Abstract
Alternative polyadenylation (APA) is a widespread mechanism for gene regulation and has been implicated in flowering, but the molecular basis governing the choice of a specific poly(A) site during the vegetative-to-reproductive growth transition remains unclear. Here we characterize HLP1, an hnRNP A/B protein as a novel regulator for pre-mRNA 3′-end processing in Arabidopsis. Genetic analysis reveals that HLP1 suppresses Flowering Locus C (FLC), a key repressor of flowering in Arabidopsis. Genome-wide mapping of HLP1-RNA interactions indicates that HLP1 binds preferentially to A-rich and U-rich elements around cleavage and polyadenylation sites, implicating its role in 3′-end formation. We show HLP1 is significantly enriched at transcripts involved in RNA metabolism and flowering. Comprehensive profiling of the poly(A) site usage reveals that HLP1 mutations cause thousands of poly(A) site shifts. A distal-to-proximal poly(A) site shift in the flowering regulator FCA, a direct target of HLP1, leads to upregulation of FLC and delayed flowering. Our results elucidate that HLP1 is a novel factor involved in 3′-end processing and controls reproductive timing via targeting APA.
Keywords: HITS-CLIP/CLIP-seq, RNA-binding protein, alternative polyadenylation, APA, flowering, FCA
Introduction
Alternative polyadenylation (APA) generates mRNAs with distinct 3′ ends and is now emerging as a pervasive mechanism for gene regulation1,2,3. More than 70% of yeast, Arabidopsis and mammalian genes4,5,6,7, ∼50% of Drosophila and zebrafish genes8,9 and ∼30% of nematode protein-coding genes10,11 have APA sites, thereby enormously magnifying the diversity and complexity of the transcriptome and proteome. The mammalian 3′-end processing machinery comprises several multi-subunit protein complexes, including cleavage and polyadenylation specificity factor (CPSF), cleavage stimulation factor (CstF), cleavage factor I (CFIm), cleavage factor II, poly(A) polymerase, symplekin and the RNA polymerase II (Pol II)2. CPSF30 and WDR33 bind to the poly(A) signal (5′-AAUAAA-3′ or the variant 5′-AUUAAA-3′) located 10-30 nucleotide (nt) upstream of the cleavage site and CstF binds to the U/GU-rich region located 30 nt downstream of the cleavage site (downstream sequence element)12,13. The choice of poly(A) site is temporally and spatially regulated by trans-acting factors such as constitutive or cell/tissue-specific RNA-binding proteins (RBPs) in combination with cis elements including the poly(A) signals, auxiliary sequences and even RNA secondary structure1,14. Misregulation of APA is associated with carcinogenesis15, pathophysiological diseases16,17 and developmental defects18.
Flowering is a critical developmental transition vital for maximum reproductive success in the angiosperms. It is tightly regulated by multiple transcriptional and posttranscriptional mechanisms19,20. RBPs represent an important class of regulators of flowering. Regulation of floral transition by APA at genes such as FCA, FPA and the floral repressor Flowering Locus C (FLC) underscores the importance of APA in developmental control. FCA and FPA are RBPs that suppress the expression of FLC to promote flowering in Arabidopsis21,22,23. APA of FCA results in two major isoforms: a full-length functional FCA-γ isoform polyadenylated at the distal site and a truncated nonfunctional FCA-β isoform polyadenylated at the proximal site within the third intron24. APA of FPA is similar to that of FCA25. Both FCA and FPA strengthen the usage of the proximal poly(A) sites of their own transcripts and enhance the usage of proximal poly(A) sites of FLC antisense transcripts25,26.
Most 3′-end processing factors are conserved in eukaryotes27,28,29; however, in some cases, the yeast and plant 3′-end processing factors differ from their mammalian counterparts in sequence preference, positioning and functionality, while a few do not have homologues in mammals28,30. For instance, the RNA recognition motif (RRM)-containing protein Hrp1, the only member of the yeast cleavage factor IB (CFIB) subunit, does not have a mammalian or plant homolog27,28. Therefore, whether plants and mammals have evolved new factors analogous to Hrp1 is unclear. Here, we describe HLP1, an evolutionarily conserved hnRNP A/B family protein, as a novel regulator of 3′-end processing. High-throughput sequencing of RNAs isolated by cross-linking immunoprecipitation (HITS-CLIP, also known as CLIP-seq) revealed that HLP1 binds to A-rich and U-rich elements around polyadenylation sites of thousands of transcripts, including those from floral genes. We further show that HLP1 controls global poly(A) site usage and to some extent regulates FLC levels by enhancing the choice of the distal over the proximal poly(A) site of the FCA transcript. Our results uncover a novel hnRNP family protein HLP1 as a 3′-end factor in the regulation of flowering by targeting APA.
Results
HLP1, an hnRNP A/B family protein, regulates flowering time in an FLC-dependent manner
The plant 3′-end processing factors were primarily characterized by sequence conservation to their mammalian or yeast counterparts27. To identify new players potentially involved in APA, we used BLAST searches with the amino acid sequence of Hrp1, the only factor identified in the yeast CFIB subunit as a query, and found a novel RBP encoded by At5g40490. Phylogenetic analysis revealed that this protein belongs to the evolutionarily conserved hnRNP A/B family with certain similarity to human hnRNP A1 and yeast Hrp1 (Supplementary information, Figure S1A). Thus, we named this Arabidopsis protein HLP1 for hnRNP A1-like protein 1 or Hrp1-like protein 1. HLP1 shares significant similarity to the two RRMs of hnRNP A1 and Hrp1, whereas the other part shows much less conservation (Supplementary information, Figure S1B).
To investigate the biological roles of HLP1, we isolated a T-DNA insertion line of HLP1, hlp1-1 (Figure 1A). RT-PCR and immunoblot results demonstrated that there was no expression of HLP1 in the hlp1-1 mutant (Figure 1B). Lesions in HLP1 result in late-flowering phenotype with increased FLC transcripts (Figure 1C and 1E). Under both long- and short-day photoperiods, the hlp1-1 mutant flowered late and showed normal responses to vernalization and GA treatments (Figure 1D). The late-flowering phenotype was suppressed when crossed to flc-3, a null allele of FLC21, and was reversed by introducing GFP-fused HLP1 full length CDS into hlp1-1 (Figure 1E). Collectively, our data suggest that HLP1 promotes floral transition in an FLC-dependent manner. As HLP1 contains two RRMs, we also investigated the impact of the RRMs on flowering time by overexpressing GFP-HLP1ΔRRM (HLP1 lacking the two RRMs) and GFP-RRM in hlp1-1 mutants, respectively. None of these transgenic plants repressed FLC transcription and rescued the delayed-flowering phenotype of hlp1-1, indicating that the RRMs are required but insufficient for flowering (Supplementary information, Figure S2).
