Abstract
Transcription and splicing are coordinated processes in mammalian cells. We have used affinity chromatography with immobilized transcription elongation factor SII to purify a protein complex that contains core RNA polymerase II (RNA Pol II), the general transcription initiation factors, and several splicing factors, including the U1, U2, and U4 small nuclear RNPs, the U2AF65, and serine/arginine-rich proteins. The splicing factors and the transcription machinery co-purify through a gel filtration column and co-immunoprecipitate in experiments using an anti-U2AF65 antibody, indicating that they are part of a unique complex. Although the RNA Pol II-containing complex does not possess splicing activity, it can complement small nuclear RNP-inactivated extracts and can promote the formation of a pre-spliceosome complex. Because interactions between components of the splicing and transcription machineries occur in the context of a complex containing a hypophosphorylated RNA Pol II capable of initiating transcription, our results suggest that the coupling between transcription and splicing begins before transcription initiation.
The splicing of pre-mRNA in mammalian cells is the nuclear process during which introns are removed from primary transcripts synthesized by RNA polymerase II (RNA Pol II).1 Splicing is achieved by the spliceosome, a large macromolecular complex, composed of small nuclear ribonucleoprotein (snRNP) particles and additional proteins including members of the serine/arginine-rich (SR) protein family (reviewed in Refs. 1 and 2). Cytological studies have revealed that splicing can occur in a co-transcriptional manner (see Refs. 3 and 4 for examples) and that factors necessary for splicing are recruited to sites of transcription (5–7). A growing body of data indicates that coupling between transcription and a variety of RNA maturation events may be achieved through the C-terminal domain (CTD) of the RPB1 subunit of RNA Pol II (reviewed in Refs. 8–11). The CTD is a seven-amino acid motif tandemly repeated 52 times in humans and 26–27 times in yeast. It is highly conserved among eukaryotic organisms and is subject to reversible phosphorylation during the transcription cycle (reviewed in Refs. 12 and 13). Two isoforms of RNA Pol II exist in vivo, namely RNA Pol IIA and IIO. RNA Pol IIA possesses a hypophosphorylated CTD and preferentially enters the preinitiation complex, whereas RNA Pol IIO harbors an extensively phosphorylated CTD and is found in the elongation complex (13, 14). Biochemical studies indicate that RNA Pol II physically interacts with components involved in capping (15–18), polyadenylation (19), and splicing (20–24). Whereas a phosphorylated CTD is essential for the interaction with capping enzymes, the polyadenylation factor CPSF is brought to the transcription complex through an interaction with TFIID and transferred to the phosphorylated CTD after transcription initiation (19, 25). The phosphorylated RNA Pol IIO isoform has been found to associate with splicing factors and has been identified in active spliceosomes (21–23). Truncation of the CTD leads to inefficient capping, polyadenylation, and splicing in cultured mammalian cells (16, 19). Both the overexpression of phosphorylated CTD peptides (26) and the use of antibodies directed against the CTD have been shown to inhibit splicing (20, 23, 24). These results suggest that the phosphorylated CTD of RNA Pol II provides an interface for the recruitment of splicing factors. Whereas the mechanisms by which the CTD presents splicing factors to the nascent pre-mRNA remain unclear, a recent study suggests that the CTD might play an active role (27).
Initiation of transcription by RNA Pol II can be achieved in vivo by large multiprotein complexes called RNA Pol II holoenzymes (reviewed in Refs. 28 and 29). Various forms of holoenzymes have been isolated from mammalian cells and all have been found to contain core RNA Pol II and a number of general transcription factors that are required for accurate transcription initiation in vitro (30). Here, we report that snRNPs, U2AF65, and SR proteins are part of a RNA Pol II holoenzyme (hereafter called RNA Pol II-containing complex) purified by affinity chromatography using transcription elongation factor SII. The splicing factors associated with this RNA Pol II-containing complex can function in splicing reactions and can promote the assembly of a pre-spliceosome complex in vitro. Our data support a model in which coordination between transcription and splicing begins prior to transcription initiation by RNA Pol II.
