Cnn and PLP directly interact at two defined sites to coordinate the cell cycle–dependent rearrangement and scaffolding activity of the centrosome to permit normal centrosome organization, cell division, and embryonic viability.
Abstract
Pericentriolar material (PCM) mediates the microtubule (MT) nucleation and anchoring activity of centrosomes. A scaffold organized by Centrosomin (Cnn) serves to ensure proper PCM architecture and functional changes in centrosome activity with each cell cycle. Here, we investigate the mechanisms that spatially restrict and temporally coordinate centrosome scaffold formation. Focusing on the mitotic-to-interphase transition in Drosophila melanogaster embryos, we show that the elaboration of the interphase Cnn scaffold defines a major structural rearrangement of the centrosome. We identify an unprecedented role for Pericentrin-like protein (PLP), which localizes to the tips of extended Cnn flares, to maintain robust interphase centrosome activity and promote the formation of interphase MT asters required for normal nuclear spacing, centrosome segregation, and compartmentalization of the syncytial embryo. Our data reveal that Cnn and PLP directly interact at two defined sites to coordinate the cell cycle–dependent rearrangement and scaffolding activity of the centrosome to permit normal centrosome organization, cell division, and embryonic viability.
Introduction
Centrosomes are composed of a pair of centrioles embedded in pericentriolar material (PCM) and function as microtubule (MT) organizing centers (MTOCs; Gould and Borisy, 1977). In mitosis, centrosomes organize the bipolar spindle, while in interphase they direct cell migration, traffic cargoes, and build cilia (Doxsey et al., 2005). These functional changes are linked to oscillations in PCM levels. Centrosomes gain MTOC activity by increasing PCM levels, or maturing, before mitosis. The process is then reversed during mitotic exit (Khodjakov and Rieder, 1999; Palazzo et al., 2000). Elucidating the regulation of PCM dynamics is critical to understanding how centrosome function is normally modulated and deregulated in disease (Nigg and Raff, 2009).
Super-resolution microscopy has revised our view of PCM from an amorphous cloud to a structured architecture (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). PCM organization into distinct zones appears conserved across taxa (Lüders, 2012; Mennella et al., 2014), and some proteins, such as Pericentrin (Pcnt)-like protein (PLP; Kawaguchi and Zheng, 2004; Martinez-Campos et al., 2004), and its mammalian orthologue, Pcnt, radially extend across zones (Lawo et al., 2012; Mennella et al., 2012). Understanding how proteins function within these subdomains is key to understanding the cell cycle dynamics, regulation, and function of PCM.
One question that emerges from the discovery of the PCM organization is the identification of the molecular glue, or scaffold, that holds the structure together. A centrosome scaffold was first proposed upon resolving Pcnt and γ-Tubulin (γTub) to a reticular, tubelike lattice (Dictenberg et al., 1998). Much of our understanding of the scaffold comes from studies of the syncytial Drosophila melanogaster embryo, where a constitutively active MTOC serves several essential functions: proper nuclear migration/spacing, actin organization, rapid progression through abridged nuclear cycles (NCs) that lack gap phases, and cellularization (Callaini and Riparbelli, 1990; Rothwell and Sullivan, 2000). Increasing evidence suggests that Centrosomin (Cnn) forms an oligomerized scaffold required to recruit other PCM proteins (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999; Zhang and Megraw, 2007; Kao and Megraw, 2009; Conduit et al., 2010, 2014a,b). Thus far, a conserved motif at the N terminus of Cnn (CM1; Zhang and Megraw, 2007), a direct interaction between Cnn and Spd2 (Conduit et al., 2014b), and phosphorylation of Cnn by Polo kinase (Conduit et al., 2014a) have all been implicated in Cnn scaffold assembly. However, how the Cnn scaffold efficiently assembles with each rapid NC remains poorly understood.
In humans, mutations in Pcnt and the Cnn orthologue, Cdk5rap2/Cep215, are associated with microcephaly (Bond et al., 2005; Rauch et al., 2008). Because a functional interaction between Pcnt and Cep215 may provide a mechanistic link between these disorders (Buchman et al., 2010), further understanding the interplay between these molecules may contribute to our understanding of disease etiology. Work in Drosophila larval neuroblasts (NBs) shows that PLP plays a minor role in organizing Cnn (Martinez-Campos et al., 2004; Galletta et al., 2014); however, a functional role for PLP has not been examined outside of NBs. Thus, it is currently unknown if PLP is required to organize the Cnn scaffold in embryos.
We use live imaging and structured illumination microscopy (SIM) to detail a cell cycle–dependent reorganization of the embryo centrosome, where PCM is expanded during interphase and compact during mitosis. We identify interphase-specific PLP satellites, novel structures that localize exclusively to the tips of Cnn flares. Furthermore, we show a Cnn and PLP direct interaction at two domains. Mutant analysis supports a role for PLP in coordinating the cell cycle–dependent rearrangement of the Cnn scaffold that is essential for normal centrosome function. Our data indicate that proper interphase centrosome organization by the PLP-Cnn scaffold is required for centrosome separation, mitotic progression, genome stability, and embryonic viability.
Results
Centrosomes remain active throughout interphase in the early Drosophila embryo
Centrosome composition, organization, and activity undergo striking cell cycle oscillations (Palazzo et al., 2000). Previous work has identified Cnn as a key regulator of centrosome dynamics through its scaffolding of PCM proteins (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999; Terada et al., 2003; Lucas and Raff, 2007; Zhang and Megraw, 2007; Dobbelaere et al., 2008; Conduit et al., 2010, 2014a,b). At embryonic centrosomes, Cnn dramatically rearranges from a compact sphere in mitosis to an expanded interphase structure that is defined by a central bolus of PCM and extended radial fibers (Li and Kaufman, 1996; Megraw et al., 2002); for clarity, we refer to these fibers as Cnn flares. In addition, Cnn particles are released into the cytoplasm primarily during interphase (Megraw et al., 2002). This leads to an attractive model whereby the cell cycle–dependent modulation of Cnn organization directly regulates centrosome activity. However, mechanisms regulating this reorganization remain poorly understood, and the physiological significance of centrosome shape changes is currently unknown.
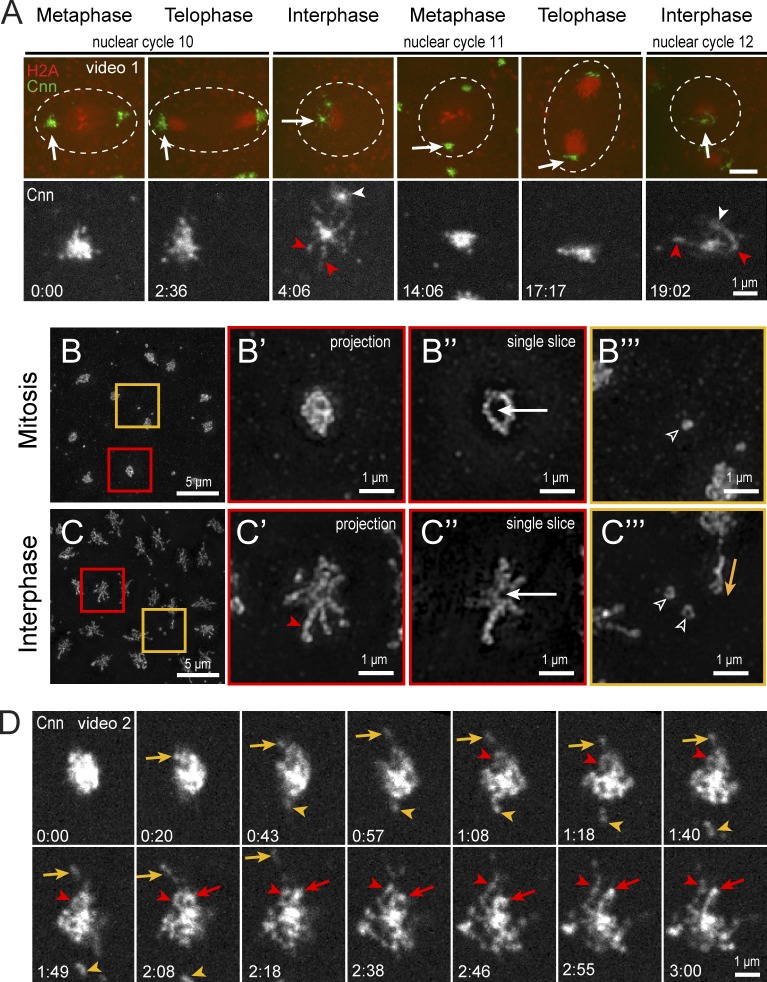
To study Cnn during the rapid early divisions, it is critical to precisely track the timing of its reorganization. By imaging recombineered Cnn in embryos coexpressing the nuclear marker H2A-RFP, we find that Cnn is a compact structure during mitosis (Fig. 1 A and Video 1). Upon mitotic exit and throughout interphase, Cnn becomes expansive and particulate, ultimately extending reticulated flares that greatly increase centrosome volume and eject particles into the cytoplasm (Fig. 1 A, 4:06). These findings are in agreement with a seminal study of Cnn dynamics (Megraw et al., 2002). To monitor the conspicuous changes to centrosome size and shape, we visualized Cnn distribution by SIM. SIM resolves mitotic Cnn as a hollow sphere with a discernable strand-like substructure (Fig. 1, B–B″) with few cytoplasmic particles (Fig. 1 B‴). In contrast, interphase centrosomes form a more elaborate shape with extended flares of uneven thickness and length (Fig. 1, C–C″). These flares are sites of Cnn particle release (Fig. 1 C‴), which appear analogous to mammalian centriole satellites (Rattner, 1992; Balczon et al., 1994; Zhang and Megraw, 2007), as both are ejected from the centrosome and bidirectionally move during interphase (Kubo et al., 1999; Megraw et al., 2002). In addition, SIM resolves Cnn particles as rings, similar to the structure of mammalian centriole satellites revealed by EM (de-Thé, 1964; Kubo et al., 1999). Although mammalian centriole satellites facilitate protein trafficking to and from centrosomes and cilia, the overall functional significance of mammalian satellites requires further study (Mahjoub and Tsou, 2013).
Figure 1.
Rearrangement of Cnn at the mitotic-to-interphase transition. (A) Live Cnn-GFP and H2A-RFP in a pseudo-cell (broken line) of a WT embryo through three cell cycles. Arrows show the enlarged regions below (Cnn, white). Cnn flares (red arrowheads) and separating daughter centrosomes (white arrowheads) in interphase are indicated. Bars in top panels, 5 µm. (B and C) SIM images of mitotic (B) and interphase (C) embryos stained for Cnn. Centrosomes (red boxes) are magnified to the right as projections (B′ and C′) and single optical sections (B″ and C″) through the centrosome center. Interphase flare (red arrowhead) and centriole position (white arrows) are shown. (B‴ and C‴) Cytoplasmic regions (orange boxes) show particles (open arrowheads) and a particle release event (orange arrow). (D) Live Cnn-GFP at mitotic exit. Released particles (orange) and unfolding flares (red) are shown. Time is given in minutes:seconds.
To observe the mitotic-to-interphase transition of Cnn in finer detail, we imaged embryos with greater temporal resolution. Unexpectedly, the compact mitotic centrosome gradually appears to unfurl as flares seemingly unfold and extend from the center mass (Fig. 1 D and Video 2), ultimately forming an interphase centrosome lattice. Retention of Cnn at the interphase centrosome is consistent with the constitutive activity of embryonic centrosomes throughout the cell cycle, which is thought to permit efficient NC progression (Callaini and Riparbelli, 1990). These findings suggest that embryonic Cnn does not follow the paradigm of PCM shedding and total centrosome dematuration during mitotic exit, and only a small amount of Cnn is released into the cytoplasm as particles. Understanding this rearrangement and how Cnn remains anchored at the centrosome is fundamental to understanding how centrosome function is modulated during the rapid divisions of early development.
Centrosomes add a flare zone in interphase
Recent studies using SIM to define the PCM structure in cultured cells revealed that the ostensibly amorphous PCM is, indeed, organized into zones (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012). To understand how interphase and mitotic centrosome shape is determined, we conducted a SIM-based survey to demarcate centrosome zones in the early Drosophila embryo.