Figure 1.
HLP1 regulates flowering. (A) Gene structure of HLP1. CDS regions are boxed in black, and the 5′ and 3′ untranslated regions are in green and grey, respectively. Introns are indicated as lines. The T-DNA insertion is indicated with a triangle. Primer pairs (CX578/CX579) for amplifying the full-length CDS of HLP1 are indicated by arrows. Bar = 500 bp. (B) RT-PCR using CX578 and CX579 primer pairs (top panel) and western blot using anti-HLP1 antibody (the third panel) show complete absence of HLP1 full-length transcript and protein in the hlp1-1 mutant. Actin and RLSU (RuBisCo Large SubUnit) were used as loading controls. (C) The hlp1-1 mutant shows late-flowering phenotype. (D) Flowering time of Col and hlp1-1 mutant under different conditions or treatments. Flowering time was assessed by the total leaf number of the plants at bolting under long-day photoperiods (LD), vernalization (LD+Ver), short-day photoperiods (SD) and gibberellin (GA) treatment (SD+GA). SD+EtOH treatment was used as a control for GA treatment. 'X' indicates that counting was terminated after the plants had produced over 100 rosette leaves before bolting. The error bars indicate standard deviation. (E) HLP1 regulates flowering in an FLC-dependent manner. Total leaf number at flowering is shown for each indicated strain. * means statistical significance by t-test. Error bars indicate SD. Expression levels of FLC sense transcripts were detected by northern blot. HLP1 protein levels were assessed by immunoblotting using anti-HLP1 antibody. Actin and HSC70 served as loading controls for the northern blot and immunoblot, respectively.
Identification of HLP1-RNA interaction in vivo by HITS-CLIP/CLIP-seq
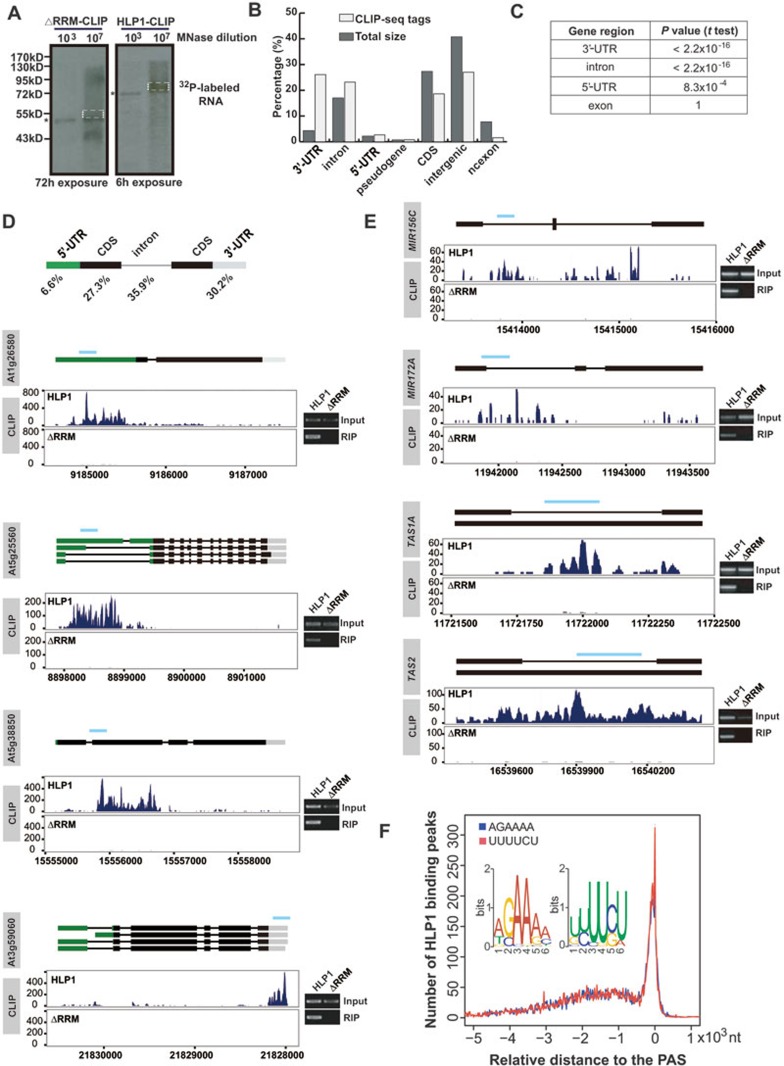
To further delineate the underlying mechanism of HLP1 in flowering regulation, we identified HLP1 in vivo targets, using a modified unbiased high-throughput sequencing of RNAs isolated by the cross-linking immunoprecipitation (HITS-CLIP)/CLIP-Seq method31,32, which was first established in animal system to capture and identify RNA target sites bound by a specific RBP. To this end, Arabidopsis seedlings were UV cross-linked to fix the protein-nucleic acid interaction. HLP1-RNA complex was then immunoprecipitated from the GFP-HLP1 transgenic plants using the GFP antibody followed by MNase digestion. A narrow smear above the HLP1 over-digestion control was separated in NuPAGE gel and was used to generate the HITS-CLIP/CLIP-Seq library for high-throughput sequencing. HITS-CLIP/CLIP-Seq reads obtained from both HLP1 and HLP1ΔRRM libraries were analyzed in parallel (Figure 2A). Overall, ∼29 million and ∼24 million raw reads were collected from the HLP1- and ΔRRM-CLIP library, respectively. After subtraction of adaptors, barcodes, multiple mapped reads and PCR duplicates, a total of 2 324 041 final tags from HLP1-CLIP were uniquely mapped to the Arabidopsis genome (TAIR10), whereas only 61 156 final tags from ΔRRM-CLIP were unique (Supplementary information, Table S1). The drastic reduction of final tags in the ΔRRM-CLIP library indicated that the vast majority of RNA binding capacity of HLP1 is contributed by the two RRMs.
Figure 2.
