Abstract
The tumor suppressor Merlin/NF2 functions upstream of the core Hippo pathway kinases Lats1/2 and Mst1/2, as well as the nuclear E3 ubiquitin ligase CRL4DCAF1. Numerous mutations of Merlin have been identified in Neurofibromatosis type 2 and other cancer patients. Despite more than two decades of research, the upstream regulator of Merlin in the Hippo pathway remains unknown. Here we show by high-resolution crystal structures that the Lats1/2-binding site on the Merlin FERM domain is physically blocked by Merlin's auto-inhibitory tail. Angiomotin binding releases the auto-inhibition and promotes Merlin's binding to Lats1/2. Phosphorylation of Ser518 outside the Merlin's auto-inhibitory tail does not obviously alter Merlin's conformation, but instead prevents angiomotin from binding and thus inhibits Hippo pathway kinase activation. Cancer-causing mutations clustered in the angiomotin-binding domain impair angiomotin-mediated Merlin activation. Our findings reveal that angiomotin and Merlin respectively interface cortical actin filaments and core kinases in Hippo signaling, and allow construction of a complete Hippo signaling pathway.
Keywords: Merlin, Nf2, angiomotin, Hippo pathway, Lats1/2, phosphorylation, PAK1/2
Introduction
The tumor suppressor gene Nf2, first identified more than two decades ago in the familial Neurofibromatosis type 2 cancer patients, encodes the protein 4.1, Ezrin, Radixin, Moesin (FERM) domain-containing protein Merlin1,2. In addition to hereditary central and peripheral nervous system tumors, somatic mutations of Nf2 are also known to be associated with many other tumors including schwannomas, meningiomas, ependymomas, mesotheliomas, melanoma, glioma, renal cancer, and thyroid cancer (3,4,5,6 and http://www.sanger.ac.uk/cosmic). The tumor suppressor activities of Nf2 are closely linked to the critical roles of Merlin in contact inhibition of cell growth and proliferation by functioning as an upstream activator of the Hippo pathway4,7,8,9,10. Merlin can initiate the Hippo signaling by directly activating Mst1/2 (Hpo in Drosophila)10,11,12, or by recruiting Lats1/2 (Wts in Drosophila) to membranes for phosphorylation by Mst1/213. Lats1/2-mediated phosphorylation of YAP/TAZ (Yki in Drosophila) inactivates its transcriptional activity via cytoplasmic sequestration of phosphor-YAP/TAZ14,15. Additionally, Merlin has also been shown to function in nuclei by inhibiting the activity of the nuclear E3 ubiquitin ligase CRL4DCAF1 via binding to its substrate adaptor DCAF116.
Despite more than two decades of extensive research on Merlin, the precise mechanism by which Merlin functions to regulate Hippo pathway kinases or any other targets are poorly understood. Most of the current working models for Merlin are inferred based on the action mechanisms of the related Ezrin, Radixin, and Moesin (ERM) family proteins and biochemical and cell biology studies of Merlin, as the family of FERM domain proteins share high amino acid sequence homology (FERM domains in particular) and similar domain organizations (Figure 1A). The C-terminal tail domain (CTD) of Merlin is assumed to bind to its FERM domain intra-molecularly, resulting in a conformationally closed and functionally dormant form of Merlin17,18,19, although a recent study suggests that mammalian Merlin adopts a partially open conformation20. Unique to Merlin, Ser518 at the beginning of the proposed inhibitory CTD of Merlin (Figure 1A), instead of Thr577 which is the equivalent of Thr558 in Moesin, can undergo p21-activated kinase (PAK)- or cyclic AMP-dependent protein kinase (PKA)-dependent phosphorylation4,21,22. It is generally accepted that phosphorylation of Ser518 acts to convert Merlin into a less active state in suppressing cell growth20,22,23,24,25,26. However, whether phosphorylation of Ser518 leads to a more closed or more open conformation of Merlin remains a topic of debate. A very recent small-angle scattering-based study has shown that the Ser518Asp substitution does not significantly alter the overall closed conformation of the full-length Merlin27. In contrast, in another study the Ser518Ala substitution has been shown to cause Merlin to be more functionally active and interpreted to release Merlin's auto-inhibited conformation20, although it is rather stretching to imagine how the subtle Ala substitution for Ser might open the auto-inhibited conformation of Merlin.
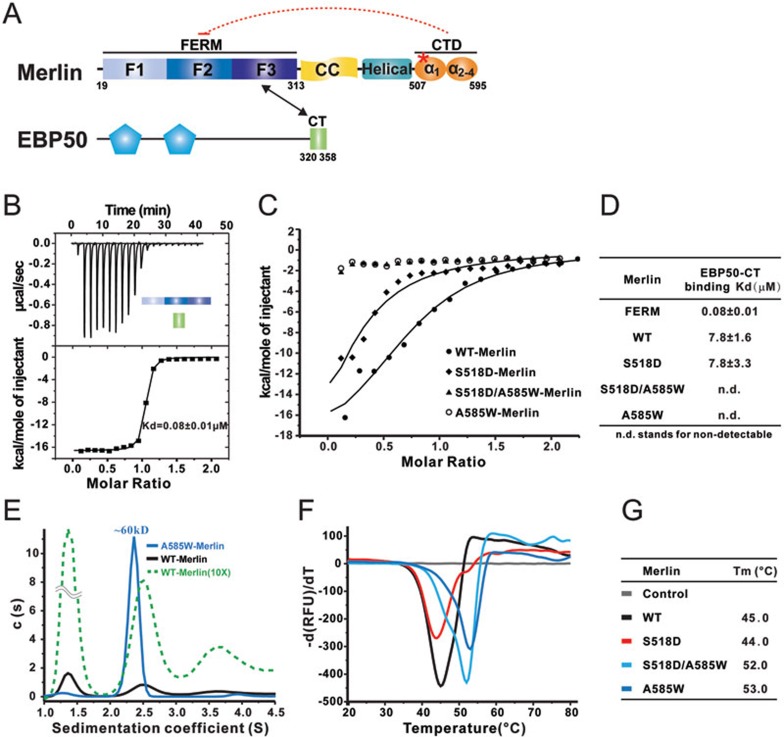
Figure 1.
Mammalian Merlin adopts a semi-open conformation. (A) Schematic diagrams showing the domain organizations of Merlin and EBP50. The Merlin-FERM/EBP50-CT interaction is indicated by a two-way arrow. The S518 site is indicated by a red asterisk and used throughout the rest of the figures. The color coding scheme of Merlin is used throughout the rest of the paper unless specified otherwise. (B-D) ITC-based measurements of the bindings between EBP50-CT and Merlin-FERM (B), WT-Merlin and various mutant forms of full-length Merlin (C, D). (E) Sedimentation velocity analysis showing the hydrodynamic properties of WT Merlin and A585W-Merlin. The fitted molecular weight of A585W-Merlin is indicated. The green dashed line scales the y-axis of the WT-Merlin profile by 10 folds to show that the broader peak at S value ∼2.5 when compared to that of A585W-Merlin. The loading concentrations of the two proteins for the experiment are the same. (F, G) Differential scanning fluorimetry-based thermal denaturation assay showing the temperature-dependent denaturation profiles of WT-Merlin, S518D-Merlin, A585W-Merlin, and S518D/A585W-Merlin.
A major question left unanswered in the Hippo pathway is which factor(s) acts directly upstream of Merlin to regulate its auto-inhibited conformation. Numerous studies have connected an array of proteins regulating cell polarity and inter-cellular junctions with Merlin28,29,30,31,32, although not all Merlin-mediated contact-dependent inhibition has to go through the Hippo pathway33,34. This is in line with Merlin's role in suppressing further growth and proliferation after cells reach confluence. However, it is not clear how the polarity/cell-cell junctional cues can be transmitted to and subsequently release the auto-inhibited conformation of Merlin. Phosphorylation of Ser518 by kinases such as PAK has been shown to modulate Merlin cellular activity21,22,25,26. However, as we show in this work, phosphorylation of Ser518 does not obviously alter the conformational status of Merlin per se. Thus, additional upstream Merlin regulatory factor(s) should exist and remains to be discovered.
In this work, we solved the structure of the Merlin FERM domain (Merlin-FERM) in complex with its own CTD (Merlin-CTD) bearing two amino acid residue substitutions. The structure provides hints to the auto-inhibition mechanism of the full-length Merlin. Unexpectedly, Ser518 from Merlin-CTD is not involved in the auto-inhibition of Merlin-FERM, indicating that Ser518 phosphorylation-mediated inactivation does not originate from direct conformational changes of Merlin. We also solved the structures of Merlin-FERM in complex with FERM-binding domains on Lats1 and Lats2 (Lats1/2-FBD). These structures reveal that the auto-inhibitory tail of Merlin needs to be released before Merlin binding to Lats1/2. We discovered that angiomotin (AMOT), a family of scaffold proteins known to regulate cell growth and polarity establishment35, binds to an elongated helical fragment of Merlin CTD containing Ser518, and subsequently releases the auto-inhibited conformation of Merlin. Importantly, Ser518 phosphorylation weakens Merlin's binding to AMOT, thereby stabilizing the auto-inhibited and functionally dormant conformation of Merlin. Therefore, AMOT can function as a cell contact-dependent upstream activator of Merlin via a Ser518 phosphorylation-regulated mechanism.
