Abstract
Many Gram-negative pathogens harbor type IV secretion systems (T4SS) that translocate bacterial virulence factors into host cells to hijack cellular processes. The pathology of the gastric pathogen Helicobacter pylori strongly depends on a T4SS encoded by the cag pathogenicity island. This T4SS forms a needle-like pilus, and its assembly is accomplished by multiple protein–protein interactions and various pilus-associated factors that bind to integrins followed by delivery of the CagA oncoprotein into gastric epithelial cells. Recent studies revealed the crystal structures of six T4SS proteins and pilus formation is modulated by iron and zinc availability. All these T4SS interactions are crucial for deregulating host signaling events and disease progression. New developments in T4SS functions and their importance for pathogenesis are discussed.
Keywords: β-catenin, CagA, CagL, cagPAI, CagY, c-Met, cortactin, EGF receptor, gastric disease, GTPase, Helicobacter pylori, integrin, MARK2/PAR1b kinase, signaling, T4SS, type IV secretion, tyrosine kinases, VirB proteins
Helicobacter pylori is a human-specific pathogen that colonizes the hostile environment of the stomach. Approximately half of the world’s population carries this Gram-negative bacterium, which is associated with a spectrum of gastric pathologies ranging from chronic gastritis to peptic ulcers and even gastric cancer in a subset of individuals [1,2]. Infection commonly appears in early childhood and, if not treated, H. pylori can persist throughout life. Evolutionary analyses indicate that H. pylori has been associated with modern humans for at least 100,000 years, possibly after the bacterium was first acquired by a host jump from an unknown source [3]. During this long time of coexistence, H. pylori developed various mechanisms of host immune evasion, and is now considered a paradigm for chronic infections [2]. The clinical outcome of infections is controlled by complex host–pathogen interactions, and influenced by factors such as genetic predisposition of the host, bacterial genotype and environmental parameters. Worldwide H. pylori strains are genetically diverse, and this variability affects virulence factors generating different degrees of pathogenicity potential. Well-known pathogenicity-associated processes include urease-triggered neutralization of acidic pH, flagella-mediated motility, expression of multiple adhesins (BabA/B, SabA, OipA, AlpA/B, HopQ, HopZ and others), cell damage caused by the vacuolating cytotoxin VacA, cleavage of E-cadherin by the protease HtrA as well as induction of apoptosis and inhibition of T-cell proliferation by γ-glutamyl transpeptidase [2,4].
One virulence factor that clearly enhances the risk of gastric cancer development is the cag pathogenicity island (cagPAI), a typical chromosomal DNA insertion element [5]. The cagPAI was discovered in 1996 and its presence discriminates highly virulent cagPAI-positive (type-1) from less virulent cagPAI-negative (type-2) strains [6]. The cagPAI is about approximately 40 kb in size and contains 27–31 genes, including cagA and others which encode proteins forming a type IV secretion system (T4SS) [7]. The cagPAI was acquired by a single horizontal DNA transfer event about 60,000 years ago, and its maintenance until today suggests that the T4SS provided H. pylori with evolutionary advantages [8]. This T4SS forms a needle-like surface appendage (T4SS pilus), which is induced upon host contact [9]. The cagT4SS exports CagA from adherent H. pylori across the bacterial and epithelial membranes into the host cell cytoplasm [10,11]. CagA itself has originally been recognized as an immunodominant antigen [12–15]. Patients that are seropositive both for H. pylori and CagA harbor a 5.8-fold higher risk of developing intestinal and diffuse gastric adenocarcinoma compared with uninfected persons, whereas individuals infected with CagA-negative type-2 strains are only at a 2.2-fold higher risk of developing distal gastric adenocarcinoma compared with uninfected persons [16]. Thus, the cagT4SS is a strong predictor of severe disease outcome. Here we review recent developments in the composition, structure and function of the cagT4SS with an emphasis on its role in H. pylori pathogenesis.
Composition of the cagT4SS
T4SSs are found in many Gram-negative as well as Gram-positive bacteria. They are evolutionarily related to DNA conjugation machineries, but are exploited by several pathogenic bacteria including Agrobacterium, Bartonella, Bordetella, Brucella, Helicobacter and Legionella to deliver DNA or protein substrates into a wide range of eukaryotic target cells [17]. A T4SS apparatus commonly consists of 11 VirB proteins, encoded by the virB1–virB11 genes in the prototypical T4SS of Agrobacterium tumefaciens. By contrast, 15–16 different components of the cagT4SS are required for its two major phenotypes, induction of IL-8 secretion from epithelial cells and translocation of the CagA protein. The cagT4SS contains proteins with detectable sequence similarity to all VirB proteins (Table 1), and additionally five essential components without any similarity to known proteins, making this system clearly distinct from canonical T4SSs in other bacteria [11].
Table 1.
Nomenclature, localization and functional importance of T4SS proteins encoded by the cagPAI of Helicobacter pylori.