EXPERIMENTAL PROCEDURES
Affinity Purification of RNA Polymerase II-containing Complexes
RNA Pol II-containing complexes were purified from HeLa whole cell extract as described previously (31). Briefly, a HeLa whole cell extract in ACB buffer (10 mM Hepes, pH 7.9, 0.1 mM EDTA, 0.1 mM dithiothreitol, and 10% glycerol) containing 50 mM NaCl was loaded on a GST-SII affinity column. The column was washed with ACB buffer containing 50 mM NaCl and eluted with ACB buffer containing 300 mM NaCl. An equivalent GST column was used as a control. The eluate from the GST-SII affinity column was further purified by chromatography through a Sepharose CL2B gel filtration column in ACB containing 50 mM NaCl and 20% glycerol (31). Samples were concentrated 5–10-fold before use in Western blot and transcription assays. The RNA Pol II-containing complexes were found to elute slightly faster than blue dextran on the CL2B column.
Transcription Assay
Transcription reactions were performed as described by Pan et al. (31) using the adenovirus major late promoter fused to a G-less cassette as template (32). Run-off transcripts were analyzed on 8% denaturating polyacrylamide gels.
Immunoprecipitation
The GST-SII column eluate (10 μg) was treated with RNase A (0.5 μg) for 30 min at room temperature and then incubated with protein A-Sepharose beads (25 μl), which had previously been bound to 5 μl of either anti-U2AF65 or preimmune rabbit serum in a final volume of 1 ml of immunoprecipitation buffer (20 mM Hepes, pH 7.9, 150 mM KOAc, 1 mM dithiothreitol, 1 mM EDTA, 20% glycerol, and 0.5% Nonidet P-40) supplemented with bovine serum albumin (625 μg). The beads were washed three times with immunoprecipitation buffer, resuspended in loading buffer, and boiled for 5 min. The proteins were separated using a 12.5% SDS gel and were transferred to polyvinylidene difluoride filters, which were then probed with specific antibodies.
S100 Complementation Assay
The 32P-labeled RNA transcripts used in the splicing assays were prepared as described (33). The S100 fraction (34) and the SR proteins (35) were prepared as previously described. Either 87.5, 175, or 350 ng of purified SR proteins, or 2.5, 5, and 10 μg of GST-SII eluate were used to complement the splicing activity of the S100 extract.
snRNA Complementation Assays
Specific snRNAs were removed from HeLa nuclear extracts and GST-SII column eluates by incubation with oligonucleotides (25 ng/μl) complementary to different snRNAs and RNase H (1 unit/μl) for 90 min at 37 °C (36). For the U4 depletion, ATP (0.5 mM) and creatine phosphate (20 mM) were also included in the depletion procedure. The sequences of the oligonucleotides used are available upon request. Splicing assays were performed as described for the S100 complementation assay, except that nuclear extracts or depleted nuclear extracts were used as sources of splicing factors and were complemented using 10 μg of either GST-SII eluates or snRNA-depleted GST-SII column eluates as described in the figure legends.
Splicing Complex Formation
Radiolabeled adenovirus transcript (~5 fmol) (33) was incubated with nuclear extract (2.5 μl), U2-depleted nuclear extract (2.5 μl), GST-SII column eluate (2.5 μg), or U2-depleted GST-SII column eluate (2.5 μg) under conditions identical to those used for the splicing reactions. Aliquots were collected at different time points and analyzed on 3.5% acrylamide:0.043% bisacrylamide native gels.