Results
Mammalian Merlin adopts a semi-open conformation
Similar to the ERM family proteins, the CTD of Merlin can directly bind to its FERM domain, thus forming a head-to-tail auto-inhibited conformation (Figure 1A and Supplementary information, Figure S1A). Consistent with earlier observations showing that the conformation of mammalian Merlin was dynamic in its native state20,36, the binding of Merlin-CTD to Merlin-FERM is significantly weaker (Kd of ∼3 μM, Supplementary information, Figure S1A) than the corresponding interaction between Moesin's FERM domain and its tail domain (Kd of ∼0.016 μM, Supplementary information, Figure S1B).
The C-terminus of ERM-binding phosphor-protein 50 (EBP50, also known as NHERF1) was shown to bind to ERM-FERM but not to the full-length ERM proteins37,38, as the full-length ERMs adopt tightly closed conformation due to their very strong head-to-tail interactions (Supplementary information, Figure S1B). We found that the binding affinity between EBP50 C-terminal tail (EBP50-CT) and Merlin-FERM is also very strong (Kd of ∼0.08 μM, Figure 1B). We therefore used the EBP50-CT binding to probe the conformational states of the full-length Merlin (WT-Merlin), as well as several of its mutants in the following study. In contrast to non-detectable binding between EBP50-CT and the full-length Moesin (data not shown), the interaction between EBP50-CT and WT-Merlin is clearly measurable (Kd of ∼8 μM), albeit that the binding is ∼100-fold weaker than that between EBP50-CT and Merlin-FERM (Figure 1C and 1D). The above result indicates that WT-Merlin indeed adopts a partially auto-inhibited conformation, and this finding is consistent with a recently published study showing partial inhibition of EBP50-CT binding to Merlin-FERM by Merlin-CT27. A full-length Merlin mutant (A585W-Merlin, in which Ala585 is substituted with Trp) has been shown to be functionally dormant and proposed as fully closed in its conformation20, and this mutant shows no detectable binding to EBP50-CT (Figure 1C and 1D). Consistent with these biochemical data, the WT-Merlin displays multiple conformations in solution, and the substitution of Ala585 with Trp converts Merlin into a highly homogeneous monomer conformation as shown by analytical ultracentrifugation analysis (Figure 1E). Finally, we demonstrated using a temperature-dependent denaturation assay that A585W-Merlin is thermally more stable than WT-Merlin (Figure 1F and 1G). Together, our biochemical and biophysical analyses indicate that WT-Merlin adopts a semi-open conformation and the A585W substitution traps Merlin in a more stable, fully-closed conformation.
Unexpectedly, substitution of Ser518 with a phosphorylation-mimicking residue Asp (S518D-Merlin) neither alters the semi-open conformation (Figure 1C and 1D) nor changes the thermal stability (Figure 1F and 1G) of the full-length Merlin. Importantly, our above conclusion (i.e., substitution of Ser518 with Asp does not alter the semi-open conformation of Merlin) is entirely consistent with a small-angle X-ray scattering-based conformation study of WT- and S518D-Merlin27. This finding is in apparent disagreement with the well-known observations that phosphorylation of Ser518 (or the S518D substitution) can convert Merlin into its functionally inactive form (i.e., inactive in suppressing cell growth), presumed via certain direct, phosphorylation-induced conformational changes to the full-length protein.
The structure of Merlin in its fully closed conformation
Identification of a fully closed conformation of Merlin (A585W-Merlin) provided us with a unique opportunity to obtain the crystal structure of the head-to-tail auto-inhibited form of Merlin, which is otherwise not feasible due to the conformational dynamics of WT-Merlin. We were able to obtain high-diffraction quality crystals of Merlin-FERM in complex with Merlin-CTD bearing S518D/A585W substitutions and solved the complex structure (Figure 2A and Supplementary information, Table S1).
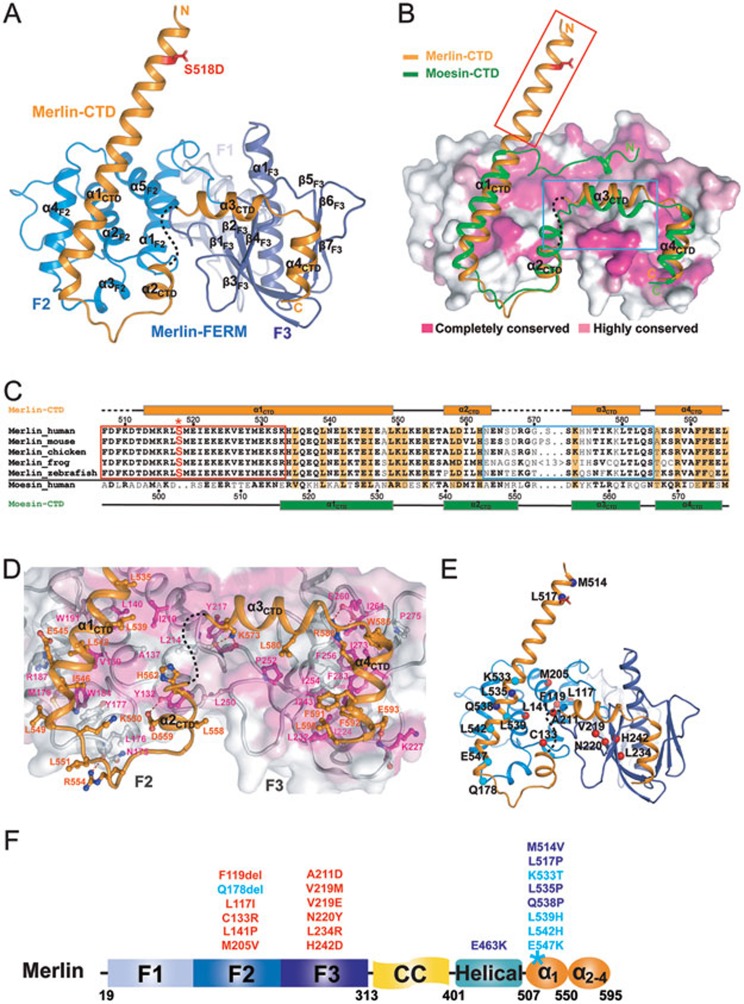
Figure 2.
The overall structure of the Merlin-FERM/Merlin-CTD complex. (A) Ribbon diagram showing the crystal structure of the stabilized, fully closed Merlin-FERM/Merlin-CTD complex. S518D is shown in the stick model and colored in red. (B) Comparison of the structures of the Merlin FERM/CTD complex and the Moesin FERM/CTD complex (PDB code: 1EF1). In this representation, the Merlin-CTD and Moesin-CTD are shown as ribbons, and the Merlin-FERM is drawn in the surface model based on the degree of their amino acid sequence conservation between Merlin and ERMs. The N-terminal half of Merlin α1CTD is boxed in red to emphasize its structural uniqueness. (C) Sequence alignment of the CTD regions from Merlin across different species and human Moesin-CTD. Secondary structural elements for Merlin-CTD and Moesin-CTD are indicated above and below the alignment, respectively. Residues involved in the auto-inhibition of Merlin are emphasized in orange and S518 is highlighted in red and with an asterisk. The residues within the red box form the first half of α1CTD highlighted in red box in B. (D) Molecular details of the Merlin FERM/CTD interaction. (E, F) Ribbon-dot model (E) and a schematic diagram (F) summarizing 21 human cancer-related missense mutations. The mutations that are predicted to interfere with the folding of FERM are shown in red; the mutations that may alter the auto-inhibition and/or AMOT binding of Merlin are colored in cyan and blue.
In the complex, the FERM domain consists of three lobes (F1, F2, and F3), forming a typical cloverleaf-like FERM domain architecture. The CTD folds into four consecutive α-helices (α1CTD-α4CTD) and extensively interacts with the F2 and F3 lobes of the FERM domain, forming a tightly closed conformation (Figure 2A). Importantly, the N-terminal half of α1CTD is projected away from the FERM domain, and Ser518 located at the beginning of α1CTD is solvent exposed and free of contact with any part of the FERM domain (Figure 2A). It is noted that the N-terminal 8 residues of α1CTD is involved in the inter-molecular crystal packing, which likely contributes to the stability of the protruding helical structure. The structure of the fully closed FERM/CTD complex also indicates that Ser518 does not directly participate in the conformational opening/closing of Merlin.
Except for their respective N-terminal halves of α1CTD, both the structures of the CTDs and the interaction modes between FERM and CTD of Merlin and Moesin17,18 are similar (Figure 2B and 2C). Additionally, the surface residues responsible for binding to their CTDs in Merlin and Moesin FERMs are highly conserved (Figure 2B). The FERM/CTD interaction in Merlin is mainly hydrophobic. Specifically, the hydrophobic face of the C-terminal half of α1CTD (composed of Leu535, Leu539, Leu542, Ile546, and Leu549) packs onto the hydrophobic groove of F2 (formed by Val139, Leu140, Tyr177, Met179, Trp184, Trp191, and Ile210; Figure 2A and 2D). The α4CTD uses Trp585, Phe592, and Leu595 in its one face to fit into a hydrophobic groove (composed of Ile224, Leu232, Ile243, Ile254, Phe256, Ile261, Ile273, Pro275, Phe283, and Phe285) between two β-sheets (β1F3-β4F3, and β5F3-β7F3) of F3 (Figure 2A and 2D). Trp585, which is an Ala in the native Merlin, inserts its indole ring into the hydrophobic cavity of F3 and presumably enhances the interaction between α4CTD and F3, thereby artificially stabilizing the fully closed conformation of Merlin. Residues from α2CTD and α3CTD form some polar and hydrophobic contacts with residues in the interface between the F2 and F3 lobes of the FERM domain (Figure 2A and 2D). Most residues located in the interface of FERM and CTD are highly conserved among Merlin and ERMs (Figure 2B-2D and Supplementary information, Figure S2), thus explaining their structural similarity. However, the residues in the N-terminal half of α1CTD and in α3CTD together with its two flanking ends are obviously different between Merlin and Moesin (Figure 2C; note that Ala585 is at the C-terminal flanking end of α3CTD). In Moesin, the region corresponding to the N-terminal half of α1CTD forms an extended loop following by a short helix and the entire segment is directly involved in binding to its FERM domain17,18. In contrast, the N-terminal half of Merlin α1CTD is not involved in the FERM/CTD interaction (Figure 2B). This structural difference, together with the relatively loose interaction of α3CTD and its two flanking ends with the F2 and F3 lobes (Figure 2D and Supplementary information, Figure S3A), provides an explanation to why WT-Merlin adopts a semi-open instead of a fully closed conformation.