| Gene name† | Protein name‡ | Size (kDa)† | Subcellular localization§ | Structure data | Phospho CagA¶ | IL-8 secretion# | T4SS pilus formation†† | Proposed function§ |
|---|---|---|---|---|---|---|---|---|
| hp0520 | Cagζ/Cag1 | 13 | IM | n.d. | +++ | +++ | n.d. | AF |
| hp0521 | Cagε/Cag2 | 10 | Cyt | n.d. | +++ | +++ | n.d. | AF |
| hp0522 | Cagδ/Cag3 | 50 | OM | n.d. | – | – | –‡‡ | OM complex |
| hp0523 | Cagγ/Cag4 | 18 | Per | n.d. | – | – | n.d. | VirB1, PGN hydrolase |
| hp0524 | Cagβ/Cag5 | 80 | IM | n.d. | – | +++ | +++‡‡ | VirD4, coupling factor |
| hp0525 | Cagα/VirB11 | 35 | Cyt, IM | Yes [18] | – | – | +++‡‡ | VirB11, NTPase |
| hp0526 | CagZ | 21 | Cyt, IM | Yes [19] | ++ | ++ | n.d. | Cagβ stabilization |
| hp0527 | CagY | 220 | IM, OM, SA | n.d. | – | – | –‡‡,§§ | VirB10, core complex, integrin binding |
| hp0528 | CagX | 55 | IM, OM, SA | n.d. | – | – | ¶¶,‡‡ | VirB9, core complex |
| hp0529 | CagW | 55 | IM | n.d. | – | – | n.d. | VirB6, IM channel |
| hp0530 | CagV | 27 | IM | n.d. | – | – | –¶¶ | VirB8, core complex |
| hp0531 | CagU | 23 | IM | n.d. | – | – | n.d. | AF |
| hp0532 | CagT | 30 | OM, SA | n.d. | – | – | –¶¶,‡‡,## | VirB7, core complex, OM lipoprotein |
| hp0534 | CagS | 21 | Cyt | Yes [20] | +++ | +++ | n.d. | AF |
| hp0535 | CagQ | 14 | IM | n.d. | +++ | +++ | n.d. | AF |
| hp0536 | CagP | 13 | IM | n.d. | +++ | +++ | n.d. | AF |
| hp0537 | CagM | 40 | OM | n.d. | – | – | –¶¶ | OM complex |
| hp0538 | CagN | 32 | Per, IM | n.d. | + | + | n.d. | Accessory factor |
| hp0539 | CagL | 26 | Per, SA | Yes [21] | – | – | –†††,## | Pilus biogenesis, VirB5-like, integrin targeting |
| hp0540 | CagI | 40 | Per, SA | n.d. | – | –# | –## | Pilus biogenesis, integrin targeting |
| hp0541 | CagH | 39 | IM | n.d. | – | – | ++++## | Pilus biogenesis |
| hp0542 | CagG | 16 | Per | n.d. | – | ++ | n.d. | AF |
| hp0543 | CagF | 29 | Cyt, IM | n.d. | – | +++ | n.d. | Chaperone of CagA |
| hp0544 | CagE | 100 | IM | n.d. | – | – | –## | VirB3/VirB4, NTPase |
| hp0545 | CagD | 22 | IM, Per, SA | Yes [22] | + | +‡‡‡ | n.d. | Accessory factor |
| hp0546 | CagC | 13 | IM, OM, SA | n.d. | – | – | +++¶¶ | VirB2, pilus subunit? |
| n.d. | CagB | 8 | Cyt | n.d. | n.d. | n.d. | n.d. | AF |
| hp0547 | CagA | 125 | Cyt, SA | Yes [23,24] | – | +++ | n.d. | Translocated effector |
Gene designations and corresponding protein sizes are according to H. pylori strain 26695 [GenBank:AE000511.1].
Cag protein designations are according to a previous study [11].
Subcellular localization of cagPAI proteins and proposed functions were reviewed by a previous article [11].
Phosphorylation of CagA and IL-8 induction in the corresponding cag gene knockout mutants of strain 26695 are listed according to a previous study [23]. Abbreviations used for the observed responses: -: Abolished; +: Weak; ++: Moderate; +++: Strong (like wild-type).
Corrected data with regard to requirement for IL-8 induction [25].
T4SS pilus formation in the corresponding knockout mutants are according to numerous previous studies listed below. Abbreviations used for pilus formation compared with wild-type H. pylori: -: Abolished; + : Weak; ++: moderate; +++: Unchanged; ++++: Hyperpiliated.
T4SS pilus formation in strain NCTC11637 and corresponding knockout mutants are according to Tanaka and coworkers [26].
Conflicting data exist with regard to T4SS pilus formation in the corresponding cagY knockout mutant in strain J166 [27].
T4SS pilus formation in strain 26695 and corresponding knockout mutants are according to Johnson and coworkers [28].
T4SS pilus formation in strain 26695 and corresponding knockout mutants are according to Shaffer and coworkers [29].
T4SS pilus formation in strain P12 and the corresponding cagL knockout mutant are according to Kwok and coworkers [9].
AF: Accessory factor with yet unknown function; Cyt: Cytoplasmic; IM: Inner membrane; n.d.: Not determined; OM: Outer membrane; Per: Periplasmic; PGN: Peptidoglycan; SA: Surface associated or secreted into the supernatant; T4SS: Type IV secretion system.
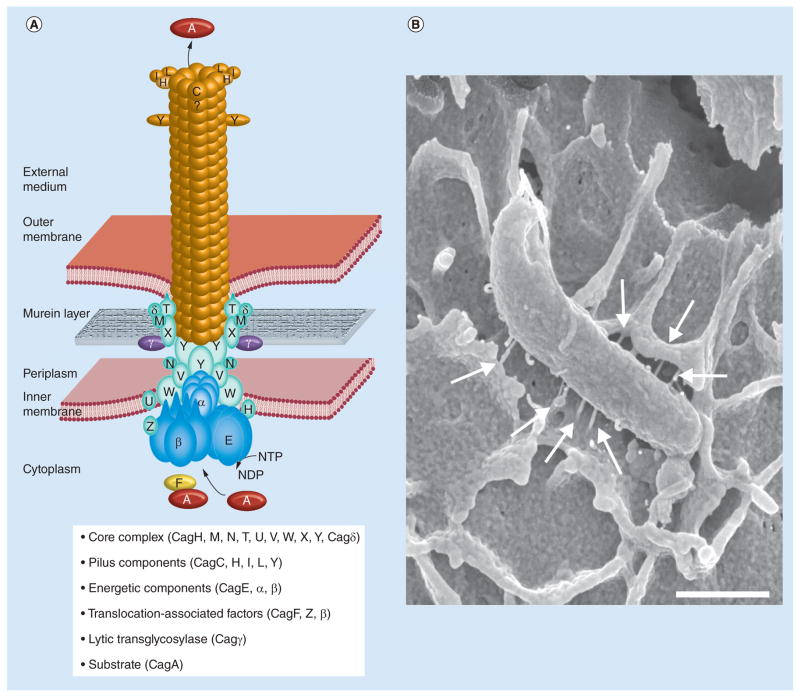
Assuming that those proteins with sequence similarity to components of other T4SSs assemble in an analogous manner to the Trw T4SS of conjugative plasmid R388, for which the structure has recently been elucidated [31], we propose that the cagPAI secretion apparatus consists of a core complex assembled by CagT (VirB7), CagX (VirB9) and CagY (VirB10), and of an inner membrane complex comprising CagE (VirB3/B4), CagW (VirB6) and CagV (VirB8) (Figure 1A). However, the outer membrane-associated part of the cagT4SS additionally contains two essential cagPAI-specific components, CagM [32] and Cag3 [33], which would have to co-assemble with CagT, CagX and CagY into an extended core complex. Likewise, the inner membrane complex might additionally contain a cagPAI-specific protein, CagU, for which no protein interactions within the system have been described so far [32]. One further essential component with a predicted transmembrane helix, CagH, is instead considered to be involved in T4SS pilus formation, which might also involve other essential proteins such as CagC [34], CagL [9,29] or CagI [29] (see below). Further essential components are the putative peptidoglycan hydrolase Cag4/Cagγ, and the cytoplasmic ATPase Cagα (VirB11) [30].