RESULTS
Affinity Purification of a Human RNA Polymerase II-containing Complex That Also Contains Splicing Factors
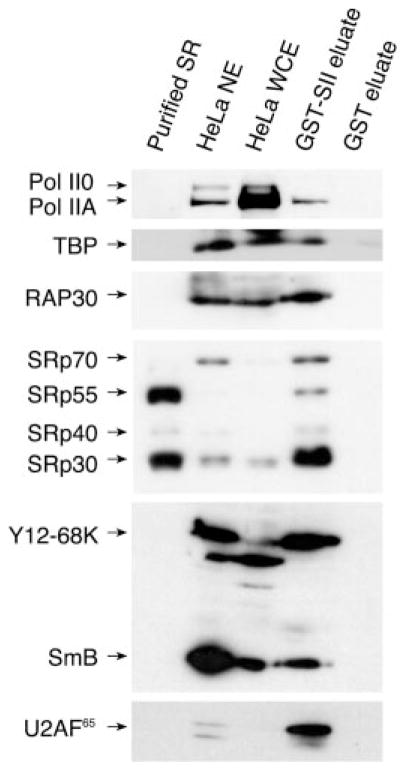
The transcription elongation factor SII (also called TFIIS) has previously been used as a ligand in protein-affinity chromatography for the purification of a RNA Pol II-containing complex that contains core RNA Pol II and the general transcription factors (31) as well as capping and polyadenylation factors (16, 19). The size of this complex is estimated to be in the 2–4-MDa range, sedimenting as a 70 S particle (31). The RNA Pol II-containing complex is sensitive to high salt concentrations, and increasing the Na+/K+ concentration above 0.3 M causes its dissociation in subcomplexes (data not shown). We show here that the eluate from a GST-SII column, but not from a GST control column, contains SR proteins, at least one Sm protein, the Y12-68K polypeptide, and the U2AF65 splicing factor (Fig. 1). Although the eluate contains the bulk of U2AF65 and is enriched for a subset of SR proteins, it contains a smaller proportion of the total Y12-68K and Sm proteins. In agreement with Greenblatt and co-workers (31), the general transcription initiation factors including TBP, TFIIB, TFIIE, TFIIF and TFIIH, and core RNA Pol II are present in the GST-SII column eluate (Fig. 1 and data not shown). As revealed by Western blot experiments with the N-20 antibody that specifically recognizes the N terminus of RPB1 (Fig. 1, first panel), the RNA Pol II in the complex corresponds to the hypophosphorylated IIA form (i.e. the form involved in transcription initiation), which we estimated to be 100-fold more abundant than the hyper-phosphorylated IIO form in the complex preparation. This observation was confirmed by Western blot experiments with antibodies that specifically recognize either the IIA (Ab 8WG16) or the IIO (Ab CC3) forms of RNA Pol II. By comparing the ratios of RNA Pol IIA over the total amount of proteins in the whole cell extract and the SII column eluate, we estimated the purification factor of the SII affinity column to be approximately 1000-fold. The association of splicing factors with a complex containing the hypophosphorylated IIA form of RNA Pol II and capable of transcription initiation (see below) contrasts with previous reports that have documented an interaction between splicing factors and RNA Pol IIO or a phosphorylated CTD (20, 22).
Fig 1. Association of splicing factors with a human RNA Pol II-containing complex.
SR proteins, the U2AF65 factor, the Y12-68K polypeptide and Sm proteins co-purify with a RNA Pol II-containing complex on a GST-SII affinity column. Western blot analysis of purified SR proteins (lane 1), HeLa nuclear extract (NE) (lane 2), HeLa whole cell extract (WCE) (lane 3), the GST-SII column eluate (lane 4), and the GST column eluate (lane 5) was performed using antibodies directed against the N terminus of the RPB1 subunit of RNA Pol II (Ab N-20) and detecting both the hypophosphorylated (IIA) and hyperphosphorylated (IIO) forms. Antibodies directed against different general transcription factors were also used (only experiments using anti-TBP and anti-RAP30 are shown). In addition, the monoclonal Ab104 and Y12 antibodies revealed the presence of SR, Sm, and Y12–68K proteins.