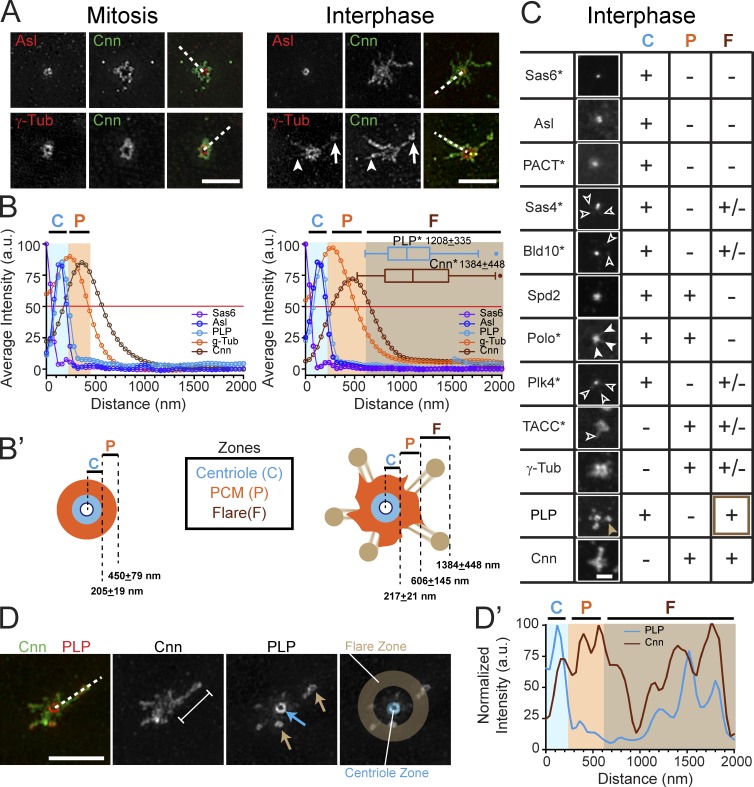
As expected, we found that the distributions of centriole proteins, such as SAS6 and Asterless (Asl), remain constant in mitosis and interphase (Fig. 2, A and B). These data were used to define a centriole zone with an ∼200-nm radius (Fig. 2 B, blue shading; and Fig. S1, A and B). In contrast, the PCM zone, defined by γTub, expands from a radius of 450 nm in mitosis to 600 nm in interphase (Fig. 2 B, orange shading; and Fig. S1, A and B). Thus, the outer edge of the PCM zone expands as embryos enter interphase. The major structural change to the interphase centrosome is the addition of extensive Cnn flares, which protrude well beyond the PCM zone. We term this zone the interphase flare zone (Fig. 2 B, brown shading; and Fig. S1, A and B). Notably, these interphase-specific flares extend ∼1,380 nm, with some reaching >2 µm, more than twice as far as the PCM zone (Figs. 2 B and S1 A). In sum, our analysis of embryonic centrosome organization shows that interphase centrosomes form a distinct flare zone in addition to the centriole and PCM zones (Fig. 2 B′). Moreover, our imaging (Fig. 1 C) reveals that the mitotic Cnn bolus appears to unfold to provide the source of interphase Cnn flares.
Figure 2.
Reorganization of the centrosome structure in interphase. (A) SIM images of WT embryos stained for the indicated proteins. The presence (arrows) and absence (arrowheads) of γTub within Cnn flares is shown. (B) Mean radial intensity distribution of centrosome proteins in mitosis (left) and interphase (right) calculated from line scans derived from n = 30–110 centrosomes (broken lines in A). Shaded areas show the centriole (C, blue), PCM (P, orange), and flare (F, brown) zones as defined by the outer edges (OE) of Asl, γTub, and Cnn, respectively (see Materials and methods). The asterisk denotes satellite or flare measurement. (B′) Diagram of centrosome zones at mitosis (left) and interphase (right). (C) Confocal projections of the indicated proteins assayed for localization to the C, P, and F zones; +, present; −, absent; and +/−, low or variable levels; *, protein detected by GFP transgene. See Fig. S1 C for contrast-enhanced versions of Sas4, Bld10, Plk4, Polo, and Spd2. Open arrowheads show low localization of protein to the flare zone; closed arrowheads show Polo extending into the PCM zone. The brown arrowhead highlights the strong localization of PLP to the flare zone. (D) SIM image of a WT interphase centrosome with a Cnn flare (bracket); arrows show PLP at the centriole (blue) and satellites (brown). Line scan (broken line, D′) shows representative distribution relative to the centriole center. Bars: (A and D) 2.5 µm; (C) 1 µm.
Interphase PLP satellites define the margin of interphase Cnn flares
Reasoning that factors enriched in the interphase flare zone might regulate or contribute to Cnn scaffold function, we assayed the localization of many other centrosome proteins (Fig. 2 C). As expected, Sas6, Asl, and the centriole marker, PACT-GFP, are restricted to the centriole zone. Sas4 and Bld10/Cep135 are also enriched in the centriole zone, but show low levels at discrete foci within the flare zone (Fig. S1 C, enhanced image). Polo, a key regulator of maturation, radially extends into the PCM zone, while low levels of PLK4/SAK, a key regulator of centriole duplication, localize to small foci within the flare zone (Figs. 2 C and S1 C). Because we detect only low levels of Sas4, Cep135, and PLK4 in the interphase flare zone upon overexpression of GFP fusion constructs, and could not verify the endogenous localization of these proteins, we cannot speculate on their function within this zone.
The defining molecule of the PCM zone, γTub, is found at low levels in the interphase flare zone (Fig. 2, A and C). However, even at a single centrosome, γTub levels vary greatly between flares (Fig. 2 A, arrows). This heterogeneity is also true for TACC (Fig. 2 C), as previously reported (Megraw et al., 2002). We find endogenous Spd2 primarily within the centriole and PCM zones (Figs. 2 C and S1 C), in agreement with GFP-tagged Spd2 localization at centrioles and PCM with only low amounts in “peripheral” areas (Conduit et al., 2014b), which we presume is the flare zone.
In contrast, we find endogenous PLP at high levels in nearly every Cnn flare (94.7%; n = 190 flares, 82 centrosomes) in addition to its known centriole localization, which suggests that PLP may coordinate centrosome shape changes with Cnn, while almost no PLP is detected in the PCM zone (Figs. 2, B–D′). More than two decades ago, Pcnt was localized to “PCM-like bodies (satellites)” in mammalian cells using immuno-EM (see Fig. 4; Doxsey et al., 1994). Subsequent work proved that Pcnt forms a biochemical complex with the canonical mammalian centriole satellite marker, PCM-1 (Li et al., 2001; Dammermann and Merdes, 2002). Although a functional orthologue of PCM-1 has not been identified in Drosophila, the discrete localization of PLP foci at flare tips is reminiscent of satellites orbiting a central body (the centriole); therefore, we refer to PLP localized within the interphase flare zone as PLP satellites.
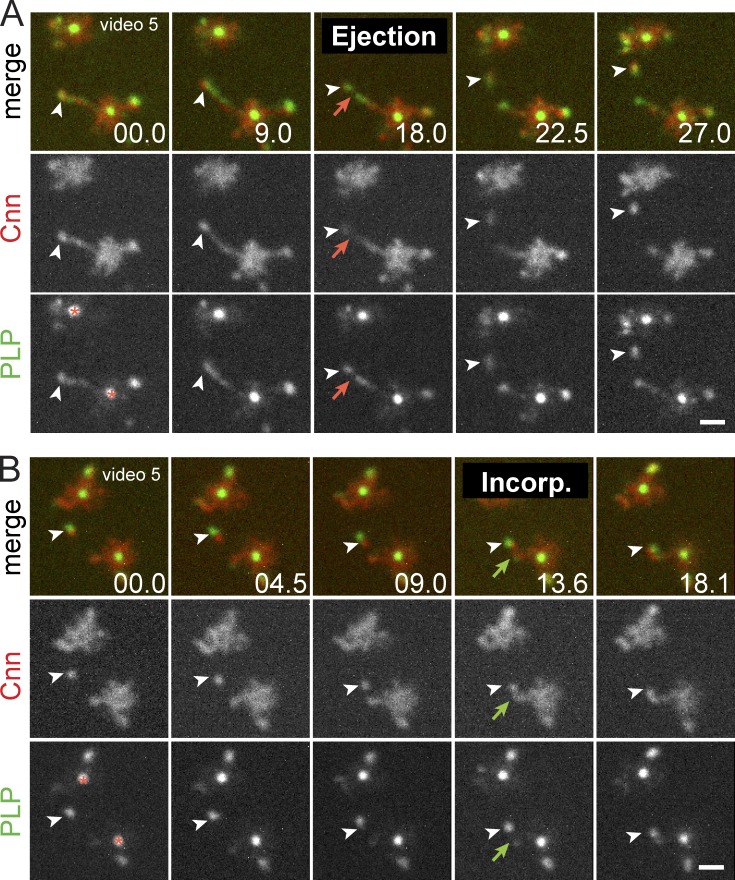
Figure 4.
Cnn and PLP are packaged together into dynamic flares. (A) Live PLPFL and Cnn-mCherry at two centrosome pairs within a single embryo show coincidence (arrowheads) at the tip of an extended flare. Red arrows show particle release. (B) Particle associates with existing PCM (green arrows). The asterisks mark the centriole. Time is given in seconds. Bars, 1 µm.
PLP satellites are found at the tips of Cnn flares (Fig. 2, D and D′) and extend ∼1,208 nm from the centriole center (Figs. 2 B and S1 A), 10-fold farther than the predicted ∼110-nm length of a PLP molecule (Mennella et al., 2012). Thus, PLP localizes to two distinct pools (centrioles and satellites) separated by a considerable distance. SIM resolves PLP satellites as rings with a diameter of 187 ± 22.9 nm (Fig. 2 D, n = 40). Based on the unique localization of PLP satellites to the tips of the extended interphase centrosome, we hypothesized that PLP functions as a molecular scaffold that stabilizes Cnn flares and contributes to centrosome shape and, in turn, centrosome function.
PLP satellites and Cnn flares share similar dynamics and coassemble
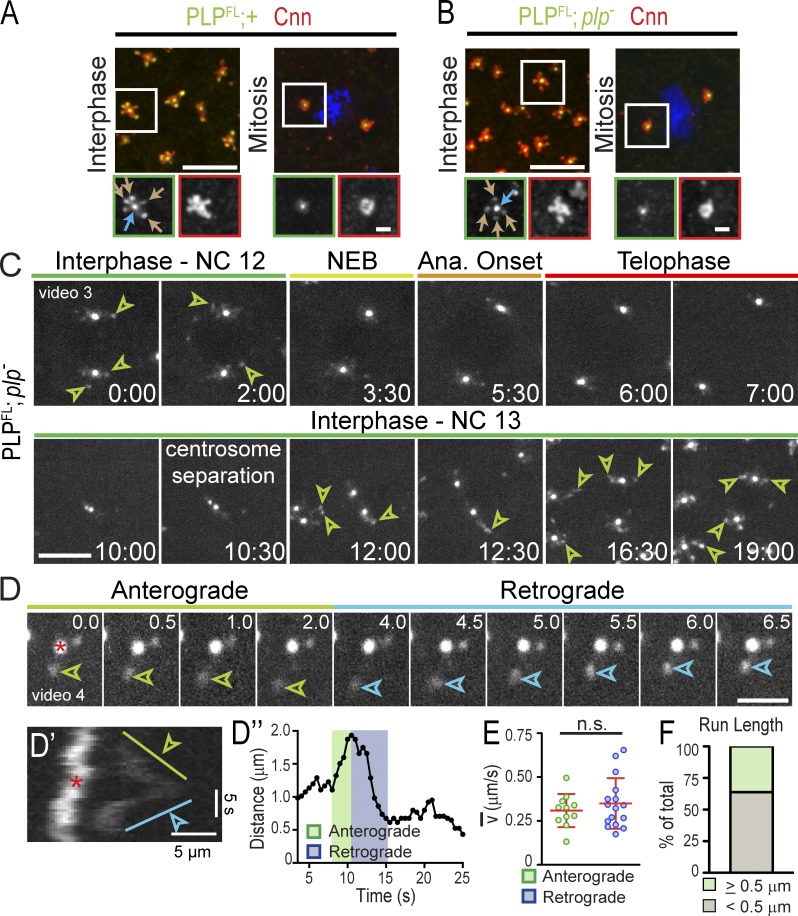
To investigate the function of PLP satellites in detail, we examined live embryos expressing full-length PLP-GFP (PLPFL), which mirrors endogenous PLP distribution (Fig. 3, A and B) and fully rescues plp− viability (Galletta et al., 2014). Live imaging shows that PLPFL remains closely apposed to centrioles throughout the cell cycle, but expands into the flare zone as PLP satellites in interphase (30/31 embryos; Fig. 3 C and Video 3). Satellites diminish upon nuclear envelope breakdown (Fig. 3 C, 3:30), are absent in mitosis (Fig. 3 C, 7:00), reemerge in interphase during centrosome separation (Fig. 3 C, 10:30), and become prominent once centrosomes fully separate (Fig. 3 C, 19:00). Therefore, PLP satellite assembly and disassembly are entrained with the cell cycle where satellite-permissive conditions are present during interphase. Moreover, the timing of PLP satellite addition and removal bears a striking resemblance to that of Cnn flares (Fig. 1 A).
