Abstract
Adaptive evolution occurs as beneficial mutations arise and then increase in frequency by positive natural selection. How, when, and where in the genome such evolutionary events occur is a fundamental question in evolutionary biology. It is possible to detect ongoing positive selection or an incomplete selective sweep in species with sexual reproduction because, when a beneficial mutation is on the way to fixation, homologous chromosomes in the population are divided into two groups: one carrying the beneficial allele with very low polymorphism at nearby linked loci and the other carrying the ancestral allele with a normal pattern of sequence variation. Previous studies developed long-range haplotype tests to capture this difference between two groups as the signal of an incomplete selective sweep. In this study, we propose a composite-likelihood-ratio (CLR) test for detecting incomplete selective sweeps based on the joint sampling probabilities for allele frequencies of two groups as a function of strength of selection and recombination rate. Tested against simulated data, this method yielded statistical power and accuracy in parameter estimation that are higher than the iHS test and comparable to the more recently developed nSL test. This procedure was also applied to African Drosophila melanogaster population genomic data to detect candidate genes under ongoing positive selection. Upon visual inspection of sequence polymorphism, candidates detected by our CLR method exhibited clear haplotype structures predicted under incomplete selective sweeps. Our results suggest that different methods capture different aspects of genetic information regarding incomplete sweeps and thus are partially complementary to each other.
Keywords: positive selection, selective sweep, composite likelihood, polymorphism
POSITIVE natural selection is one of the most fundamental driving forces for biological evolution. However, it is known that mutations conferring higher relative fitness to carriers, or beneficial mutations, do not occur frequently at a given gene or genomic region of interest in most natural populations of plants and animals. Even if a beneficial allele is currently under strong directional selection, its direct identification at the sequence level is not easy since the allele frequency change is likely to be too slow to follow over time in typical population genetic surveys unless the generation time is very short and a large amount of serially sampled sequences are available. Therefore, it is extremely difficult to directly follow the random occurrence of beneficial mutations and their spread under selective environments in nature. For this reason, the investigation depends heavily on detecting the signature of past episodes of positive selection, whether the beneficial mutation is already fixed in the population or still on the way to fixation (i.e., ongoing selection for a mutation that occurred in the past but still segregating in the population), from the present-day patterns of within- and between-species genetic variation (reviewed in Nielsen 2005; Sabeti et al. 2006; Akey 2009; Stephan 2010). Such signatures of positive selection provide information for reconstructing evolutionary events that happened in the population’s history. In addition, signals of positive selection imply functional importance of the loci and thus can be used to identify genetic variation that contributes to phenotypic diversity or annotate the genome functionally (Biswas and Akey 2006).
One of the basic methods for detecting positive selection is to search for the distinct pattern of within-species genetic variation left by a “selective sweep.” A selective sweep occurs when a new advantageous mutation increases in frequency quickly in the population and results in a great reduction in variation, a temporary increase in linkage disequilibrium, and a skew in allele frequency distribution in the nearby region of a recombining chromosome (Maynard Smith and Haigh 1974; Kaplan et al. 1989; Fay and Wu 2000; Kim and Nielsen 2004). A selective sweep may be “complete” when the advantageous mutation goes to fixation and all local variation is removed except those that escaped the sweep by recombination. This type of selective sweep has drawn much attention and a number of statistical tests, mostly based on summary statistics such as Tajima’s D, Fu and Li’s D and F, and Fay and Wu’s H test, were proposed to detect mainly complete positive selection from sequences sampled shortly after the fixation of a beneficial mutation (Tajima 1989; Fu and Li 1993; Fay and Wu 2000). More advanced statistical tests based on composite likelihood were also proposed (Kim and Stephan 2002; Meiklejohn et al. 2004; Nielsen et al. 2005).
Hudson et al. (1994) first observed evidence of an ongoing selective sweep—a subgroup of sampled sequences harboring very low variation due to linkage to the putative beneficial allele that reached an intermediate frequency—at the Sod locus in Drosophila melanogaster. However, as the availability of population genomic data was limited and discovering rare episodes of recent selective sweeps was considered very difficult in natural populations, capturing such “incomplete” or ongoing selective sweeps must have been considered even more difficult. Therefore, theoretical work mainly focused on inferring selective sweeps that were already completed in the past (Kaplan et al. 1989; Barton 1998; Fay and Wu 2000; Kim and Stephan 2002; Przeworski 2002). However, Sabeti et al. (2002), in one of the first large-scale population genomic surveys for detecting recent positive selection, showed that the human genome harbors a number of loci with clear signatures of incomplete selective sweeps. Since then, detecting this type of selective sweep soon became an important topic in both empirical and theoretical population genetics (Quesada et al. 2003; Meiklejohn et al. 2004; Sabeti et al. 2006; Saunders et al. 2006; Voight et al. 2006).
Sabeti et al. (2002) introduced a long-range haplotype test based on extended haplotype homozygosity (EHH) that quantifies the residual association between an allele at the core locus and its genetic background (i.e., the linked haplotype at the time of the allele’s mutational origin). Under neutrality, a haplotype associated with an allele at higher frequency extends to a shorter distance, thus yielding smaller EHH, since the allele is older (Toomajian et al. 2003). A significantly large EHH for a given allele frequency at the focal locus then suggests the hitchhiking effect driven by positive selection. If the ancestral vs. derived alleles of a polymorphic site can be distinguished, positive selection is expected to generate a much larger EHH for the derived allele than that for the ancestral allele. This is the rationale of the iHS statistic in Voight et al. (2006) that is now routinely used in population genomic studies. Recently, Ferrer-Admetlla et al. (2014) proposed a new statistic, nSL, that is similar to iHS but is robust to recombination rate variation and exhibits improved power to detect sweeps.
The success and popularity of discovering incomplete sweeps may be attributable to unique haplotype structures that can be relatively easily and reliably captured by a rather simple test statistic such as iHS. If the local mutation rate fluctuates, it may create a random region of severely reduced variation that might be taken as a candidate for a complete selective sweep (Kim and Stephan 2002). With an incomplete sweep, the pattern of polymorphism in the haplotype block containing the ancestral allele of the focal locus reflects genetic variation that existed before the start of the selective sweep. Then, this haplotype block is effectively a negative control for the selective sweep that would alleviate the problem of local fluctuation in mutation rate. In the case of local adaptation, the inclusion of sequences from a neighboring deme, where positive selection did not take place, into analysis was shown to increase the statistical power of detecting positive selection (Innan and Kim 2008). The ancestral haplotype block in an incomplete sweep is expected to play a similar role in increasing statistical power to detect selection to that of the neighboring deme for complete sweeps.
The analysis of incomplete selective sweeps therefore provides a great opportunity for understanding positive natural selection in nature. However, as the current methods are not built on an explicit model of selection, information regarding the process of selection underlying the incomplete sweeps was limited. In this study, we obtain an approximate formula for sampling probabilities in a model of an incomplete selective sweep and then build a composite-likelihood-ratio (CLR) test for formal hypothesis testing and parameter estimation. Previously, Meiklejohn et al. (2004) proposed a CLR test for detecting an incomplete sweep by extending the sampling probabilities under complete selective sweeps of Kim and Stephan (2002) into cases where the final frequency of the beneficial allele in the population is less than one. However, in this approach the probability of sampling a neutral variant from the entire set of samples was obtained without explicitly specifying the polymorphic site causing an incomplete sweep or the joint configuration of polymorphism in the neutral and the putative selected loci. While a key parameter in their composite-likelihood ratio is the final frequency, β, of a beneficial mutation in the population, the frequency spectrum of the total data contains only a limited amount of information, yielding a very broad peak of the composite-likelihood ratio over the parameter space. Therefore, the joint estimation of β and the location/strength of selection was not accurate and the statistical power to detect selection was much lower compared to that of the iHS test. To overcome this difficulty, this study uses an approach to take each single-nucleotide polymorphism (SNP) in data as a putative locus under selection, essentially identical to the iHS method above. Namely, the derived alleles at all SNPs are tested to find whether they increased to the current frequencies by strong directional selection, by jointly analyzing the pattern of linked polymorphism surrounding the derived allele of each SNP and that surrounding the ancestral allele. This test is aimed at detecting selection in large-scale population genomic data generated by next-generation sequencing (NGS) methods, which inevitably contain occasional low-quality or missing base calls. The composite-likelihood approach can be straightforwardly applied to such data with missing information. By applying this method to simulated data and a population genomic data set in D. melanogaster, we demonstrate that this approach improves our ability to detect clear signatures of incomplete selective sweeps.