The auto-inhibited structure of Merlin is also valuable for us to understand the mechanisms governing many human disease-related mutations identified in Merlin (39 and http://www.hgmd.org). All, except one (“Q178del”), of the 12 mutations found in the F2 and F3 lobes are expected to cause folding defects of the FERM domain (Figure 2E and 2F, labeled in red), and thus impair Merlin's function (see40 for detailed discussions of this category of Merlin mutations). Gln178 is located in the so-called “blue box” (BB) region, and mutation or deletion of the BB region is known to result in an over-proliferation phenotype in Drosophila41. In the closed Merlin structure, the BB region (residues176-183) directly contacts the loop between α1CTD and α2CTD (Figure 2D and 2E, and Supplementary information, Figure S3B). A shortened BB region resulted from the deletion of Gln178 may perturb a series of interactions between the α1CTD/α2CTD loop and the BB region (Supplementary information, Figure S3B), thus weakening the auto-inhibited conformation of Merlin. Counterintuitively, the BB mutant of Drosophila Merlin is known to be inactive in suppressing cell proliferation41, yet our structure-based analysis indicates that the mutant Merlin should adopt a more open conformation and thus more active in suppressing cell growth. This apparent contradiction can be nicely explained by the structure of Merlin-FERM in complex with Lats1/2 presented in the next section. The BB region of Merlin-FERM forms a large part of the Lats1/2-binding surface (Figure 3G). The deletion of the BB region is expected to weaken or disrupt Lats1/2 (or Wts in fly) binding capacity of Merlin, thus preventing Lats1/2 from phosphorylating YAP13.
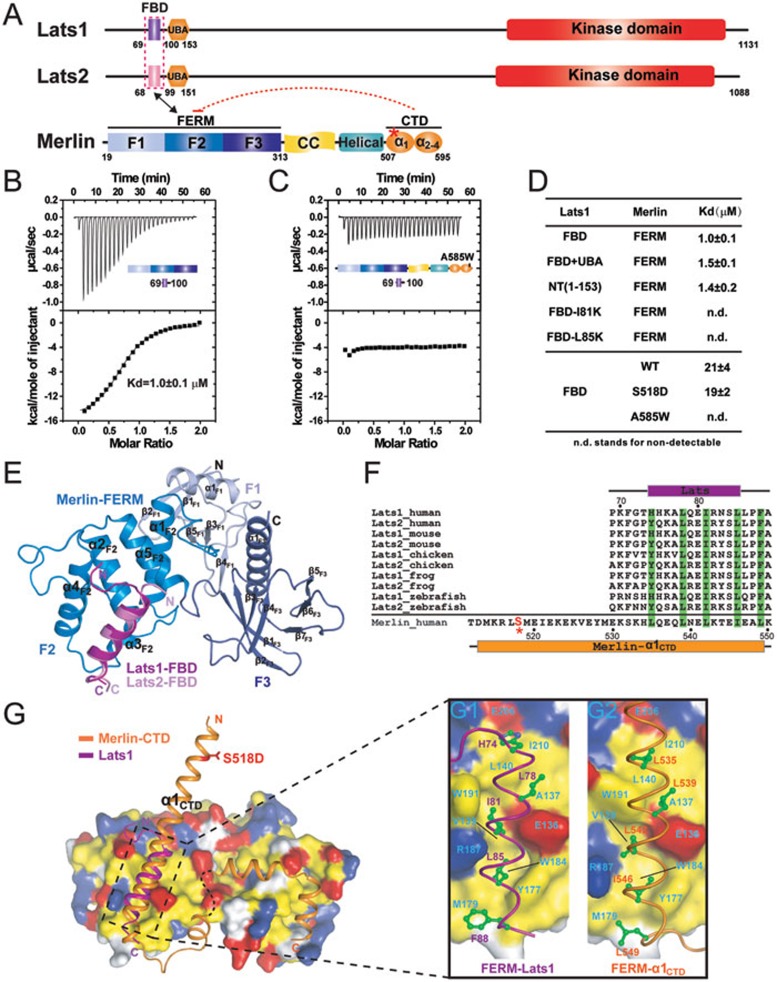
Figure 3.
Lats1/2 bind to the F2 lobe of Merlin-FERM. (A) Domain organizations of Lats1/2 and Merlin. The Merlin-FERM/Lats1/2 interaction is indicated by a two-way arrow. (B, C) ITC-based measurements show the binding of Lats1-FBD to Merlin-FERM (B) or A585W-Merlin (C). (D) A table summarizing the binding affinities between Merlin-FERM and various forms of Lats1, and between Lats1-FBD and various forms of full-length Merlin. (E) The crystal structures of Merlin-FERM in complex with Lats1- and Lats2-FBD drawn in ribbon diagram representation. (F) Sequence alignment of Lats1/2-FBD across different species and human Merlin-α1CTD fragment. The hydrophobic residues of Lats1/2-FBD and Merlin-α1CTD responsible for their respective binding to the Merlin-FERM F2 lobe are highlighted in green. (G) Surface representations showing the comparison of Merlin-FERM/Lats1-FBD (G1) and Merlin-FERM/Merlin-CTD (G2). Residues in Merlin-FERM that are hydrophobic, positively charged and negatively charged are colored in yellow, blue, and red, respectively.
Another frequently mutated region of Merlin is in α1CTD, where 8 missense mutations have been identified (Figure 2E and 2F). Among these mutations, L539H and L542H may impact the hydrophobic interaction between α1CTD and the F2 lobe. Mutations of Lys533 and Glu547, which form salt bridges with Glu537 and Lys550, respectively, are likely to destabilize α1CTD (Figure 2E). The two mutations in the N-terminal end of α1CTD (M514V and L517P; Figure 2E) are not likely to be directly related to the auto-inhibition of Merlin, as these two residues are far away from the contact sites between CTD and FERM. As we will show later on in this study, most of the mutations in α1CTD cause Merlin to be defective in responding to its upstream regulatory factor and thus alter its cell growth suppression activity.
Lats1/2 bind to the FERM domain of Merlin in open conformation
As an upstream regulator of the Hippo pathway, Merlin was shown to interact with Lats1/2 through its FERM domain (Figure 3A), and physically position Lats1/2 for being phosphorylated by Mst1/213. We verified the interaction between Merlin-FERM and Lats1 using purified recombinant proteins. Quantitative binding assays showed that the N-terminus of Lats1 (Lats1-NT) binds to Merlin-FERM with a Kd of ∼1.4 μM (Figure 3D). Further boundary mapping revealed that the minimal FERM-binding domain of Lats1 (Lats1-FBD) binds to Merlin-FERM with essentially the same affinity as Lats1-NT does (Figure 3B and 3D). The Merlin-binding sequences of Lats1 and Lats2 are highly similar (Figure 3F), indicating that Lats2 uses the same minimal fragment to bind to Merlin-FERM.
To elucidate the molecular mechanism governing the Merlin and Lats1/2 interaction, we determined the crystal structures of Merlin-FERM in complex with the FBDs from both Lats1 and Lats2 (Figure 3E). The two complex structures are essentially identical (overall RMSD of 0.6 Å, Supplementary information, Figure S4). Thus, we only describe the Merlin-FERM/Lats1-FBD interaction in details here. In the complex, Lats1-FBD forms an amphipathic α-helix and uses its hydrophobic phase to bind to the F2 lobe of the FERM domain (Figure 3E). The hydrophobic residues (Leu78, Ile81, Leu85, and Phe88) from Lats1-FBD that are critical for binding to Merlin-FERM are extremely conserved both in Lats1 and Lats2 (Figure 3F and 3G). Correspondingly, the hydrophobic residues forming the Lats1-FBD-binding surface in the F2 lobe of Merlin-FERM (Val139, Leu140, Tyr177, Met179, Trp184, Trp191, and Ile210) are also highly conserved (Figure 3G and Supplementary information, Figure S2; note that the BB region of Merlin is critical for Lats1/2 binding). Consistently, ITC-based measurements showed that substitution of either Ile81 or Leu85 of Lats1-FBD with Lys completely abolished the binding of Lats1 to Merlin-FERM (Figure 3D). The interaction mode revealed by the structures of Merlin-FERM in complex with Lats1/2-FBD nicely explains the over-proliferation phenotype of the MerΔBB and Mer3 (M177I) mutant alleles observed in Drosophlia41, as these Merlin mutants are expected to be defective in binding to Wts.