Figure 3.
PLPFL localizes to dynamic satellite structures. (A and B) Embryos were stained for the indicated proteins. Arrows show PLP at centrioles (blue) and satellites (brown). The boxed sections are enlarged below. (C) Live PLPFL shows interphase PLP satellites (arrowheads) in NC 12 and 13. Time is given in minutes:seconds. (D) Anterograde (green) and retrograde (blue) satellite run relative to centriole (asterisk). Time is given in seconds. (D′) Corresponding kymograph and plot of distance over time (D″). (E) Average velocity of directed runs; n.s., not significant; n = 27 runs. (F) Frequency of satellites with directed (≥0.5 µm) runs; n = 58 centrosomes. Bars: (A and B, top) 5 µm; (A and B, bottom) 1 µm; (C) 5 µm; (D) 2 µm.
Imaging individual embryos progressing from NC 10 or 11 (early blastoderm) through NC 14 (cellularization) reveals that PLPFL satellites become brighter and more structured with each NC, reaching a peak in NC 14 (n = 5/5 embryos; Fig. 3 C, 0:00 vs. 19:00; Fig. S2 A, 97:30). PLP satellites are no longer detected after cellularization (Fig. S2 A′), once the interphase centrosomes are inactivated (Harris and Peifer, 2007). Thus, PLP satellite formation is also developmentally regulated, suggesting that their function may be critical during the increasingly prolonged interphases of later syncytial NCs (Foe and Alberts, 1983).
To examine PLP satellite dynamics in detail, we imaged PLPFL at higher temporal resolution. This reveals that PLP satellites undergo bidirectional, linear runs (Fig. 3, D–D″; and Video 4) with an average velocity of 0.33 ± 0.13 µm/s (n = 27; Fig. 3 E), which is in agreement with velocities reported for cytoplasmic Cnn flare particles (Megraw et al., 2002). More than 35% (n = 21/58) of PLP satellites undergo directed runs ≥0.5 µm (Fig. 3 F), with an average run length of 0.77 ± 0.22 µm and some runs exceeding 1.5 µm or interspersed with pauses. We conclude that PLP satellites share many of the same dynamic properties as Cnn.
A model where a PLP–Cnn complex can assemble into a stable scaffold is supported by experiments that show that overexpression of either mammalian Cep215 or Pcnt is sufficient to form a super scaffold that recruits high levels of the other (Lawo et al., 2012; Pagan et al., 2015) and by the biochemical association of PLP and Cnn within embryonic extracts (Conduit et al., 2010; Gopalakrishnan et al., 2011). To investigate whether PLP satellites and Cnn are assembled into a complex, we performed live imaging of PLP-GFP and Cnn-mCherry, which revealed an enrichment of Cnn at flare tips that is coincident with PLP satellites (Figs. 4 A and 2 D). Visualizing the release of a Cnn particle shows that Cnn and PLP are packaged together and co-traffic within the cytoplasm (Fig. 4 A and Video 5). Likewise, mobile cytoplasmic particles containing both Cnn and PLP can associate with existing flares (Fig. 4 B and Video 5). In contrast, live imaging of embryos expressing PLP-GFP and another dynamic PCM component, TACC-RFP, reveals an uneven distribution within PLP satellites (Fig. S2 B). Therefore, Cnn and PLP share a specific relationship within the centrosome and likely function together to direct local changes to its size and activity.
Given these coordinated movements, we sought to test if Cnn flare extension is required for PLP satellite formation. Previous studies show that treatment with the MT drug colchicine causes Cnn flares to collapse into a compact, mitotic-like configuration (Megraw et al., 2002). Injecting embryos expressing PLP-GFP and Cnn-mCherry with colchicine results in the concomitant compaction of Cnn and PLP (Fig. S2 C). Thus, while MTs are dispensable for the localization of Cnn to the PCM zone (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999) and PLP to the centriole zone (Kawaguchi and Zheng, 2004), we find that MTs are required for the extension of Cnn and PLP into the interphase flare zone. Another correlation is observed upon examination of mother and daughter centrosomes. It was previously noted that higher levels of Cnn accumulate on the mother as compared with the daughter (Conduit et al., 2010). Similarly, we note that greater amounts of PLP associate with the larger, Cnn-rich mother centrosome than the smaller daughter centrosome (Fig. S2 D). These data further suggest that Cnn and PLP are coregulated and may cooperate to determine centrosome size and activity.
PLP organizes the Cnn scaffold
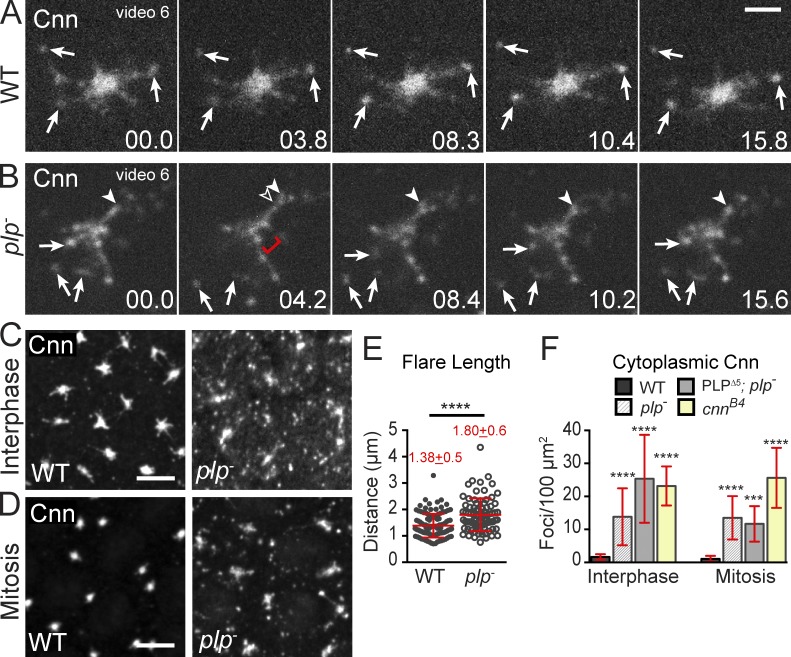
To date, a functional role for PLP has not been examined in the Drosophila embryo. However, a recently reported mouse model shows that Pcnt organizes Cep215 and is required for cardiovascular and neural development (Chen et al., 2014). To investigate whether PLP regulates centrosome size or function, and whether this regulation is important for early Drosophila development, we generated plp− null embryos (Fig. S3 A). Loss of PLP leads to 100% lethality; 32% die as embryos and 68% as first instar larva (see Materials and methods), which indicates that PLP is essential for viability. To determine if PLP is required to maintain the Cnn interphase lattice, we visualized Cnn in live wild-type (WT) and plp− embryos. Compared with WT embryos expressing Cnn-GFP (Fig. 5 A and Video 6), plp− centrosomes show Cnn disorganization and dispersal (Fig. 5 B). In plp− embryos, Cnn appears to ooze away from the centrosome, forming extended flares that eject an increased number of cytoplasmic particles. In addition, large voids interrupt the normally uniform distribution of Cnn within the PCM zone (Fig. 5 B, bracket).
Figure 5.
PLP organizes the Cnn scaffold. (A and B) Live Cnn-mCherry in WT (A) and plp− (B) embryos in interphase. Released particles (arrows), extending flare (arrowheads), and disrupted PCM (bracket) are shown. Time is given in seconds. (C and D) Embryos stained for Cnn. (E) Cnn flare length in interphase embryos; n > 80 centrosomes. Mean ± SD is indicated. (F) Cytoplasmic Cnn particles in a 100-µm2 area (interphase embryos: WT n = 87, plp− n = 18, PLPΔ5 n = 24, cnnB4 n = 21; mitotic embryos: WT n = 40, plp− n = 18, PLPΔ5 n = 9, cnnB4 n = 15). Data are mean ± SD (error bars). ***, P < 0.001; ****, P < 0.0001 by a Student’s two-tailed t test relative to WT. Data shown are from a single representative experiment out of two or more repeats. Bars: (A and B) 2.5 µm; (C and D) 5 µm.
To quantify these abnormalities, we imaged endogenous Cnn in plp− embryos (Fig. 5, C and D). Strikingly, Cnn flares in plp− mutants are significantly longer by ∼30% (1.80 µm vs. 1.38 µm in WT; Fig. 5 E) with some flares reaching >3 µm. The presence of long Cnn flares and the degree of Cnn disorganization at plp− centrosomes made it difficult to accurately measure Cnn within the PCM zone. Therefore, we also investigated endogenous γTub behavior in plp− embryos, which confirmed the significant PCM disorder (Fig. S3 B, brackets). Live imaging of GFP-γTub throughout the cell cycle further shows that PCM dispersal in plp− mutants is most evident in interphase (Fig. S3, C and D, green boxes; and Video 7); γTub during mitosis appears less disrupted and more similar to controls (Fig. S3, B–D).
Our analysis also revealed a significant ninefold increase in cytoplasmic Cnn particles in interphase plp− embryos (13.8/100 µm2 vs. 1.6/100 µm2 in WT; Fig. 5, C, D, and F). Moreover, these flares remain in the cytoplasm and fail to consolidate into the centrosome in mitosis (13.5/100 µm2 vs. 1.0/100 µm2 in WT). Our data show that PLP is required for normal Cnn lattice arrangement and for Cnn retention at interphase centrosomes (Fig. 5, B–D). Our findings that γTub dispersal is more severe in interphase plp− embryos (Fig. S3, B and D), and that PLP satellites are only present in interphase hint that PLP satellites might function as structural elements that physically scaffold and anchor the PCM to properly determine interphase centrosome shape. This positive regulatory role for PLP-mediated regulation of interphase centrosome activity agrees with results from mammalian and Drosophila cultured cells (Loncarek et al., 2008; Kim and Rhee, 2011; Lawo et al., 2012; Mennella et al., 2012; Wang et al., 2013; Pagan et al., 2015), as well as mitotic Drosophila neuroblasts (Martinez-Campos et al., 2004; Galletta et al., 2014), but contrasts with a negative regulatory role for PLP in interphase neuroblasts (Lerit and Rusan, 2013). Thus, modulation of centrosome activity by PLP is differentially regulated in various cellular contexts.
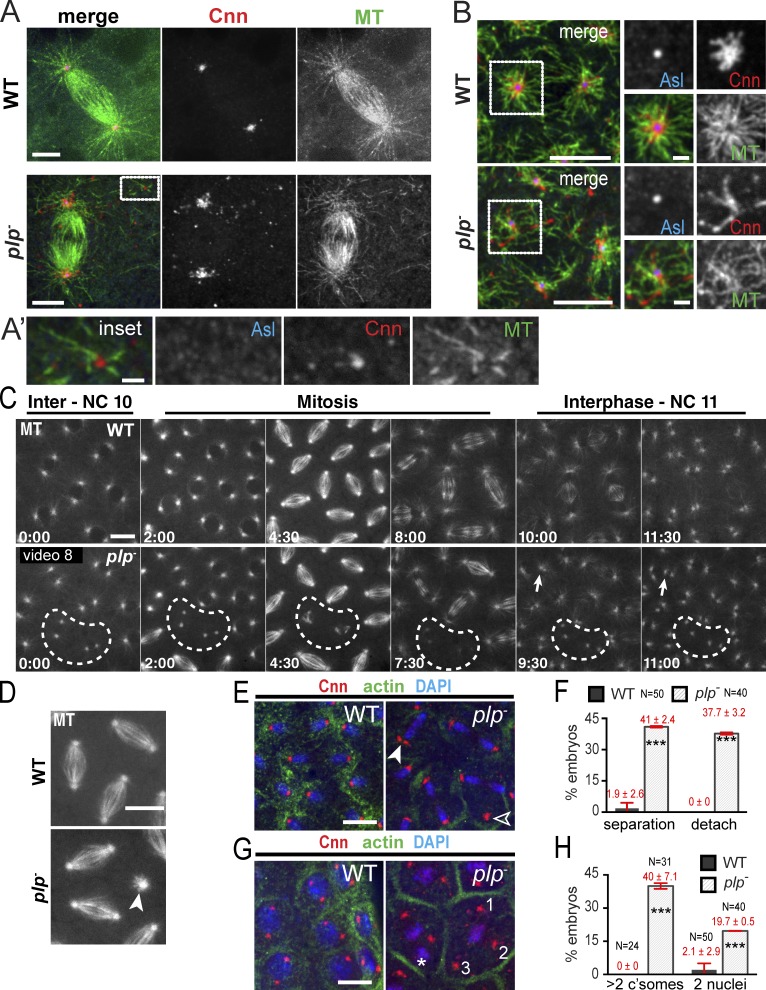
PLP mediates efficient MT radial organization and centrosome segregation
Expanded localization of Pcnt to centrosomes has previously been correlated with MT organization in mouse epithelial cells (Mogensen et al., 1997), and knockdown of Pcnt is associated with MT disorganization in cultured cells (Zimmerman et al., 2004), indicating that Pcnt contributes to proper MT organization. While immunodepletion experiments previously suggested that PLP plays a role in MT nucleation (Kawaguchi and Zheng, 2004), mutant analysis within NBs indicates that plp− cells are efficient MT nucleators (Martinez-Campos et al., 2004; Lerit and Rusan, 2013; Singh et al., 2014). Indeed, similar to cnn− mutants (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999), we found that embryonic plp− centrosomes maintain robust MTOC activity comparable to WT (Fig. 6 A). In addition, the cytoplasmic Cnn particles found in plp− mutants serve as a platform for MT organization independent of the centrosome (80%; n = 33/40 of cytoplasmic foci; Fig. 6 A′), which is consistent with ectopic cytoplasmic Cnn foci organizing MT asters within unfertilized eggs (Conduit et al., 2014a). In interphase, it was difficult to determine if these cytoplasmic particles altered the density of the MT network, but there is a clear reduction in radial MT symmetry in plp− embryos, as evident by MT crossovers and more randomized MTs (55%; n = 11/20 embryos; Fig. 6 B). Similarly, disorganized astral MTs were recently reported in a novel Pcnt mouse model (Chen et al., 2014), which suggests that MT organization is a conserved function of PLP. Live imaging of MTs shows that plp− mutants (n = 3/5 embryos) display MTOC inactivation followed by failed and abortive spindle formation (Fig. 6 C, broken line), indicating that PLP is required to maintain MTOC activity throughout the cell cycle. Further, loss of PLP also disrupts mitotic spindle orientation (Fig. 6 D), and collisions of neighboring nuclei result in uneven nuclear spacing and association of more than two MTOCs with a single nucleus (Fig. 6 C, arrows; and Video 8).