Materials and Methods
Sampling probability under an incomplete selective sweep
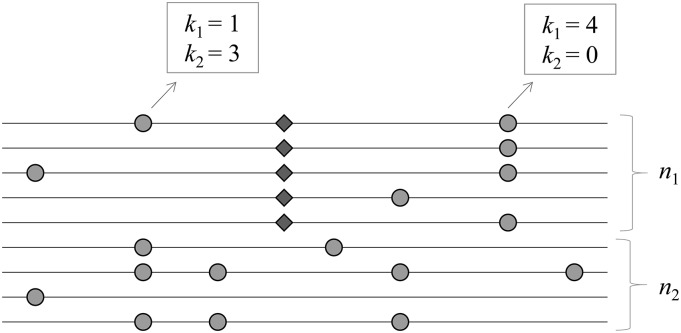
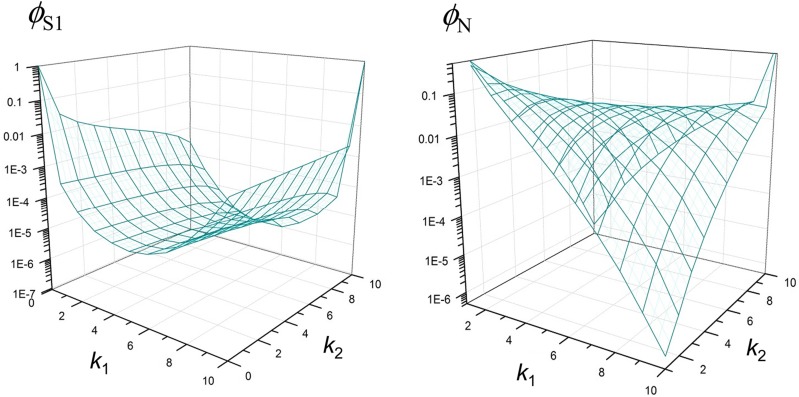
We aim to detect the signature of an incomplete selective sweep in which a beneficial allele originating from a single event of a point mutation (thus a hard selective sweep) reaches an intermediate frequency in a population. Consider multisite polymorphism observed in the alignment of n randomly sampled homologous chromosomes (Figure 1). It is assumed that neutral alleles segregate at these polymorphic sites, except one under selection (denoted the “S locus”) with n1 copies of the beneficial allele and n2 (= n – n1) copies of the ancestral allele. The strength of selection for the beneficial allele is given by α = 2Ns, where N is the number of diploid individuals in the population and s is the selection coefficient [the relative fitness of the beneficial over the ancestral allele is 1 + s, assuming codominance (h = 0.5)]. At a neutral site that is d nucleotides away from the S locus, let k1 (k2) be the count of the derived allele in the subsample of n1 (n2) chromosomes carrying the beneficial (ancestral) allele. If d is small enough to generate the hitchhiking effect of the beneficial allele, an increased or decreased frequency of the derived neutral allele due to hitchhiking is reflected by k1/n1, while its frequency before hitchhiking is estimated by k2/n2, assuming that the frequency of the neutral allele among chromosomes carrying the ancestral allele at the S locus does not change during the sweep (see Appendix). Therefore, the hypothesis of an incomplete sweep acting on the (putative) S locus predicts a very distinct joint probability distribution of k1 and k2, compared to an alternative (i.e., neutral) hypothesis (Figure 2). Our goal is to build a parametric test based on this joint sampling probability, denoted by for detecting an incomplete selective sweep (i.e., identifying the S locus in DNA sequence polymorphism).
Figure 1.
Pattern of DNA sequence polymorphism under an incomplete sweep. Lines indicate individual sequences and circles indicate derived alleles at neutral sites. Diamonds indicate new advantageous mutations spreading in the population.
Figure 2.
Joint sampling probability as a function of k1 and k2 (= 0, . . . , 10) for n1 = n2 = 10 and N = 105, under the incomplete sweep model with r/s = 0.04 (left) and the neutral model (right).
We obtained two approximate solutions to such a joint sampling probability, and by modifying the equivalent solution for complete sweeps in Nielsen et al. (2005) and that in Etheridge et al. (2006), respectively (Appendix ). The corresponding probability under the null hypothesis (no selection), can also be obtained. The primary parameter that determines and is where r = rnd (rn = recombination rate per nucleotide per generation) is the recombination rate between the S locus and the neutral site and R = 4Nr. In comparison against simulated data generated by msms (Ewing and Hermisson 2010) under the model of an incomplete sweep, approximates the sampling probability much better than for small recombination rates (Supporting Information, Figure S1). However, is not applicable for larger recombination rates () (Etheridge et al. 2006).
CLR test
Let x be the position of the putative S locus, which is assumed to be one of the polymorphic sites in the sequence alignment and thus partition the data into subsamples of n1 and n2 chromosomes carrying the derived and ancestral alleles at the locus, respectively. n1 is also denoted as n1(x) to emphasize that the position x uniquely determines the derived allele frequency of the putative S locus. This partition by the S locus also determines the counts of the derived neutral allele at nucleotide site i in the two subsamples, and Then, for a given x, a maximum-composite-likelihood estimate of the strength of selection, , is obtained as a value of α (if R per site is externally given) that maximizes the CLR,
| (1) |
where
| (2) |
and
| (3) |
are composite likelihoods under the hypotheses of incomplete selective sweep and neutrality, respectively. In the following, unless stated otherwise we use only for Equation 2 despite its error for small r/s. The impact of this error on the performance of our likelihood test is addressed below. Unless stated otherwise, multiplication above is done across all sites in the data, including monomorphic sites It is also possible to multiply probabilities over polymorphic sites only (analogous to L2 and L4 in Kim and Nielsen 2004), which leads to a composite-likelihood test for detecting selection based on the joint allele frequency spectrum only but not on the patterning of polymorphic sites along the sequence.
It is straightforward to calculate the CLR given in Equation 1 in the presence of missing values in sequence data, for example due to low-quality base calls that are common in NGS data sets. If missing base calls are made at a site on chromosomes in the sample, the sampling probability for this site is calculated after n1 and/or n2 are reduced accordingly. If base calls are missing at the core SNP (the putative S locus), entire chromosomes carrying the missing base calls are excluded from the calculation of composite likelihoods.
Next, the maximum-composite-likelihood estimate of the locus under selection is obtained by calculating for all polymorphic sites in a given chromosomal region and then identifying the site (at position ) that maximizes This procedure also implies that a test statistic for hypothesis testing would be given by
| (4) |
and we may reject the null hypothesis (neutrality) if T0 is larger than a certain cutoff value. The null distribution of T0 is determined by applying the above calculation to polymorphic sites in a large number of data sets simulated under the neutral model. Imposing fixed polymorphic sites (-s option in ms) or fixed scaled mutation rate (-t option) but conditioning on the similar number of polymorphic sites in simulated data led to almost identical distributions (data not shown). However, it was observed that the maximum CLR for a given focal site () is negatively correlated with , most likely because a derived allele with smaller allele frequency originated more recently and is thus associated with a longer extended haplotype. If not corrected, this will bias the estimated locus of selection to be a polymorphic site with a lower frequency of the derived (putatively beneficial) allele. A solution to this problem would be to transform to remove its correlation with We tried and evaluated various forms of the normalized test statistic. The following procedure yielded the most optimal performance of parameter estimation (see below). Let and be the mode and the 1 − ε quantile of the distribution of obtained from polymorphic sites whose derived allele frequency is f in simulated neutral data sets. Then, we define a new statistic
| (5) |
Then, the estimated location of the S locus, is the value of x that achieves the maximum in the above formula. This also leads to the final estimate of the strength of selection, for a given set of sequences. For a given ε, the null distribution of T1 is obtained by applying the above procedure to a large number of sequence samples, with an equivalent number of polymorphic sites and a scaled recombination rate, that are generated by neutral simulation. Unless stated otherwise, we reject the null hypothesis of neutrality (no selection) with significance level P = 0.001, which resulted in an optimal range of statistical powers with varying parameter values chosen below.
Analysis of D. melanogaster population genomic data
We used 22 primary core genomes in the Rwanda (RG) sample of D. melanogaster described in Pool et al. (2012), available for download from the DPGP2 project (http://www.dpgp.org/). As the violation of the random sample from unrelated individuals may generate spurious occurrence of long-range haplotype homozygosity, we removed identical-by-descent (IBD) tracts detected by Pool et al. (2012): in the sample of 22 sequences, if any pair of chromosome segments are IBD, we treated one of them as a missing observation, replacing it by a sequence of “N” characters. Then, we extracted a phased table of polymorphic sites with their physical locations. Next, the ancestral and derived alleles were inferred using the syntenic assembly of D. melanogaster and D. simulans (available at www.dpgp.org) and designating the allele observed in simulans as ancestral or the table of ancestral allele probability for polymorphic sites calculated for DPGP1 RAL sequences (Chan et al. 2012). This procedure could assign the ancestral/derived states for ∼85% of polymorphic sites obtained above. The remaining polymorphic sites were not included in input files. We also excluded from analysis the telomeric and centromeric regions of each chromosome arm with low recombination rates: from the midpoint of a chromosome arm we moved toward the telomere and toward the centromere until the points over which the mean genetic distance per megabase first becomes <1 cM, using the best-fitting equations for crossing-over rates on 100-kb windows obtained by Comeron et al. (2012).