HLP1 preferentially binds to the A- and U-rich elements in the 3′-UTR and introns of targeted transcripts. (A) Immunoprecipitation of the HLP1-RNA complex. Positions of the 32P-labeled HLP1ΔRRM-RNA complex (left panel) and the HLP1-RNA complex (right panel) after MNase over-digestion (*) are indicated. The protein-RNA complex in dashed box was purified and used to prepare the library for high-throughput sequencing. (B) Percentages of 5′-UTR, 3′-UTR, intron, CDS, pseudogene, ncexon and intergenic region in Arabidopsis genome (dark grey) and HLP1 CLIP-seq tags (light grey). (C) HLP1 CLIP tags are significantly enriched in 3′-UTR and intron. (D) Distribution and percentage of HLP1 binding sites in genes. Binding sites are shown as wiggle plots on the left (blue for the HLP1 library and dark grey for the ΔRRM library). CDS regions are boxed in black. The 5′-UTR and 3′-UTR are boxed in green and grey, respectively. Introns are indicated as lines. Blue line above gene structure indicates RIP-RT-PCR amplified region. The x axis indicates genome location in chromosome. The y axis indicates normalized HITS-CLIP/CLIP-seq abundance. HITS-CLIP/CLIP-seq tag counts were normalized to tag per 10 million (TP10M) to adjust for differences of two HITS-CLIP/CLIP-seq libraries in sequencing depth. Right panels show validation of binding by RIP-RT-PCR. (E) HLP1 binds to transcripts of small RNA genes. Binding sites are shown as wiggle plots on the left (blue for the HLP1 library and grey for the ΔRRM library). CDS regions are boxed in black. Introns are indicated as lines. Blue line above gene structure indicates RIP-RT-PCR amplified region. The x axis indicates genome site in chromosome. The y axis indicates normalized HITS-CLIP/CLIP-seq abundance. HITS-CLIP/CLIP-seq tag counts were normalized to tag per 10 million (TP10M) to adjust for differences of two HITS-CLIP/CLIP-seq libraries in sequencing depth. Right panels show validation of binding by RIP-RT-PCR. (F) Distribution of the two overrepresented binding motifs (insert) relative to the poly(A) site (PAS, indicated as 0) are indicated by blue and red curves, respectively.
The uniquely mapped HLP1-CLIP tags showed a broad range of binding sites to sense targets (1 934 562), encompassing 5′-UTR (2.69%), coding sequences (CDS, 18.61%), introns (23.18%), 3′-UTR (26.11%), intergenic regions (27.03%), pseudogene (0.80%) and noncoding gene exons (1.58%; Figure 2B). We also observed additional 389 479 antisense tags bound by the HLP1. Markedly, these tags are significantly enriched in 3′-UTR (P < 2.2 × 10−16, t-test) and introns (P < 2.2 × 10−16, t-test), compared with 5′-UTR (P = 8.3 × 10−4, t-test) and CDS (P = 1, t-test; Figure 2C). Final HLP1 tags were further normalized and clustered as binding sites. Overall, we identified 9 031 binding sites from the sense transcripts of 5 569 genes (∼16.6% of Arabidopsis genes) and 1 192 binding sites from the antisense transcripts of 866 genes (∼2.6% of Arabidopsis genes; Supplementary information, Figure S3A). As mature mRNAs do not contain introns, by systematically investigating the distribution of HLP1 binding peaks along pre-mRNAs versus mature mRNAs, we found that HLP1 was significantly enriched (from <400 binding peaks in mRNAs to >600 peaks in pre-mRNAs) in the introns and 3′-UTR of its targeted sense transcripts, suggesting that HLP1 may play various roles in regulating pre-mRNA splicing and 3′-end formation (Supplementary information, Figure S3B). Cases of HLP1 binding to each gene region were either experimentally validated (Figure 2D), or shown as snapshots (Supplementary information, Figure S4). Transcripts of some small RNA genes are also targeted by HLP1, indicating that HLP1 may regulate microRNA processing or tasi-RNA biogenesis (Figure 2E). Using all the HITS-CLIP/CLIP-seq tags obtained from HLP1 binding sites, unbiased search for motifs enriched in HLP1 binding sites revealed over-represented A-rich (5′-AGAAAA-3′) and U-rich (5′-UUUUCU-3′) sequences predominantly near the poly(A) sites (Figure 2F). The top 10 motifs were listed in Supplementary information, Figure S5.
To confirm that HLP1 preferentially binds to 3′-UTR and intronic regions, we generated a second set of HLP1-CLIP libraries using HLP1 antibody to immunoprecipitate the endogenous HLP1-RNA complex directly from the wild-type plant (Col-0) and the hlp1-1 mutant, respectively (Supplementary information, Figure S6A). Similarly, HLP1 showed enriched binding tags at 3′-UTR and introns (Supplementary information, Figure S6B). Cases of HLP1 binding to each gene region were shown as snapshots (Supplementary information, Figure S7A) or experimentally validated by RIP-qPCR from two biological replicates (Supplementary information, Figure S7B). Most importantly, the HLP1 binding targets from the two sets of HLP1-CLIP libraries share significant overlap (P = 0, Supplementary information, Figure S6C), suggesting the reproducibility of the CLIP experiment.
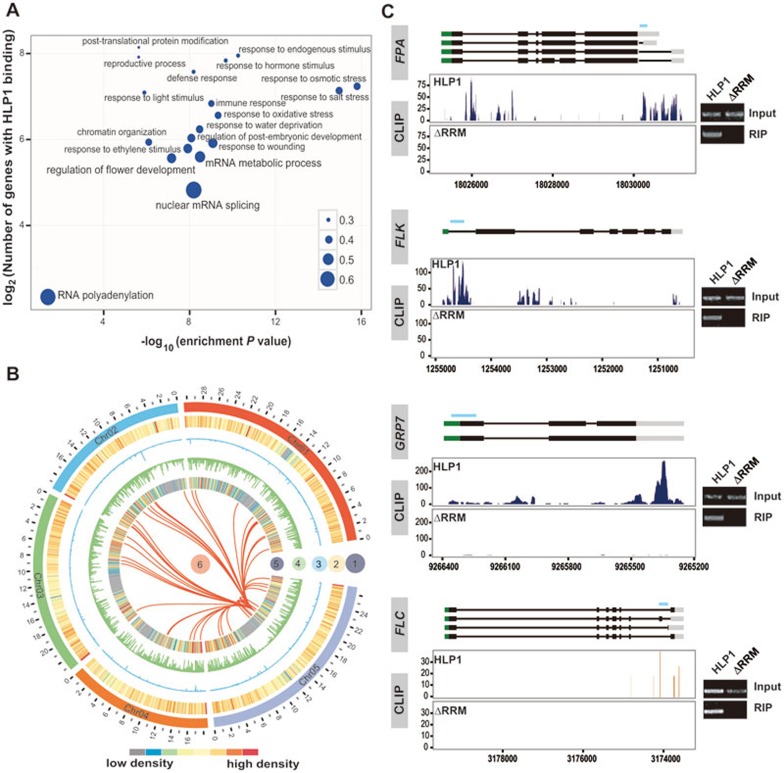
Gene Ontology analysis of the HLP1 sense binding targets revealed significantly enriched terms in RNA metabolism, flower development, various stimuli and stress responses, suggesting that HLP1 may have profound regulatory roles in these physiological processes (Figure 3A). Consistent with its role in floral transition, HLP1 binds to many transcripts of flowering-related genes, some of which encode RNA-processing factors or RBPs involved in RNA processing (Figure 3B, 3C and Supplementary information, Table S2). We showed that HLP1 binds to FLC antisense transcripts around the proximal polyadenylation site, suggesting a role in FLC antisense processing (Figure 3C, bottom panel).