Figure 1. Model for the assembled T4SS complex in Helicobacter pylori. (A).
The T4SS encoded by the cag pathogenicity island is a multicomponent protein complex spanning the inner and outer membranes and exhibits homology to the VirB/VirD4 T4SS machinery of Agrobacterium and other Gram-negative bacteria. The Helicobacter pylori T4SS assembly and cellular localization of the proteins are shown in a simplified manner and according to current knowledge. Pilus components, core complex proteins, energetic components and other factors are highlighted in the box as indicated. The reported substrates for this T4SS are CagA and possibly peptidoglycan. We have chosen the cag nomenclature to highlight the various factors, see also Table 1. (B) Scanning electron microscopy of H. pylori infection of AGS gastric epithelial cells. Typical T4SS pili are shown, connecting the bacterium with the host cell membrane (arrows).
Scale bar represents 1 μm. For more details see text.
T4SS: Type IV secretion system.
(A) Adapted with permission from [10] © John Wiley and Sons.
(B) Kindly provided by Manfred Rohde (HZI Brausnschweig, Germany) [35] with kind permission from © Nature Publishing. For color figures, please see online at www.futuremedicine.com/doi/full/10.2217/FMB.15.32
For translocation of the CagA protein, the VirD4-like coupling protein Cag5/Cagβ and the cagPAI-specific translocation factors CagF, CagZ (and possibly CagD), are required. Cagβ is thought to be responsible, probably together with its interaction partner CagZ, for recognition of the CagA T4SS signal sequence [11]. CagF is considered as a CagA secretion chaperone, and recent evidence shows that CagF forms a high-affinity, 1:1 complex with CagA, in which different domains of CagA contribute to binding [36].
T4SS pilus formation
Most of the well-characterized T4SSs (particularly the DNA-transporting systems) produce surface pili that are typically composed of a major and a minor pilin subunit [17]. Therefore, it was no surprise that pilus-like surface structures associated with the cagT4SS could be identified by electron microscopy studies (Figure 1B). These structures are probably induced by contact with host cells and seem to be more irregular than classical pili, and there is some evidence that they are sheathed [9,26,29,37]. The cagT4SS pili could be stained with antibodies directed against several components, including CagY, CagX, CagT and CagL [9,26,37]. For CagY, CagX and CagT, this finding seems to contradict their involvement in the putative core complex, but it should be noted that all three proteins are considerably different from their canonical orthologs and probably contain additional domains with as yet unknown functions. Since CagL has been detected on T4SS pili and since it interacts with CagI and CagH [25,29], it is possible that in fact all three proteins are associated with T4SS pili, although neither CagI nor CagH localization to pili has been experimentally demonstrated. Deletion of cagI or cagL genes results in a loss of pilus formation, whereas a hyperpiliated phenotype with longer and thicker pili has been described for the cagH mutant [29]. Finally, immunogold stainings also showed the presence of CagA at the tips of the T4SS pili, providing first direct evidence that CagA can be delivered through these surface appendages [9].
Despite the association of different proteins with cagT4SS pili, the major subunit of these pili is still unknown. Since one essential component of the cagT4SS, CagC, is an ortholog of the canonical major pilin VirB2, this protein has been assumed to fulfil a similar function, and immunogold stainings showed its presence at the bacterial surface [34]. However, a recent study in which a cagC mutant was found to produce similar amounts of pili as the wild type, has challenged the notion that CagC is the major pilin [28]. While pilus production has been reported to be independent of CagY as well [27], Cag3, CagX, CagT, CagV and CagM were described to be necessary for pilus formation [28]. These findings cannot be accommodated in a standard model of T4SS pilus composition and assembly. Further studies are therefore required to elucidate the constituents and the assembly of cagT4SS pili.
Formation of pili seems to be regulated by the host and environmental conditions. For instance, iron limitation induces the cagT4SS to produce more pili and to translocate more CagA [38]. Interestingly, injection of CagA into polarized cells by an active T4SS has been shown to facilitate apical surface colonization by recruiting iron [39]. On the contrary, a recent study has shown that depletion of zinc ions, as it would occur in the presence of the neutrophil effector calprotectin, reduces the number of T4SS pili and thus inhibits CagA translocation [40].