Importantly, comparison of the Merlin-FERM/Merlin-CTD and Merlin-FERM/Lats1/2-FBD complex structures reveals that Lats1/2-FBD and the auto-inhibitory α1CTD of Merlin bind to the same region on the F2 lobe of Merlin-FERM (Figure 3G and the enlarged view shown in panels G1 and G2). Additionally, the FERM-binding residues of Lats1/2-FBD and Merlin-α1CTD are highly similar and can be aligned very well with each other at both the primary and tertiary structure levels (Figure 3F and 3G). The above analysis indicates that the bindings of Lats1/2-FBD and Merlin-CTD to the FERM domain are mutually exclusive, and thus the auto-inhibited conformation of Merlin is expected to prevent Lats1/2 from binding to Merlin. Fully fitting with our prediction, the Lats1-FBD shows a much weaker but detectable binding to WT-Merlin with semi-open conformations (Figure 3D). The fully closed A585W mutant Merlin has no detectable binding to Lats1-FBD (Figure 3C and 3D).
Phosphorylation of Ser518 neither alters the conformational opening/closing, nor directly affects target bindings of Merlin-FERM
The Merlin-FERM/Merlin-CTD and Merlin-FERM/Lats1/2-FBD complex structures presented here predict that the phosphorylation of Ser518 should neither affect its auto-inhibitory conformation nor interfere Merlin's binding to Lats1/2, as Ser518 is located far away from the overlapping site between Lats1/2-FBD and α1CTD (Figure 3G). Fully fitting with the above prediction, the WT Merlin-CTD and its Ser518 phosphorylation mimetic mutant (Merlin-CTD-S518D) display essentially identical dissociation constant (Kd of ∼4 μM) in binding to Merlin-FERM (Supplementary information, Figure S1D and S1E), indicating that substitution of Ser518 with Asp does not alter its semi-open conformation (also see27). Additionally, WT-Merlin and S518D-Merlin display similar thermal stabilities (Figure 1F and 1G). Finally, the WT-Merlin and S518D-Merlin share similar binding affinities to either EBP50-CT or Lats1-FBD (Figures 1C, 1D and 3D). The opposite roles of Ser518 phosphorylation in Merlin's conformation/activity status observed from previous cellular and genetic investigations and from our biochemical and structural studies here can be reconciled if an additional regulatory factor(s) exists to sense the conformational changes originating from Ser518 phosphorylation. If such factor indeed exists, it should be able to bind to Merlin-CTD and thus release the auto-inhibitory conformation of Merlin, leading to Merlin-mediated activation of the Hippo pathway kinases. It is further predicted that the binding of this hypothetical factor to Merlin-CTD can be disrupted or at least weakened by Ser518 phosphorylation, which thus converts the phosphorylated Merlin back to its functionally dormant form.
AMOT binds to and activates Merlin specifically
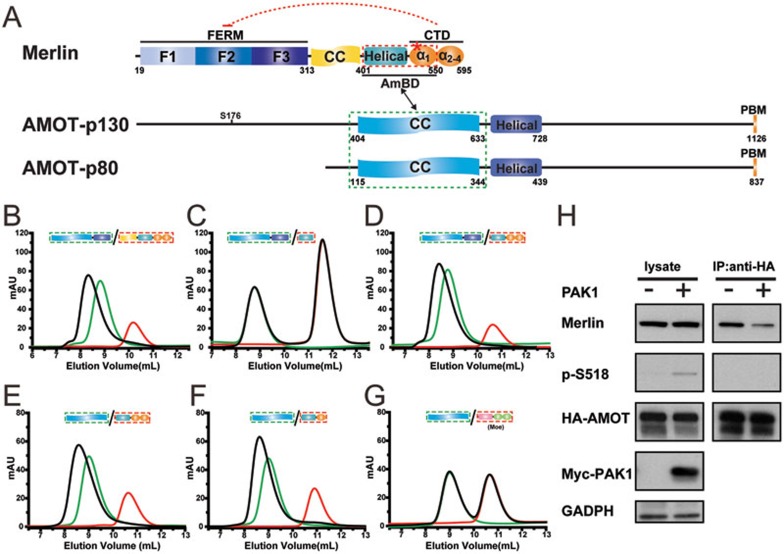
AMOT, a recently identified Hippo pathway component, was proposed to act downstream of Merlin and regulate Ras-MAPK activities30,42. The relationship between AMOT and Merlin in the Hippo pathway is unclear. Recently, the predicted coiled-coil (CC) domain of AMOT was shown to interact with the helical region between the FERM domain and the inhibitory CTD of Merlin30. We reasoned that AMOT might function as a candidate of our hypothesized, Ser518 phosphorylation-modulated Merlin activity regulatory factor. To test this hypothesis, we first verified the AMOT/Merlin interaction and mapped their precise binding domains using high-quality, purified recombinant proteins. Consistent with the reported finding30, an elongated α-helix-rich region of AMOT (residues 404-728 of AMOT-p130, Figure 4A) robustly binds to the entire C-terminal half of Merlin, forming a stable complex with 1:1 stoichiometry (Figure 4B). Further truncation experiments demonstrated that the predicted CC domain of AMOT-p130 (AMOT-CC, residues 404-633) is sufficient for binding to Merlin (Figure 4E and 4F). Interestingly, the helical region of Merlin previously predicted to interact with AMOT shows weak binding to AMOT (Figure 4C and Supplementary information, Figure S5) and the CC domain N-terminal to this helical region is not required for AMOT binding (Figure 4D). The above biochemical data indicate that at least part of the inhibitory Merlin-CTD is also required for AMOT binding. Further boundary mapping revealed that truncation of the α2CTD-α4CTD portion of Merlin-CTD has no impact on the Merlin/AMOT binding (Figure 4E and 4F), indicating that, in addition to the helical region, the α1CTD segment is also critical for Merlin's binding to AMOT. Therefore, the minimal and complete AMOT-binding domain of Merlin (Merlin-AmBD) encompasses residues 401-550 (Figure 4A). Importantly, the AMOT-CC fails to bind to the corresponding “Helical-α1CTD” region in Moesin (Figure 4G), indicating that the AMOT/Merlin interaction is very specific. It is important to note that the amino acid sequences of the “Helical-α1CTD” regions are the most diverse ones between Merlin and ERMs (Supplementary information, Figure S2), providing a possible mechanistic explanation to the specificity of the AMOT/Merlin interaction.
Figure 4.
AMOT specifically binds to Merlin. (A) Domain organizations of Merlin, AMOT-p130, and AMOT-p80. The interaction between Merlin and AMOT-CC is indicated by a two-way arrow. (B-F) Analytical gel filtration chromatography-based mapping of the minimal binding domains of AMOT and Merlin for their mutual interaction. The elution profiles of various forms of Merlin, AMOT, and their mixtures are shown in red, green, and black, respectively. (G) Analytical gel filtration chromatography showing that AMOT-CC does not bind to a Moesin fragment containing both the helical region and CTD. (H) Immunoprecipitation of overexpressed HA-tagged AMOT and endogenous Merlin showing that the AMOT/Merlin interaction occurs in cultured HEK293 cells, and this interaction is weakened by PAK1-mediated phosphorylation (right panel). The inputs are shown on the left. It is noted that in the AMOT-bound fraction of Merlin, no phosphor-Merlin is detected.
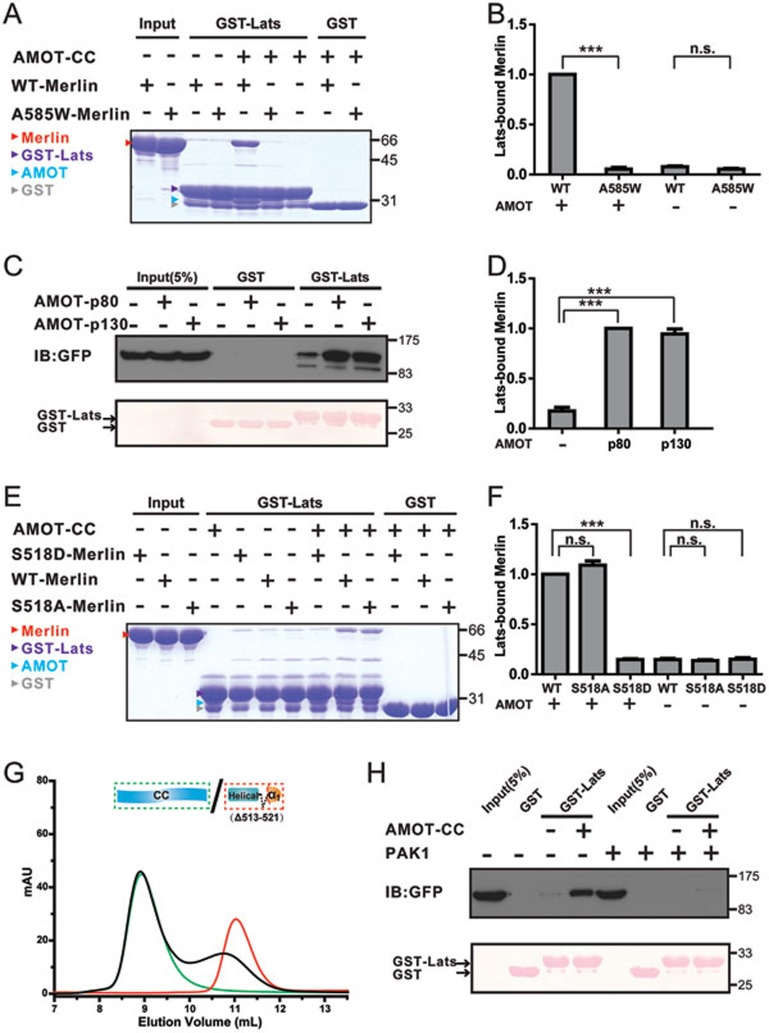
The FERM domain-mediated target bindings of Merlin require the release of the CTD-mediated auto-inhibition (e.g., a frequently used but rather physiologically artificial approach is truncating a part of CTD, see13 for an example). The α1CTD segment, a core element of Merlin-AmBD, is central for the Merlin head-to-tail auto-inhibition, as well as for directly blocking the binding of Lats1/2-FBD to Merlin-FERM (Figure 3G). Therefore, the binding of AMOT-CC to Merlin-AmBD may release the inhibitory CTD from Merlin-FERM, thereby facilitating Merlin to bind to its downstream targets such as Lats1/2. Consistent with this prediction, addition of AMOT-CC fragment or the full-length AMOT (either the p80 or the p130 isoform) can significantly potentiate the binding of Lats1-FBD to WT-Merlin (Figure 5A-5D). This biochemical result qualifies AMOT as an upstream activator of Merlin in the Hippo pathway. Interestingly, the fully closed mutant Merlin (A585W-Merlin) can no longer be activated by AMOT-CC (Figure 5A and 5B), pointing to the functional importance of the semi-open conformation of Merlin.