Figure 6.
PLP is required for MT organization. (A and B) WT and plp− mitotic (A) and interphase (B) embryos stained for the indicated proteins. (A’) Acentriolar Cnn particle (inset) organizes MTs. Boxes in B show radial MT array, enlarged in the insets on the right. (C and D) Live GFP-MT in embryos. Broken circles show MTOC inactivation. Arrows show nuclear collisions and the arrowhead shows an orthogonal spindle. (E) Centrosome separation (closed arrowhead) and detachment (open arrowhead) defects are quantified in F. (G) Centrosome and nucleus positioning defects with two nuclei (asterisk) and more than two centrosomes (numerals) per pseudo-cell are quantified in H and F, and show mean ± SD (error bars). ***, P < 0.0001. Data shown are from a single representative experiment out of two repeats. Bars: (A and B, main panels) 5 µm; (A′ and B, right) 1 µm; (C–G) 10 µm.
Given this MT disorganization, we reasoned that centrosome separation, a process known to require properly arranged anti-parallel MTs, would be impaired. Analysis confirms that 41% of plp− embryos show centrosome separation defects (vs. 1.9% in WT), resulting in two centrosomes at some mitotic spindle poles (Fig. 6, E and F). In addition, 38% of plp− embryos show centrosomes detached from nuclei/chromosomes, an event never seen in WT (Fig. 6, E and F). Similar centrosome segregation and detachment defects occur in embryos deficient in dynein (Robinson et al., 1999), Polo (Archambault et al., 2008), and Cnn (Lucas and Raff, 2007; Zhang and Megraw, 2007). These centrosome separation and detachment defects, coupled with the abnormal mitotic spindles observed in our live imaging, may account for the elevated centrosome segregation defects, where 40% (n = 14/31) of plp− embryos contain pseudo-cells with more than two centrosomes (Fig. 6, G and H).
Defects in centrosome positioning and interphase MT arrangement may contribute to aberrant nuclear spacing and cell cycle length. Indeed, polyploid pseudo-cells were observed in ∼20% of plp− embryos (Fig. 6, G and H). Additional analysis shows that cell cycle progression is altered in some cells, as nuclei in plp− mutants lose the stereotyped mitotic synchrony characteristic of WT syncytial divisions (Fig. 7, F and F′). Together, these data support a role for PLP in organizing the symmetric, compact PCM necessary for proper MT organization, centrosome separation, and efficient cell cycle progression. Furthermore, the extensive similarities observed in plp− and cnn− loss-of-function studies further support a model where PLP and Cnn function in a common pathway to modulate centrosome size and activity.
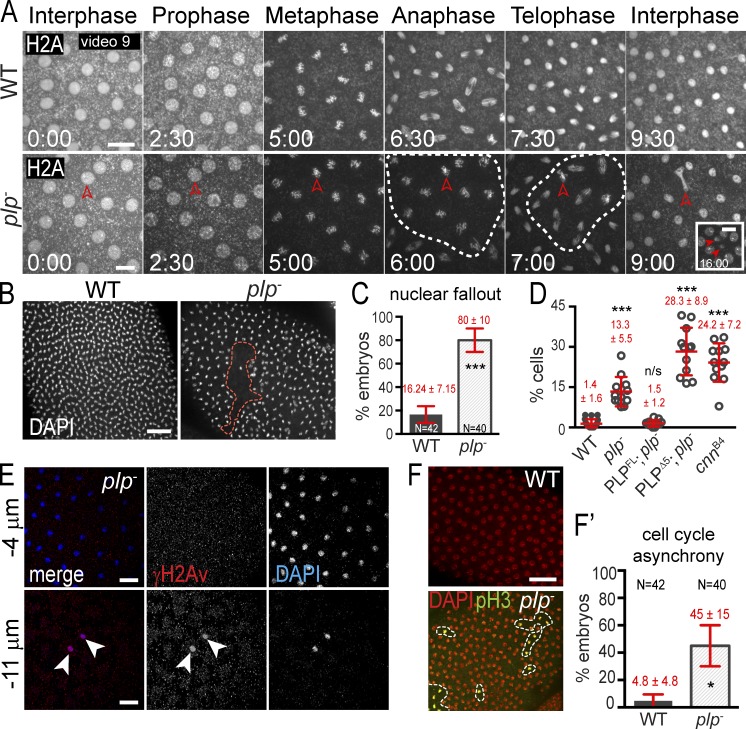
Figure 7.
PLP maintains genome stability. (A) Live H2A-RFP in embryos. Broken circles show mitotic asynchrony. Arrowheads show lagging chromosomes (9:00) followed by NUF (16:00). (B) NUF (broken circle) detected by DAPI. (C and D) The frequency (C) and amount (D) of NUF is quantified. (E) γH2Av (red) labels nuclei (DAPI, blue) ejected from the cortex. Arrowheads show nuclei that have undergone nuclear fallout and stain positive for γ-H2A.The negative sign indicates distance below the embryo surface. (F) Embryos stained with DAPI (red; all nuclei) and pH3 (green; mitotic nuclei) to detect mitotic asynchrony. Results are quantified in F′. Data are mean ± SEM (error bars) for C, all other data are mean ± SD. Time is given in minutes:seconds. *, P < 0.01; ***, P < 0.0001; n.s., not significant. Data shown are from a single representative experiment out of two or more repeats. Bars: (A) 10 µm; (B, E, and F) 20 µm.
PLP is required for genome stability
Drosophila embryos have evolved a mechanism, nuclear fallout (NUF), to eject damaged nuclei from the cortex in order to protect the developing embryo from propagating aberrant chromosomes (Sullivan et al., 1993; Rothwell et al., 1998; Takada et al., 2003). In cnn− mutant embryos, the accumulation of progressive mitotic failures leads to nuclear collisions, irregular nuclear spacing, and NUF, which prohibit cellularization and lead to embryonic lethality (Megraw et al., 1999; Vaizel-Ohayon and Schejter, 1999; Zhang and Megraw, 2007). To detect chromosome segregation defects and NUF in plp− mutants, we imaged successive cell cycles in embryos expressing H2A-RFP. While nuclei synchronously divided without chromosome segregation errors in WT embryos, plp− embryos revealed lagging chromosomes followed by NUF (Fig. 7 A and Video 9). Fixed analysis showed that NUF is significantly increased in plp− embryos (80% of embryos vs. 16% in WT; Fig. 7, B and C) and is more severe, as more nuclei per embryo are removed from the cortex (13% vs. <2% in WT; Fig. 7 D). Further analysis of ejected nuclei revealed the accumulation of γH2Av, a marker of double-stranded DNA breaks (Fig. 7 E), which suggests that chromosome segregation errors trigger DNA damage, leading to NUF. Similarly, NUF is evident in 100% of hypomorphic cnn− embryos (cnnB4; n = 25) with nearly 25% of nuclei removed from the cortex (Fig. 7 D), indicating that both PLP and Cnn function to ensure chromosome fidelity. We speculate that the accumulation of DNA damage and resultant NUF contributes to embryonic lethality.
Previous studies show nuclei that have experienced chromosome missegregation or DNA damage fail to progress through the cell cycle efficiently (Hayashi and Karlseder, 2013; Poulton et al., 2013). In cultured mammalian cells, Pcnt disruption is linked to aberrant mitotic progression (Tibelius et al., 2009; Wang et al., 2013). Quantification of this defect in fixed embryos indicates there is a 10-fold increase in mitotic asynchrony in plp− mutants (45% of embryos vs. <5% in WT; Fig. 7, F and F′). We suspect that these patches of mitotic asynchrony correspond to the patches of NUF observed in plp− mutants (Fig. 7 B), as they are of roughly the same size and occur during the same embryonic stages. These data indicate that PLP ensures efficient progression through the cell cycle to confer genomic stability.
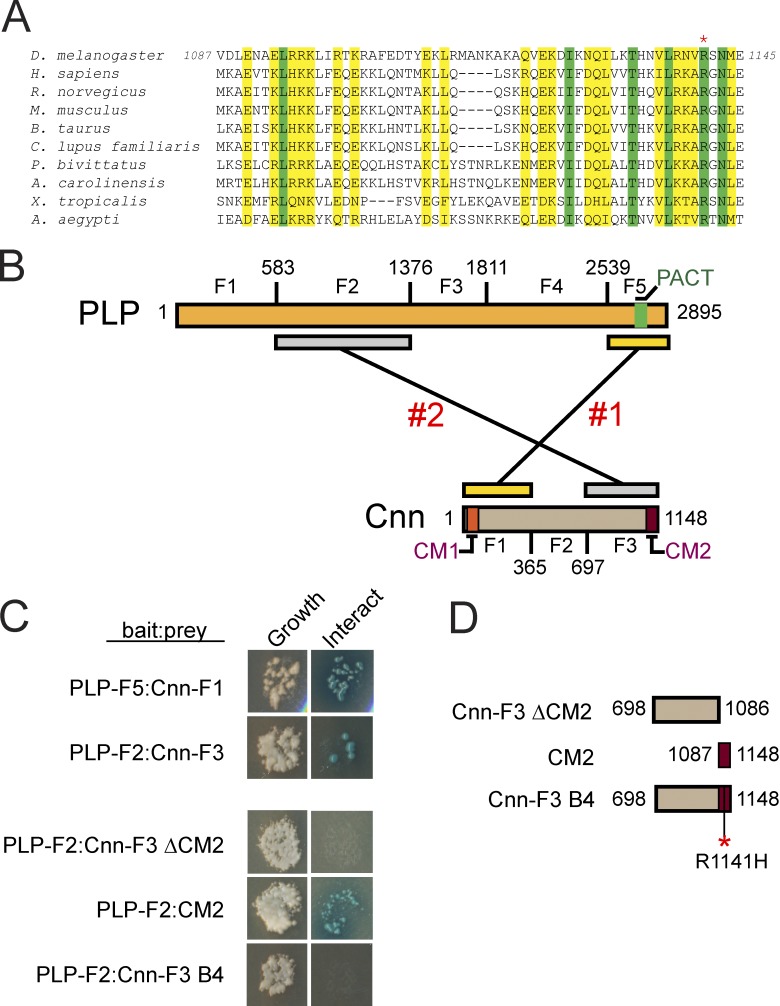
Cnn and PLP directly interact at two distinct domains
Our collective work strongly suggests that Cnn and PLP function within a complex to regulate centrosome size and activity. To date, a direct interaction between Cep215/Cnn and Pcnt/PLP has not been reported. Immunoprecipitation experiments from mammalian and Drosophila extracts, however, indicate that these proteins comprise a biochemical complex through a conserved motif located in the C terminus of Cep215/Cnn (Fig. 8 A), termed Conserved Motif 2 (CM2; Kao and Megraw, 2009; Buchman et al., 2010; Conduit et al., 2010; Wang et al., 2010; Gopalakrishnan et al., 2011; Kim and Rhee, 2014). To test whether Cnn and PLP interact directly, we truncated Cnn and PLP proteins into a series of fragments (Fig. 8 B) and conducted yeast two-hybrid (Y2H) analysis (Fig. 8 C). These studies identified two sites likely to mediate direct interaction. One occurs between Cnn fragment 1 (Cnn-F1) and PLP fragment 5 (PLP-F5; Fig. 8, B and C). A second was detected between Cnn fragment 3 (Cnn-F3) and PLP fragment 2 (PLP-F2; Fig. 8, B and C). Previous work indicates that CM2 interacts directly with Calmodulin (Wang et al., 2010) and Centrocortin (Cen; Kao and Megraw, 2009), which suggests that this conserved motif mediates several protein interactions. Further Y2H analysis confirms that the CM2 domain within Cnn-F3 is necessary and sufficient to mediate the direct interaction with PLP-F2 (Fig. 8, C and D).