Figure 3.
Enriched HLP1 binding to floral genes. (A) Gene Ontology (GO) analysis of HLP1 binding targets. Significantly enriched GO terms of genes with HLP1 binding were identified using the BiNGO software (hypergeometric test with Benjamini and Hochberg false discovery rate correction). The x axis indicates the enrichment P-value on a −log10 scale; the y axis indicates number of genes with HLP1 binding on a log2 scale. The size of each point is proportional to the ratio of HLP1-bound genes associated with one GO term to all genes associated with this GO term. (B) Circos diagram shows enriched HLP1 binding sites at flowering-related transcripts: circle 1, Arabidopsis chromosomes (indicated as chr01-05 with different colors); circle 2, heat map displaying all of Arabidopsis genes; circle 3, binding density of HLP1ΔRRM showing very few binding peaks of the truncated HLP1; circle 4, binding density of HLP1 indicating all the HLP1 binding sites across the transcriptome; circle 5, heat map view of genes with HLP1 binding; circle 6, red link lines indicating flowering-related transcripts with HLP1 binding. (C) Examples and validations of flowering associated genes bound by HLP1. Binding sites are shown as wiggle plots on the left. CDS regions are boxed in black and the 5′-UTR and 3′-UTR are boxed in green and grey, respectively. Introns are indicated as lines. Blue line above gene structure indicates RIP-RT-PCR amplified region. The x axis indicates genome site in chromosome. The y axis indicates normalized HITS-CLIP/CLIP-seq abundance. HITS-CLIP/CLIP-seq tag counts were normalized to tag per 10 million (TP10M) to adjust for differences of two HITS-CLIP/CLIP-seq libraries in sequencing depth. Blue for binding peaks at FPA, FLK, GRP7 sense transcripts and orange for peaks at FLC antisense transcripts. Right panels show validation of binding by RIP-RT-PCR.
HLP1 is required for APA
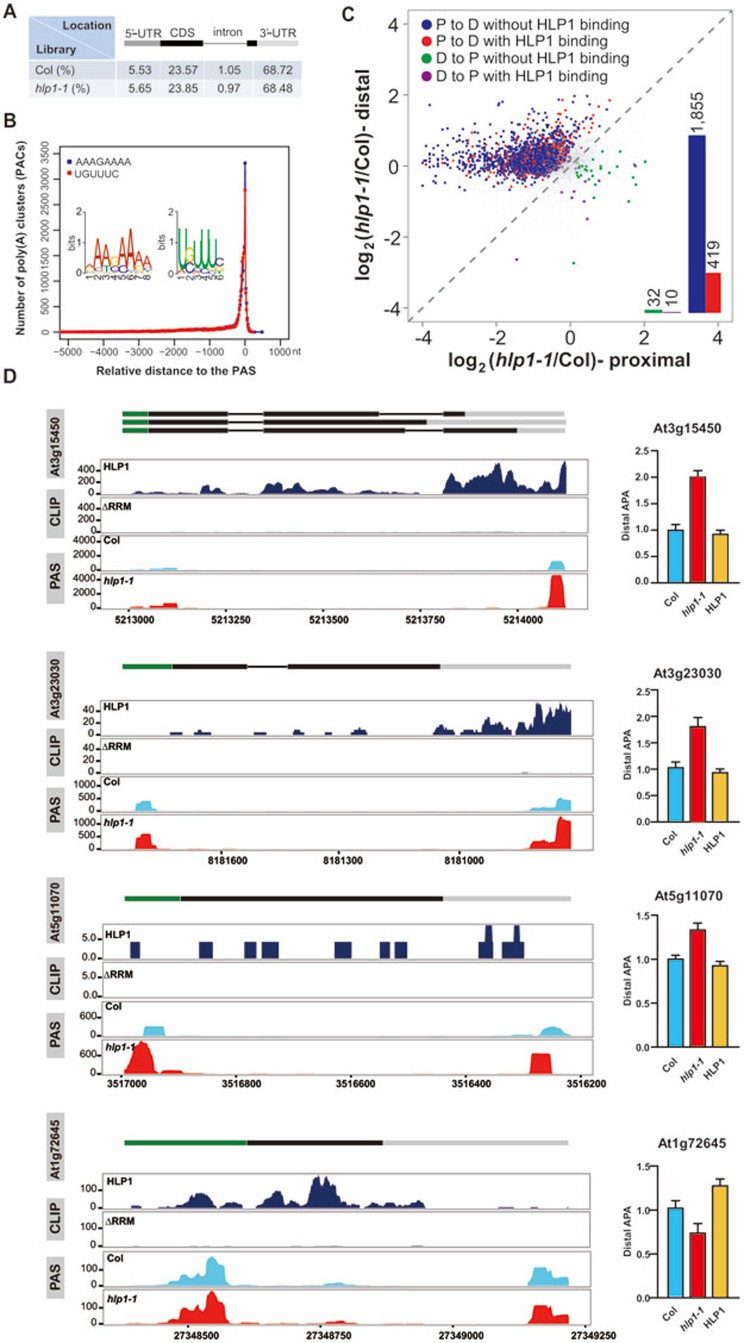
Although HLP1 shares low similarity to the yeast Hrp1, which has been shown to bind the AU-rich efficiency element (EE) and has been implicated in correct positioning of the cleavage site and regulating 3′-end formation of pre-mRNAs33,34, it binds to the U-rich far upstream element (FUE) and A-rich near upstream element (NUE), the plant cis-element equivalent to the yeast EE and A-rich positioning element (PE), respectively. Therefore, HLP1 may function as the Hrp1 ortholog determining the ploy(A) site in plants. To address this, poly(A) site sequencing (PAS-Seq) was used to quantitatively profile poly(A) site usage35,36 in hlp1-1 mutant and the wild-type plant Col (Supplementary information, Figure S8 and Table S3). As expected, the poly(A) clusters (PACs) are predominantly located in the 3′-UTR (∼70%), but also were found in CDS (∼23%), 5′-UTR (∼5%) and introns (∼1%), suggesting APA in CDS, and to a lesser extent in 5′-UTR, as potential regulatory mechanisms (Figure 4A). However, we cannot exclude the possibility that the high percentage of PACs in CDS could be artifacts caused by internal priming11,37. The overrepresented 5′-AAAGAAAA-3′ and 5′-UGUUUC-3′ motifs surrounding the poly(A) site are very similar to the HLP1-binding motifs (Figure 4B). Notably, out of 2 691 HLP1 binding sites at the 3′-UTR, 78% (2 088/2 691) overlap with 1 777 PACs at the 3′-UTR (∼13% of PACs at this region) in wild-type plant (standard score Z = 82), further supporting the role of HLP1 in 3′-end formation (Supplementary information, Figure S9A). By analyzing overlapping APA profiles from two APA biological replicates, we found that HLP1 mutation caused proximal-to-distal poly(A) site shifts in 2 274 transcripts compared with Col (P < 0.02, Fisher's exact test), suggesting that HLP1 is a 3′-end factor predominantly suppressing the usage of distal poly(A) sites (Figure 4C). The single-molecule direct RNA sequencing (DRS) is a newly developed method and is believed to have less or no artifacts for PAS-seq analysis37. Comparison of transcripts with poly (A) site shifts in our data with the DRS data show that 94% switched PACs were also detected by DRS, suggesting that these switched PACs are reliable (Supplementary information, Figure S9B). Both proximal-to-distal and distal-to-proximal polyadenylation shifts in genes were validated by q-PCR or shown as wiggle plots (Figure 4D and Supplementary information, Figure S10).