Figure 5.
AMOT potentiates WT but not the Ser518-phosphorylated Merlin to bind to Lats1. (A) Binding of the semi-open WT-Merlin, but not the closed A585W-Merlin, to Lats1-FBD can be significantly enhanced in the presence of AMOT-CC. (B) Lats1 binding to WT-Merlin and A585W-Merlin in the presence or absence of AMOT-CC was quantified. Values are mean ± SD from three independent experiments (as with the rest of the binding experiments shown in this figure and Figure 7). ***P < 0.001, and n.s. stands for non-significant. (C) Addition of the full-length AMOT (AMOT-p80 or AMOT-p130) potentiates Merlin's binding to Lats1. (D) Quantification of the binding experiments shown in C. (E) Phosphorylation-mimic S518D-Merlin shows only a background level of binding to Lats1 both in the presence and absence of AMOT-CC. In contrast, S518A-Merlin displays an AMOT-CC-dependent binding to Lats1 as WT-Merlin does. (F) Quantification of the binding experiments shown in E. (G) Analytical gel filtration-based assay shows that deletion of a 9-residue fragment surrounding Ser518 (Δ513-521) of Merlin-AmBD essentially disrupts its binding to AMOT-CC. (H) Phosphorylation at Ser518 of Merlin by PAK1 prevents its AMOT-CC-potentiated binding to Lats1. In this assay, we co-transfected GFP-tagged full-length Merlin and Myc-tagged constitutively active form of PAK1 (empty Myc-tagged vector as the control) into the HEK293 cells, and compared AMOT-potentiated binding between Merlin and Lats1. In the upper panel, the presence of PAK1 eliminated AMOT's capacity in potentiating Merlin/Lats1 interaction. The bottom panel shows the input of GST and GST-Lats1 by Ponceau S staining.
Ser518 phosphorylation weakens Merlin's binding to AMOT
Ser518 is located at the center of Merlin-AmBD (Figure 4A). Thus, phosphorylation of Ser518 may disrupt or weaken Merlin's binding to AMOT, thereby converting Merlin back into its conformationally auto-inhibited and biologically less active form. To test this hypothesis, we compared the bindings of S518D-Merlin, S518A-Merlin, and WT-Merlin to Lats1 with and without the presence of AMOT-CC using purified recombinant proteins. In contrast to WT-Merlin, S518D-Merlin has only the background level of binding to Lats1 both in the presence and absence of AMOT-CC. Importantly, S518A-Merlin displays AMOT-CC-dependent binding to Lats1 as WT-Merlin does (Figure 5E and 5F). The above biochemical data strongly indicate that the phosphorylation of Ser518 disrupts/weakens Merlin's binding to AMOT. An Nf2 in-frame deletion leading to the loss of a 9-residue fragment of Merlin (Δ513-521) was identified in vestibular schwannoma patients39. The deletion includes the critical Ser518 but is outside of the auto-inhibitory region of Merlin-CTD (Figure 3F and 3G), therefore the resulted Merlin mutant is expected to be incapable of binding to AMOT, but such deletion does not directly alter the auto-inhibitory conformation of Merlin. Indeed, the Merlin-AmBD bearing this 9-residue deletion (Δ513-521) shows no detectable binding to AMOT-CC (Figure 5G).
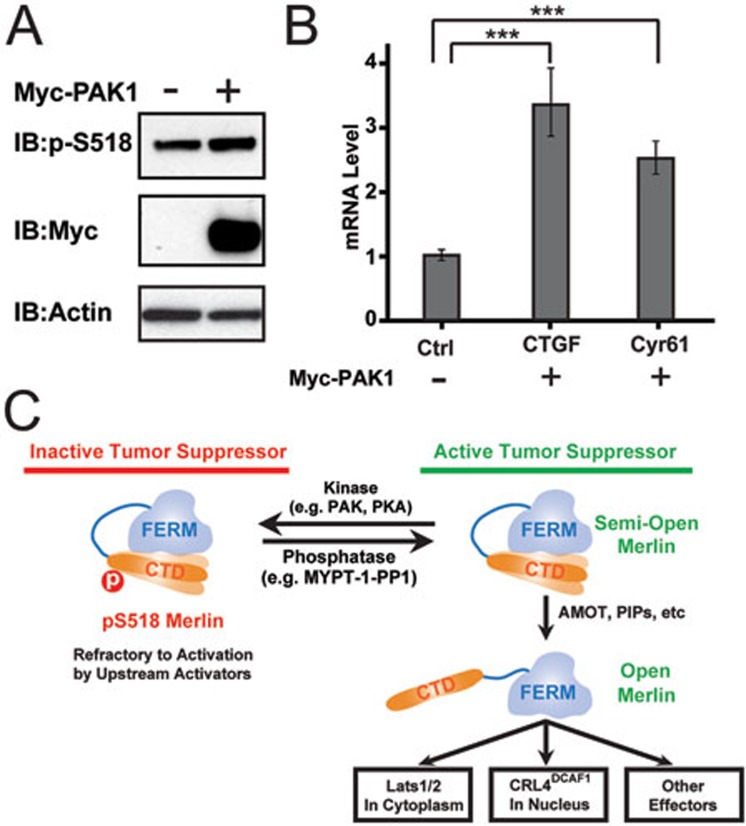
Finally, we directly tested the effect of Ser518 phosphorylation of Merlin on its binding to AMOT. As PAK1/2 have been shown to specifically phosphorylate Ser518 of Merlin22,26 (Supplementary information, Figure S6), we assessed the impact of co-expression of a constitutively active form of PAK1 on the interaction between AMOT and Merlin in cultured HEK293 cells. Consistent with our in vitro interaction data, expression of PAK1 led to a decreased association between AMOT and Merlin (Figure 4H). We also assessed the role of the PAK1-mediated phosphorylation of Merlin on AMOT-potentiated Merlin/Lats1/2 interaction. Upon co-expression of PAK1 with GFP-Merlin, the interaction between Merlin and Lats1 was largely diminished even in the presence of an excess amount of AMOT-CC (Figure 5H), indicating that the activation of Merlin by AMOT can be reversed by PAK1-mediated phosphorylation of Merlin on Ser518. Consistent with the above biochemical studies, overexpression of active PAK1 in HEK293 cells led to an increased level of Ser518-phosphorylated Merlin and a concomitant increase of YAP transcriptional activity (Figure 6A and 6B), presumably at least in part due to the weakened AMOT/phosphor-Merlin interaction. We noticed that PAK1-induced Merlin phosphorylation at Ser518 is clearly observable but rather modest, indicating that additional kinases or other regulatory mechanisms may exist to modulate Merlin phosphorylation in vivo. Taken together, our biochemical and structural data presented above not only identify AMOT as an upstream activator of Merlin, but also elucidate a mechanism by which Merlin phosphorylation at Ser518 regulates its activity in the Hippo pathway (Figure 6C).
Figure 6.
A model depicting the phosphorylation-modulated activity regulation of Merlin. (A, B) PAK1 can enhance phosphorylation of Merlin (A), and cause the inactivation of the Hippo-YAP signaling pathway as evidenced by increased expression levels of two YAP target genes CTGF and Cyr61 (B), presumably due to the weakened AMOT/phosphor-Merlin interaction. (C) A model depicting the phosphorylation-mediated activity regulation of Merlin. In this model, WT-Merlin adopts a semi-open conformation via an intra-molecular head-to-tail interaction. Upstream regulators such as AMOT can bind to Merlin-CTD, fully expose the FERM domain and thus activate Merlin. Phosphorylation of Ser518 can weaken AMOT's binding to Merlin-CTD and thus maintain Merlin in its inactive state.