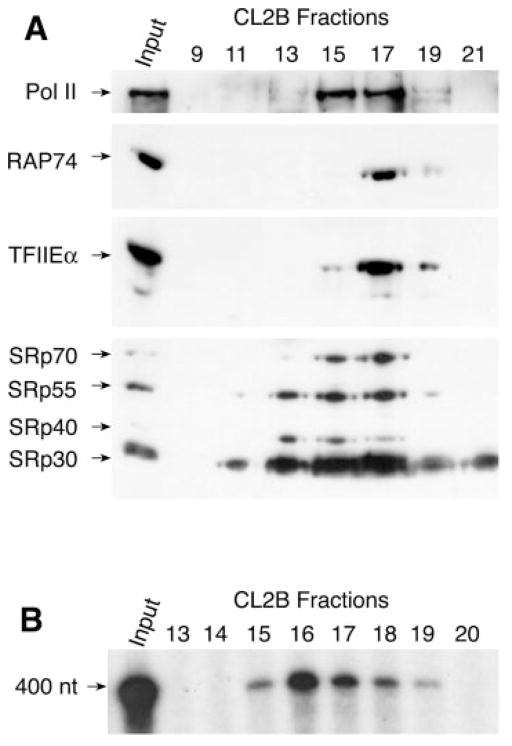
To confirm that the splicing and transcription factors are part of the same multiprotein complex, we submitted the GST-SII column eluate to a gel filtration column and an immunoprecipitation experiment. Following a Sepharose CL2B gel filtration column, the splicing factors continue to co-purify with core RNA Pol II and the general transcription factors RAP74 and TFIIEα (Fig. 2A), indicating that all these splicing and transcription factors are either part of a unique complex or are distributed in a collection of complexes of similar molecular masses. Virtually all the polypeptides that had bound to the GST-SII column co-migrated on the Sepharose CL2B column (data not shown), again indicating that the proteins that had bound to SII are part of a single complex or a set of complexes of similar molecular weight. As determined by the synthesis of a 400-nucleotide run-off transcript accurately initiated from the adenovirus major late promoter in vitro, the transcriptional activity found in the GST-SII column eluate also co-purifies with the splicing factors, core RNA Pol II, and the general initiation factors (Fig. 2B). In addition, and notably, core RNA Pol II is recovered from both the GST-SII column eluate and the crude whole cell extract in immunoprecipitation experiments using an anti-U2AF65 antibody but not a control antibody (Fig. 3). Extensive treatment with RNase A does not dissociate U2AF65 from RNA Pol II in co-immunoprecipitation assays (Fig. 3), clearly demonstrating that the splicing machinery is primarily associated with the complex via protein-protein interactions. Together, these results provide compelling evidence that several components of the splicing machinery are tightly associated with a RNA Pol II-containing complex that is fully capable of initiating transcription in vitro, suggesting that coordination between splicing and transcription begins before transcription initiation.
Fig. 2. Co-migration of the SR splicing factors with the RNA Pol II-containing complex and transcriptional activity on a gel filtration column.
A, co-purification of SR proteins with the general initiation factors and core RNA Pol II. The GST-SII column eluate was chromatographed through a Sepharose CL2B column, and the resulting concentrated fractions were analyzed by Western blot using antibodies directed against RPB1 (form IIA), RAP74, TFIIEα, and SR proteins. B, the RNA Pol II-containing complex is active in in vitro transcription. The GST-SII column eluate was chromatographed through a Sepharose CL2B gel filtration column, and the resulting fractions were tested in transcription assays in vitro. The position of the run-off transcript (400 nt) is indicated.
Fig. 3. Immunoprecipitation of the RNA Pol II-containing complex using anti-U2AF65 before, or following, an extensive treatment with RNase A.
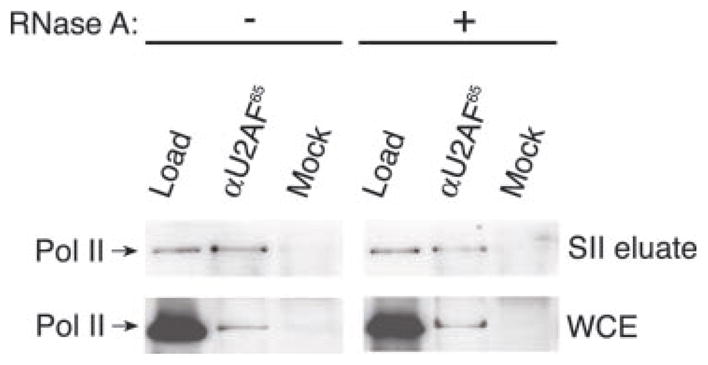
The SII-column eluate (top panel) and the crude whole cell extract (WCE; bottom panel) were either treated (+) or not treated (−) with RNase A and then immunoprecipitated using either an anti-U2AF65 (αU2AF65) or a pre-immune (Mock) antibody. A proportion (5%) of the loading reactions (Load) and the whole pellets were then analyzed by Western blot using an anti-Pol IIA antibody (Ab 8WG16).