Figure 4.
HLP1 regulates global APA. (A) Distribution of poly(A) clusters. (B) Distribution of A-rich and U-rich PAC motifs relative to the poly(A) site (PAS) are indicated by blue and red curves, respectively. (C) Scatter plots show poly(A) site shifts in hlp1-1 mutant. The x axis and y axis show the ratio of PAC counts on a log2 scale between hlp1-1 and Col at proximal and distal poly(A) sites, respectively. Genes with significant proximal-to-distal poly(A) site shifts (P-to-D) are indicated by blue dots and red dots; genes with significant distal-to-proximal shifts (D-to-P) are colored in green and purple. Red and purple dots represent APA shifts with HLP1 binding. Bar graphs indicate the number of these dots. Grey dots represent genes without significant changes in APA. The poly(A) site shifts were evaluated using Fisher's exact test (P < 0.02). (D) Case studies of transcripts with APA shift. Wiggle plots on the left panels show P-to-D shift at At3g15450, At3g23030, At5g11070 and D-to-P shift at At1g72645. CDS regions are boxed in black. The 5′-UTR and 3′-UTR are boxed in green and grey, respectively. Introns are indicated as lines. The x axis indicates genome site in chromosome. The y axis indicates normalized HITS-CLIP/CLIP-seq or PAS-seq abundance. HITS-CLIP/CLIP-seq or PAS-seq tag counts were normalized to tag per 10 million (TP10M) to adjust for differences of two libraries (wild-type and mutant) in sequencing depth. Right panels show RT-qPCR assessments of the shift.
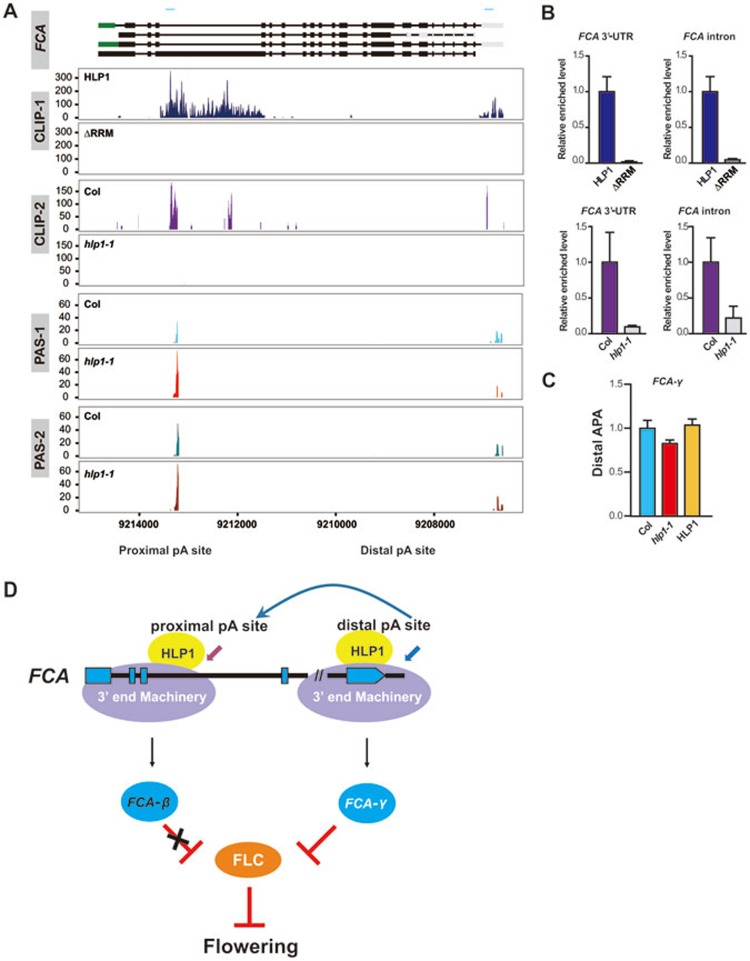
Among the genes whose APA is regulated by HLP1, we found that HLP1 binds directly to the 3′-UTR and the third intron of FCA transcripts (Figure 5A and 5B). Lesion in HLP1 caused upregulated retention of the third intron, which contains the proximal poly(A) site (Figure 5A). PAS-Seq analysis showed decreased polyadenylation at the distal poly(A) site in FCA transcripts in hlp1-1 mutant, resulting in reduced FCA-γ functional isoform, and this shift was rescued in a GFP-HLP1 complementary line (Figure 5A and 5C). This reduction releases the suppression of the floral repressor FLC in hlp1-1 mutant, which leads to delayed flowering (Figures 1E and 5D).
Figure 5.
HLP1 directs poly(A) site choice at the FCA transcripts. (A) The genomic structures of FCA transcripts are shown in the top panel. CDS regions are boxed in black. The 5′-UTR and 3′-UTR are boxed in green and grey, respectively. Introns are indicated as lines. Blue line above gene structure indicates RIP-qPCR amplified region. Wiggle plots of two HITS-CLIP/CLIP-seq (CLIP-1 and CLIP-2) and two PAS-seq (PAS-1 and PAS-2) replicates are shown below the gene structure. HLP1 binding peaks in the third intron and 3′-UTR of FCA transcripts are indicated by blue and purple wiggle plots. The x axis indicates genome site in chromosome. The y axis indicates normalized HITS-CLIP/CLIP-seq or PAS-seq abundance. HITS-CLIP/CLIP-seq or PAS-seq tag counts were normalized to tag per 10 million (TP10M) to adjust for differences of two libraries (wild-type and mutant) in sequencing depth. PAS-Seq analyses show decreased PAC numbers at the distal 3′-UTR poly(A) site in hlp1-1 mutant (red plots in PAS-1 and brown plots in PAS-2) compared with Col (light blue plots in PAS-1 and green plots in PAS-2). (B) RIP-qPCR validation of HLP1 binding to the third intron and 3′-UTR of FCA transcripts. (C) Quantitative PCR results show decreased APA at the distal poly(A) site of FCA in hlp1-1 mutant. (D) Working model for HLP1-directed APA of FCA in the regulation of flowering.