Merlin-AmBD is frequently mutated in Neurofibromatosis type 2 and other cancer patients
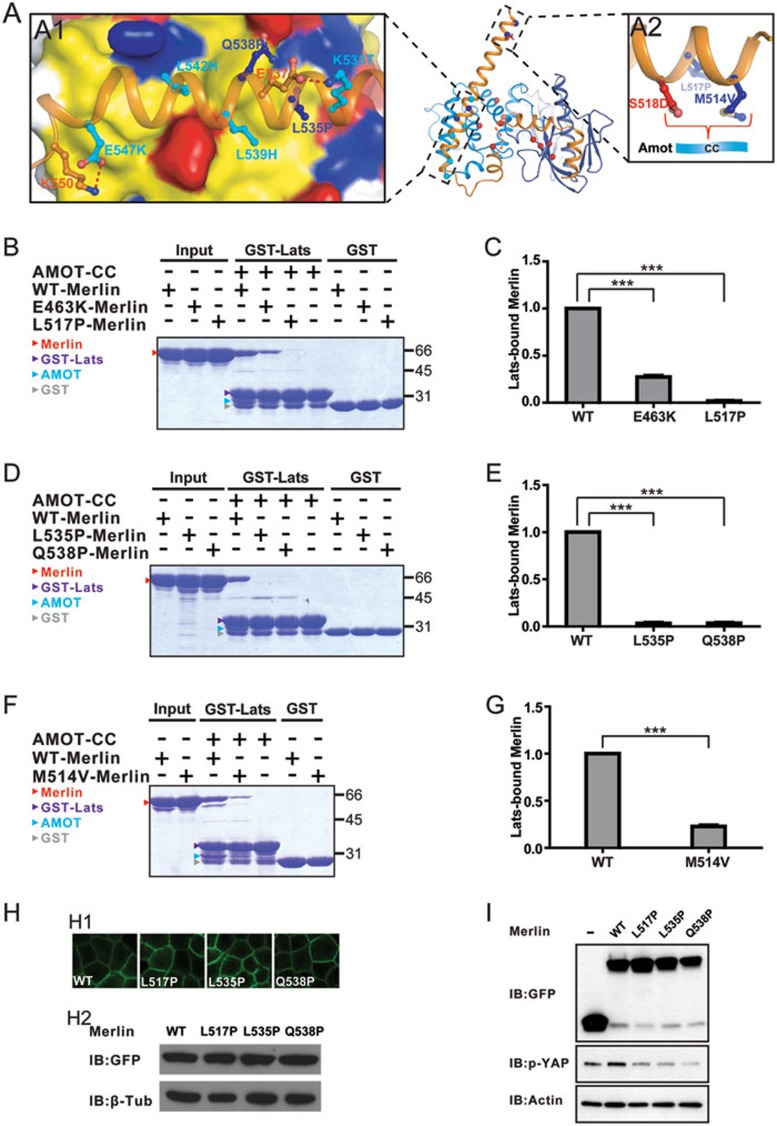
Interestingly, Merlin-AmBD is enriched with missense mutations found in Neurofibromatosis type 2 patients39. It is further noted that α1CTD (part of Merlin-AmBD), but not the rest of auto-inhibitory CTD (α2CTD-α4CTD), contains at least 8 different mutations (Figures 2F and 7A). All these mutations are not expected to affect Merlin-FERM-mediated binding to Lats1/2, and thus we tested the impact of these mutations on AMOT-mediated activation of Merlin. Among the five of these eight Merlin mutants characterized in this study, four (M514V, L517P, L535P, and Q538P; Figure 7A) failed to bind to Lats1 even in the presence of AMOT (Figure 7B-7G), indicating the critical role of α1CTD for Merlin to bind to AMOT. Based on the structure shown in Figure 7A, these mutations may disrupt/destabilize the helical structure of α1CTD, and thus compromise Merlin's binding to AMOT. One Neurofibromatosis type 2 mutation (E463K) is located in the “Helical” region N-terminal to α1CTD, and this mutation also significantly weakens AMOT-potentiated Lats1 binding to Merlin (Figure 7B and 7C), further supporting that the “Helical” region and “α1CTD” together form the Merlin-AmBD.
Figure 7.
Merlin-AmBD is rich in disease-related mutations found in Neurofibromatosis 2 and other cancer patients. (A) Combined surface and stick representation of disease-related missense mutations located in Merlin-AmBD, with (A1) showing the positions of the mutation sites and their interaction networks for the C-terminal half of α1CTD, and (A2) showing that the two mutation sites at the beginning of α1CTD are spatially close to Ser518 and thus expected to disturb Merlin's binding to AMOT. (B, D, F) Even with the presence of an excess amount of AMOT-CC, the cancer-related mutations such as L517P (B), L535P and Q538P (D) of Merlin have only the background level of binding to Lats1, and mutations like E463K (B) and M514V (F) have much weakened binding to Lats1. To ensure that the binding data are directly comparable and quantifiable, all binding experiments in each panel were performed in parallel with freshly purified proteins and ran in one single SDS-PAGE. The bindings of WT-Merlin to GST-Lats1 in each panel serve as the cross-panel control for the quantifications. (C, E, G) Quantifications of the bindings of various Merlin mutants to Lats1 shown in B, D and F. *** P < 0.001. (H) Membrane localization and protein stability of WT-Merlin and several of its mutants when expressed in polarized MDCK cells. (I) HEK293 cells overexpressing Merlin mutants with weakened AMOT binding abilities displayed increased YAP activation (i.e., decreased phosphorylation of YAP).
We next performed cell-based assays to assess the impact of some of the above Merlin mutations (L517P-, L535P-, and Q538P-Merlin) on Hippo pathway activity. First, we demonstrated that, like WT-Merlin, these three mutants are properly localized to cell cortex when expressed in polarized Madin-Darby canine kidney (MDCK) cells (Figure 7H). Expression of WT-Merlin in HEK293 cells leads to increased level of phosphor-YAP, presumably due to Merlin-mediated enhancement of Lats1/2 activation. In contrast, none of the three Merlin mutants could induce phosphor-YAP increase. In fact, the expression of each of these Merlin-mutants led to decreased levels of phosphor-YAP (Figure 7I).
Discussion
Actin cytoskeleton is a major regulator of the Hippo pathway, as essentially all Hippo-mediated cellular processes (e.g., cell growth and differentiation, cell-cell, and cell-matrix contact-induced tissue morphogenesis/homeostasis, cell migrations, etc.) involve changes in the F-actin structures43,44,45,46,47. A series of recent studies have shown that AMOT can relay the signals initiated by F-actin structural changes to YAP activity by either Hippo-dependent or Hippo-independent mechanisms48,49,50. A converging picture in the Hippo-dependent F-actin/AMOT signaling is that AMOT can activate the core Hippo kinase activity when AMOT is released from F-actin induced either by decrease in the amount of F-actin fibers (e.g., in tissues with densely packed cells and inner cells of mammalian preimplantation embryos;32,49) or by phosphorylation of actin-binding motif in AMOT50,51,52. However, the exact role of AMOT after being released from F-actin is not clear. We demonstrated in this study that AMOT can directly bind to and concomitantly release the auto-inhibited conformation of Merlin, thereby promoting Merlin's binding to Lats1/2 and activation of the Hippo pathway. As such, a complete Hippo signaling pathway initiated from cortical F-actin cytoskeleton can be constructed. In this pathway, AMOT and Merlin interface the cortical actin filaments and the core Hippo pathway kinases, respectively. It is important to note that the scaffolding roles of AMOT and Merlin in the Hippo pathway are intimately linked with their phosphorylation statuses. It has been shown that phosphorylation of AMOT by kinases such as Lats1/2 can cause AMOT to be released from cortical F-actin structures, and consequently promotes Merlin's binding to Lats1/2 and Hippo pathway activity (Figures 5 and 6C). The activated Lats1/2 will in return enhance phosphorylation of AMOT, thus forming a positive feedback Hippo signaling activation circuit. Importantly, the above AMOT-mediated Hippo signaling activation circuit can be effectively inhibited by phosphorylation of Merlin at Ser518, as pS518-Merlin can no longer bind to and be activated by AMOT (Figure 6C). It should be noted that Merlin has been reported to interact directly with Mst1/2, resulting in subsequent phosphorylation and activation of Lats1/210,53. AMOT may also directly activate Hippo pathway via physical binding to and subsequent activation of Lats1/2 kinase54. AMOT has also been reported to bind directly to YAP/TAZ in cytoplasm, and thereby negatively regulate YAP/TAZ activity in a Hippo-independent manner42,55. Therefore, the functions of AMOT and Merlin in cell growth control are likely to be diverse and cell/tissue-type dependent. Finally, it is likely that additional factors other than AMOT exist in cells that can also regulate conformational opening/closing of Merlin. One such Merlin activity regulatory factor may be phosphatidylinositol phospholipids, as phosphatidylinositol 4,5-bisphosphate was found to be able to promote conformational opening of WT- but not S518D-Merlin27.
In the Hippo signaling pathway shown in Figure 6C, Merlin functions as a critical hub and rheostat in integrating and interpreting cell growth/proliferations signals. Consistent with the critical roles of Merlin in the Hippo pathway, numerous Nf2 mutations have been identified in Neurofibromatosis 2 or other cancer patients (http://www.sanger.ac.uk/cosmic). The majority of these disease-causing missense mutations of Merlin can be divided into two categories. One group is mutations that occur in the folding core of Merlin-FERM40 (Figure 2E and 2F), and these mutations are expected to affect the FERM domain-mediated bindings of Merlin to its various targets such as Lats1/2 and CRL4DCAF1. The mutations in the other group are clustered in α1CTD of the inhibitory tail and overlapped with the AMOT-binding region of Merlin (Figure 2E). These mutations can abolish or weaken AMOT's binding to Merlin (Figure 7), and thus prevent the Hippo pathway from being activated properly upon AMOT's release from cortical actin filaments. Mutations in the FERM domain of Merlin (e.g., the Q178del mutant) can directly interfere Merlin's binding to Lats1/2, thereby impairing Merlin's tumor suppressor activity. Mutations in the AMOT-binding tail region of Merlin can also impair Merlin's tumor suppressor activity, though via a very different and indirect mechanism (i.e., by weakening AMOT-potentiated interaction between Merlin and Lats1/2). The detailed structural and biochemical studies presented in our work not only uncover the cell growth activity regulatory mechanism of Merlin, but also set a solid foundation for understanding many cancer-causing mutations found in Merlin. Finally, the findings presented in this work are expected to be valuable for finding possible therapeutic methods for Nf2 mutation-caused cancers in humans.