The Splicing Factors Associated with the RNA Polymerase II-containing Complex Can Function in Splicing Reactions
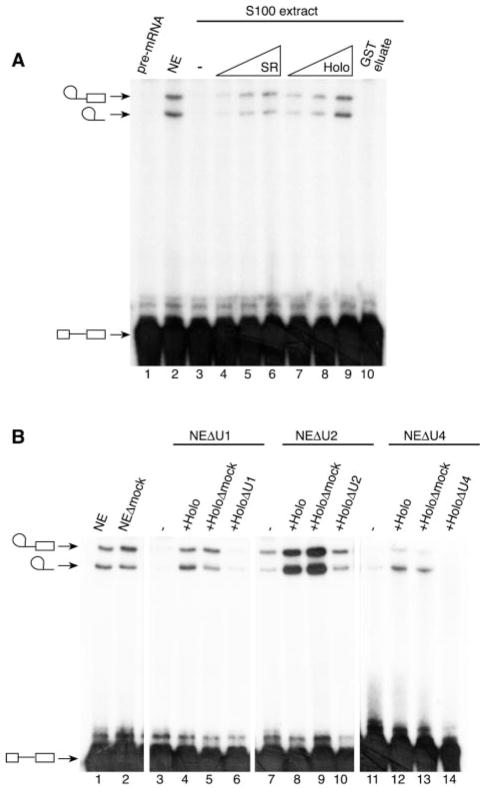
Although the RNA Pol II-containing complex displays transcriptional activity and contains many splicing factors, it failed to splice a simple pre-mRNA in vitro (data not shown), suggesting that at least one essential splicing factor is missing and/or that some factors in the preparation are inactive. To assess the activity of the SR proteins present in the RNA Pol II-containing complex, we used the GST-SII column eluate to complement splicing reactions performed with a HeLa S100 extract. S100 extracts are known to contain all the factors required for accurate splicing of a pre-mRNA in vitro except the SR proteins (37). Fig. 4A shows the human RNA Pol II-containing complex complementing the S100 extract (lanes 7–9) with an efficiency comparable with that of purified SR proteins (lanes 4–6), whereas the eluate from a GST control column does not (lane 10). Clearly, the SR proteins that are associated with the RNA Pol II-containing complex can function in pre-mRNA splicing in vitro.
Fig. 4. Functional SR proteins and U1, U2, and U4 snRNPs associated with the RNA Pol II-containing complex.
A, the RNA Pol II-containing complex (Holo) can complement the splicing activity of an S100 extract. Splicing reactions were performed using either a HeLa nuclear extract (lane 2) or an S100 extract alone (lane 3), or supplemented with increasing amounts of purified SR proteins (lanes 4–6), the GST-SII column eluate (lanes 7–9), or the GST column eluate (lane 10). Lane 1 contained the pre-mRNA alone. B, the RNA Pol II-containing complex (Holo) can complement the splicing activity of nuclear extracts depleted of U1, U2, and U4 snRNAs. Splicing assays were performed using either an untreated nuclear extract (NE; lane 1) or nuclear extracts depleted using a mock oligonucleotide (NEΔmock; lane 2), the U1-specific oligonucleotide (NEΔU1; lanes 3–6), the U2-specific oligonucleotide (NEΔU2; lanes 7–10), or the U4-specific oligonucleotide (NEΔU4; lanes 11–14). Some reactions were complemented using the RNA Pol II-containing complex (lanes 4, 8, and 12) or the RNA Pol II-containing complex depleted using the mock oligonucleotide (lanes 5, 9, and 13), the U1-specific oligonucleotide (lane 6), the U2-specific oligonucleotide (lane 10), or the U4-specific oligonucleotide (lane 14). The positions of the splicing products are indicated.
The presence of the Y12-68K and the Sm proteins in the GST-SII column eluate, as detected by Western blot analysis (see Fig. 1), suggests that at least the U1 snRNP particle is associated with the RNA Pol II-containing complex. To identify the snRNPs present in the complex, we used a splicing assay performed with nuclear extracts that had specific snRNAs inactivated by treatment with RNase H and specific antisense oligonucleotides (36). Fig. 4B shows that the RNA Pol II-containing complex preparation can stimulate the splicing activity of nuclear extracts depleted of U1, U2, or U4 snRNAs (lanes 4, 8, and 12). In each case, restoration of splicing activity was due to the presence of U1, U2, or U4 snRNAs in the RNA Pol II-containing complex, since GST-SII column eluates depleted of U1, U2, or U4 did not restore the splicing activity of nuclear extracts depleted of the same snRNA (compare lane 3 with lane 6, lane 7 with lane 10, and lane 11 with lane 14 in Fig. 4B). Because the bulk of U4 snRNA exists in association with U5 and U6 in the form of a tri-snRNP complex (2), these data suggest that the human RNA Pol II-containing complex is associated with all the spliceosomal snRNPs.