Figure 8.
Identification of two sites of direct interaction between Cnn and PLP. (A) ClustalW multiple sequence alignment of the Cnn CM2 motif; similar (yellow) and identical (green) residues are shown. The asterisk shows an invariant arginine mutated in cnnB4 mutants. (B) Graphic showing PLP and Cnn truncations used in Y2H. Two distinct interaction sites are shown. (C) Y2H assays for growth (left) and interaction (right; Materials and methods). (D) Graphic showing truncations of Cnn-F3 used for interaction refinement. The asterisk shows the R1141H mutation that mimics the cnnB4 mutation.
Cnn and PLP are mutually required for proper localization and function
The power of Drosophila genetics allows us to test the significance of these interactions in the context of an intact organism and to test whether the interactions between Cnn and PLP are required to influence centrosome activity and regulate the cell cycle–dependent rearrangement of the centrosome. To investigate a role for the PLP(F5)–Cnn(F1) interaction in PLP satellite and Cnn flare formation, we generated animals expressing a GFP-tagged PLP transgene that lacks the F5 region. Fixed and live studies of this construct showed it localizes to centrioles and satellites in both WT and plp− backgrounds (PLPΔ5; plp−, hereafter PLPΔ5; Fig. S4, A–C; and Video 10). The centriole localization was unexpected because PLP-F5 contains the highly conserved PACT domain that is sufficient for centriole targeting (Gillingham and Munro, 2000; Kawaguchi and Zheng, 2004; Martinez-Campos et al., 2004). Our data show that motifs outside of PLP-F5 are sufficient to generate PLP satellites and maintain colocalization with Cnn within flares and cytoplasmic particles.
Notably, PLPΔ5 satellites are less structured and extend farther from the centriole (1.5 ± 0.5 µm vs. 1.2 ± 0.3 µm for PLPFL and 1.2 ± 0.3 µm for endogenous PLP; n = 20 centrosomes). PLPΔ5 also exacerbates the release of Cnn cytoplasmic particles found in plp− mutants (25/100 µm2 vs. 13.8/100 µm2 for plp− and 1.6/100 µm2 for WT; Figs. 5 F and S4 D, arrows). Surprisingly, live imaging reveals that PLPΔ5 satellites persist throughout the cell cycle (n = 6/6 embryos; Fig. S4 C and Video 10), which is unlikely due to overexpression of PLP, as PLPFL (Fig. 3 A) or PLPΔ5 in the WT background do not perturb the normal centrosome structures (Fig. S4 A). Importantly, the PLPΔ5 gain-of-function phenotype in mitosis lends support to the hypothesis that PLP satellites act to organize Cnn, because the abnormal mitotic PLPΔ5 satellites also organize abnormal interphase-like Cnn flares (Fig. S4, B and E). Even when only one mitotic centrosome at a nucleus displays PLPΔ5 satellites, it also extends Cnn flares (Fig. S4 D, solid box), whereas the PLP satellite-free centrosome shows compact Cnn that is characteristic of normal mitotic centrosomes (Fig. S4 D, broken box), implicating local regulation of centrosome organization by the PLP–Cnn complex. Overall, these data suggest that PLP-F5 is dispensable for PLP satellite and Cnn flare formation, but is required for their proper organization and mitotic compaction. Furthermore, we conclude that the PLP(F5)–Cnn(F1) interaction is dispensable for PLP–Cnn colocalization, and possibly complex formation.
These data raise the intriguing possibility that the second interaction defined between Cnn CM2 and PLP-F2 may be important for the normal formation of Cnn flares and PLP satellites in interphase. Unfortunately, despite testing 16 truncations of PLP-F2, our efforts to narrow the interaction region were unsuccessful (Fig. S5 A). Likewise, despite screening many candidate transgenic lines, we were unable to generate an animal expressing a PLP transgene that lacks the F2 region (see Materials and methods). Therefore, to abrogate the interaction between Cnn CM2 and PLP-F2, we used a previously published allele of cnn, cnnB4, which is defined by a single point mutation at an invariant arginine residue (R1141H; Fig. 8 A) within CM2 (Vaizel-Ohayon and Schejter, 1999; Kao and Megraw, 2009). Remarkably, introduction of R1141H into Cnn-F3 is sufficient to abolish the interaction with PLP-F2 (Fig. 8, C and D), which suggests that cnnB4 mutants are particularly useful to probe the significance of the Cnn(CM2)–PLP(F2) interaction in vivo.
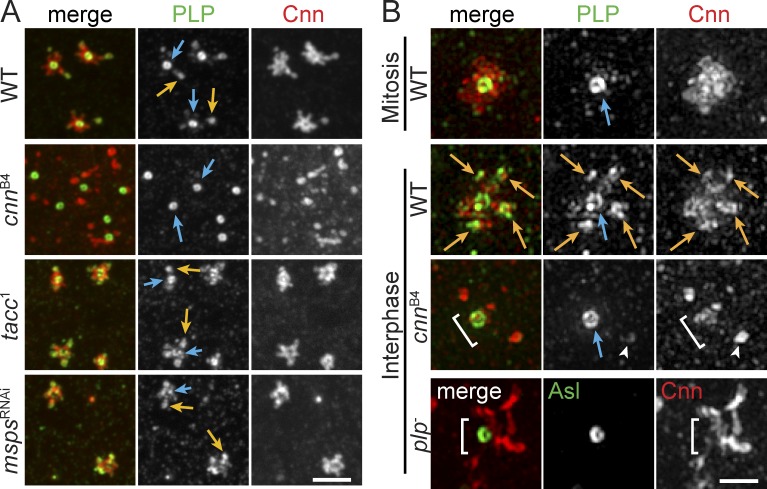
Despite producing normal levels of Cnn protein, prior detailed analysis of cnnB4 embryos indicates pronounced similarities in the PCM dispersion and NUF phenotypes we observe in plp− mutants (Kao and Megraw, 2009). Consistent with these studies, all interphase cnnB4 embryos display highly disordered Cnn and γTub (Figs. 9 A and S5 B; n = 34). PLP localization was not previously examined in cnnB4 mutants. Thus, to assay the significance of the Cnn CM2 interaction with PLP-F2, we examined PLP distribution in control and cnnB4 embryos. Significantly, localization of PLP to the satellites is eliminated in interphase cnnB4 embryos, whereas PLP at centrioles is unperturbed (Fig. 9 A; n = 34). The effect of Cnn on PLP satellite formation is specific, as disruption of other PCM factors, such as TACC and MSPS, previously localized to flare-like particles (Lee et al., 2001), did not alter PLP satellite formation (Fig. 9 A).
Figure 9.
Localization of PLP to satellites requires Cnn CM2. Embryos were stained for the indicated proteins and imaged by confocal microscopy (A) or SIM (B). PLP satellites (orange arrows) are present in all genotypes but cnnB4 mutants, which resemble mitotic centrosomes. The PLP centriole pool is present in all genotypes (blue arrows). plp− and cnnB4 mutants do not properly assemble PCM around the centriole (brackets). Arrowheads show a cytoplasmic particle or rare PLP satellite in cnnB4 mutant. Bars: (A) 2.5 µm; (B) 1 µm.
SIM confirms that interphase centrosomes in cnnB4 embryos lack PLP satellites but maintain PLP at centrioles (Fig. 9 B), which signifies that the two PLP pools are regulated by distinct mechanisms. Furthermore, we conclude that the Cnn(CM2)–PLP(F2) interaction is dispensable for PLP centriole localization but is essential for PLP satellite formation. The lack of PLP satellites in cnnB4 embryos corresponds with a loss of Cnn flares and an increase in Cnn cytoplasmic particles, similar to the plp− phenotype (Figs. 5 F and 9 B). While it remains formally possible that the cnnB4 mutation abrogates the structure of the Cnn molecule and/or its overall function, mitotic cnnB4 centrosomes are much less disrupted (Kao and Megraw, 2009), suggesting that the pathway that organizes Cnn during mitosis remains intact and that the Cnn molecule is not completely disrupted. Therefore, we propose that PLP satellites function to form a Cnn scaffold in the interphase flare zone that organizes and confines Cnn to interphase centrosomes (Fig. 10). Our data demonstrate that Cnn and PLP are mutually required for the proper localization and function of the other, thereby demonstrating the presence of a positive feedback loop that ensures the proper formation of the interphase centrosome scaffold.
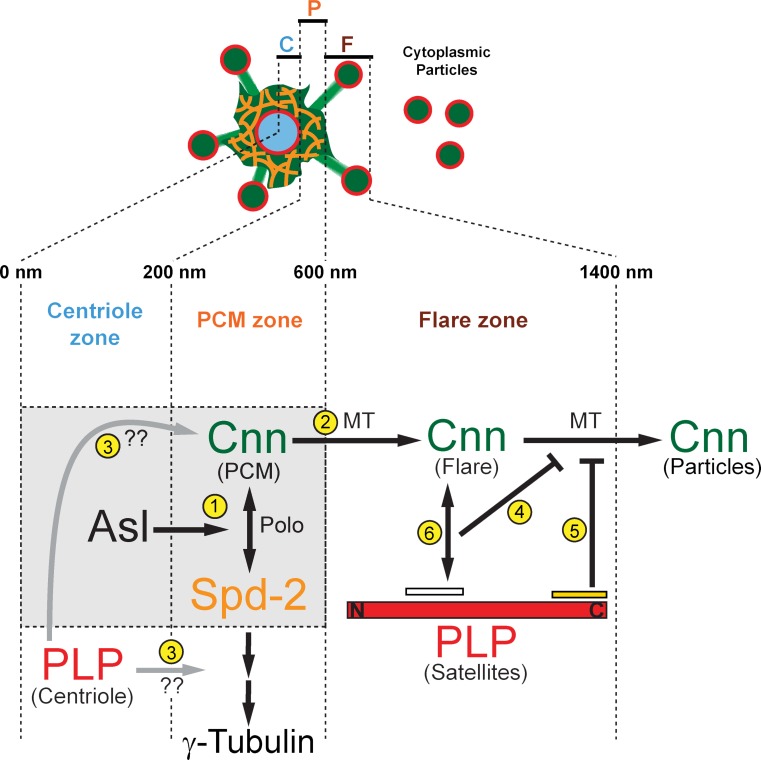
Figure 10.
Model of PLP-Cnn coregulation at interphase centrosomes. Diagram depicting centrosome scaffold formation during interphase. Our data support an interphase-specific Cnn scaffold in the interphase flare zone that is organized by PLP satellites. See text for details.
Discussion
Our identification of a positive feedback loop between Cnn and PLP that coordinates the cell cycle–dependent reorganization of the centrosome structure adds to the growing recognition that the centrosome is subject to sophisticated layers of regulation to ensure proper function. The interdependence of three major PCM components (Pcnt/PLP, Cep215/Cnn, and Cep192/Spd2) and two kinases (Plk1/Polo and Aurora A) in generating a fully functional mitotic centrosome is well established (for a recent review see Mennella et al., 2014; Fu et al., 2015). However, we demonstrate a novel role for PLP in building an elaborate interphase centrosome scaffold in the early embryo. Our data show that PLP undergoes a major rearrangement that is entrained to the cell cycle and is critical for proper Cnn dynamics, distribution, and functions upstream of γTub.
Interestingly, our imaging of Cnn during mitotic exit is highly suggestive of Cnn forming a reticular, strand-like structure that is compact in mitosis, but unravels as the cell enters interphase to form a giant centrosome with constitutive MTOC activity. The mechanisms that control these Cnn dynamic movements will require further studies, including the use of fluorescence photoconversion methods. What mediates this cell cycle–dependent transition in centrosome architecture? One compelling candidate is Polo, which is known to phosphorylate Cnn and promote its lattice formation (Conduit et al., 2014a). What is the role and significance of this cell cycle–dependent transition in centrosome architecture? Our collective data support a model where this temporal regulation of PCM expansion and compaction likely ensures normal MT organization to avoid erroneous MT attachments, collisions with neighboring nuclei, and the ensuing DNA damage and embryonic lethality.