Materials and Methods
Protein expression and purification
WT-Merlin (residues 20-595), Merlin-FERM (residues 1-320), helical region (residues 401-485), Merlin-CTD (residues 507-595), and Merlin-AmBD (residues 401-550) were amplified by PCR using the human Merlin cDNA as the template. Mouse AMOT-CC (residues 404-633), helical region (residues 634-728), Lats1-NT (residues 1-153), Lats1-FBD (residues 69-100), human Lats2 FERM-binding domain (Lats2-FBD, residues 68-99), and EBP50-CT (residues 320-358) were amplified by PCR using the mouse or human library as the template, respectively. Each PCR product was individually cloned into a modified pET32a vector or the PGEX-4T-1 vector. Various mutants were created using standard two-step PCR-based methods and confirmed by DNA sequencing. Recombinant proteins with N-terminal Trx-His6-tagged or GST-tagged were transformed to E. coli BL21(DE3) cells, cultured at 37 °C to OD ∼0.6 and induced with 0.2 mM IPTG at 16 °C overnight. The expressed proteins were purified by a Ni2+-NTA agarose affinity chromatography or GSH-Sepharose affinity chromatography followed by a size-exclusion chromatography. During purification, all protein samples were detected and analyzed by SDS-PAGE coupled with Coomassie blue staining.
Isothermal titration calorimetry (ITC) assay
ITC was carried out on a MicroCal VP-ITC at 25 °C. All proteins were dissolved in a buffer containing 50 mM Tris pH 7.5, 250 mM NaCl, 1 mM EDTA, and 1 mM DTT. The titration processes were performed by injecting 2-10 μl aliquots of protein samples in syringe (concentration of 200 μM) into protein samples in cell (concentration of 20 μM) at time intervals of 120s to ensure that the titration peak returned to the baseline. The data were analyzed using the Origin 7.0 and fitted by the one-site binding model.
Analytical gel filtration chromatography
Analytical gel filtration chromatography was carried out on an AKTA FPLC system (GE Healthcare). Recombinant proteins with N-terminal Trx- and His6-tagged were concentrated to 40-50 μM and loaded onto a Superose12 10/300 GL column (GE Healthcare) equilibrated with the assay buffer (50 mM Tris pH 7.5, 250 mM NaCl, 1 mM EDTA, and 1 mM DTT). The data were analyzed using the Origin 7.0.
GST pull-down assay
For GST pull-down assays, 2 nmol GST-tagged Lats1-FBD (or GST as the negative control) was loaded to 30 μl GSH-Sepharose 4B slurry beads in an assay buffer (50 mM Tris pH7.5, 250 mM NaCl, 1 mM EDTA, and 1 mM DTT). The lysates of HEK293 cells expressing GFP-tagged WT-Merlin co-transfected with constitutively active PAK1 or purified proteins of WT-Merlin and several of its mutants were first incubated with/without AMOT-CC protein for 1 h at 4 °C. The GST fusion protein-loaded beads were then incubated with the mixture for 30 min at 4 °C. After three times washing, the captured proteins were eluted by boiling, resolved by 10% SDS-PAGE, and detected by western blot with specific antibodies or Coomassie blue staining.
Analytical ultracentrifugation
The sedimentation velocity and equilibrium experiments were performed on a Beckman/Coulter XL-I analytical ultracentrifuge. All protein samples were dissolved in the assayed buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT. The rotor speed for velocity and equilibrium was 42 000 rpm and 9 000 rpm, respectively. The buffer composition (density and viscosity) and protein partial specific volume (V-bar) were obtained using the program SEDNTERP (http://www.rasmb.bbri.org/). The data were analyzed using the programs SEDFIT and SEDPHAT56,57. Global data fitting for different protein concentrations were performed under a heterodimer association model and simulated annealing algorithms.
Differential scanning fluorimetry (DSF)
The DSF experiment was performed with a CFX96 real-time PCR instrument (Bio-Rad). A typical DSF solution is composed of 0.4 mg/ml of protein mixed with 9× Sypro Orange (Invitrogen) in an assay buffer containing 50 mM, Tris pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT. During DSF assays, all samples were scanned from 20 °C to 95 °C at a rate of 0.5 °C per minute. Protein denaturation was monitored by increased fluorescence signal of Sypro Orange, which captures exposed hydrophobic residues during thermal unfolding of proteins. The recorded curves were analyzed by the software CFX-Manager (Bio-Rad). The temperature corresponding to the inflection point was defined as the melting temperature (Tm).
Crystallization
For crystallization, the tagged proteins were treated by a small amount of HRV 3C protease at 4 °C overnight to cleave the fusion tags and further purified by a size-exclusion chromatography. Crystals of the Merlin-FERM/Lats1-FBD, Merlin-FERM/Lats2-FBD, and Merlin-FERM/Merlin-CTD complex were obtained by hanging drop vapor diffusion method at 16 °C within 3-5 days. To set up a hanging drop, 1 μl of concentrated protein mixture (∼20 mg/ml) at 1:1 stoichiometric ratio was mixed with 1 μl of crystallization solution with 0.1 M Sodium malonate pH 7.0, 10% PEG 3350 for Merlin-FERM/Lats1-FBD, 18% iso-Propanol, 0.1 M Tris, pH 7.5, 5% PEG 8000 for Merlin-FERM/Lats2-FBD, and 0.1 M Bis-Tris, pH 6.0, 21% PEG 3000 for Merlin-FERM/Merlin-CTD. Before diffraction experiments, crystals were soaked in crystallization solution containing 10%-30% glycerol for cryoprotection. The diffraction data of Merlin-FERM/Merlin-CTD was collected at the beamline NW3A at the Photon Factory at Tsukuba, Japan, and the data for Merlin-FERM/Lats1/2-FBD were collected at the Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U. All data were processed and scaled using HKL200058 (Supplementary information, Table S1).
Structure determination
The initial phase for all structures was determined by molecular replacement using the apo form of Merlin-FERM (PDB code: 1ISN) as the searching model. The models were refined in Phenix59 against the 2.3 Å data set for Merlin-FERM/Lats1-FBD and Merlin-FERM/Merlin-CTD, and 2.7 Å data set for Merlin-FERM/Lats2-FBD. The Lats1-FBD, Lats2-FBD, and Merlin-CTD were built subsequently in COOT60. In the final stage, an additional TLS refinement was performed in Phenix. The final model was further validated using MolProbity61. The refinement statistics are listed in Supplementary information, Table S1. All structure figures were prepared using PyMOL (http://pymol.sourceforge.net/). The sequence alignments were prepared using ClustalW62. Atomic coordinates and structure factors have been deposited in the Protein Data Bank with PDB codes of 4ZRJ, 4ZRK, and 4ZRI for the structures of Merlin-FERM in complex with CTD, Lats1, and Lats2, respectively.
Cell culture, immunoprecipitation, and immunoblotting
About 1 million HEK293 cells were seeded into 6-cm dishes a day before transfection. The cells were co-transfected with HA-AMOT either with vector or Myc-PAK. The transfected cells were first seeded into 15-cm dishes and grown for 24 h, and then replenished with fresh complete media and kept for another 4 days. The cells were washed once with ice-cold PBS and subsequently lysed in ice-cold lysis buffer (50 mM Tris, pH7.4, 150 mM NaCl, 0.1% NP40, and protease inhibitors cocktail). After clearance by a spin (10 000g, 15 min), 1 mg whole-cell lysates in lysis buffer was incubated with monoclonal anti-HA agarose at 4 °C overnight. The beads were then washed 2 times with lysis buffer with 500 mM NaCl and 0.5% NP40, and 3 times with lysis buffer. The immunoprecipitated proteins were resolved on SDS-polyacrylamide gels and blotted onto the nitrocellulose membrane. The filters were blocked in 5% skimmed milk in PBS and probed with the indicated primary antibodies followed by horseradish peroxidase-conjugated secondary antibody (Pierce). Signals were visualized using Supersignal (Pierce).
For EGFP-Merlin and mutants experiments, 70% confluent HEK293 cells in 6-cm dishes were transfected with the indicated plasmids. Twenty-four hours after transfection, the cells were washed once with ice-cold PBS and subsequently lysed in ice-cold lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.3, 0.25 mM EDTA, pH 8.0, 1% sodium deoxycholate, 1% Triton X-100, 0.2% sodium fluoride, and 0.1% sodium orthovanadate supplemented with protease inhibitor cocktail). The immunoblotting described above was then performed.
RNA isolation and real-time PCR
Total RNA was isolated from culture cells using the RNeasy Mini Kit (Qiagen). cDNA was synthesized by reverse transcription using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and subjected to real-time PCR using specific probes and reagents from Applied Biosystems. Relative abundance of mRNA was calculated using program provided by Applied Biosystems.