Composite likelihood is calculated by taking a SNP as the putative S locus (“core SNP”): thus the sample is partitioned into n1 and n2 sequences as described above. The sample frequency of the focal derived allele is therefore f = n1/(n1 + n2). Note that, as this SNP may contain missing values (N in data) and the corresponding chromosomes are excluded from calculating the composite likelihood, n1 + n2 can be <22. For computational convenience, we assumed scaled recombination rate 4Nrn = 0.012 per site in the calculation of likelihood for all chromosome arms. As sampling probability under selection is primarily a function of but only slightly modified by α alone, a deviation of actual recombination rate from the above assumption would lead to a corresponding error in the estimate of α, without affecting the location and value of the maximum-composite-likelihood ratio. Local fluctuation in the scaled mutation rate, θ, was also ignored: we estimated mean θ for each chromosome arm and used it in the calculation of likelihoods for any region within the chromosome arm. Incorrect assumptions of θ were shown to affect minimally the performance of our test (see below).
Joint sampling probabilities were obtained using the approximation proposed in Nielsen et al. (2005), i.e., assuming that the ancestral pattern of polymorphism at the time of the beneficial mutation follows either standard neutral equilibrium (test option A) or the currently observed genome-wide empirical frequency spectrum (test option B) (Appendix). The significance of the CLR, maximized with respect to α and then normalized for the derived allele frequency, is assessed as described for T1 above, however, using the site-wise null distribution of CLR obtained from individual polymorphic sites (5 × 105 SNPs) generated by msms under neutrality, with parameters adjusted to match sample size, mean recombination rate, and the mean density of polymorphic sites to those of Drosophila genome data. Namely, multiple-test correction, as implemented above by the null distribution of the local maximum of test statistic (T1) in a window of defined sequence length, is not performed here. Therefore, a P-value determined this way cannot be compared to that used for analyzing simulated incomplete sweeps above. We consider sites that yield large normalized CLR, corresponding to P < 0.001, as candidate loci under selection. This level of significance is rather arbitrary. However, rather questionable candidates of incomplete sweeps (with unclear haplotype structure upon visual inspection; see Results below) are already detected at this level and, therefore, a less stringent level will likely increase the number of such loci.
Haplotype homozygosity tests
We applied two haplotype homozygosity tests, iHS (Voight et al. 2006) and nSL (Ferrer-Admetlla et al. 2014), to detect incomplete sweep in simulated data as well as D. melanogaster data. For the analysis of simulated data, unstandardized iHS (log[iHHA/iHHD]) was calculated for individual polymorphic sites according to Voight et al. (2006), using the rehh R package (Gautier and Vitalis 2012) (http://cran.r-project.org/web/packages/rehh/index.html), and unstandardized nSL was calculated through the program provided by Ferrer-Admetlla et al. (2014) at http://cteg.berkeley.edu/∼nielsen/. Using the same set of simulated neutral samples as used above for CLR analysis, the 1 − ε quantile and mode of the distribution of the unstandardized iHS were obtained for each derived allele frequency and these values were used to define standardized iHS by applying the procedure of obtaining T1 by Equation 5. The test statistic for detecting an incomplete selective sweep in a replicate of a 100-kb sequence sample is therefore the most negative standardized iHS among sites and the procedure of obtaining the null distribution and assessing the significance of this statistic is identical to that of T1 by the CLR method. The same normalization procedure was applied for the nSL statistic. We also tried the standardization procedure based on the assumption of normal distribution described in Voight et al. (2006) for both statistics and discovered that our standardization procedure leads to slightly increased statistical power (data not shown).
For the genomic scan of D. melanogaster data below, we first obtained standardized iHS and associated P-values for individual polymorphic sites in the data according to the procedure described by Voight et al. (2006) performed by the rehh package. In the calculation of iHHA and iHHD by this package haplotype homozygosity for sequences does not extend from the core SNP if missing base calls are encountered in a subset of the sequences. Namely, missing bases (N) are treated as an allele distinct from A, C, G, or T. We found that this frequently generates very small iHHA and thus erroneously very negative iHS (i.e., false detection of selection). To correct this problem, we wrote our own code that calculates iHHA and iHHD while skipping positions of missing bases in extending haplotype homozygosity. We also used the full data including all polymorphic sites rather than the input data used above for the CLR test, in which ∼15% of polymorphic sites were excluded as their ancestral/derived alleles could not be determined. Excluding these sites caused frequent false positives since the excluded sites are often clustered to make the region to falsely appear monomorphic and thus inflate iHHD. These corrections led to detection of clearer signatures of incomplete sweeps (upon visual inspection of haplotype structures). For the nSL statistic, since we find it less sensitive to missing data than iHS, we used the same input data as used for the CLR test and then performed standardization according to Ferrer-Admetlla et al. (2014).
Simulated data under different demographic assumptions
To explore the robustness of the CLR test to demographic assumptions, we generated neutral data sets, using msms (Ewing and Hermisson 2010) under three different scenarios: population bottleneck, exponential population growth, and population subdivision. All data sets were generated with equal sequence length (100 kb) and number of polymorphic sites (3000). For the bottleneck model, we simulated a population bottleneck lasting from 0.4N to 0.2N generations in the past with different severities c = 0.4, 0.2, and 0.1 (c = Nb/N, where Nb is the population size during the bottleneck). In the case of the exponential growth model, populations start growing exponentially from a population size of 0.4N, with three different growth rates g = 10, 100, and 500. For the population subdivision model, we simulated a two-island model with symmetric, constant migration rates M = 0.1, 1, 10 and then drew all sequences for each sample from one island. We also varied the recombination rate [R (per 10 kb) = 4000, 6000, 8000, 10,000, 12,000] in each model to study the effect of altered linkage disequilibrium on the null distribution. For each parameter set, at least 1000 replicates with a sample size of 20 chromosomes were obtained.
Codes and scripts
All source codes developed here for analyzing simulated and actual data are available upon request. Command line scripts for simulations performed above using ms and msms are provided in File S1.
Results
Statistical power of the composite-likelihood test
To evaluate the performance of the composite-likelihood method described above, we applied it to simulated data sets generated by msms (Ewing and Hermisson 2010) under the model of incomplete selective sweeps. In simulation, the beneficial allele of the S locus, located in the middle of the 100-kb sequence, reaches frequency β = 0.5 in the population and a sample of 20 sequences is generated. Then, test statistic T1 in Equation 5 was determined after the maximum CLRs were calculated over all SNPs in the sample with derived allele frequency ≥3 and ≤17. The null hypothesis of neutrality was rejected if T1 > the 99.9th percentile of the null distribution, which was obtained from data sets simulated under the model of neutral equilibrium with the same sequence length and recombination rate. This cutoff value (P = 0.001) of T1 is a function of ε. When various values of ε (0.0006, 0.001, 0.0016, 0.002, and 0.003) were tried, the statistical power fluctuated moderately (∼5%) while ε = 0.002 resulted in the best performance in parameter estimation (the largest proportion of replicates in which the correct site, at position 50 kb, yielded the largest T1). We thus use ε = 0.002 in the following analyses.
The statistical power of the test increased as the final frequency of the beneficial mutation, β, in the population increased: with R = 2000, α = 2000, and β increasing from 0.3 to 0.7 by 0.1, the statistical powers were 0.45, 0.71, 0.87, 0.95, and 0.98, respectively. This test performed better with larger β presumably because as a larger proportion of individuals (thus sequences in a sample) are affected by selection, the pattern of polymorphism becomes more distinctive from the neutrality, and also because the component of sampling probability was obtained from the solution obtained for a complete selective sweep. For a fixed value of β, the statistical power increased with increasing strength of selection, as expected (Table 1).
Table 1. Accuracy of parameter estimates using the composite-likelihood, iHS, and nSL methods.
| Parameters | Composite likelihood | iHS | nSL | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ra | α | Statistical power (%)b | mean ± SD | deviation mean (median)c | Exact site detected (%)d | Statistical power (%)b | deviation mean (median)c | Exact site detected (%)d | Statistical power (%)b | deviation mean (median)c | Exact site detected (%)d |
| Using both polymorphic and monomorphic sites | |||||||||||
| 2000 | 1000 | 57 (17) | 1160 ± 660 | 3.68 (1.73) | 9.23 [10.9] | 24 | 5.67 (2.91) | 4.92 [5.29] | 41 | 4.72 (2.23) | 11.1 [13.1] |
| 2000 | 86 (36) | 2070 ± 920 | 4.11 (2.43) | 10.6 [12.7] | 63 | 6.18 (4.05) | 3.54 [3.22] | 63 | 5.01 (2.95) | 9.4 [11.4] | |
| 4000 | 97 (30) | 3850 ± 1780 | 6.03 (3.92) | 9.07 [10.7] | 89 | 8.58 (6.09) | 2.61 [2.23] | 76 | 7.10 (4.45) | 6.9 [7.0] | |
| 4000 | 1000 | 35 (16) | 1290 ± 840 | 2.92 (0.95) | 15.6 [20.4] | 9.6 | 5.71 (1.80) | 7.37 [9.00] | 42 | 3.36 (1.21) | 18.6 [24.5] |
| 2000 | 75 (40) | 2290 ± 1000 | 2.51 (1.39) | 15.9 [19.2] | 41 | 3.98 (2.30) | 5.46 [6.58] | 73 | 2.77 (1.62) | 16.3 [17.1] | |
| 4000 | 94 (48) | 4030 ± 1610 | 3.56 (2.20) | 13.8 [16.7] | 78 | 5.12 (3.45) | 3.90 [4.29] | 87 | 3.66 (2.35) | 12.6 [12.4] | |
| Using only polymorphic sites | |||||||||||
| 4000 | 1000 | 4.0 | 860 ± 950 | 3.62 | 13.3 | ||||||
| 2000 | 36 | 1630 ± 1010 | 3.21 | 13.3 | |||||||
| 4000 | 82 | 3290 ± 1870 | 4.91 | 10.3 | |||||||
Scaled recombination rate (4Nr) across the 100-k-long simulated sequence.