Discussion
Precise regulation of APA relies on the crosstalk between cis-elements that recruit specific subunits of the processing complex and certain RBPs that recognize the specific sequence elements. Recent studies have revealed that this process is much more complex than previously thought and involves multiple protein-RNA interactions12,13. Using integrated high-throughput technologies, we identified HLP1, an hnRNP A/B family protein, as a novel regulator of APA. Hrp1, the yeast CFIB and the counterpart of HLP1, binds AU-rich EE and regulates correct positioning of the cleavage site and 3′-end formation of pre-mRNAs33,34,38. Interestingly, Hrp1 interacts directly with Rna14 (the yeast counterpart of CstF77)38, suggesting a possible link between HLP1 and CstF77, or other components in the mRNA 3′-end processing machinery. Although it is not clear if HLP1 itself is a direct cleavage factor, and the mechanism of selecting a specific poly(A) site remains elusive, we postulate that HLP1 is the functional ortholog of Hrp1 and binds to A-rich or U-rich FUE, NUE or CE (cleavage element), and helps the 3′-end machinery to select the appropriate poly(A) site for subsequent cleavage events.
HLP1 binds to transcripts of thousands of genes, many of which have general roles in plants. Among the 5 569 genes with HLP1 binding sites at their sense transcripts, 429 genes show poly(A) site shift in hlp1-1 mutant, suggesting HLP1 may be directly responsible for APA of these transcripts. Remarkably, HLP1 also binds to sequences other than the poly(A) sites, suggesting additional roles for HLP1. These non-poly(A) binding events may affect the processing (splicing), stability or transport of target transcripts. We found only 21.3% (217/1 018) of differentially expressed genes in hlp1-1 mutant were associated with HLP1 binding, suggesting that a large portion of altered gene expression is indirectly caused by HLP1 mutation (Supplementary information, Figure S11).
The cellular concentration of CstF64 has been reported to affect the selection of poly(A) site in B cells39. Therefore, the expression levels of CstF64 and other 3′-end factors in Col and hlp1-1 mutant were analyzed using RNA-Seq (Supplementary information, Figure S12). Most of these factors show very slight changes in abundance, except for CPSF73-II, which is reduced in hlp1-1 mutant, suggesting that HLP1 may regulate APA mainly through modulating protein-protein interactions or composition of the 3′-end processing complex instead of directly affecting abundance of these factors. However, we can not exclude the possibility that many of these altered APA can result primarily from the reduced level of CPSF73-II in hlp1-1 mutants.
The specific binding of HLP1 to FCA transcripts around the two poly(A) sites suggests HLP1 may be involved in poly(A) site choice of FCA. Previously, the RRM-type RBP FCA was shown to physically interact with the polyadenylation factor FY to regulate its own expression by alternative splicing and polyadenylation mechanisms, and such interaction is required for the regulation of flowering40. Later, FCA and another RRM-type RBP FPA were shown to repress FLC expression22,24,41. Most recently, FCA (probably works together with the mRNA 3′-end processing factor CstF64 and CstF77) and FPA have been shown to function independently in promoting the 3′-end processing of the FLC antisense transcripts at the proximal site, which could silence FLC sense transcripts25,26. We show that HLP1 promotes polyadenylation at the distal site of FCA, which leads to decreased FLC transcripts. HLP1 also directly binds to the proximal poly(A) site of FLC antisense, indicating that HLP1 not only affects FCA poly(A) site choice, but may also play a role in FLC antisense processing together with other 3′-end processing factors. We propose that the late-flowering phenotype could be a net result of altered gene expression upon HLP1 mutation.
Unlike HLP1 and Hrp1, hnRNP A1, the human counterpart of HLP1 has been implicated in regulating pre-mRNA splicing42 and microRNA maturation43. It would not be surprising if hnRNP A1 is also involved in 3′-end formation since increasing number of splicing factors, including Nova32, U1 snRNP44 and hnRNP H45 have been shown to regulate cleavage and polyadenylation.
Materials and Methods
Plant materials and growth conditions
All of the Arabidopsis thaliana lines were in the Col background. Plant growth, flowering time analysis, and plant transformation were performed as previously reported46. The flc-3 mutant was described previously21. The hlp1-1 mutant was isolated from the SALK collection (SALK_021452). Seedlings were grown on Murashige and Skoog (MS) plates containing 3% sucrose at 23 °C under long day conditions for 12 days.
DNA constructs
For the 35S::GFP-HLP1 construct, HLP1 full-length CDS was PCR-amplified with primers CX578 and CX579 and digested with BglII and XbaI, then cloned into BamHI and XbaI sites of the pCAMBIA1300-GFP vector. For the 35S::GFP-HLP1ΔRRM construct, the deletion form of HLP1ΔRRM was PCR-amplified using primers CX7157/CX7158 from the 35S::GFP-HLP1 construct.
Antibodies used in western blotting and HITS-CLIP/CLIP-seq
The rabbit polyclonal HLP1 antibody used in this study was generated against the full-length HLP1 and affinity purified. CX578 and HX1443 primers were used to amplify the coding sequence of HLP1. HSC70 antibody (SPA-818, Enzo Life Sciences) was used as a loading control.
Quantitative PCR and northern blot
Total RNAs were extracted from 12-day-old seedlings using TRNzol reagent (Tiangen). FLC northern blot was performed as previously described46. First-strand cDNA was reverse-transcribed using SuperScript Reverse Transcriptase (Invitrogen). Quantitative PCR was performed using the CFX96 Real-Time System (Biorad) and SYBR Green I (Invitrogen). Primers used for PCR are listed in Supplementary information, Table S4.