Interaction of T4SS proteins with host cell receptors
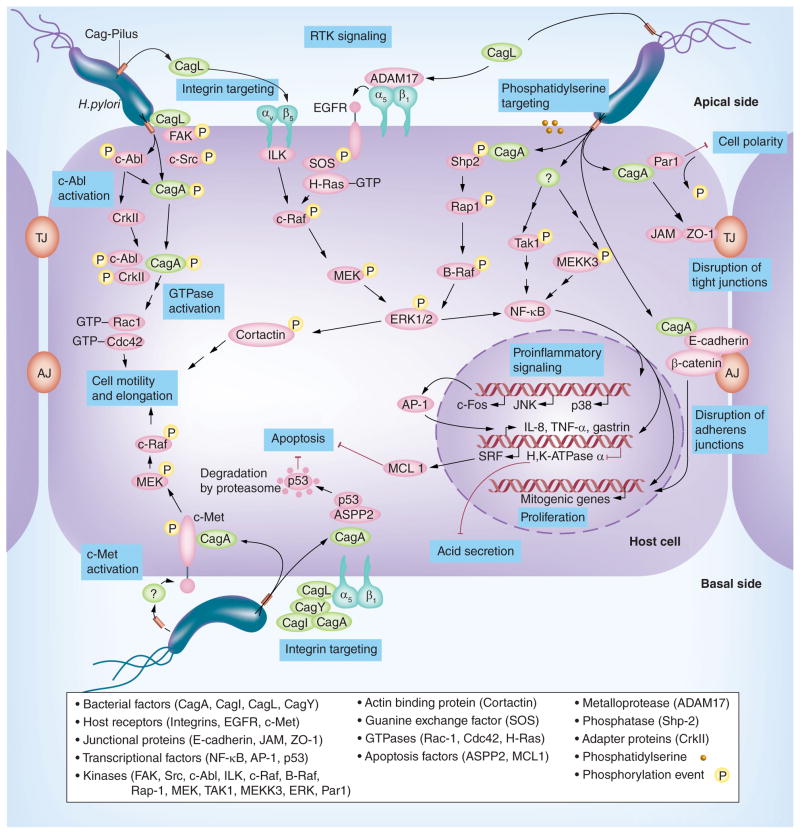
Comparative analysis of cagPAI sequences from different geographic H. pylori populations showed a high overall gene conservation, but suggested that some genes (cagY, cagQ, cagN, cagL, cagI, cagC and cagA) have most likely evolved under diversifying selection [8], suggesting a surface localization of the corresponding proteins and their recognition by the immune system. Such a selective pressure, possibly exerted by the immune response, could be more directly demonstrated for recombination events in CagY with impact on its proinflammatory (IL-8 inducing) activity [27]. Interestingly, some of the cagPAI proteins under evolutionary pressure, including CagL [9,41], CagY, CagI and CagA [42], have also been identified as proteins capable of interacting with integrin molecules as receptors on host cells (Figure 2). Specifically, CagL has been shown to bind to integrins α5β1 and αVβ5 [9,43]; for CagY, CagI and CagA, interactions with integrin α5β1 have been demonstrated [42]. For binding to integrin α5β1 and to epithelial cells, an RGD motif within CagL is necessary [9,21], but the requirement of this motif for T4SS functionality is controversial. Consequences of integrin binding by CagL are an activation of the nonreceptor host cell kinases FAK and c-Src in the case of integrin α5β1 [41] or an upregulation of gastrin expression in the case of integrin αVβ5 (Figure 2) [43].
Figure 2. Model for the role of Helicobacter pylori T4SS effector proteins in host cell interaction and disease-associated signal transduction.
T4SS effectors alter different cellular processes in polarized gastric epithelial cells as illustrated. CagA is translocated into the cytoplasm of infected cells and modulates various signaling cascades associated with cell proliferation, inflammation, motility, cytoskeletal rearrangements, disruption of tight and adherens junctions (AJ and TJ) and suppression of apoptosis or acid secretion, as shown. Receptor tyrosine kinases such as EGFR and c-Met, transcription factors NF-κB and AP-1 as well as small Rho GTPases Rac1 and Cdc42 can also be activated by the T4SS. However, because it is still unclear if integrin-mediated delivery of CagA into the cells occurs at apical and/or basolateral sites, the integrin receptors were indicated at both locations. The various bacterial effectors and host signaling molecules are explained in the bottom box. For more details see text.
T4SS: type IV secretion system.
Adapted with permission from [44] © John Wiley and Sons.
The finding of CagA at the bacterial surface or at the pilus tips [9,42] suggested that surface-associated CagA might be a translocation intermediate, although the CagA protein does not enter cells autonomously in the absence of H. pylori. In support of this notion, recombinant CagA fragments, which bind to integrin, can block CagA translocation by H. pylori when added to bacteria-epithelial cell cocultures [23]. As a further aspect, CagA has also been shown to bind to phosphatidylserine exposed to the outer leaflet of the target cell membrane upon infection, and site-specific CagA mutants that are unable to bind this phospholipid are translocated at very low levels [45]. It remains to be determined, however, how integrin binding and phospholipid interaction contribute to translocation of CagA across the target cell membrane.
Structural analyses of cagT 4SS proteins
A major contribution to our current understanding of T4SS transporters came from unraveling the structure of a T4SS core assembly from conjugative plasmid pKM101, a large approximately 1-MDa complex composed of three structural VirB/D proteins, which are embedded in the bacterial double membrane [46]. Moreover, recent determination of the structure of approximatey 3-MDa assembly of eight T4SS components from plasmid R388 shows intriguing details and revolutionized our view of molecular T4SS architecture [31]. In addition to these major findings, the crystal structures of six H. pylori T4SS proteins have been reported (Table 1). Cagα (VirB11) forms a hexameric ring in conjunction with the HP 1451 regulator [18]. This complex operates as a gating factor in the inner bacterial membrane, which cycles between ATP binding/hydrolysis resulting in closed and open VirB11 conformations [18]. The crystal structure of CagD unraveled a covalent dimer in which each monomer folded as a single domain comprising three α-helices and five β-strands [22]. The structures of CagZ, a 23-kDa protein involved in the translocation of CagA, and CagS, a 23-kDa protein encoded by a conserved cagPAI gene whose function remains elusive, have also been solved [19,20]. In addition, CagL forms an elongated four-α-helix bundle, but revealed substantial flexibility in seven crystallographically independent structures [21]. The RGD-motif is surface exposed but located within a long α-helix, which is unprecedented as previously characterized integrin binding RGD motifs are found within extended or flex-ible loops. Intramolecular disulfide bonds engineered to reduce CagL flexibility resulted in a more stable protein, but were unable to support cell adhesion, suggesting that CagL may partly unfold for integrin α5β1 receptor binding [21].