Moreover, we uncovered PLP localized to the tip of each Cnn flare. These PLP satellites are distinct from a PCM pool of PLP predicted by FRAP analysis of PACT-GFP (a GFP fusion with a motif from the C terminus of PLP; Martinez-Campos et al., 2004), as we find that PACT-GFP is absent from PLP satellites (Fig. 2 C). Thus, PACT is not an accurate predictor of PLP localization, at least not beyond the centriole pool (see also Fu and Glover, 2012). More recently, FRAP of PLP-GFP also showed nonuniform recovery within the centrosome, but a second major peak corresponding to PLP satellites at distances beyond the PCM zone at 650 nm was not reported (Conduit et al., 2014b). Thus, our work is the first to describe the dynamic behavior of PLP satellites, which define the end of Cnn flares and the outer margin of the interphase centrosome.
Several recent studies support a model of Cnn scaffold formation in Drosophila. Polo phosphorylates Cnn at sites within Cnn-F2 near the centriole wall, which is required for the formation of higher-order Cnn structures (Conduit et al., 2014a). The Cnn scaffold then migrates into the PCM zone (between 200 and 600 nm; Fig. 10). Spd2 is critical for this process (Fu and Glover, 2012; Conduit et al., 2014b), where it functions in a positive feedback loop with Cnn to form a Spd2–Cnn PCM scaffold (Fig. 10, gray box, 1), which is consistent with earlier studies showing that Cep192/Spd2 recruits PCM (Dix and Raff, 2007; Gomez-Ferreria et al., 2007; Giansanti et al., 2008; Haren et al., 2009; Fu and Glover, 2012). Our work suggests that PLP satellites and Cnn form a second, spatially distinct scaffold (Fig. 10, outside of gray box) located in the interphase centrosome flare zone (Fig. 10, 600–1,400 nm). These two scaffolds are likely interactive, as our live imaging suggests that Cnn emerges from the PCM zone and extends into the flare zone in an MT-dependent manner (Fig. 10, 2). Additionally, a conserved motif at the N terminus of Cnn, CM1, is required for the extension of Cnn flares (Zhang and Megraw, 2007). We speculate that CM1 may link Cnn to MTs and drive scaffold expansion in embryos and/or centrosome dematuration in other cell types.
Although related, the two scaffolds are likely functionally unique. The Spd2–Cnn scaffold is the main anchor of γTub within the PCM zone, whereas the PLP–Cnn scaffold functions to anchor Cnn within the flare zone, effectively preventing interphase centrosome dematuration by resisting MT-dependent particle release forces (Fig. 10, 4 and 5). Critically, unlike the Spd2–Cnn scaffold, the PLP–Cnn scaffold is exclusive to interphase centrosomes. We conclude that each architectural subdomain that comprises the centrosome structure is subject to multiple layers of regulation, some being zone specific. This stratification of regulatory organization defines the function of each zone.
Our Y2H studies show that PLP and Cnn likely interact at two defined domains. Analysis of PLPΔ5 suggests that the Cnn(F1)–PLP(F5) interaction is required to properly organize Cnn flares and PLP satellites, and is required to dampen Cnn particle release (Fig. 10, 5). However, this large truncation includes the PACT domain and may disrupt other PLP interactions. Nevertheless, we can conclude that the Cnn(F1)–PLP(F5) interaction is not essential for PLP–Cnn scaffold formation, because Cnn and PLP still colocalize at flares and cytoplasmic particles. The second interaction between Cnn-F3 and PLP-F2 requires the Cnn CM2 domain (Fig. 10, 6) and is disrupted by the cnnB4 mutation. However, this mutation also disrupts an interaction with Cen required for actin furrow assembly (Kao and Megraw, 2009). Formally, we cannot rule out the possibility that the cnnB4 mutation may disrupt Cnn function due to protein destabilization. In addition, because gross actin aberrations were not observed in plp− mutants (unpublished data), it is likely that Cnn CM2 mediates several interactions involved in distinct pathways that are disrupted in the cnnB4 mutant background. Nevertheless, the cnnB4 mutation does lead to a loss of PLP satellites, with no effect on the PLP centriole pool (Fig. 10, 3). This intriguing result supports a model where PLP satellites and Cnn flares are functionally interdependent within the interphase flare zone, analogous to the interdependence of Spd2 and Cnn within the PCM zone. Moreover, the cnnB4 mutation demonstrates a specific separation of PLP function at satellites versus centrioles. Our data show the disordered localization of Cnn and γTub, as well as NUF and embryonic lethality, is more severe in cnnB4 than in plp− mutants. These data suggest that Cnn functions to organize interphase centrosome structure through multiple pathways, with only one requiring PLP activity.
Previous work on the regulation of Cnn by Polo and Spd2, combined with our functional analysis of direct interactions with PLP, paints a complex portrait of the Cnn molecule. We propose that the coregulation of PLP satellites and Cnn flares is required for their anchorage to the centrosome, preventing interphase centrosome inactivation, and maintaining robust MT asters throughout the cell cycle. Disruption of this interphase PLP–Cnn scaffold leads to increased cytoplasmic PCM particles that act as platforms for MTs, leading to the formation of an aberrant interphase MT array in the embryo. These effects on MTs then cause defects in centrosome separation, nuclear spacing, and spindle assembly, culminating in chromosome missegregation, DNA damage, and embryonic lethality. Together, our data emphasize the fundamental importance of regulating interphase centrosome form and function for viability.
Materials and methods
Fly stocks
The following mutant strains and transgenic lines were used: the plp2172 allele is a P-element insertion that interrupts the PLP sequence (Spradling et al., 1999) and was recombined onto the FRT2A chromosome to generate plp2172; FRT2A chromosomes were used for the generation of germline clones (plp− embryos) by the FLP/ovoD method (Chou and Perrimon, 1996); the tacc1 allele is a strong hypomorph (Gergely et al., 2000); and Df(3R)Excel6142 (Bloomington Stock Center), mspsRNAi (Vienna Drosophila Resource Center transformant 21982), and maternal-Tubulin GAL4 expressed GAL4 under the control of the maternal Tubulin promoter (Bloomington Stock Center); cnnB4 is a hypomorphic allele caused by an R1141H single amino acid mutation (Vaizel-Ohayon and Schejter, 1999), Ubi-GFP-γ-Tub23C expresses GFP-γ-Tub23C under the Ubiquitin promoter, and PBac-GFP-SAS6 is a recombineered GFP-SAS6 construct (Lerit and Rusan, 2013); UASp-Bld10-GFP expresses Cep135/Bld10-GFP under UAS (upstream activating sequence) regulator elements (Mottier-Pavie and Megraw, 2009); Ubi-GFP-PACT is a fusion of GFP with the PACT domain of PLP expressed under the Ubiquitin promoter (Martinez-Campos et al., 2004); Ubi-GFP-SAK expresses GFP-SAK/Plk4 under the Ubiquitin promoter (Basto et al., 2008); GFP-Polo expresses GFP-Polo with a Polo promoter (Moutinho-Santos et al., 1999); Ubi-D-TACC-RFP expresses TACC-RFP under the Ubiquitin promoter (Conduit et al., 2010); Ubi-GFP-SAS4 expresses GFP-SAS4 under the Ubiquitin promoter (Dix and Raff, 2007); H2AvD-mRFP expresses H2A-RFP under endogenous regulatory elements (Pandey et al., 2005); Ubi-GFP-α-Tubulin expresses GFP-α-Tubulin under the Ubiquitin promoter (Rebollo et al., 2004); and mCherry-Cnn (a gift from P. Singh and C. Cabernard, University of Basel, Basel, Switzerland), unpublished reagent generated by tagging Cnn with mCherry at the endogenous locus as described in Singh et al., 2014. PLPFL-GFP expresses full-length PLP fused to GFP under the Ubiquitin promoter (Galletta et al., 2014) and rescues the uncoordinated phenotype of plp2172/Df(3L)Brd15 (Martinez-Campos et al., 2004) adults. y−w− were used as WT controls unless otherwise noted. For live imaging, PLPFL-GFP was expressed in plp2172/Df(3L)Brd15 mutants, and all other transgenes (GFP-γTub, H2A-RFP, Cnn-mCherry, and PLPΔ5-GFP) were expressed in plp− germline clones unless otherwise noted.
Construction of transgenic animals
All transgenic flies were generated by BestGene, Inc., using standard P-element transformation. For PLPΔ5-GFP, a C′-terminal truncation (deleting amino acids 2,539–2,895, which include the PACT domain) of PLPFL was PCR amplified using the primers 5′-CACCATGGCCATTAATATTGCTTTATTTACG-3′ and 5′-TTCATTGAAGTGTTCCAACTCTGTTTCGGC-3′, directionally cloned into the pENTR-D vector (Invitrogen), and recombined into the P-element destination vector pUWG (Ubiquitin promoter, C’-terminal GFP; Drosophila Genomics Resource Center) via the Gateway cloning system (Invitrogen). PBac-GFP-Cnn was generated from the bacterial artificial chromosome clone CH322-10I24 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute) using the following primers to amplify the N-terminal GFP-Kanamycin cassette from the N-EGFP (N-terminal EGFP) template vector (Venken et al., 2008): 5′-TCAAGTGTTAGAATTATTGTGTGCGAAAGTTAACTATTTGAGGACCTCCCATGGTGAGCAAGGGCGAGGAG-3′ and 5′-GTACCATTGCCGTCGCCGCAATAGTCCCGCAAAACCTGTTTAGACTGGTCACTAGTGGATCCCCTCGAGGGAC-3′. This modified bacterial artificial chromosome was directionally integrated into the genome using the PhiC31 system at site VK33 (Chromosome 3). PLPΔ2-GFP was generated by PCR amplification of PLP-F1 flanked by 5′-pEntr and 3′-PLP-F3 homology arms using the primers 5′-GCAGGCTCCGCGGCCGCCCCCTTCACCAGGATGAATCTGTACACTATATACGATTGGATC-3′ and 5′-GCATTTCCCGCATGCTCTTGAAGATCGGCGGATCCTGCTCCTCTTC-3′. A second fragment consisting of PLP-F3 through PLP-F5 flanked by 5′-PLP-F1 and 3′-pEntr homology arms was PCR amplified using the primers 5′-GAAGAGGAGCAGGATCCGCCTGATCTTCAAGAGCATGCGGGAAATGC-3′ and 5′-AGCTGGGTCGGCGCGCCCACCCTTGTCTAGATGATGCCGCGCATGCGCTC-3′. The two fragments were enzymatically combined by a one-step isothermal DNA assembly (Gibson et al., 2009) into the pENTR-D vector, and then recombined into the P-element destination vector pUWG (Ubiquitin promoter, C’-terminal GFP; Drosophila Genomics Resource Center) via the Gateway cloning system. More than 32 independent PLPΔ2-GFP transgenic lines were screened for fluorescence using confocal microscopy. None of these lines showed GFP localization in cycling embryos.
Assessment of zygotic lethality and hatch rate analysis
Because males zygotically homozygous for the plp2172 mutation are uncoordinated and sterile (Martinez-Campos et al., 2004), we crossed males heterozygous for plp2172 (over the TM6B,Tb balancer chromosome) to females with germlines homozygous mutant for plp, thus 50% of the analyzed embryos received a paternal WT plp gene and were paternally rescued. Eggs were collected at 25°C and the percentage of hatched larvae was divided by the total number of fertilized eggs. Embryonic lethality for plp− was 16% (n = 64/404 embryos) and is significantly increased compared with WT (2.4%; n = 458 embryos; P < 0.0001 by Fisher’s exact test). Because 50% of the embryos were paternally rescued, lethality for plp− maternal/zygotic mutant embryos may be estimated to be 32% (64/202). The remaining 68% (138/202) maternal/zygotic mutant embryos hatched, but die as first instar larvae, as we do not detect any second instar larvae that are non-Tubby (Tb). We conclude that all of the detected viable animals had received a WT copy of plp from the balancer chromosome. PLPΔ5 flies showed significantly greater embryonic lethality (54.9%; n = 89/162 embryos) than either controls or plp− (P < 0.0001). Because 50% of embryos were paternally rescued, this suggests that PLPΔ5 maternal/zygotic mutants are 100% embryonic lethal. The fact that embryonic lethality of PLPΔ5 exceeded plp− suggests it may exert dominant-negative effects.