Acknowledgments
We thank the SSRF (Shanghai, China) and the Photon Factory (Tsukuba, Japan) for data collection, Dr Dongyan Jin for the PAK1 construct, and Dr Fei Ye for assistance with the analytical ultracentrifuge analysis. This work was supported by the Hong Kong Research Grants Council (RGC; 663811, 663812, 664113, AoE/M09/12, and T13-607/12R), the Asia Fund for Cancer Research to MZ, the National Basic Research Program of China (973 Program; 2014CB910201 to JL and MZ), and the National Natural Science Foundation of China (31270815 to JL). MZ is a Kerry Holdings Professor in Science and a Senior Fellow of IAS at HKUST.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Head-to-tail auto-inhibition of Merlin and ERMs.
Sequence alignment of the full length Merlin across different species and with human Moesin.
Detailed interactions between Merlin FERM and CTD.
Comparison of the structures of the Merlin-FERM/Lats1 and Merlin-FERM/Lats2 complexes.
ITC-based assay of the binding between the Merlin helical region (residues 401-485) to AMOT-CC showing that the Merlin helical region indeed binds to AMOT, albeit with a relatively weak affinity.
Ser518 of Merlin can be specifically phosphorylated by PAK1.
Statistics of data collection and model refinement
References
- Rouleau GA, Merel P, Lutchman M, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- Bianchi AB, Hara T, Ramesh V, et al. Mutations in transcript isoforms of the neurofibromatosis 2 gene in multiple human tumour types. Nat Genet. 1994;6:185–192. doi: 10.1038/ng0294-185. [DOI] [PubMed] [Google Scholar]
- Cooper J, Giancotti FG. Molecular insights into NF2/Merlin tumor suppressor function. FEBS Lett. 2014;588:2743–2752. doi: 10.1016/j.febslet.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder RD, Angelo LS, Kurzrock R. NF2/merlin in hereditary neurofibromatosis 2 versus cancer: biologic mechanisms and clinical associations. Oncotarget. 2014;5:67–77. doi: 10.18632/oncotarget.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli AM, Fernandez-Valle C.Role of Merlin/NF2 inactivation in tumor biology Oncogene 2015 Apr 20. doi: 10.1038/onc.2015.125 [DOI] [PMC free article] [PubMed]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, et al. The tumour-suppressor genes NF2/Merlin and expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway – an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Li W, You L, Cooper J, et al. Merlin/NF2 suppresses tumorigenesis by inhibiting the E3 ubiquitin ligase CRL4(DCAF1) in the nucleus. Cell. 2010;140:477–490. doi: 10.1016/j.cell.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson MA, Reczek D, Bretscher A, Karplus PA. Structure of the ERM protein moesin reveals the FERM domain fold masked by an extended actin binding tail domain. Cell. 2000;101:259–270. doi: 10.1016/s0092-8674(00)80836-3. [DOI] [PubMed] [Google Scholar]
- Li Q, Nance MR, Kulikauskas R, et al. Self-masking in an intact ERM-merlin protein: an active role for the central alpha-helical domain. J Mol Biol. 2007;365:1446–1459. doi: 10.1016/j.jmb.2006.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Agosti C, Wiederhold T, Herndon ME, Gusella J, Ramesh V. Interdomain interaction of merlin isoforms and its influence on intermolecular binding to NHE-RF. J Biol Chem. 1999;274:34438–34442. doi: 10.1074/jbc.274.48.34438. [DOI] [PubMed] [Google Scholar]
- Sher I, Hanemann CO, Karplus PA, Bretscher A. The tumor suppressor merlin controls growth in its open state, and phosphorylation converts it to a less-active more-closed state. Dev Cell. 2012;22:703–705. doi: 10.1016/j.devcel.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Paez JG, Curto M, et al. The Nf2 tumor suppressor, merlin, functions in Rac-dependent signaling. Dev Cell. 2001;1:63–72. doi: 10.1016/s1534-5807(01)00009-0. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Beeser A, Chernoff J, Testa JR. p21-activated kinase links Rac/Cdc42 signaling to merlin. J Biol Chem. 2002;277:883–886. doi: 10.1074/jbc.C100553200. [DOI] [PubMed] [Google Scholar]
- Kissil JL, Wilker EW, Johnson KC, Eckman MS, Yaffe MB, Jacks T. Merlin, the product of the Nf2 tumor suppressor gene, is an inhibitor of the p21-activated kinase, Pak1. Mol Cell. 2003;12:841–849. doi: 10.1016/s1097-2765(03)00382-4. [DOI] [PubMed] [Google Scholar]
- Surace EI, Haipek CA, Gutmann DH. Effect of merlin phosphorylation on neurofibromatosis 2 (NF2) gene function. Oncogene. 2004;23:580–587. doi: 10.1038/sj.onc.1207142. [DOI] [PubMed] [Google Scholar]
- Rong R, Surace EI, Haipek CA, Gutmann DH, Ye K. Serine 518 phosphorylation modulates merlin intramolecular association and binding to critical effectors important for NF2 growth suppression. Oncogene. 2004;23:8447–8454. doi: 10.1038/sj.onc.1207794. [DOI] [PubMed] [Google Scholar]
- Kissil JL, Johnson KC, Eckman MS, Jacks T. Merlin phosphorylation by p21-activated kinase 2 and effects of phosphorylation on merlin localization. J Biol Chem. 2002;277:10394–10399. doi: 10.1074/jbc.M200083200. [DOI] [PubMed] [Google Scholar]
- Ali Khajeh J, Ju JH, Atchiba M, et al. Molecular conformation of the full-length tumor suppressor NF2/Merlin — a small-angle neutron scattering study. J Mol Biol. 2014;426:2755–2768. doi: 10.1016/j.jmb.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand D, Curto M, Saotome I, Giovannini M, McClatchey AI. NF2 deficiency promotes tumorigenesis and metastasis by destabilizing adherens junctions. Genes Dev. 2003;17:1090–1100. doi: 10.1101/gad.1054603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladden AB, Hebert AM, Schneeberger EE, McClatchey AI. The NF2 tumor suppressor, Merlin, regulates epidermal development through the establishment of a junctional polarity complex. Dev Cell. 2010;19:727–739. doi: 10.1016/j.devcel.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C, Troutman S, Fera D, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin's regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin ME, Kruger GM, Slocum KL, et al. The Nf2 tumor suppressor regulates cell-cell adhesion during tissue fusion. Proc Natl Acad Sci USA. 2007;104:3261–3266. doi: 10.1073/pnas.0700044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirate Y, Hirahara S, Inoue K, et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol. 2013;23:1181–1194. doi: 10.1016/j.cub.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J Cell Biol. 2007;177:893–903. doi: 10.1083/jcb.200703010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamouche S, Curto M, Saotome I, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moleirinho S, Guerrant W, Kissil JL. The Angiomotins – from discovery to function. FEBS Lett. 2014;588:2693–2703. doi: 10.1016/j.febslet.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen R, Reczek D, Bretscher A. Hierarchy of merlin and ezrin N- and C-terminal domain interactions in homo- and heterotypic associations and their relationship to binding of scaffolding proteins EBP50 and E3KARP. J Biol Chem. 2001;276:7621–7629. doi: 10.1074/jbc.M006708200. [DOI] [PubMed] [Google Scholar]
- Bretscher A, Edwards K, Fehon RG. ERM proteins and merlin: integrators at the cell cortex. Nat Rev Mol Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Ahronowitz I, Xin W, Kiely R, Sims K, MacCollin M, Nunes FP. Mutational spectrum of the NF2 gene: a meta-analysis of 12 years of research and diagnostic laboratory findings. Hum Mutat. 2007;28:1–12. doi: 10.1002/humu.20393. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Seto A, Maita N, et al. Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain. J Biol Chem. 2002;277:10332–10336. doi: 10.1074/jbc.M109979200. [DOI] [PubMed] [Google Scholar]
- LaJeunesse DR, McCartney BM, Fehon RG. Structural analysis of Drosophila merlin reveals functional domains important for growth control and subcellular localization. J Cell Biol. 1998;141:1589–1599. doi: 10.1083/jcb.141.7.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, et al. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011;30:2325–2335. doi: 10.1038/emboj.2011.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mana-Capelli S, Paramasivam M, Dutta S, McCollum D. Angiomotins link F-actin architecture to Hippo pathway signaling. Mol Biol Cell. 2014;25:1676–1685. doi: 10.1091/mbc.E13-11-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Commun. 2013;4:2251. doi: 10.1038/ncomms3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Guo F, Tan I, Leung T, Hong W. Actin-binding and cell proliferation activities of angiomotin family members are regulated by Hippo pathway-mediated phosphorylation. J Biol Chem. 2013;288:37296–37307. doi: 10.1074/jbc.M113.527598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler JJ, Johnson DE, Heller BL, et al. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci USA. 2013;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, et al. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration, and angiogenesis. J Biol Chem. 2013;288:34041–34051. doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D. Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell. 2010;18:288–299. doi: 10.1016/j.devcel.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam M, Sarkeshik A, Yates JR III, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W. Hippo pathway-independent restriction of TAZ and YAP by angiomotin. J Biol Chem. 2011;286:7018–7026. doi: 10.1074/jbc.C110.212621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Head-to-tail auto-inhibition of Merlin and ERMs.
Sequence alignment of the full length Merlin across different species and with human Moesin.
Detailed interactions between Merlin FERM and CTD.
Comparison of the structures of the Merlin-FERM/Lats1 and Merlin-FERM/Lats2 complexes.
ITC-based assay of the binding between the Merlin helical region (residues 401-485) to AMOT-CC showing that the Merlin helical region indeed binds to AMOT, albeit with a relatively weak affinity.
Ser518 of Merlin can be specifically phosphorylated by PAK1.
Statistics of data collection and model refinement