The Splicing Factors Associated with the RNA Polymerase II-containing Complex Can Form a Pre-spliceosomal Complex
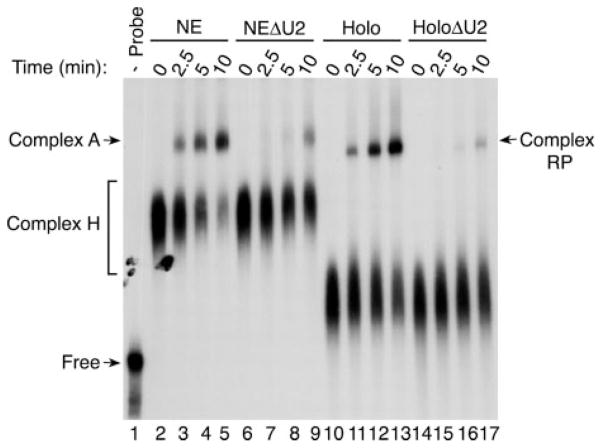
To determine whether the splicing factors associated with the RNA Pol II-containing complex can support the assembly of splicing complexes, the GST-SII column eluate was incubated with a radiolabeled transcript containing an adenovirus splicing cassette. Complex formation was analyzed on native polyacrylamide gels. Fig. 5 shows that the factors in the RNA Pol II-containing complex can support the assembly of complexes with an efficiency nearly comparable with that of a nuclear extract (compare lanes 2–5 with lanes 10–13). One of these complexes (RNA polymerase) nearly co-migrates with spliceosomal complex A, and its formation is severely compromised by inactivation of U2 snRNA (compare lanes 6–9 with 14–17). These results indicate that the splicing factors present in the RNA Pol II-containing complex preparation can be efficiently utilized to promote the assembly of a pre-spliceosomal complex.
Fig. 5. Efficient formation of a U2-dependent complex by splicing factors associated with the RNA Pol II-containing complex.
Nuclear extracts (lanes 2–5) and the RNA Pol II-containing complex (Holo; lanes 10–13) were incubated with an adenovirus pre-mRNA containing a splicing cassette. A nuclear extract (NEΔU2; lanes 6–9) and the RNA Pol II-containing complex (HoloΔU2; lanes 14–17) depleted of snRNA U2 were used as controls. Splicing complex formation was monitored over time as indicated. The positions of the H and A complexes formed in nuclear extracts are indicated. The RNA Pol II-containing complex also leads to the assembly of a U2-dependent complex (RP) that nearly migrates with the A complex.
DISCUSSION
Because the physical interaction between RNA Pol II and splicing factors was shown initially to occur with a phosphorylated CTD (20–23), coupling between transcription and splicing was thought to begin during transcription elongation. The fact that a RNA Pol II-containing complex exists in association with various components of the splicing machinery and that the CTD in the RNA Pol II-containing complex is hypophosphorylated implies that coupling can be initiated before transcription begins. Thus, a phosphorylated CTD is not absolutely required for the interaction of splicing factors with transcription complexes, suggesting that splicing factors can associate with an as yet unidentified component of the transcription machinery. Interactions between splicing factors and the general transcription machinery, including the transcription initiation factors TBP (38) and TFIIH,2 have been reported. The splicing components that interact with the RNA Pol II-containing complex include SR proteins and snRNPs, factors that also associate with hyperphosphorylated RNA Pol II (20, 22, 23). In addition, we document for the first time the interaction of U2AF65 with transcription complexes. Thus, as shown by the capacity of the RNA Pol II-containing complex to support splicing complex formation, all the components required for committing a pair of splice sites are part of the complex. However, the complex preparation cannot support splicing of a pre-mRNA, as if it is lacking one (or some) component(s) of the spliceosome. The identity of the splicing factor(s) that is lacking from the complex remains unknown. As is the case for the polyadenylation factor CPSF (25), we propose that CTD phosphorylation may trigger the transfer of splicing factors from the non-CTD site to the phosphorylated CTD (see Fig. 6 for a model). Splicing factors may then escort the elongating RNA Pol II until they are loaded onto the nascent pre-mRNA when appropriate RNA sites emerge from the elongating enzyme.