Microscopy
Embryos were imaged using 40×, 1.3 NA or 100×, 1.49 NA oil immersion objective lenses on a microscope (Eclipse Ti; Nikon) fitted with a stage incubator (20/20 Technology, Inc.) heated to 25°C, a CSU-22 spinning-disk confocal head (Yokagawa Electric Corporation), a cooled CCD camera (Clara; Andor Technology), and the Perfect Focus System (Nikon) all run by an automated controller (MAC6000; Ludl Electronic Products) using MetaMorph software (Molecular Devices). Laser excitation was supplied by a laser merge module equipped with 491 nm, 561 nm, and 642 nm solid-state lasers (VisiTech International). For super-resolution SIM, immuno-labeled samples were mounted in Vectashield (Vector Laboratories, Inc.) and imaged with the DeltaVision OMX4 SIM Imaging System (Applied Precision).
Embryos (1–2 h old) were prepared for live imaging as described previously (Lerit and Gavis, 2011) with the following modifications: series 700 halocarbon oil (Sigma-Aldrich) was used to cover embryos adhered to a sticky #1.5 coverslip layered with glue extracted from double-sided Scotch tape (3M). The embryos and coverslip were inverted onto a 50-mm gas-permeable lumox dish (Sarstedt) fitted with two halves of a broken #1 coverslip used as spacers. Images were collected at 0.25-µm intervals over a 5–10 µm volume at 30-s time intervals or imaged at 500-ms intervals in a single optical plane as noted. Images were assembled using ImageJ (National Institutes of Health), Photoshop (Adobe), and QuickTime Player Pro 7 (Apple) software to crop regions of interest, adjust brightness and contrast, separate or merge channels, and generate maximum-intensity projections.
Image analysis
Total fluorescence intensity projections of the entire centrosome/centriole volume were used to measure the peak (P) and outer edge (OE) position of Sas6, Asl, and PLP, and the OE position of γTub and Cnn. Single optical sections through the middle of the centrosome were used for P measurements of γTub and Cnn. All line scans were single-pixel-wide line scans of 5 µm generated by the Plot Profile tool in ImageJ. Line scans were converted to a −2.50 to 2.50 scale such that the 0.0 µm position marked the center of the centriole. Average background fluorescence was subtracted from each pixel along the line scan. GraphPad Software was then used to fit each line scan to a Gaussian curve from which the mean (μ) and SD (Gaussian RMS) was calculated. μ represents the position of the Gaussian peak (P), which is the peak fluorescence intensity. The full width at half maximum (FWHM) was calculated for each curve using 2.35482×SD. The protein’s OE was calculated as μ + (0.5×FWHM). The mean P and OE for each measured centrosome protein is reported in Fig. S1 A. For Sas6, the entire −2.5 to 2.5 line scan was used to fit the Gaussian curve, for all other proteins two independent curves were generated along the −2.5 to 0.0 and 0.0 to 2.5 positions. For the γTub and Cnn within the PCM zone, a Gaussian Blur tool in ImageJ was used to smooth the edges of the PCM such that the fitted Gaussian produced an R2 >0.90 (Fig. S1 B). Comparisons of individual native and blurred line scans indicate a negligible impact of the blurring function on position measurements. For OE measurements of PLP satellites and Cnn flares, a half-Gaussian curve was fitted to the distal edge of the flare. A mirror image of this curve was added to generate the full-Gaussian from which μ and SD were calculated (Fig. S1 B, OE flare).
Cytoplasmic Cnn flares (not continuous with the centrosome or satellites) were manually counted from a 20 × 20-µm (400 µm2) box from three random areas in at least five different embryos for each genotype and cell cycle stage. Flare density is reported per 100 µm2. To analyze PLP satellite dynamics, kymographs along a single pixel line scan were generated using ImageJ software, and satellites that remained visible for at least five consecutive frames were manually tracked using the mTrackJ plugin (Meijering et al., 2012). Mean velocities were determined for individual satellites by dividing the time of transit by the distance between the first and last monitored positions. NUF was quantified as described previously (Poulton et al., 2013); in brief, embryos were stained for actin to label cortical boundaries and DAPI to label DNA. Empty (devoid of nucleus) actin cages were counted as sites of NUF. γH2Av labels double-stranded DNA breaks and was observed in all nuclei undergoing NUF. Data were plotted and statistical analysis was performed using Excel (Microsoft) and Prism software (GraphPad Software). To calculate significance, the normality of the distributions was confirmed with the D’Agnostino and Pearson normality test. Data were then subjected to Student’s two-tailed t test or the Wilcoxon rank-sum test and are displayed as mean ± SD unless otherwise noted.
Immunofluorescence
Embryos were fixed in a 1:4 solution of 4% paraformaldehyde/heptane for 20 min before devitellinization in methanol. Samples were rehydrated stepwise into PBST (PBS with 0.1% Tween-20) then blocked in BBT (PBST with 0.1% BSA) for 2 h before an overnight incubation with nutation at 4°C in BBT with primary antibody. Samples were washed, further blocked in BBT supplemented with 2% normal goat serum (NGS), then incubated for 2 h at room temperature in secondary antibody. For visualization of γTub, embryos were blocked in PBST with 1% BSA. For visualization of MTs, embryos were prepared as described previously (Lerit and Gavis, 2011) according to the method of Theurkauf (1994). In brief, embryos were fixed for 3 min in 1:1 37% paraformaldehyde/heptane, rinsed three times in fresh PBS, and hand devitellinized. After staining, samples were washed in PBST and mounted in Aqua-Poly/Mount (Polysciences, Inc.) and imaged on a spinning disk confocal microscope (Eclipse Ti; Nikon).
Primary antibodies used were as follows: rabbit and guinea pig anti-PLP (made against amino acids 8–351; 1:3,000; Brownlee et al., 2011), guinea pig anti-Asl (made against full-length Asl; 1:3,000; gift from G. Rogers, University of Arizona Cancer Center, Tucson, AZ), rabbit anti-Cnn (made against amino acids 271–1,034; 1:2,000; gift from T. Megraw, Florida State University, Tallahassee, FL); anti–phospho-Histone H3 Ser10 (pH3; 1:1,000; EMD Millipore), mouse anti-γTub ascites GTU-88 (1:50; Sigma-Aldrich), mouse anti–α-Tubulin DM1α (1:500; Sigma-Aldrich), rabbit anti-γH2Av made against QPDQRKGNVILSQAY (1:500; gift from K. McKim, Rutgers University, Piscataway, NJ), and rabbit anti–Spd-2 (1:2,000; Rodrigues-Martins et al., 2007). Secondary antibodies and counterstains were as follows: Alexa Fluor 488, 568, or 627 (1:500; Molecular Probes); DAPI (4’6-diamidino-2-phenylindole; 10 ng/ml); and Alexa Fluor 488 phalloidin (1:500; Life Technologies).
Y2H analysis
Y2H was performed as described in Galletta et al. (2014). In brief, fragments of PLP and Cnn were introduced into pDEST-pGADT7 and pDEST-pGBKT7 using Gateway technology (Invitrogen), transformed into Y187 or Y2HGold yeast strains (Takara Bio Inc.), and grown in −leu or −trp media to select for plasmids. After mating of the two strains, yeast were grown on −leu, −trp (DDO) plates, then replica plated onto plates of increasing stringency: DDO; −ade, −leu, −trp, −ura (QDO); −leu, −trp plates supplemented with Aureobasidin A and X-α-Gal (DDOXA; Takara Bio Inc.); and −ade, −leu, −trp, −ura plates supplemented with Aureobasidin A and X-α-Gal (QDOXA). Interactions were scored based on growth and the presence of blue color. For simplicity, only DDO (growth) and the most stringent condition, QDOXA (interaction), are shown. All plasmids were tested for the ability to drive reporter activity in the presence of an empty vector (autoactivation). Plasmids that conferred autoactivity were omitted from further analysis (see Fig. S5 A).
Online supplemental material
Fig. S1 shows analysis of centrosome protein distribution. Fig. S2 shows the dynamics of PLP satellites. Fig. S3 shows disruption of γTub in plp− embryos. Fig. S4 shows that PLP-F5 is required for compaction of the mitotic centrosome. Fig. S5 shows interaction refinement of PLP-F2. Video 1 shows Cnn-GFP and H2A-RFP through multiple NCs in a control embryo. Video 2 shows Cnn-GFP during the mitosis-to-interphase transition in a control embryo. Video 3 shows PLPFL in a cycling plp− embryo. Video 4 shows PLP satellite dynamics. Video 5 shows that PLPFL and Cnn-mCherry are packaged as dynamic particles. Video 6 shows Cnn-mCherry in a control and plp− embryo. Video 7 shows GFP-γTub through multiple NCs in a control and plp− embryo. Video 8 showsGFP-MT in a control and plp− embryo. Video 9 shows H2A-RFP in a control and plp− embryo. Video 10 shows PLPΔ5 in a cycling plp− embryo. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201503117/DC1.
Supplementary Material
Acknowledgements
We thank T. Megraw, C. Cabernard (for the unpublished mCherry-Cnn), and the Bloomington Drosophila Stock Center for fly stocks; K. Plevock for generating the CM2 alignment; R. Guillen for generating PLPΔ5; A. Zajac for Fig. 1 D data; R. Fischer and C. Waterman for microinjection help; the Drosophila Genomics Resource Center for DNA; M. Bettencourt-Dias, K. McKim, T. Megraw, and G. Rogers for antibodies; the NHLBI Light Microscopy Core; and K. O’Connell and A. Kelly for critical comments.