Moreover, two groups reported crystal structures of an approximately 100-kDa amino-terminal fragment of the effector protein CagA [23,24]. Three distinct domains were identified, giving insights into intramolecular and intermolecular interactions. Domain-I constitutes a mobile N-terminus, whereas domain-II tethers CagA to the plasma membrane through binding of phosphatidylserine [24] or facilitates integrin α5β1 engagement [23]. Domain-III inter-acts intramolecularly with the intrinsically disordered C-terminal domain, and this interaction potentiates the pathogenic scaffold/hub function of CagA [24]. In addition, the co-crystal structure of a subdomain of CagA with the human kinase MARK2/PAR1b was solved, showing that a CagA peptide (amino acids 885–1005) mimics substrates of this kinase family, resembling eukaryotic protein kinase inhibitors [47]. Further studies showed that CagA-MARK2 binding is necessary to induce cell polarity defects [48]. Co-crystals were also obtained for the association of CagA with apoptosis-stimulating protein of p53–2 (ASPP2), a human tumor suppressor [49]. A specialized three-helix bundle at the N-terminus of CagA binds to a 7-kDa proline-rich sequence of ASPP2 and disruption of this interaction decreased survival of H.pylori- infected cells [49]. These reports underscore that structural data are crucially important for understanding the functions of CagA and other T4SS proteins (Figure 2).
Delivery & function of T4SS effectors
Clinical studies and animal infection models have shown that the presence of cagT4SS and CagA is associated with the development of gastric disease [1–2,16]. Interestingly, Mongolian gerbils infected with wild-type H. pylori exhibit gastric pathology similar to infected humans and approximately 75% of the animals developed gastric adenocarcinomas after only 8 weeks of infection, while this was not the case with isogenic cagA or T4SS mutants [38,50]. In addition, the first direct causal link between CagA and oncogenesis was reported in CagA-expressing transgenic mice, which developed polyps and adenocarcinomas in the stomach after 72 weeks [51]. Thus, CagA and the T4SS play central roles during H. pylori pathogenesis in vivo. Most of the functional studies, however, were performed with infected cultured gastric cells in vitro. These studies revealed profound T4SS activities that were either CagA dependent or CagA independent, and provided fundamental insights in disease-associated signaling events [4,52]. Some of these important signaling pathways are highlighted in Figure 2.
First, upon delivery, CagA localizes to the plasma membrane and undergoes phosphorylation (CagAPY) by c-Src and c-Abl tyrosine kinases targeting EPIYA-sequence motifs [53]. Intracellular CagA can then interact with at least 24 host proteins in phosphorylation-dependent or -independent manners [10,54]. Besides the two binding partners (ASPP2 and MARK2/PAR1b) discussed above, other studies have shown that CagAPY also binds to and activates phosphatase SHP-2, triggering gastric epithelial cell elongation by stimulating the Rap1→B-Raf→ERK1/2 signaling pathway [55] and serine-phosphorylation of the actin-binding protein cortactin [56]. CagA can also recruit the tight junction proteins ZO-1 and JAM leading to disturbed cell junctions [57]. Injected CagA has been also found to bind the c-Met receptor which is activated in this scenario, and triggers cell scattering involving the small Rho GTPases Rac1 and Cdc42 [58]. This signaling also requires the activity of c-Abl, the adapter protein CrkII and CagAPY which form a trimeric complex upstream of the GTPases [59,60]. Furthermore, CagA has been reported to promote proliferative and antiapoptotic gene regulation via activation of growth factor receptors, β-catenin, and other signaling molecules (Figure 2). Consequently, the uncontrolled cellular growth in combination with the disruption of epithelial integrity by CagA can potentiate pre-cancerous changes in the stomach and explains the high incidence of gastric adenocarcinoma in patients infected with CagA-positive H. pylori.
Second, the T4SS-dependent but CagA-independent effects mainly comprise the induction of transcription factor NF-κB [61]. H. pylori induces a transient activation of the inhibitor of NF-κB (IκBα) kinase (IKK) complex [62]. IKKα and IKKβ expression stabilizes the regulatory IKK complex subunit NF-κB essential modulator, and this is tightly controlled by two kinases, MEKK3 and TAK1 [62]. The activation of NF-κB finally results in the induction of proinflammatory cytokines such as IL-8 and TNF-α (Figure 2). However, the activation of NF-κB also suppresses gastric acid secretion. The mechanism of the latter involves CagL binding to integrin α5β1 which induces the dissociation of the metalloprotease ADAM-17 from this receptor complex [63]. This resulted in the activation of ADAM-17 and EGFR, followed by NF-κB-dependent transcriptional suppression of H, K-ATPase (α-subunit), the central enzyme controlling gastric acid secretion (Figure 2). Indeed, hypochlorhydria is commonly diagnosed during acute phase of infections, and thus, CagL-triggered cell signaling significantly contributes to H. pylori pathogenesis. Together, these reports validate that H. pylori subvert multiple cellular key processes by structural T4SS proteins and/or yet unknown effector(s).