HITS-CLIP/CLIP-seq and RIP-RT-PCR validation
The HITS-CLIP/CLIP-seq experiment was performed as described before31,32 with modifications. Briefly, 12-day-old seedlings (from MS plates containing 3% sucrose grown under LD at 23 °C) expressing either GFP-HLP1 (designated as HLP1-CLIP) or GFP-HLP1ΔRRM (designated as ΔRRM-CLIP) were soaked in ice-cold PBS buffer and irradiated twice at 400 mJ/cm2 in a Hoefer UVC 500 Ultraviolet Crosslinker (GE). After UV irradiation, Arabidopsis seedlings were lysed and the cross-linked RNA-protein complexes (RNPs) were immunoprecipitated using anti-GFP antibody (Clontech). The RNPs were digested by micrococcal nuclease (Fermentas) partially (107 dilution) or completely (103 dilution). 3′ RNA adapters were ligated to the immunoprecipitated RNAs, which were 32P-labeled and separated by PAGE gel. Subsequently, protein was digested with proteinase K and purified RNAs were ligated to 5′ RNA adapter. In our CLIP assay, four random nucleotides (barcode) were added to the 3′-terminus of the 5′ adapter to distinguish PCR duplications. After reverse transcription and PCR amplification, high-throughput sequencing was performed on an Illumina platform. For RIP-RT-PCR, RNAs were extracted from GFP-HLP1-RNAs and GFP-HLP1ΔRRM-RNAs complexes using TRNzol (Tiangen), and digested with DNase I (Ambion) before reverse transcription by SuperScript III using random hexamers (Invitrogen). For HLP1-CLIP library replicates and RIP-qPCR validation, HLP1 antibody was used to immunoprecipitate the endogenous HLP1-RNA complex directly from the wild-type plant (Col-0) and the hlp1-1 mutant. Adapters and primers for HITS-CLIP/CLIP-seq and RIP-RT-PCR are listed in Supplementary information, Table S4.
PAS-Seq
PAS-Seq was performed as described previously35,36 with modifications at the HITS-3′ adaptor and sequencing primer. Briefly, poly(A) RNAs were purified using an mRNA purification kit (Invitrogen), and fragmented by heating at 95 °C for 30 min. Reverse transcription (Superscript, Invitrogen) was carried out using our modified HITS-3′ adaptor at 42 °C for 30 min, then the HITS-5′ adaptor (a SMART oligo) was added and incubated for an additional 30 min. The cDNAs were purified using a Qiagen PCR Cleanup kit and the second strand cDNAs were synthesized by three cycles of PCR using Phusion DNA polymerase (NEB) and the PE1.0 and PE2.0 primers. PCR products were separated on a 2% agarose gel and 200-300 bp bands were excised and purified. Gel-extracted DNAs were amplified for additional 13 cycles. PCR products were purified using a Qiagen PCR Cleanup kit. TA-cloning was performed before Illumina sequencing. Oligos for PAS-Seq are listed in Supplementary information, Table S4.
Bioinformatics analysis
Analysis of RNA-seq data RNA-seq libraries for Col and hlp1-1 were constructed following the mRNA sequencing Sample Preparation Guide (Illumina, Cat#RS-930-1001). All raw RNA-seq reads were aligned to the Arabidopsis genome (TAIR10) using TopHat (1.3.0)47 with no more than six mismatches. Reads with unique locations were kept for subsequent analyses. Expression levels of each gene were calculated by counting the number of sequenced tags mapped to the gene and normalized by mapped reads per kilo base of exon per million mapped reads (RPKM) based on a previous method48. Genes with a RPKM fold change > 1.5 and P < 0.01 were regarded as differentially expressed genes.
Analysis of HITS-CLIP/CLIP-seq data HITS-CLIP/CLIP-seq libraries were sequenced on an Illumina platform. 3′-end adaptors and 5′-end 4 nt barcodes were removed using a FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit). Overall, ∼29 million and ∼24 million raw reads (36 nt) were collected from the HLP1 and ΔRRM-CLIP libraries, respectively (Supplementary information, Table S1). Tags ≥ 14 nt in length (about 66% of the total recovered tags) were kept for further analysis, while the shorter tags were discarded, which were usually aligned to multiple loci in the genome. Tags ranging from 14-19 nt (no mismatch allowed) and tags ≥ 20 nt (two mismatches allowed) were aligned to the Arabidopsis genome (TAIR10) using Bowtie (0.12.7)49. Only tags with a unique genomic location were kept for further analysis. Identical sequences with the same 4 nt random barcode were regarded as one tag to exclude PCR duplication. Regions with at least two overlapping tags were grouped and subjected to in silico random CLIP simulations to discard transient and nonspecific HLP1-RNA interactions31. In brief, observed tags were randomly aligned to a given gene region and the maximum number of overlapping tags was calculated. This simulation was repeated 500 times. P-value was calculated by counting the frequency of maximum cluster height from the 500 repeats. Peak heights with P < 0.01 were used as thresholds to identify significant HLP1 binding sites/peaks.
Analysis of PAS-seq data Tags less than 20 nt after removal of the 3′ linker and 4 nt random barcodes were discarded. All tags were reverse-complemented because the tags were sequenced from 3′-end of transcripts. Trimmed tags were then mapped to the Arabidopsis genome (TAIR10) using Bowtie (0.12.7)49, allowing two mismatches. Only uniquely mapped tags were kept for downstream analyses. Uniquely located tags with six or more continuous adenines downstream of the poly(A) junction in a 10-nt window were considered as internal priming tags and discarded36. About 9-12 million comparable clean reads, namely poly(A) tags (PATs), were obtained from each library (Supplementary information, Table S3). Due to intrinsic heterogeneity, poly(A) sites located within a 24-nt window with TPM (tag per million) > 3 in the same gene were pooled and defined as a poly(A) site cluster as described7. Overall, > 21 000 PACs in the sense orientation were identified from each library (Tag Per Million > 3; Supplementary information, Table S3). To ensure data reliability, two biological replicates were performed for wild-type Col and hlp1-1 mutant. Both data sets were highly consistent (Supplementary information, Figure S8B and S8C). For statistical analysis of APA shifts, genes with at least two PACs (TPM > 3) were selected for analysis of differential usage. We used Fisher's exact test to compare the APA shift events between the two most predominant PACs as described7. A shift with P < 0.02 was regarded as significant different shifts. Overlapping APA shift events from two biological replicates were used for downstream analysis.
Motif analysis HLP1 binding motifs were identified using MEME as previously described50. Range of motif widths was set to 4 and 20 as the minimum motif width and maximum motif width, respectively. Poly(A) site cluster motifs were identified by extracting PAC sequences with TPM > 3.
Phylogenetic tree and domain analysis HLP1 orthologs were retrieved from the InParanoid database51. A phylogenetic tree of HLP1 and related proteins was then constructed by MEGA4 using the Neighbor-joining method with a bootstrap value of 1 000.
Accession numbers
Sequencing data sets described in this study have been deposited in GEO database under accession number GSE39051.