Percentages of simulated samples that yield P < 0.001. In parentheses: using an adjusted recombination rate in neutral simulations to yield mean LD identical to that of data to be analyzed.
Mean (median) deviation in kilobases of the estimated location of selection from the true location (x = 50,000):
Percentages of simulated samples for which = 50,000. Those of samples in which the frequency of the beneficial allele is exactly 10 are given in brackets.
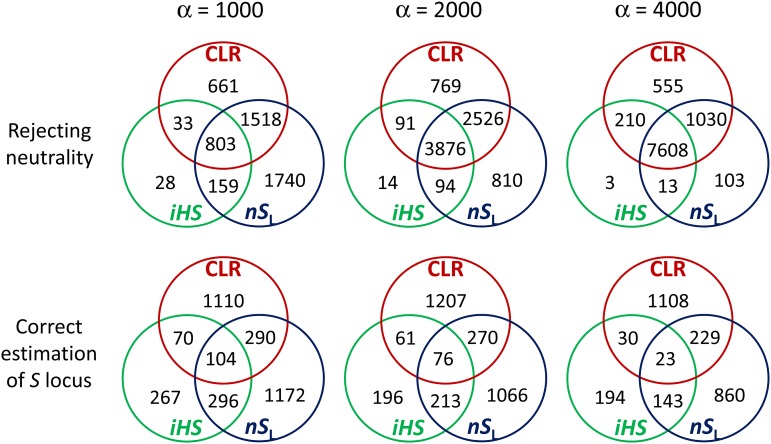
This performance of the composite-likelihood test was compared to that of long-range haplotype methods that use iHS and nSL statistics (Voight et al. 2006; Ferrer-Admetlla et al. 2014). Instead of using the normal distribution-based standardization of iHS and nSL, we applied the normalization procedures that were used to obtain T1 above (see Materials and Methods), which made it possible to directly compare the performance of the CLR, iHS, and nSL methods. In all parameter sets tested, the statistical power of our composite-likelihood method is higher than that of iHS but only slightly better than that of the nSL method [Table 1; note that results here were obtained assuming that the correct scaled recombination rate of the sequence is available (see below)]. Interestingly, there are a relatively large number of simulated incomplete sweeps detected by either CLR or nSL only, particularly with weaker strength of selection (Figure 3), suggesting that the CLR and nSL methods capture different aspects of data as signatures of incomplete sweeps and thus are largely complementary to each other.
Figure 3.
Numbers of simulation replicates (of 10,000) from which incomplete selection sweeps were detected (rejection of null hypothesis at P < 0.001) and the correct site under selection was inferred, individually or jointly by the CLR, iHS, and nSL methods. R = 4000.
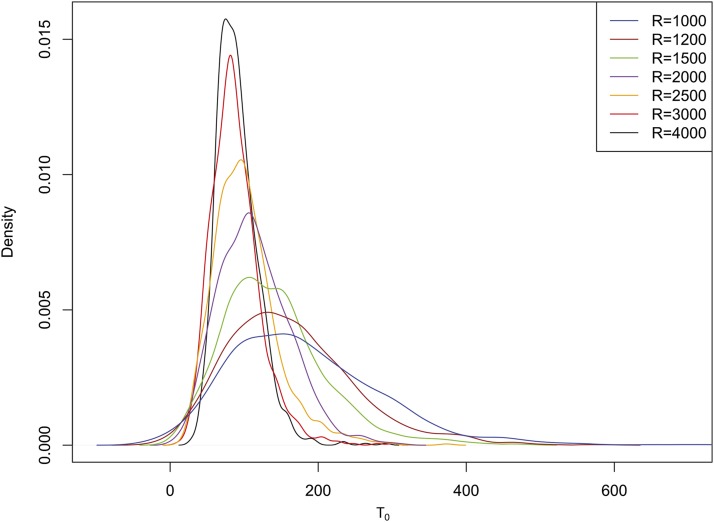
Effect of recombination rate and linkage disequilibrium
The above result is based on the null distribution of the test statistic obtained from neutral simulations that used recombination rates identical to those used in the simulation of incomplete sweeps. However, in practice, the correct rate of recombination, scaled or unscaled, for a given genomic region may not be available. This turns out to be a serious problem for our CLR method, as we found that the null distribution of the likelihood ratio is highly sensitive to the scaled recombination rate (Figure 4). It appears that, with decreasing recombination rate, linkage disequilibrium (LD) between adjacent polymorphic sites increases and this inflates the likelihood of an incomplete sweep (LIS) relative to that of neutral evolution (LN). Therefore, one approach to control the false-positive rate of detection might be to adjust the recombination rate during neutral simulation until the average LD among sites matches that of data under examination (see below for the case of demographic complications). As an incomplete selective sweep generates a high level of LD around the S locus (Stephan et al. 2006), to generate samples with an equivalent level of LD [measured by the average ρ2 over all pairs of sites, where ρ is the normalized LD as measured by correlation of allele frequencies between two loci (Hill and Robertson 1968)] the recombination rate needs to be greatly reduced in the neutral simulation. When we obtained the null distribution of T1 from such low-recombination simulation, the statistical power of our CLR method decreased dramatically (Table 1). In contrast, the null distribution of nSL was affected minimally by recombination rate variation (data not shown), as it was originally proposed to cope with uncertainty in recombination rates (Ferrer-Admetlla et al. 2014).
Figure 4.
Distribution of maximum CLR, where the maximum was obtained over the set of polymorphic sites with n1 = 10 (S[10] for each replicate), for samples generated under standard neutral simulation with varying recombination rate.
Inferring the strength of selection and the position of the S locus
Because sequences are randomly sampled from a population, the copy number of the beneficial allele, nB = n1(50,000), in a sample is variable (binomial): with β = 0.5, nB < 3 or >17 in <0.5% of replicates, which makes it impossible to detect the true locus under selection. In the other replicates, the exact locus is detected if the maximum T1 is obtained at the correct site (i.e., = 50,000). Compiling results from all replicates (regardless of whether the correct site is inferred or not), we find that the estimate of the strength of selection is unbiased, although the variance of the estimate is large (Table 1). More than half of replicates yielded within ∼1–3 kb from the target of selection. The proportion of replicates in which the exact site is inferred ranges from 9 to 16%, more accurate estimates occurring with higher recombination rates. If the sample frequency of the beneficial allele matches the population frequency (0.5), this proportion significantly increases (Table 1).
In the iHS and nSL methods the estimated location of the S locus, , is given as the polymorphic site from which the most negative normalized statistic is obtained. Applied to the same sets of simulated data, by iHS was less accurate than by either CLR or nSL (Table 1). The exact position of the S locus was correctly inferred about three times more often by CLR than by iHS but roughly as often as by nSL. CLR also yielded the smallest mean deviation of from the true location. Overall, the accuracies of estimates are similar between the CLR and nSL methods. Surprisingly, however, the three methods are weakly correlated with respect to estimating the exact location of selection (Figure 3): for example, applied to 10,000 replicates of simulation with α = 4000, the CLR and nSL methods detected the correct site under selection in 1390 and 1255 replicates, respectively. However, in only 252 replicates the correct site was detected by both methods. Again, this result suggests that the CLR and iHS/nSL methods capture slightly different information in multisite polymorphism to detect incomplete sweeps and estimate the position of the putative S locus. When we define a new estimate as the average over those by the CLR and nSL methods, its mean deviation from the correct site in kilobases [i.e., ] is 2.74, 2.42, and 3.25 for α = 1000, 2000, and 4000, respectively (with R = 4000), which is smaller than the deviation obtained by an individual method (Table 1). Therefore, small improvements in the accuracy of position estimates are made by combining the two methods.
Modification of composite likelihoods
So far, sampling probability based on approximation by Nielsen et al. (2005), was used for obtaining composite likelihoods. For small recombination rates (), we may replace by more accurate approximation, based on Etheridge et al. (2006). This, however, did not lead to a significant change in the profile of the CLR (Figure S2). We also examined the effect of not including monomorphic sites in the data. When the CLR is calculated by multiplying joint sampling probabilities over only polymorphic sites in the data, it leads to lower statistical power to detect selection and larger errors in estimating the strength and position of selection than when multiplication was done over all sites (Table 1). This result suggests that not only the (joint) frequency spectrum of polymorphism but also the spatial distribution or density of polymorphic sites contains information regarding incomplete selective sweeps.