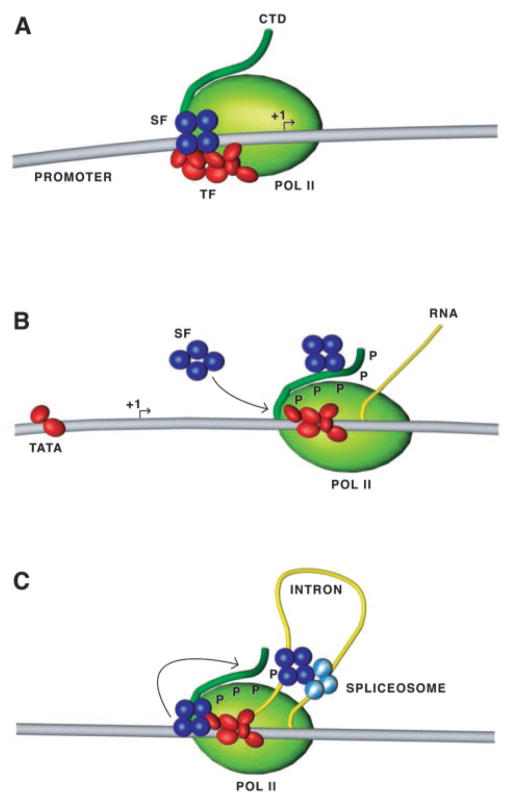
Fig. 6. A model for transcription-coupled splicing.
A, splicing factors (SF) involved in pre-spliceosomal complex formation are part of the RNA Pol II-containing complex in association with a non-CTD site located either on a transcription factor (TF) or core RNA Pol II (Pol II). B, after transcription initiation, the CTD of RNA Pol II is phosphorylated, and the splicing factors are transferred to the hyperphosphorylated CTD. The non-CTD site then becomes available for the recruitment of a new set of splicing factors. C, the position of the CTD near the RNA exit channel of RNA Pol II allows binding and unloading of the splicing factors to the nascent transcript and formation of the spliceosome. Splicing factors at the non-CTD site could be transferred to the phosphorylated CTD in preparation for splicing of the next intron.
While our results suggest that the coupling between transcription and splicing occurs before transcription is initiated, it is difficult to envision how the initial loading of splicing factors can sustain the splicing of multiple introns on a single pre-mRNA. An attractive possibility is that the non-CTD portion of the RNA Pol II-containing complex that interacts with splicing factors facilitates the continuous recruitment of splicing factors to the elongating enzyme as previous sets of splicing factors are transferred to the CTD (see Fig. 6). Because the CTD and the RNA exit channel are closely juxtaposed in the initiation complex (39, 40), transit through the CTD may facilitate their unloading to the nascent pre-mRNA. If not loaded rapidly after transcription, secondary structure formation on the pre-mRNA may compromise the recognition of splicing signals by splicing factors.
The association of splicing factors with a RNA Pol II-containing complex capable of initiating transcription also suggests that transcription initiation may play a critical role in the control of splicing. Consistent with this view, Cramer et al. (41, 42) have shown that the identity of the promoter can influence the SR-dependent alternative splicing of a fibronectin pre-mRNA. Although recent studies have linked such effects to differences in the processivity of polymerase complexes (43, 44), it remains possible that various forms of RNA Pol II-containing complexes with different subsets of splicing factors may be recruited to different promoters in preparation for the accurate regulation of downstream splicing events. However, regulatory splicing factors that are recruited in initiation complexes may be used in the constitutive splicing of introns before the transcription machinery reaches the alternative splicing unit. The continuous recruitment of the same subset of regulatory splicing factors may be achieved if the assembly of transcription initiation complexes localizes transcription sites near regions that are specifically enriched in such factors. Thus, the model posits that the identity of the splicing regulators would be determined by interactions with transcription factors bound to the promoter. During transcription elongation, the phosphorylated CTD portion of RNA Pol II would act as a transit platform, essential for the efficient downloading of splicing factors to the nascent pre-mRNA.
Acknowledgments
We are grateful to Jack Greenblatt and members of our laboratories for helpful discussions. We thank J. Patton for kindly providing anti-U2AF65 antibody. We thank Will Home for comments on the manuscript and Diane Bourque for help with the figures.