Acknowledgments
We thank all the members in Cao group for discussions. And we are grateful to Drs Haofeng Chen, Xingyu Hu and Ms Fei Meng at the genomics core facility at IGDB, CAS for deep sequencing. This work was supported by the National Basic Research Program of China (2014CB943400 to CL, 2011CB915400 to XC, and 2013CB967303 to XD), the National Natural Science Foundation of China (31330020 and 31210103901 to XC, 31171184 and 90919033 to CL, and 31200900 to XD), the Transgenic Project (2014ZX08010-002 to XC), the Chinese Academy of Sciences (KSCX2-EW-Q-24 to CL) and the State Key Laboratory of Plant Genomics (2014B0227-01 and 2015B0129-01 to XC).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
HLP1 (At5g40490) belongs to the Arabidopsis hnRNP A/B family.
The RRMs of HLP1 are required but insufficient for promoting floral transition.
An overview of HLP1 binding tags and binding sites.
A snapshot of HLP1 binding to different regions of transcripts.
The top 10 motifs of HLP1 binding tags analyzed using MEME software.
HLP1-CLIP replicate using HLP1 antibody.
Replicates of CLIP-seq and RIP-qPCR validation of HLP1 binding.
Poly(A) site sequencing.
Statistical analysis of PAS-seq.
Case studies of transcripts with APA shift in two biological repeats.
Correlation between HLP1 binding and differential gene expression.
Expression levels of RNA 3′ end processing factors in hlp1-1 mutant compared to Col determined by RNA-Seq.
Summary of HLP1 HITS-CLIP from two biological replicates
Flower-related genes with HLP1 binding
Summary of PAS-Seq reads from two biological replicates
Primers and oligos
References
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43:853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22:1173–1183. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, et al. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods. 2013;10:133–139. doi: 10.1038/nmeth.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, et al. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell. 2010;143:1018–1029. doi: 10.1016/j.cell.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Liu M, Downie B, et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA. 2011;108:12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, et al. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep. 2012;1:277–289. doi: 10.1016/j.celrep.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, et al. Extensive alternative polyadenylation during zebrafish development. Genome Res. 2012;22:2054–2066. doi: 10.1101/gr.139733.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, et al. The landscape of C. elegans 3′UTRs. Science. 2010;329:432–435. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. Formation, regulation and evolution of Caenorhabditis elegans 3UTRs. Nature. 2011;469:97–101. doi: 10.1038/nature09616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonemann L, Kuhn U, Martin G, et al. Reconstitution of CPSF active in polyadenylation: recognition of the polyadenylation signal by WDR33. Genes Dev. 2014;28:2381–2393. doi: 10.1101/gad.250985.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Huppertz I, Yao C, et al. CPSF30 and Wdr33 directly bind to AAUAAA in mammalian mRNA 3 processing. Genes Dev. 2014;28:2370–2380. doi: 10.1101/gad.250993.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Brunkow ME, Ramsdell F, et al. A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA-->AAUGAA) leads to the IPEX syndrome. Immunogenetics. 2001;53:435–439. doi: 10.1007/s002510100358. [DOI] [PubMed] [Google Scholar]
- Comstock CE, Augello MA, Benito RP, et al. Cyclin D1 splice variants: polymorphism, risk, and isoform-specific regulation in prostate cancer. Clin Cancer Res. 2009;15:5338–5349. doi: 10.1158/1078-0432.CCR-08-2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal M, Elkon R, Loayza-Puch F, et al. The poly(a)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Terzi LC, Simpson GG. Regulation of flowering time by RNA processing. Curr Top Microbiol Immunol. 2008;326:201–218. doi: 10.1007/978-3-540-76776-3_11. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell. 1999;11:949–956. doi: 10.1105/tpc.11.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, et al. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Schomburg FM, Patton DA, Meinke DW, Amasino RM. FPA, a gene involved in floral induction in Arabidopsis, encodes a protein containing RNA-recognition motifs. Plant Cell. 2001;13:1427–1436. doi: 10.1105/tpc.13.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Duroux M, Laurie R, Dijkwel P, Simpson G, Dean C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell. 2002;14:877–888. doi: 10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG. The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell. 2010;18:203–213. doi: 10.1016/j.devcel.2009.12.009. [DOI] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327:94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- Hunt AG. Messenger RNA 3' end formation in plants. Curr Top Microbiol Immunol. 2008;326:151–177. doi: 10.1007/978-3-540-76776-3_9. [DOI] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3' end processing regulation. Nucleic Acids Res. 2010;38:2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3'-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Li QQ. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip Rev RNA. 2011;2:445–458. doi: 10.1002/wrna.59. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MM, Henry MF, Shen E, et al. Hrp1, a sequence-specific RNA-binding protein that shuttles between the nucleus and the cytoplasm, is required for mRNA 3'-end formation in yeast. Genes Dev. 1997;11:2545–2556. doi: 10.1101/gad.11.19.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Guisbert KS, Li H, Guthrie C. Alternative 3′ pre-mRNA processing in Saccharomyces cerevisiae is modulated by Nab4/Hrp1 in vivo. PLoS Biol. 2007;5:e6. doi: 10.1371/journal.pbio.0050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, et al. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 2011;21:741–747. doi: 10.1101/gr.115295.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17:761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A, Duc C, Cole C, et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol. 2012;19:845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S, Moore C. Five subunits are required for reconstitution of the cleavage and polyadenylation activities of Saccharomyces cerevisiae cleavage factor I. Proc Natl Acad Sci USA. 2001;98:6080–6085. doi: 10.1073/pnas.101046598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–771. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C. FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell. 2003;113:777–787. doi: 10.1016/s0092-8674(03)00425-2. [DOI] [PubMed] [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson GG. Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J. 2003;22:3142–3152. doi: 10.1093/emboj/cdg305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Krainer AR. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Niu L, Lu F, et al. Mutations in the Type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007;144:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund G, Schmitt T, Forslund K, et al. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HLP1 (At5g40490) belongs to the Arabidopsis hnRNP A/B family.
The RRMs of HLP1 are required but insufficient for promoting floral transition.
An overview of HLP1 binding tags and binding sites.
A snapshot of HLP1 binding to different regions of transcripts.
The top 10 motifs of HLP1 binding tags analyzed using MEME software.
HLP1-CLIP replicate using HLP1 antibody.
Replicates of CLIP-seq and RIP-qPCR validation of HLP1 binding.
Poly(A) site sequencing.
Statistical analysis of PAS-seq.
Case studies of transcripts with APA shift in two biological repeats.
Correlation between HLP1 binding and differential gene expression.
Expression levels of RNA 3′ end processing factors in hlp1-1 mutant compared to Col determined by RNA-Seq.
Summary of HLP1 HITS-CLIP from two biological replicates
Flower-related genes with HLP1 binding
Summary of PAS-Seq reads from two biological replicates
Primers and oligos