Effect of complex demography
Next, to evaluate the robustness of the CLR method to complex demography and population structure, we examined how the null distribution of the test statistic (T1) changes if it is obtained from data sets simulated under the models of population bottleneck, expansion, and subdivision (see Materials and Methods). In each model parameters were chosen to produce a significant deviation of the frequency spectrum from that under neutral equilibrium. The number of polymorphic sites (3000) per sample remained constant for varying models and parameters. First, with a population bottleneck that lasted from 0.4N to 0.2N generations ago, decreasing the size of the bottlenecked population (c = NB/N decreasing from 0.2, 0.1, to 0.05) dramatically shifted the distribution of the CLR upward (Figure S3A). This shift appears to be explained by a reduction in scaled (population-level) recombination rate due to the bottleneck, which leads to increased LD: when the recombination rate was increased to reduce LD (quantified by mean pairwise ρ2), the distribution of the CLR shifted back downward (Figure S4A). With matching LD, distributions obtained under the bottleneck (4Nrn = 0.1; mean ρ2 = 0.0543) and under the standard neutral model (a constant-sized panmictic population; 4Nrn = 0.04; mean ρ2 = 0.0543) are very similar (Figure S4A). However, the right tail of the distribution is still slightly larger than that of neutral equilibrium.
Similarly, the null distribution of the CLR shifts upward due to rapid exponential growth of population size (g > 100) in the expansion model and limited migration (M < 1) in the subdivision model (Figure S3, B and C). Again, by increasing the recombination rate in the simulation, thus reducing the average level of LD among sites, these distributions are shifted downward. Similar distributions of the CLR (right tails) are obtained from simulations under the standard and complex demography if the levels of LD match (Figure S4). These results suggest that, in the analysis of a genomic region for which underlying population demography and/or correct recombination rate are not known, the false-positive rate of detecting incomplete sweeps by CLR can be greatly reduced, if not completely, by generating samples with matching LD by standard neutral simulation.
Results above were obtained by calculating the likelihood of incomplete sweeps, assuming the standard neutrality at the time of beneficial mutation [f0(p) = θ/p; test option A]. We can replace f0(p) with the empirical distribution of the derived allele frequency observed in the simulations of these demographic models (test option B). The latter option is essentially the approach by Nielsen et al. (2005) to minimize the compounding effect of complex demography in detecting the signature of selection. However, it had little effect in correcting the null distribution and did not prevent the inflation of the CLR with increasing LD between segregating sites (Figure S3).
Application to D. melanogaster genomic data
The composite-likelihood method described above was applied to population genomic data of D. melanogaster to detect incomplete sweeps. We used 22 haploid genome sequences from Rwanda (the RG sample) described in Pool et al. (2012). As the species’ ancestral range is known to lie within southern and eastern Africa, the RG sample is likely to satisfy the assumption of equilibrium demography (constant-sized random-mating population before the start of the sweep in our model) better than any other available genomic data sets in D. melanogaster. However, when we examined the genome-wide distribution of derived allele frequency, a slight but clear deviation (excess of rare alleles) from the standard neutrality was observed (Figure S5). This is likely due to nonequilibrium demography (mild population bottleneck and recent population growth) that may have affected the RG sample (Pool et al. 2012) but might also be due to errors in base calling and ancestral/derived state inference.
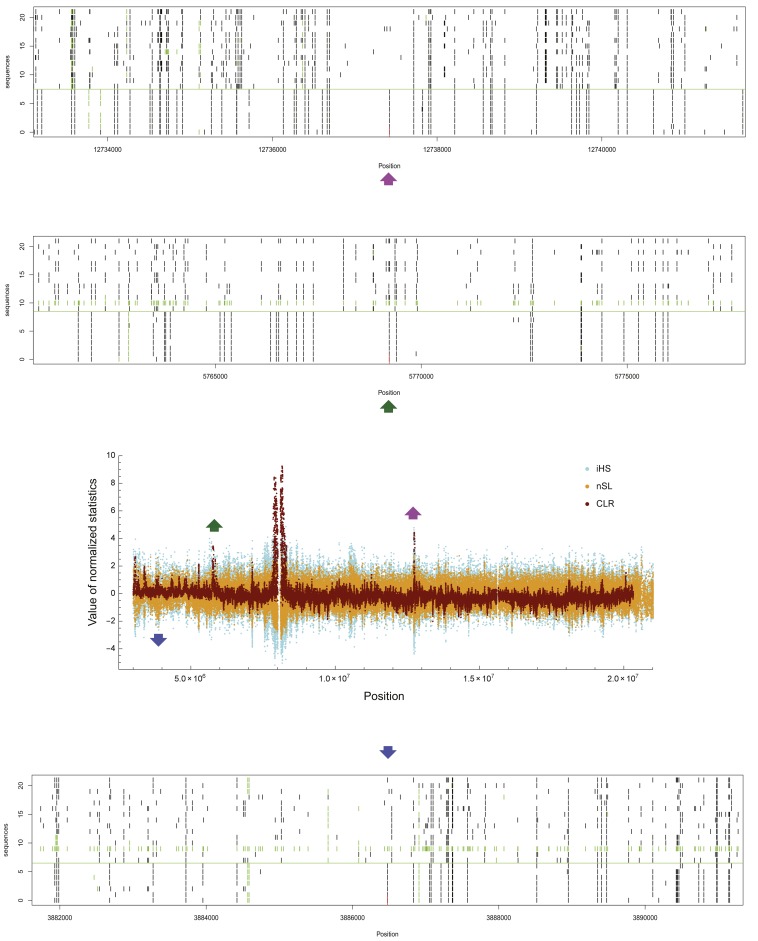
A genome scan was conducted by sequentially taking all polymorphic sites in the data with derived allele frequencies satisfying 0.35 < f < 0.8 as core SNPs and calculating composite likelihoods. We observed clear clustering of SNPs yielding a large CLR (Figure 5 for chromosome arm 2R), corresponding to P < 0.001 (see Materials and Methods), scattered over the five major chromosome arms. We consider each cluster as a footprint of a single episode of an incomplete selective sweep. (Other scattered and isolated sites that yield P < 0.001 but do not form clusters were not considered.) A SNP with the largest CLR within a cluster (the “peak”) is therefore a candidate position of ongoing selection. There are 42 clusters in total, using test option A, and we identified an annotated gene in FlyBase (version FB2014_03) containing or closest to the peak in each cluster (Table 2). Test options A and B generated very similar profiles of the CLR along the chromosome (Figure S6) and thus led to the detection of almost identical sets of candidate loci in each chromosome arm. When clusters are ranked according to T1 within each chromosome arm, ranks by options A and B are strongly correlated (Table 2). Upon visual inspection of aligned and sorted sequences, we observed clear segregating patterns of SNPs indicative of incomplete sweeps—far fewer polymorphisms and high linkage disequilibrium among chromosomes containing the derived allele compared to those containing the ancestral allele at the core SNP—at the majority of these candidate loci (Figure 5 and Figure S7).
Figure 5.
Plots of normalized maximum-composite-likelihood ratio (T1x; dark red) and standardized iHS (light blue) and nSL (orange) along chromosome 2R. Haplotype structures around three putative positions under selection (positions 5,769,223 and 12,737,423 detected by the CLR method and position 3,886,479 detected by the nSL method) are shown. Aligned sequences were sorted by the position of the putative site, with those carrying ancestral alleles on the top and derived alleles on the bottom (divided by a horizontal green line). Derived alleles and missing base calls at polymorphic sites are marked by black and green bars, respectively.
Table 2. List of putative loci under incomplete selective sweeps in D. melanogaster Rwanda population detected by the CLR method.