Conclusion & future perspective
Research on the H. pylori T4SS has provided us with fascinating insights into its biology and function, however, many open questions remain. It was established that the cagPAI plays a crucial role in gastric disease development and various involved mechanisms have been clarified over the years. The structure of the cagT4SS pilus has been visualized and recombination in genes like cagY was shown to ‘turn on’ or ‘turn off’ T4SS functions [27]. However, the composition of the cag pilus is still not clear. The surprising finding that cagC mutants still produce T4SS pili like wild-type H. pylori [28] is not compatible with current T4SS models in other bacteria [17]. Thus, the molecular assembly of the cagT4SS pilus needs to be rigorously reinvestigated. In addition, several T4SS-associated adhesins for integrin α5β1 were discovered, including CagL and CagY. It will be important to examine how these proteins are transported to the pilus surface and in which way the various integrin α5β1 interactions are regulated. Furthermore, much remains to be learned about when and how CagA is delivered into cells. The discovery of a T4SS receptor, suggests the existence of a sophisticated control mechanism for CagA injection. Importantly, integrins are normally present at basal cell surfaces and not apical sites. Thus, we need to investigate how H. pylori can make contact with integrins in polarized cells. The situation becomes even more complex considering very recent reports indicating that various factors encoded outside of the cagPAI (such as hopQ and LPS genes) are also necessary for T4SS-dependent delivery of CagA or NF-κB activation [64]. How this works is fully unclear yet and requires further investigation. It will also be interesting to find out if the cagT4SS translocates substrates other than CagA. Delivery of peptidoglycan is an intriguing possibility [65], but follow-up studies on T4SS-dependent peptidoglycan transportation are required. Finally, the evolutionary advantage of the T4SS for H. pylori needs to be uncovered. Potentially, reduction of gastric epithelial cell turnover by CagA and getting access to iron could be adaptive advantages for H. pylori , but this should be further studied. It therefore appears that the cagT4SS will continue to be an attractive research topic in future.
EXECUTIVE SUMMARY.
The type IV secretion system encoded by the cag pathogenicity island of Helicobacter pylori represents a major disease-associated factor involved in gastric cancer development.
The cagT4SS forms a specific T4SS pilus structure which is induced upon host cell contact.
T4SS assembly is accomplished by multiple cag pathogenicity island protein–protein interactions.
Delivery of CagA in host target cells requires the expression of a host cell receptor, which was identified as the integrin member α5β1
Several pilus-associated proteins (CagA, CagI, CagL and CagY) were identified to interact with integrin α5β1, which is required for delivery of CagA into target cells.
Translocated CagA is phosphorylated and activated by oncogenic c-Src and c-Abl tyrosine kinases of the host.
Intracellular CagA can interact with at least 24 host cell proteins in a phosphorylation-dependent and -independent fashion.
In this way, CagA mimics several host cell proteins to hijack intracellular signaling cascades.
Studies in transgenic mice showed that CagA is the first bacterial oncoprotein.
The route of CagA delivery in polarized gastric epithelial cells remains unclear.
More work needs to be done to clarify if CagA is delivered at apical and/or basolateral surfaces and the evolutionary advantage of the T4SS.
Acknowledgments
The authors apologize to all their colleagues whose important work could not be directly cited.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
Work in the laboratory of S Backert was supported by grants of the German Science Foundation (project B10 in CRC-796) and National Institutes of Health (1R011AM087). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136(6):1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2••.Salama NR, Hartung ML, Mueller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat Rev Microbiol. 2013;11(6):385–399. doi: 10.1038/nrmicro3016. Broad overview of Helicobacter pylori lifestyle in the stomach and pathogenicity mechanisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moodley Y, Linz B, Bond RP, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Backert S, Clyne M, Tegtmeyer N. Molecular mechanisms of gastric epithelial cell adhesion and injection of CagA by Helicobacter pylori. Cell Commun Signal. 2011;9:28. doi: 10.1186/1478-811X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8(11):1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 6•.Censini S, Lange C, Xiang Z, et al. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc Natl Acad Sci USA. 1996;93(25):14648–14653. doi: 10.1073/pnas.93.25.14648. Describes the original discovery of the cag pathogenicity island and first functional studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pacchiani N, Censini S, Buti L, et al. Echoes of a distant past: the cag pathogenicity island of Helicobacter pylori. Cold Spring Harb Perspect Med. 2013;3(11):a010355. doi: 10.1101/cshperspect.a010355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olbermann P, Josenhans C, Moodley Y, et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6(8):e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9••.Kwok T, Zabler D, Urman S, et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature. 2007;449(7164):862–866. doi: 10.1038/nature06187. First study on the identification of a host cell receptor (β1-integrin) and corresponding adhesin necessary for type IV secretion system (T4SS) function. [DOI] [PubMed] [Google Scholar]
- 10.Tegtmeyer N, Wessler S, Backert S. Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 2011;278(8):1190–1202. doi: 10.1111/j.1742-4658.2011.08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer W. Assembly and molecular mode of action of the Helicobacter pylori cag type IV secretion apparatus. FEBS J. 2011;278(8):1203–1212. doi: 10.1111/j.1742-4658.2011.08036.x. [DOI] [PubMed] [Google Scholar]
- 12.Cover TL, Dooley CP, Blaser MJ. Characterization of and human serologic response to proteins in Helicobacter pylori broth culture supernatants with vacuolizing cytotoxin activity. Infect Immun. 1990;58(3):603–610. doi: 10.1128/iai.58.3.603-610.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crabtree JE, Taylor JD, Wyatt JI, et al. Mucosal IgA-recognition of Helicobacter 120 kDa protein, peptic ulceration, and gastric pathology. Lancet. 1991;338(8763):332–335. doi: 10.1016/0140-6736(91)90477-7. [DOI] [PubMed] [Google Scholar]