Footnotes
This work was supported by grants from the Canadian Institutes of Health Research (to B. Coulombe and B. Chabot), the Cancer Research Society, Inc. (to B. Coulombe), and the National Cancer Institute of Canada (to B. Chabot).
The abbreviations used are: Pol II, polymerase II; SR, serine/arginine-rich; snRNP, small nuclear ribonucleoprotein; CTD, C-terminal domain; GST, glutathione S-transferase; Ab, antibody; TF, transcription factor.
J. M. Egly, personal communication.
References
- 1.Kramer A. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 2.Moore MJ, Query CC, Sharp PA. In: The RNA World: the Nature of Modern RNA Suggests a Prebiotic RNA World. Gesteland RF, Atkins JF, editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 303–358. [Google Scholar]
- 3.Bauren G, Wieslander L. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 4.Beyer AL, Yvonne N, Osheim YN. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 5.Misteli T, Spector DL. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 6.Misteli T, Caceres JF, Spector DL. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 7.Zhang G, Taneja KL, Singer RH, Green MR. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 8.Corden JL, Patturajan M. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 9.Lewis JD, Tollervey D. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- 10.Proudfoot N. Trends Biochem Sci. 2000;25:290–293. doi: 10.1016/s0968-0004(00)01591-7. [DOI] [PubMed] [Google Scholar]
- 11.Steinmetz EJ. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 12.Corden JL. Trends Biochem Sci. 1990;10:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 13.Dahmus ME. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 14.O’Brien T, Hardin S, Greenleaf A, Lis JT. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 15.Cho EJ, Takagi T, Moore CR, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Proc Natl Acad Sci U S A. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komarnitsky P, Cho EJ, Buratowski S. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 20.Chabot B, Bisotto S, Vincent M. Nucleic Acids Res. 1995;23:3206–3213. doi: 10.1093/nar/23.16.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E, Du L, Bregman DB, Warren SL. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. Proc Natl Acad Sci U S A. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vincent M, Lauriault P, Dubois MF, Lavoie S, Bensaude O, Chabot B. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. Proc Natl Acad Sci U S A. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dantonel JC, Murthy KGK, Manley JL, Tora L. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 26.Du L, Warren SL. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose Y, Tacke R, Manley JL. Genes Dev. 1999;13:1234–1239. doi: 10.1101/gad.13.10.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenblatt J. Curr Opin Cell Biol. 1997;9:310–331. doi: 10.1016/s0955-0674(97)80002-6. [DOI] [PubMed] [Google Scholar]
- 29.Myer VE, Young RA. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- 30.Coulombe B, Burton ZF. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pan G, Aso T, Greenblatt J. J Biol Chem. 1997;272:24563–24571. doi: 10.1074/jbc.272.39.24563. [DOI] [PubMed] [Google Scholar]
- 32.Coulombe B, Killeen M, Liljelund P, Honda B, Xiao H, Ingles CJ, Greenblatt J. Gene Expr. 1992;2:99–110. [PMC free article] [PubMed] [Google Scholar]
- 33.Chabot B. In: RNA Processing: a Practical Approach. Higgins SJ, Hames BD, editors. Vol. 1. Oxford University Press; Oxford, UK: 1994. pp. 1–29. [Google Scholar]
- 34.Dignam JD, Lebovitz RM, Roeder RG. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zahler AM, Lane WS, Stolk JA, Roth MB. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 36.Black DL, Chabot B, Steitz JA. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 37.Fu XD. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshida T, Makino Y, Tamura T. FEBS Lett. 1999;457:251–254. doi: 10.1016/s0014-5793(99)01048-0. [DOI] [PubMed] [Google Scholar]
- 39.Douziech M, Forget D, Greenblatt J, Coulombe B. ) J Biol Chem. 1999;274:19868–19873. doi: 10.1074/jbc.274.28.19868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 41.Cramer P, Caceres CF, Cazalla D, Kadener S, Muro AF, Baralle FE, Kornblihtt AR. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 42.Cramer P, Pesce CG, Baralle FE, Kornblihtt AR. Proc Natl Acad Sci, U S A. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CWJ. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kadener S, Cramer P, Nogues G, Cazalla D, de La Mata M, Fededa JP, Werbajh SE, Srebrow A, Kornblihtt AR. EMBO J. 2001;20:5759–5768. doi: 10.1093/emboj/20.20.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]