| Chr. arm | Cluster start to enda | Max T1e | Site of max T1f | Sample DAFg | Rank by option A (B)h | iHSi | nSL | Closest gene | |
|---|---|---|---|---|---|---|---|---|---|
| 2L | 1,517,366–1,529,788b | 1.71 | 1,527,302 | 8,000 | 13/19 | 11 (12) | −0.662 | −0.42 | halo |
| 5,803,333–5,815,486b,c,d | 2.04 | 5,805,001 | 8,000 | 9/22 | 5 (3) | −1.87 | −1.44 | CG11034 | |
| 6,650,567–6,657,389b,d | 1.92 | 6,652,011 | 8,000 | 17/22 | 7 (13) | −0.577 | −0.46 | Tango1 | |
| 7,409,201–7,413,132 | 1.81 | 7,409,825 | 2,828 | 11/22 | 9 (6) | −0.577 | −1.96 | CG5181 | |
| 14,094,223–14,102,937b | 1.72 | 14,100,158 | 8,000 | 11/21 | 10 (9) | −1.81 | −2.29 | nAChRalpha5 | |
| 16,001,993–16,020,004c | 2.05 | 16,005,369 | 16,000 | 8/21 | 4 (5) | −2.52 | −1.88 | Beat-Ic | |
| 17,221,968–17,339,080c,d | 6.66 | 17,271,945 | 64,000 | 8/21 | 1 (1) | −2.23 | −1.58 | CG6380 | |
| 17,602,170–17,624,875c,d | 2.28 | 17,616,351 | 11,313 | 11/21 | 3 (4) | −1.42 | −1.69 | Sytalpha | |
| 18,446,487–18,454,519 | 1.55 | 18,453,145 | 11,313 | 8/20 | 12 (11) | −0.892 | −1.30 | bsf | |
| 18,993,669–18,999,601 | 1.93 | 18,996,657 | 11,313 | 13/20 | 6 (10) | −1.23 | −0.97 | CG10650 | |
| 19,480,924–19,546,720 | 1.91 | 19,493,563 | 22,627 | 8/21 | 8 (8) | −1.32 | −0.94 | swm | |
| 19,729,511–19,790,297c,d | 3.13 | 19,756,197 | 22,627 | 9/21 | 2 (2) | −2.14 | −1.08 | CG10631 | |
| 2R | 3,058,650–3,079,356d | 2.67 | 3,073,701 | 45,255 | 8/20 | 4 (5) | −2.88 | −2.06 | didum |
| 5,269,193–5,278,463 | 2.12 | 5,271,741 | 11,313 | 11/20 | 6 (6) | −2.38 | −2.14 | CG13954 | |
| 5,756,581–5,788,311 | 3.45 | 5,769,223 | 32,000 | 9/21 | 3 (3) | −0.44 | −0.95 | Sec24AB | |
| 7,126,724–7,131,721c,d | 2.27 | 7,127,281 | 5,656 | 11/22 | 5 (4) | −3.87 | −2.40 | CG13215 | |
| 7,882,689–8,178,854c,d | 9.24 | 8,157,979 | 11,313 | 9/22 | 1 (1) | −3.88 | −2.08 | otk | |
| 12,727,369–12,759,779b,c,d | 4.43 | 12,737,423 | 22,627 | 8/22 | 2 (2) | −2.40 | −1.81 | IntS8 | |
| 20,061,659–20,075,219b | 1.53 | 20,073,016 | 5,656 | 17/22 | 7 () | −1.11 | −0.48 | Nop60B | |
| 3L | 3,173,086–3,190,811b | 2.01 | 3,175,908 | 8,000 | 15/22 | 5 (5) | −1.48 | −0.97 | Girdin |
| 4,478,135–4,479,071b,c | 1.38 | 4,478,135 | 2,828 | 11/22 | 8 (8) | −2.44 | −2.91 | CG7465 | |
| 6,100,579–6,157,140b,c,d | 2.09 | 6,146,679 | 11,313 | 11/21 | 3 (3) | −0.179 | −0.62 | Lcp65Ag2 | |
| 6,550,102–6,557,978b,c,d | 1.81 | 6,551,837 | 8,000 | 9/21 | 6 (4) | −3.92 | −3.52 | CG18769 | |
| 11,825,913–11,831,926b,d | 1.35 | 11,829,615 | 2,000 | 17/22 | 9 (9) | −2.11 | −2.38 | CG43064 | |
| 13,425,802–13,431,380b,d | 1.70 | 13,430,186 | 5,656 | 11/22 | 7 (7) | −2.72 | −3.18 | CG10713 | |
| 16,084,427–16,136,825c,d | 2.86 | 16,106,542 | 16,000 | 17/22 | 2 (2) | −1.79 | −2.45 | Taf4 | |
| 17,733,967–17,741,172 | 2.04 | 17,735,433 | 8,000 | 16/22 | 4 (6) | 0.0795 | −0.49 | CG7460 | |
| 19,211,270–19,237,070c,d | 2.89 | 19,220,338 | 22,627 | 8/22 | 1 (1) | −1.85 | −2.74 | fz2 | |
| 3R | 3,697,516–3,769,886 | 2.90 | 3,727,631 | 45,255 | 9/19 | 5 (7) | −0.147 | 0.08 | mRpS9 |
| 4,155,075–4,182,535 | 2.68 | 4,158,518 | 64,000 | 11/20 | 7 (9) | −1.06 | −1.03 | CG9601 | |
| 5,530,419–5,688,202 | 3.91 | 5,548,751 | 90,510 | 8/20 | 2 (5) | −1.15 | −1.36 | CG8478 | |
| 8,486,190–8,497,516 | 3.32 | 8,497,516 | 32,000 | 11/19 | 3 (4) | 1.63 | 0.53 | CG14395 | |
| 9,040,956–9,111,809c | 7.03 | 9,057,704 | 64,000 | 8/19 | 1 (1) | −2.87 | −1.53 | Ace | |
| 10,380,467–10,391,240c,d | 2.73 | 10,386,839 | 8,000 | 10/21 | 6 (3) | −2.99 | −2.56 | Pde6 | |
| 12,060,488–12,066,143c,d | 2.48 | 12,066,090 | 16,000 | 17/22 | 8 (10) | −3.09 | −3.01 | tara | |
| 16,575,106–16,577,277b,d | 2.10 | 16,575,113 | 11,313 | 16/22 | 9 (14) | −1.91 | −2.34 | CG42322 | |
| 17,406,332–17,432,203b | 3.03 | 17,414,532 | 16,000 | 15/22 | 4 (2) | −1.67 | −0.54 | InR | |
| 18,232,347–18,251,667b | 2.09 | 18,245,938 | 11,313 | 12/22 | 10 (6) | −1.33 | −1.58 | lqfR | |
| X | 525,809–1,798,033c | 2.75 | 1,350,182 | 32,000 | 17/22 | 1 (1) | −1.06 | −0.97 | MED18 |
| 2,817,759–2,835,897c,d | 1.73 | 2,828,033 | 5,656 | 10/22 | 2 (2) | −4.89 | −4.64 | kirre | |
| 14,156,405–14,160,294b,d | 1.21 | 14,157,513 | 2,828 | 16/21 | 4 (4) | −3.86 | −4.50 | CG1461 | |
| 15,607,843–15,628,927b | 1.43 | 15,620,351 | 5,656 | 14/21 | 3 (3) | −1.40 | −1.90 | CG8184 |
Positions of the first (start) and last (end) sites of significant CLR (P < 0.001) within the cluster.
Overlap with a candidate region of complete selective sweep.
Overlap with a cluster detected by the iHS test.
Overlap with a cluster detected by the nSL test.
Maximum CLR (T1) within the cluster.
The location of maximum T1 or the putative nucleotide site under selection within the cluster.
The derived allele frequency (DAF) in the data at the putative site under selection.
The rank within the chromosome arm of the maximum T1 when option A (B) is used for calculating composite likelihoods.
The value of iHS and nSL calculated at the putative site under selection detected by the CLR method.
The calculations above were performed using a uniform value of scaled mutation rate, θ, for each chromosome arm. To examine whether local variation in θ has an effect on the accuracy of inference, we performed the CLR test with the local value of θ calculated from a 10-kb window surrounding each core SNP. This procedure yielded almost the same profile of composite likelihood along the chromosome and the same list of selection candidates (data not shown), presumably because the ratio of composite likelihoods depends weakly on θ: change in θ appears to affect LIS and LN in Equation 1 to a similar degree.
Patterns similar to the outcome of incomplete selective sweeps may arise by a complete selective sweep: at an appropriate recombination distance from the position of the beneficial mutation that reached fixation, low variation and high frequency of derived alleles would be observed among chromosomes whose linkages to beneficial mutation were not broken by recombination. However, a normal level of variation will be observed among chromosomes that recombined away from the beneficial mutation. We therefore checked whether our candidate regions of incomplete selective sweeps overlap with those of complete selective sweeps in the RG sample detected by Pool et al. (2012) (343 regions listed in their table S13). Seventeen of our 42 clusters overlap with the candidate regions of complete sweeps (Table 2).
Next, we calculated iHS and nSL statistics for the same data set for which CLR was obtained above. Even though corrections were made to address the complexity of data (missing base calls and incomplete inference of ancestral/derived alleles; see Materials and Methods), many sites yielding very negative iHS appear to be false positives because clear haplotype structures predicted under incomplete sweeps are not observed at those loci (Figure S8). On the other hand, sites yielding very negative nSL are associated with a much clearer haplotype pattern. However, there are still cases of very unclear haplotype patterns detected by nSL (Figure S8). We could identify clusters of negative iHS and those of negative nSL, similar to clusters of large CLR above. However, the overall pattern of clustering for negative iHS or nSL is not clear, whereas very distinct clusters of large CLR were observed (Figure 5). Many sites generated large negative iHS or nSL by themselves without belonging to any cluster and we did not consider them as candidate loci under selection. We found that these isolated occurrences of large negative iHS/nSL and other sites with large negative iHS/nSL but without clear haplotype structure of incomplete sweep are associated with unusually small iHHA. Namely, stochastic fluctuation in haplotype structure surrounding the ancestral allele appears to frequently generate false-positive signatures of selection captured by iHS or nSL.