- 14.Covacci A, Censini S, Bugnoli M, et al. Molecular characterization of the 128-kDaimmunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90(12):5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D’Elios MM, Manghetti M, De Carli M, et al. T helper 1 effector cells specific for H. pylori in the gastric antrum of patients with peptic ulcer disease. J Immunol. 1997;158(2):962–967. [PubMed] [Google Scholar]
- 16.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA-positive or CagA-negative Helicobacter pylori infection. Gut. 1997;40(3):297–301. doi: 10.1136/gut.40.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17•.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol Mol Biol Rev. 2009;73(4):775–808. doi: 10.1128/MMBR.00023-09. Comprehensive overview on T4SS transporters and effectors in various bacterial pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hare S, Fischer W, Williams R, et al. Identification, structure and mode of action of a new regulator of the Helicobacter pylori H. pylori 0525 ATPase. EMBO J. 2007;26(23):4926–4934. doi: 10.1038/sj.emboj.7601904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cendron L, Seydel A, Angelini A, et al. Crystal structure of CagZ, a protein from the Helicobacter pylori pathogenicity island that encodes for a Type IV Secretion System. J Mol Biol. 2004;340(4):881–889. doi: 10.1016/j.jmb.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Cendron L, Tasca E, Seraglio T, et al. The crystal structure of CagS from the Helicobacter pylori pathogenicity island. Proteins. 2007;69(2):440–443. doi: 10.1002/prot.21576. [DOI] [PubMed] [Google Scholar]
- 21.Barden S, Lange S, Tegtmeyer N, et al. A helical RGD motif promoting cell adhesion: crystal structures of the Helicobacter pylori type IV secretion system pilus protein CagL. Structure. 2013;21(11):1931–1941. doi: 10.1016/j.str.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Cendron L, Couturier M, Angelini A, et al. The Helicobacter pylori CagD (H. pylori 0545, Cag24) protein is essential for CagA translocation and maximal induction of interleukin-8 secretion. J Mol Biol. 2009;386(1):204–217. doi: 10.1016/j.jmb.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 23••.Kaplan-Türkoz B, Jiménez-Soto LF, Dian C, et al. Structural insights into H. pylori oncoprotein CagA interaction with β1 integrin. Proc Natl Acad Sci USA. 2012;109(36):14640–14645. doi: 10.1073/pnas.1206098109. Presents, together with [44], the first structure of the CagA N-terminus revealing details on intramolecular and intermolecular interactions involved in CagA functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi T, Senda M, Morohashi H, et al. Tertiary structure-function analysis reveals the pathogenic signaling potentiation mechanism of Helicobacter pylori oncogenic effector CagA. Cell Host Microbe. 2012;12(1):20–33. doi: 10.1016/j.chom.2012.05.010. [DOI] [PubMed] [Google Scholar]
- 25.Pham KT, Weiss E, Jiménez Soto LF, et al. CagI is an essential component of the Helicobacter pylori Cag type IV secretion system and forms a complex with CagL. PLoS ONE. 2012;7(4):e35341. doi: 10.1371/journal.pone.0035341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 2003;5(6):395–404. doi: 10.1046/j.1462-5822.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 27.Barrozo RM, Cooke CL, Hansen LM, et al. Functional plasticity in the type IV secretion system of Helicobacter pylori. PLoS Pathog. 2013;9(2):e1003189. doi: 10.1371/journal.ppat.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson EM, Gaddy JA, Voss BJ, et al. Genes required for assembly of pili associated with the Helicobacter pylori cag type IV secretion system. Infect Immun. 2014;82(8):3457–3470. doi: 10.1128/IAI.01640-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer CL, Gaddy JA, Loh JT, et al. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011;7(9):e1002237. doi: 10.1371/journal.ppat.1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer W, Püls J, Buhrdorf R, et al. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol Microbiol. 2001;42(5):1337–1348. doi: 10.1046/j.1365-2958.2001.02714.x. [DOI] [PubMed] [Google Scholar]
- 31••.Low HH, Gubellini F, Rivera-Calzada A, et al. Structure of a type IV secretion system. Nature. 2014;508(7497):550–553. doi: 10.1038/nature13081. Presents, together with [39], the structure of a T4SS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.Kutter M, Buhrdorf R, Haas J, et al. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J Bacteriol. 2008;190(6):2162–2171. doi: 10.1128/JB.01341-07. Performed a comprehensive sequence analysis of cag pathogenicity island components and yeast two-hybrid screening for potential protein–protein interactions. Immunoprecipitation studies resulted in identification of a T4SS subassembly complex at the outer membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinto-Santini DM, Salama NR. Cag3 is a novel essential component of the Helicobacter pylori Cag type IV secretion system outer membrane subcomplex. J Bacteriol. 2009;191(23):7343–7352. doi: 10.1128/JB.00946-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrzejewska J, Lee SK, Olbermann P, et al. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J Bacteriol. 2006;188(16):5865–5877. doi: 10.1128/JB.00060-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hauck CR. Microbiology: preparing the shot. Nature. 2007;449(7164):798–799. doi: 10.1038/449798a. [DOI] [PubMed] [Google Scholar]
- 36.Bonsor DA, Weiss E, Iosub-Amir A, et al. Characterization of the translocation-competent complex between the Helicobacter pylori oncogenic protein CagA and the accessory protein CagF. J Biol Chem. 2013;288(46):32897–32909. doi: 10.1074/jbc.M113.507657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohde N, Püls J, Buhrdorf R, et al. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49(1):219–234. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 38.Noto JM, Gaddy JA, Lee JY, et al. Iron deficiency accelerates Helicobacter pylori-induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123(1):479–492. doi: 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Tan S, Noto JM, Romero-Gallo J, et al. Helicobacter pylori perturbs iron trafficking in the epithelium to grow on the cell surface. PLoS Pathog. 2011;7(5):e1002050. doi: 10.1371/journal.ppat.1002050. Described, together with [30], that a major target of CagA and the T4SS is the acquisition of iron from the host, playing an important role in disease development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaddy JA, Radin JN, Loh JT, et al. The host protein calprotectin modulates the Helicobacter pylori cag type IV secretion system via zinc sequestration. PLoS Pathog. 