To examine whether the CLR, iHS, and nSL methods detect common candidate loci under selection, we adjusted the P-value cutoff of iHS or nSL for each chromosome arm so that the numbers of iHS or nSL clusters match that of the CLR in the same chromosome arm (Table S1 and Table S2). If a CLR cluster and an iHS or nSL cluster are not >50 kb away from each other, they are defined as overlapping candidates of selection. Of 25 CLR clusters that do not overlap with candidates of complete sweeps, 13 overlap with iHS clusters (Table 2). Ten of those 13 iHS clusters are also nSL clusters, reflecting a very high level of overlap between the iHS and nSL methods. There is only one case of coincidence between CLR and nSL peaks not being an iHS peak (excluding those overlapping with complete sweeps). Therefore, less than half of CLR peaks were detected also by the nSL method. Visual inspection of haplotype structures indicates that such candidate loci detected by all three methods tend to exhibit a much clearer pattern of incomplete sweeps than others (Figure S7). However, there are also loci detected by the CLR method only but with clear haplotype patterns (for example, near position 5,770,000 in 2R; Figure 5). We also identified a few peaks of negative nSL with clear haplotype patterns not overlapping with CLR or iHS peaks (for example, near position 3,886,000 in 2R; Figure 5). However, such cases are exceptional: if an nSL peak is not overlapping with the CLR or iHS peaks, it is more likely to show unclear than clear haplotype patterns (Figure S8).
Discussion
We developed a composite-likelihood method for detecting incomplete selective sweeps and inferring the location and strength of positive selection from DNA sequence polymorphism. As this method is built on analytic approximations to sampling probabilities under an explicit model of the evolutionary process, hypothesis testing and parameter estimation can be performed systematically, for example, allowing the estimation of the strength of selection. This approach also has the potential to be extended to incorporate more complex scenarios of incomplete sweeps if the sampling probabilities can be obtained as functions of additional parameters. On the other hand, statistical methods aiming to capture the extended haplotype such as the iHS and nSL tests (Voight et al. 2006; Ferrer-Admetlla et al. 2014) have an advantage of requiring fewer assumptions about the evolutionary process to be inferred (i.e., how directional selection occurs) and are also easier to implement the procedure and to interpret the result. We thus compared the performance of our CLR method and the extended haplotype method, using both simulated and actual sequence data.
Analysis of simulated data showed that our CLR approach achieves statistical power and accuracy in estimating the location of selection similar to those by the nSL method (Table 1), however, under the assumption that the true scaled recombination rate of the genomic region is known when generating the null distribution by neutral simulation. If a falsely lower estimate of the scaled recombination rate is used for a genomic region under test, which is likely true if an incomplete selective sweep left a polymorphism with long-range LD, it will greatly reduce the statistical power to detecting it as the cutoff value in the null distribution becomes larger. Such a large sensitivity of the CLR to the recombination rate (the level of linkage disequilibrium) is a major problem that needs to be addressed in future improvement of our approach. However, if local recombination rate or map distance is well estimated in advance over a large genomic region (much larger than typical sizes of sweep-affected areas), scaled recombination at a particular locus might be correctly inferred from observed polymorphism in the neighboring regions, given that LD over a large region is much less affected by local fluctuation, for example by selection. Namely, generating the null distribution with neutral simulation that yields the observed level of LD in data under test, as we suggested to correct the effect of unknown recombination rate, might be an unnecessarily conservative test, if the observed LD is definitely unusual (i.e., higher) compared to that in neighboring regions.
A related problem due to the sensitivity of our statistic to the level of linkage disequilibrium is the increased chance of detecting false-positive incomplete sweeps in the presence of nonstandard demography (Figure S4). Because various demographic processes can inflate the level of LD throughout the genome, which upwardly shifts the distribution of T1 in the absence of selection, obtaining the null distribution under the assumption of the standard neutral model can lead to erroneous detections of sweeps. Again, if the nature of (complex) demography affecting the data is not known, the false-positive detection might be controlled by the null distribution from simulated samples under the standard neutral model but adjusted to exhibit the level of LD observed in the data.
A more important result in the comparison between the CLR and iHS/nSL tests is that their performances are rather complementary to each other, as their outcomes are not so strongly correlated, especially for weak selection (α = 1000; Figure 3). It is probably because the two methods are designed to detect slightly different footprints of incomplete selective sweeps. Our method primarily captures joint frequency spectra at linked neutral loci for the two subsamples divided according to the S locus (Figure 2), whereas the iHS and nSL methods target the extended haplotype homozygosity, although these two signatures are obviously closely related through the reduction of polymorphism surrounding the putative beneficial allele.
As it was not as feasible to evaluate statistical significance of CLR tests by generating appropriate null distributions for a large number of genomic regions in D. melanogaster, we applied the CLR and iHS/nSL methods as outlier detection approaches. We evaluated the relative performance of the three methods by obtaining similar numbers of outliers (candidate loci) for each chromosome arm and visually inspecting haplotype structures surrounding the putative sites under selection. In general, the clearest haplotype patterns of incomplete selective sweeps were obtained when the loci were detected by all three methods. Candidates detected only by our CLR method exhibited relatively clean patterns compared to those detected by the iHS or nSL method (Figure 5, Figure S7, and Figure S8). Again this can be attributed to the gain of additional information from DNA sequence polymorphism in the CLR approach. Visual inspection also suggests that many false positives are detected by iHS because extended homozygosity surrounding the ancestral allele of the core SNP can be randomly reduced to very small values. Namely, while iHHD captures the hitchhiking effect of the beneficial allele, stochastic fluctuation of iHHA greatly increases the variance of iHHA/iHHD. In addition, if a small number, say n′, of sequences containing the derived allele of focal SNP are highly homozygous (e.g., hidden identity by descent) by chance while the other n1 − n′ sequences are heterozygous at the normal level, it can lead to a very large iHHD. Our approach is not affected by such problems, as our CLR does not simply depend on differences in the levels of variation between the two subsamples of data but compares neutral vs. selective scenarios as potential explanations for the subdivided pattern of polymorphism. The stochastic fluctuation of SNP density in the ancestral block appears to be less of a problem for nSL than for iHS, given that much clearer haplotype structures are detected by nSL than by iHS, probably because it does not use genetic map distance but the number of intervening SNPs for measuring the size of the extended haplotype.
As population genomic data are obtained predominantly by NGS platforms, missing or low-quality base calls in data may greatly affect the performance of evolutionary inferences from DNA sequence polymorphism. It is straightforward to calculate sampling probability under both neutral and selective hypotheses given the configuration of missing bases at each site in the data. Therefore, our CLR approach can be applied to data with an arbitrary frequency of missing bases without systematic problems. On the other hand, it is not clear how to handle missing bases in quantifying the extended homozygosity for the iHS or nSL test. We skipped the site containing a missing base in calculating the extension of homozygosity for a pair of sequences because clear haplotype structure of an incomplete sweep could not be identified otherwise. It is not clear how this procedure would affect the performance of the iHS test.
In conclusion, we proposed a composite-likelihood method for detecting incomplete selective sweeps and demonstrated that it achieves improvements in parameter estimation and ability to capture clear haplotype patterns compatible with incomplete sweeps compared to long-range haplotype tests. Although it has a disadvantage in not being robust to uncertainty in scaled recombination rates and complex demography, our composite-likelihood ratio provides information that is not captured by an advanced haplotype-based method using nSL. We thus recommend that both CLR and nSL be used together to maximize the chance of detecting true targets of selection. As incomplete selective sweeps provide excellent opportunities to estimate the strength and location of selection, due to the presence of ancestral polymorphism in the data, compared to complete sweeps, these methods will contribute to broadening our understanding of adaptive evolution in nature. In the framework of the likelihood-ratio test, we may conceive extension of this approach to study further details of incomplete selective sweeps beyond simple confirmation of positive selection and basic parameter estimation. For example, recent analysis predicted that many beneficial mutations are likely to stall at intermediate frequencies due to heterozygote advantage (Sellis et al. 2011). If this process generates sampling probabilities distinct from that left by simple directional selection with incomplete dominance, we may detect it under the current framework of the composite-likelihood test.
Supplementary Material
Acknowledgments
This research was supported by the Global Top5 Grant of Ewha Womans University 2013 and the National Research Foundation of Korea grants 2012R1A1A2004932 (to Y.K.).
Appendix
Derivation of and
We consider a constant-sized population of N diploid individuals that reproduce in discrete generations according to the Wright–Fisher model, thus equivalent to a population of 2N haploid individuals. Assume that mutation to a beneficial allele occurred at position x of a chromosome at time T = τ (generations counted backward in time) in the past. At the time of sampling (T = 0), this mutant allele reaches an intermediate frequency β in the population. A random sample of n chromosomes is assumed to contain n1 and n2 = n − n1 copies of the beneficial and the ancestral allele, respectively, that define the corresponding partition of the sample into two subsamples as illustrated in Figure 1. Let k1 and k2 be the counts of the derived allele in respective subsamples at a neutrally evolving site at position x − d or x + d. The probability of observing k1 and k2 jointly is given by
| (A1) |
where is the probability density of the derived allele frequency at the time of beneficial mutation (T = τ). is the probability of sampling k2 derived alleles in a sample of n2 chromosomes in a neutrally evolving population in which frequency of the allele drifted for τ generations starting from p. During the course of a selective sweep, the deterministic change of the linked neutral allele frequency among chromosomes carrying the ancestral allele at the S locus (frequency pA in the “ancestral background”) is predicted to be small (Stephan et al. 1992; Meiklejohn et al. 2004). A moderate deterministic change in pA occurs while β < 0.8, the range to which our method applies (Figure S9). We, however, ignore this change. We also ignore the change of allele frequency by genetic drift in the ancestral background, assuming and obtain
| (A2) |
Namely, we assume that the subsample of n2 chromosomes effectively captures the ancestral polymorphism at the time of beneficial mutation. Next, is the probability of observing k1 copies of the derived allele at position d in the subsample of n1 sequences that carry the beneficial allele. Strictly, this probability must be a function of the frequency of the beneficial allele at the time of sampling. However, as the frequency of the neutral allele among chromosomes carrying the beneficial allele (i.e., in the “beneficial background”) is known to change drastically only at the early stage of hitchhiking when the frequency of the beneficial allele is low and then change little until the fixation of the beneficial allele (Stephan et al. 1992), we approximate by sampling probability for the case of the complete selective sweep. We multiply and inside the integral of (A1), assuming that the frequency of linked neutral alleles in the beneficial background is distributed independently of possible stochastic change in allele frequency in the ancestral background and that chromosomes are sampled independently in the two genetic backgrounds. In reality, the “migration” of lineages by recombination during the selective sweep may cause correlated stochastic changes of allele frequencies in the two backgrounds. However, we ignore such complications, as the stochastic fluctuation of p in the ancestral background by genetic drift is ignored in the first place (see above).
Nielsen et al. (2005) and Etheridge et al. (2006) provided approximate solutions that allow the derivation of the above sampling probability as a function of neutral allele frequency, p, at the time of the beneficial mutation. Using a star-like genealogy approximation, Nielsen et al. (2006) obtained the probability of observing k1 derived alleles at the neutral locus from the sample of n1 chromosomes after a selective sweep,
| (A3) |
where is the probability that k of n distinct ancestral lineages at T = τ carry the derived mutant alleles and is the probability that k of n lineages at T = 0 escape the sweep by recombining away from the beneficial allele, with the escaping probability per lineage given by
Replacing (A2) and (A3) into (A1), we obtain
| (A4) |
where is the probability of having k derived alleles in a sample of n chromosomes at time τ . can be given by assuming the population at this time is under neutral equilibrium, or by the proportion of polymorphic sites with k derived alleles in the data, namely assuming that the distribution of the derived allele frequency at time τ is identical to that observed at present. The latter approach of using the empirical frequency spectrum was suggested by Nielsen et al. (2005) to correct for nonequilibrium demography. These two approximations are bases of CLR test options A and B, respectively.
Alternatively, we may derive the sampling probability from the work of Etheridge et al. (2006), which showed that n lineages at a linked neutral locus sampled at the time of a beneficial allele’s fixation are divided into three parts: l late recombinants, e early recombinants, and n − l − e nonrecombinants. Given the selection coefficient s and recombination rate r, the joint distribution of l and e, P(l, e), follows equation 2.7 of Etheridge et al. (2006). However, this result in terms of genealogical structure needs to be translated into sampling probability by considering the transmission of mutant alleles along the lineages. The probability of sampling k derived alleles can be obtained separately in the following four cases.
First, consider the case in which the beneficial allele appears on a chromosome carrying the derived allele at the neutral locus. In addition, the ancestor of early recombinants carries the ancestral allele. Therefore, the sample contains at least n − l − e derived alleles and at least e ancestral alleles. In addition, assume that in l late recombinants, there are ld derived alleles and l − ld ancestor alleles. Then, the total number of derived allele in the sample is k = n – e – (l – ld). Since ld = l – (n – e – k), the probability for this case is
| (A5) |
where p is the initial frequency of the derived allele before hitchhiking. In the case that the ancestor of early recombinants carries the derived allele,
| (A6) |
Next, the beneficial mutation is now assumed to appear on a chromosome carrying the ancestral allele of the neutral locus. Probabilities that there are k derived alleles in the sample if the ancestor of early recombinants carries the ancestral and the derived allele are, respectively,
| (A7) |
and
| (A8) |
Since these cases are mutually exclusive, the final solution for sampling probability for a complete selective sweep is after the above probabilities are weighted accordingly:
| (A9) |
Using Equations A2 and A9, Equation A1 is now turned into our second approximation:
| (A10) |
Footnotes
Communicating editor: R. Nielsen
Supporting information is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.175380/-/DC1.
Literature Cited
- Akey J. M., 2009. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Res. 19: 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton N. H., 1998. The effect of hitch-hiking on neutral genealogies. Genet. Res. 72: 123–133. [Google Scholar]
- Biswas S., Akey J. M., 2006. Genomic insights into positive selection. Trends Genet. 22: 437–446. [DOI] [PubMed] [Google Scholar]
- Chan A. H., Jenkins P. A., Song Y. S., 2012. Genome-wide fine-scale recombination variation in Drosophila melanogaster. PLoS Genet. 8: e1003090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J. M., Ratnappan R., Bailin S., 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8: e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge A., Pfaffelhuber P., Wakolbinger A., 2006. An approximate sampling formula under genetic hitchhiking. Ann. Appl. Probab. 16: 685–729. [Google Scholar]
- Ewing G., Hermisson J., 2010. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics 26: 2064–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay J. C., Wu C. I., 2000. Hitchhiking under positive Darwinian selection. Genetics 155: 1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Admetlla A., Liang M., Korneliussen T., Nielsen R., 2014. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 31: 1275–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-X., Li W.-H., 1993. Statistical tests of neutrality of mutations. Genetics 133: 693–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier M., Vitalis R., 2012. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 28: 1176–1177. [DOI] [PubMed] [Google Scholar]
- Hill W. G., Robertson A., 1968. Linkage disequilibrium in finite populations. Theor. Appl. Genet. 38: 473–485. [DOI] [PubMed] [Google Scholar]
- Hudson R. R., Bailey K., Skarecky D., Kwiatowski J., Ayala F. J., 1994. Evidence for positive selection in the superoxide dismutase (Sod) region of Drosophila melanogaster. Genetics 136: 1329–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kim Y., 2008. Detecting local adaptation using the joint sampling of polymorphism data in the parental and derived populations. Genetics 179: 1713–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N. L., Hudson R. R., Langley C. H., 1989. The “hitchhiking effect” revisited. Genetics 123: 887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Nielsen R., 2004. Linkage disequilibrium as a signature of selective sweeps. Genetics 167: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Stephan W., 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160: 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J., Haigh J., 1974. The hitch-hiking effect of a favorable gene. Genet. Res. 23: 23–35. [PubMed] [Google Scholar]
- Meiklejohn C. D., Kim Y., Hartl D. L., Parsch J., 2004. Identification of a locus under complex positive selection in Drosophila simulans by haplotype mapping and composite-likelihood estimation. Genetics 168: 265–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., 2005. Molecular signatures of natural selection. Annu. Rev. Genet. 39: 197–218. [DOI] [PubMed] [Google Scholar]
- Nielsen R., Williamson S., Kim Y., Hubisz M. J., Clark A. G., et al. , 2005. Genomic scans for selective sweeps using SNP data. Genome Res. 15: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool J. E., Corbett-Detig R. B., Sugino R. P., Stevens K. A., Cardeno C. M., et al. , 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8: e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M., 2002. The signature of positive selection at randomly chosen loci. Genetics 160: 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada H., Ramírez U. E., Rozas J., Aguadé M., 2003. Large-scale adaptive hitchhiking upon high recombination in Drosophila simulans. Genetics 165: 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti P. C., Reich D. E., Higgins J. M., Levine H. Z., Richter D. J., et al. , 2002. Detecting recent positive selection in the human genome from haplotype structure. Nature 419: 832–837. [DOI] [PubMed] [Google Scholar]
- Sabeti P. C., Schaffner S. F., Fry B., Lohmueller J., Varilly P., et al. , 2006. Positive natural selection in the human lineage. Science 312: 1614–1620. [DOI] [PubMed] [Google Scholar]
- Saunders M. A., Good J. M., Lawrence E. C., Ferrell R. E., Li W.-H., et al. , 2006. Human adaptive evolution at myostatin (GDF8), a regulator of muscle growth. Am. J. Hum. Genet. 79: 1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellis D., Callahan B. J., Petrov D. A., Messer P. W., 2011. Heterozygote advantage as a natural consequence of adaptation in diploids. Proc. Natl. Acad. Sci. USA 108: 20666–20671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan W., 2010. Detecting strong positive selection in the genome. Mol. Ecol. Res. 10: 863–872. [DOI] [PubMed] [Google Scholar]
- Stephan W., Wiehe T. H. E., Lenz M. W., 1992. The effect of strongly selected substitutions on neutral polymorphism: analytic results based on diffusion theory. Theor. Popul. Biol. 41: 237–254. [Google Scholar]
- Stephan W., Yun Y. S., Langley C. H., 2006. The hitchhiking effect on linkage disequilibrium between linked neutral loci. Genetics 172: 2647–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomajian C., Ajioka R. S., Jorde L. B., Kushner J. P., Kreitman M., 2003. A method for detecting recent selection in the human genome from allele age estimates. Genetics 165: 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight B. F., Kudaravalli S., Wen X., Pritchard J. K., 2006. A map of recent positive selection in the human genome. PLoS Biol. 4: e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.