2014;10(10):e1004450. doi: 10.1371/journal.ppat.1004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tegtmeyer N, Hartig R, Delahay RM, et al. A small fibronectin-mimicking protein from bacteria induces cell spreading and focal adhesion formation. J Biol Chem. 2010;285(30):23515–23526. doi: 10.1074/jbc.M109.096214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Jiménez-Soto F, Kutter S, Sewald X, et al. Helicobacter pylori type IV secretion apparatus exploits beta1 integrin in a novel RGD-independent manner. PLoS Pathog. 2009;5(12):e1000684. doi: 10.1371/journal.ppat.1000684. Performed yeast two-hybrid screening and identified novel cag pathogenicity island components interacting with β1-integrin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiedemann T, Hofbaur S, Tegtmeyer N, et al. Helicobacter pylori CagL dependent induction of gastrin expression via a novel αvβ5- integrin-integrin linked kinase signaling complex. Gut. 2012;61(7):986–996. doi: 10.1136/gutjnl-2011-300525. [DOI] [PubMed] [Google Scholar]
- 44.Backert S, Selbach M. Role of type IV secretion in Helicobacter pylori pathogenesis. Cell Microbiol. 2008;10(8):1573–1581. doi: 10.1111/j.1462-5822.2008.01156.x. [DOI] [PubMed] [Google Scholar]
- 45.Murata-Kamiya N, Kikuchi K, Hayashi T, et al. Helicobacter pylori exploits host membrane phosphatidylserine for delivery, localization, and pathophysiological action of the CagA oncoprotein. Cell Host Microbe. 2010;7(5):399–411. doi: 10.1016/j.chom.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Chandran V, Fronzes R, Duquerroy S, et al. Structure of the outer membrane complex of a type IV secretion system. Nature. 2009;462(7276):1011–1015. doi: 10.1038/nature08588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47•.Nešić D, Miller MC, Quinkert ZT, et al. Helicobacter pylori CagA inhibits PAR1- MARK family kinases by mimicking host substrates. Nat Struct Mol Biol. 2010;17(1):130–132. doi: 10.1038/nsmb.1705. Reveals (together with [46]) that a CagA motif mimics substrates of a host kinase family, resembling eukaryotic protein kinase inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saadat I, Higashi H, Obuse C, et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447(7142):330–333. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- 49.Nešić D, Buti L, Lu X, Stebbins CE. Structure of the Helicobacter pylori CagA oncoprotein bound to the human tumor suppressor ASPP2. Proc Natl Acad Sci USA. 2014;111(4):1562–1567. doi: 10.1073/pnas.1320631111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Franco AT, Israel DA, Washington MK, et al. Activation of beta-catenin by carcinogenic Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102(30):10646–10651. doi: 10.1073/pnas.0504927102. Describes a highly carcinogenic H. pylori strain targeting β-catenin and inducing rapid gastric cancer development in the Mongolian gerbil model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51••.Ohnishi N, Yuasa H, Tanaka S, et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA. 2008;105(3):1003–1008. doi: 10.1073/pnas.0711183105. The results provide first direct evidence for the role of CagA as a bacterial oncoprotein and the importance of CagA tyrosine phosphorylation in disease development in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glowinski F, Holland B, Thiede C, et al. Analysis of T4SS-induced signaling by H. pylori using quantitative phosphoproteomics. Front Microbiol. 2014;5:356. doi: 10.3389/fmicb.2014.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mueller D, Tegtmeyer N, Brandt S, et al. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in western and east Asian Helicobacter pylori strains. J Clin Invest. 2012;122(4):1553–1566. doi: 10.1172/JCI61143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selbach M, Paul FE, Brandt S, et al. Host cell interactome of tyrosine-phosphorylated bacterial proteins. Cell Host Microbe. 2009;5(4):397–403. doi: 10.1016/j.chom.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 55•.Higashi H, Tsutsumi R, Muto S, et al. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori CagA protein. Science. 2002;295(5555):683–686. doi: 10.1126/science.1067147. Describes the first intracellular interaction partner of CagA, the phosphatase SHP-2. [DOI] [PubMed] [Google Scholar]
- 56.Tegtmeyer N, Wittelsberger R, Hartig R, et al. Serine phosphorylation of cortactin controls focal adhesion kinase activity and cell scattering induced by Helicobacter pylori. Cell Host Microbe. 2011;9(6):520–31. doi: 10.1016/j.chom.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 57•.Amieva MR, Vogelmann R, Covacci A, et al. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science. 2003;300(5624):1430–1434. doi: 10.1126/science.1081919. Describes first evidence that CagA targets tight junctions and disrupts junction-mediated functions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Churin Y, Al-Ghoul L, Kepp O, et al. Helicobacter pylori CagA protein targets the c-Met receptor and enhances the motogenic response. J Cell Biol. 2003;161(2):249–255. doi: 10.1083/jcb.200208039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poppe M, Feller SM, Römer G, Wessler S. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene. 2007;26(24):3462–3472. doi: 10.1038/sj.onc.1210139. [DOI] [PubMed] [Google Scholar]
- 60.Tammer I, Brandt S, Hartig R, et al. Activation of Abl by Helicobacter pylori: a novel kinase for CagA and crucial mediator of host cell scattering. Gastroenterology. 2007;132(4):1309–1319. doi: 10.1053/j.gastro.2007.01.050. [DOI] [PubMed] [Google Scholar]
- 61.Backert S, Naumann M. What a disorder: proinflammatory signaling pathways induced by Helicobacter pylori. Trends Microbiol. 2010;18(11):479–486. doi: 10.1016/j.tim.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 62.Sokolova O, Maubach G, Naumann M. MEKK3 and TAK1 synergize to activate IKK complex in Helicobacter pylori infection. Biochim Biophys Acta. 2014;1843(4):715–724. doi: 10.1016/j.bbamcr.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 63.Saha A, Backert S, Hammond CE, et al. Helicobacter pylori CagL activates ADAM17 to induce repression of the gastric H, K-ATPase alpha subunit. Gastroenterology. 2010;139(1):239–248. doi: 10.1053/j.gastro.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belogolov E, Bauer B, Pompaiah M, et al. Helicobacter pylori outer membrane protein HopQ identified as a novel T4SS-associated virulence factor. Cell Microbiol. 2013;15(11):1896–1912. doi: 10.1111/cmi.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Viala J, Chaput C, Boneca IG, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5(11):1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]