M. Peifer is supported by NIH R01GM67236, N.M. Rusan is supported by the Division of Intramural Research at the National Institutes of Health/NHLBI (1ZIAHL006126), and D.A. Lerit is supported by the Lenfant Biomedical Postdoctoral Fellowship.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- Asl
- Asterless
- Cnn
- Centrosomin
- γTub
- γ-Tubulin
- MT
- microtubule
- MTOC
- MT organizing center
- NB
- neuroblast
- NC
- nuclear cycle
- NUF
- nuclear fallout
- PCM
- pericentriolar material
- Pcnt
- Pericentrin
- PLP
- Pcnt-like protein
- SIM
- structured illumination microscopy
- Y2H
- yeast two-hybrid
- WT
- wild type
References
- Archambault V., D’Avino P.P., Deery M.J., Lilley K.S., and Glover D.M.. 2008. Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis. Genes Dev. 22:2707–2720. 10.1101/gad.486808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R., Bao L., and Zimmer W.E.. 1994. PCM-1, A 228-kD centrosome autoantigen with a distinct cell cycle distribution. J. Cell Biol. 124:783–793. 10.1083/jcb.124.5.783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., and Raff J.W.. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042. 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., et al. . 2005. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 37:353–355. (published erratum appears in Nat. Genet. 2005. 37:555) 10.1038/ng1539 [DOI] [PubMed] [Google Scholar]
- Brownlee C.W., Klebba J.E., Buster D.W., and Rogers G.C.. 2011. The protein phosphatase 2A regulatory subunit twins stabilizes Plk4 to induce centriole amplification. J. Cell Biol. 195:231–243. 10.1083/jcb.201107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman J.J., Tseng H.C., Zhou Y., Frank C.L., Xie Z., and Tsai L.H.. 2010. Cdk5rap2 interacts with pericentrin to maintain the neural progenitor pool in the developing neocortex. Neuron. 66:386–402. 10.1016/j.neuron.2010.03.036 [DOI] [PubMed] [Google Scholar]
- Callaini G., and Riparbelli M.G.. 1990. Centriole and centrosome cycle in the early Drosophila embryo. J. Cell Sci. 97:539–543. [DOI] [PubMed] [Google Scholar]
- Chen C.T., Hehnly H., Yu Q., Farkas D., Zheng G., Redick S.D., Hung H.F., Samtani R., Jurczyk A., Akbarian S., et al. . 2014. A unique set of centrosome proteins requires pericentrin for spindle-pole localization and spindle orientation. Curr. Biol. 24:2327–2334. 10.1016/j.cub.2014.08.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.B., and Perrimon N.. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., Brunk K., Dobbelaere J., Dix C.I., Lucas E.P., and Raff J.W.. 2010. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 20:2178–2186. 10.1016/j.cub.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Conduit P.T., Feng Z., Richens J.H., Baumbach J., Wainman A., Bakshi S.D., Dobbelaere J., Johnson S., Lea S.M., and Raff J.W.. 2014a The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell. 28:659–669. 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B., Dix C.I., Novak Z.A., Dobbie I.M., Schermelleh L., and Raff J.W.. 2014b A molecular mechanism of mitotic centrosome assembly in Drosophila. eLife. 3:e03399 10.7554/eLife.03399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., and Merdes A.. 2002. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J. Cell Biol. 159:255–266. 10.1083/jcb.200204023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de-Thé G. 1964. Cytoplasmic microtubules in different animal cells. J. Cell Biol. 23:265–275. 10.1083/jcb.23.2.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dictenberg J.B., Zimmerman W., Sparks C.A., Young A., Vidair C., Zheng Y., Carrington W., Fay F.S., and Doxsey S.J.. 1998. Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141:163–174. 10.1083/jcb.141.1.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix C.I., and Raff J.W.. 2007. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 17:1759–1764. 10.1016/j.cub.2007.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N., and Raff J.. 2008. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6:e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S.J., Stein P., Evans L., Calarco P.D., and Kirschner M.. 1994. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 76:639–650. 10.1016/0092-8674(94)90504-5 [DOI] [PubMed] [Google Scholar]
- Doxsey S., McCollum D., and Theurkauf W.. 2005. Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21:411–434. 10.1146/annurev.cellbio.21.122303.120418 [DOI] [PubMed] [Google Scholar]
- Foe V.E., and Alberts B.M.. 1983. Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61:31–70. [DOI] [PubMed] [Google Scholar]
- Fu J., and Glover D.M.. 2012. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2:120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Hagan I.M., and Glover D.M.. 2015. The centrosome and its duplication cycle. Cold Spring Harb. Perspect. Biol. 7:a015800 10.1101/cshperspect.a015800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta B.J., Guillen R.X., Fagerstrom C.J., Brownlee C.W., Lerit D.A., Megraw T.L., Rogers G.C., and Rusan N.M.. 2014. Drosophila pericentrin requires interaction with calmodulin for its function at centrosomes and neuronal basal bodies but not at sperm basal bodies. Mol. Biol. Cell. 25:2682–2694. 10.1091/mbc.E13-10-0617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F., Kidd D., Jeffers K., Wakefield J.G., and Raff J.W.. 2000. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 19:241–252. 10.1093/emboj/19.2.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M.G., Bucciarelli E., Bonaccorsi S., and Gatti M.. 2008. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr. Biol. 18:303–309. 10.1016/j.cub.2008.01.058 [DOI] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Gillingham A.K., and Munro S.. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1:524–529. 10.1093/embo-reports/kvd105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria M.A., Rath U., Buster D.W., Chanda S.K., Caldwell J.S., Rines D.R., and Sharp D.J.. 2007. Human Cep192 is required for mitotic centrosome and spindle assembly. Curr. Biol. 17:1960–1966. 10.1016/j.cub.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan J., Mennella V., Blachon S., Zhai B., Smith A.H., Megraw T.L., Nicastro D., Gygi S.P., Agard D.A., and Avidor-Reiss T.. 2011. Sas-4 provides a scaffold for cytoplasmic complexes and tethers them in a centrosome. Nat. Commun. 2:359 10.1038/ncomms1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould R.R., and Borisy G.G.. 1977. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J. Cell Biol. 73:601–615. 10.1083/jcb.73.3.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Stearns T., and Lüders J.. 2009. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE. 4:e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris T.J., and Peifer M.. 2007. aPKC controls microtubule organization to balance adherens junction symmetry and planar polarity during development. Dev. Cell. 12:727–738. 10.1016/j.devcel.2007.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M.T., and Karlseder J.. 2013. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 32:4593–4601. 10.1038/onc.2012.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao L.R., and Megraw T.L.. 2009. Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr. Biol. 19:937–942. 10.1016/j.cub.2009.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi S., and Zheng Y.. 2004. Characterization of a Drosophila centrosome protein CP309 that shares homology with Kendrin and CG-NAP. Mol. Biol. Cell. 15:37–45. 10.1091/mbc.E03-03-0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., and Rieder C.L.. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. 10.1083/jcb.146.3.585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., and Rhee K.. 2011. The pericentriolar satellite protein CEP90 is crucial for integrity of the mitotic spindle pole. J. Cell Sci. 124:338–347. 10.1242/jcs.078329 [DOI] [PubMed] [Google Scholar]
- Kim S., and Rhee K.. 2014. Importance of the CEP215-pericentrin interaction for centrosome maturation during mitosis. PLoS ONE. 9:e87016 10.1371/journal.pone.0087016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Sasaki H., Yuba-Kubo A., Tsukita S., and Shiina N.. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–980. 10.1083/jcb.147.5.969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawo S., Hasegan M., Gupta G.D., and Pelletier L.. 2012. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat. Cell Biol. 14:1148–1158. 10.1038/ncb2591 [DOI] [PubMed] [Google Scholar]
- Lee M.J., Gergely F., Jeffers K., Peak-Chew S.Y., and Raff J.W.. 2001. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat. Cell Biol. 3:643–649. 10.1038/35083033 [DOI] [PubMed] [Google Scholar]
- Lerit D.A., and Gavis E.R.. 2011. Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr. Biol. 21:439–448. 10.1016/j.cub.2011.01.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerit D.A., and Rusan N.M.. 2013. PLP inhibits the activity of interphase centrosomes to ensure their proper segregation in stem cells. J. Cell Biol. 202:1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., and Kaufman T.C.. 1996. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 85:585–596. 10.1016/S0092-8674(00)81258-1 [DOI] [PubMed] [Google Scholar]
- Li Q., Hansen D., Killilea A., Joshi H.C., Palazzo R.E., and Balczon R.. 2001. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J. Cell Sci. 114:797–809. [DOI] [PubMed] [Google Scholar]
- Loncarek J., Hergert P., Magidson V., and Khodjakov A.. 2008. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10:322–328. 10.1038/ncb1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas E.P., and Raff J.W.. 2007. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 178:725–732. 10.1083/jcb.200704081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J. 2012. The amorphous pericentriolar cloud takes shape. Nat. Cell Biol. 14:1126–1128. 10.1038/ncb2617 [DOI] [PubMed] [Google Scholar]
- Mahjoub M.R., and Tsou M.F.. 2013. The AmAZI1ng roles of centriolar satellites during development. PLoS Genet. 9:e1004070 10.1371/journal.pgen.1004070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., and Raff J.W.. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw T.L., Li K., Kao L.R., and Kaufman T.C.. 1999. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development. 126:2829–2839. [DOI] [PubMed] [Google Scholar]
- Megraw T.L., Kilaru S., Turner F.R., and Kaufman T.C.. 2002. The centrosome is a dynamic structure that ejects PCM flares. J. Cell Sci. 115:4707–4718. 10.1242/jcs.00134 [DOI] [PubMed] [Google Scholar]
- Meijering E., Dzyubachyk O., and Smal I.. 2012. Methods for cell and particle tracking. Methods Enzymol. 504:183–200. [DOI] [PubMed] [Google Scholar]
- Mennella V., Keszthelyi B., McDonald K.L., Chhun B., Kan F., Rogers G.C., Huang B., and Agard D.A.. 2012. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nat. Cell Biol. 14:1159–1168. 10.1038/ncb2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V., Agard D.A., Huang B., and Pelletier L.. 2014. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 24:188–197. 10.1016/j.tcb.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen M.M., Mackie J.B., Doxsey S.J., Stearns T., and Tucker J.B.. 1997. Centrosomal deployment of γ-tubulin and pericentrin: evidence for a microtubule-nucleating domain and a minus-end docking domain in certain mouse epithelial cells. Cell Motil. Cytoskeleton. 36:276–290. [DOI] [PubMed] [Google Scholar]
- Mottier-Pavie V., and Megraw T.L.. 2009. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell. 20:2605–2614. 10.1091/mbc.E08-11-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Santos T., Sampaio P., Amorim I., Costa M., and Sunkel C.E.. 1999. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol. Cell. 91:585–596. 10.1111/j.1768-322X.1999.tb01104.x [DOI] [PubMed] [Google Scholar]
- Nigg E.A., and Raff J.W.. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678. 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Pagan J.K., Marzio A., Jones M.J., Saraf A., Jallepalli P.V., Florens L., Washburn M.P., and Pagano M.. 2015. Degradation of Cep68 and PCNT cleavage mediate Cep215 removal from the PCM to allow centriole separation, disengagement and licensing. Nat. Cell Biol. 17:31–43. 10.1038/ncb3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo R.E., Vogel J.M., Schnackenberg B.J., Hull D.R., and Wu X.. 2000. Centrosome maturation. Curr. Top. Dev. Biol. 49:449–470. [DOI] [PubMed] [Google Scholar]
- Pandey R., Heidmann S., and Lehner C.F.. 2005. Epithelial re-organization and dynamics of progression through mitosis in Drosophila separase complex mutants. J. Cell Sci. 118:733–742. 10.1242/jcs.01663 [DOI] [PubMed] [Google Scholar]
- Poulton J.S., Mu F.W., Roberts D.M., and Peifer M.. 2013. APC2 and Axin promote mitotic fidelity by facilitating centrosome separation and cytoskeletal regulation. Development. 140:4226–4236. 10.1242/dev.094425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J.B. 1992. Ultrastructure of centrosome domains and identification of their protein components. In The Centrosome. Kalnins V.I., editor. Academic Press, San Diego, CA: 45–68. 10.1016/B978-0-12-394770-3.50007-3 [DOI] [Google Scholar]
- Rauch A., Thiel C.T., Schindler D., Wick U., Crow Y.J., Ekici A.B., van Essen A.J., Goecke T.O., Al-Gazali L., Chrzanowska K.H., et al. . 2008. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science. 319:816–819. 10.1126/science.1151174 [DOI] [PubMed] [Google Scholar]
- Rebollo E., Llamazares S., Reina J., and Gonzalez C.. 2004. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2:E8 10.1371/journal.pbio.0020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T., Wojcik E.J., Sanders M.A., McGrail M., and Hays T.S.. 1999. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 146:597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., and Glover D.M.. 2007. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17:1465–1472. 10.1016/j.cub.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Rothwell W.F., and Sullivan W.. 2000. The centrosome in early Drosophila embryogenesis. Curr. Top. Dev. Biol. 49:409–447. [DOI] [PubMed] [Google Scholar]
- Rothwell W.F., Fogarty P., Field C.M., and Sullivan W.. 1998. Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organization. Development. 125:1295–1303. [DOI] [PubMed] [Google Scholar]
- Singh P., Ramdas Nair A., and Cabernard C.. 2014. The centriolar protein Bld10/Cep135 is required to establish centrosome asymmetry in Drosophila neuroblasts. Curr. Biol. 24:1548–1555. 10.1016/j.cub.2014.05.050 [DOI] [PubMed] [Google Scholar]
- Sonnen K.F., Schermelleh L., Leonhardt H., and Nigg E.A.. 2012. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol. Open. 1:965–976. 10.1242/bio.20122337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A.C., Stern D., Beaton A., Rhem E.J., Laverty T., Mozden N., Misra S., and Rubin G.M.. 1999. The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics. 153:135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W., Fogarty P., and Theurkauf W.. 1993. Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development. 118:1245–1254. [DOI] [PubMed] [Google Scholar]
- Takada S., Kelkar A., and Theurkauf W.E.. 2003. Drosophila checkpoint kinase 2 couples centrosome function and spindle assembly to genomic integrity. Cell. 113:87–99. 10.1016/S0092-8674(03)00202-2 [DOI] [PubMed] [Google Scholar]
- Terada Y., Uetake Y., and Kuriyama R.. 2003. Interaction of Aurora-A and centrosomin at the microtubule-nucleating site in Drosophila and mammalian cells. J. Cell Biol. 162:757–764. 10.1083/jcb.200305048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E. 1994. Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 44:489–505. [DOI] [PubMed] [Google Scholar]
- Tibelius A., Marhold J., Zentgraf H., Heilig C.E., Neitzel H., Ducommun B., Rauch A., Ho A.D., Bartek J., and Krämer A.. 2009. Microcephalin and pericentrin regulate mitotic entry via centrosome-associated Chk1. J. Cell Biol. 185:1149–1157. 10.1083/jcb.200810159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaizel-Ohayon D., and Schejter E.D.. 1999. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9:889–898. 10.1016/S0960-9822(99)80393-5 [DOI] [PubMed] [Google Scholar]
- Venken K.J., Kasprowicz J., Kuenen S., Yan J., Hassan B.A., and Verstreken P.. 2008. Recombineering-mediated tagging of Drosophila genomic constructs for in vivo localization and acute protein inactivation. Nucleic Acids Res. 36:e114 10.1093/nar/gkn486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wu T., Shi L., Zhang L., Zheng W., Qu J.Y., Niu R., and Qi R.Z.. 2010. Conserved motif of CDK5RAP2 mediates its localization to centrosomes and the Golgi complex. J. Biol. Chem. 285:22658–22665. 10.1074/jbc.M110.105965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Dantas T.J., Lalor P., Dockery P., and Morrison C.G.. 2013. Promoter hijack reveals pericentrin functions in mitosis and the DNA damage response. Cell Cycle. 12:635–646. 10.4161/cc.23516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., and Megraw T.L.. 2007. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell. 18:4037–4049. 10.1091/mbc.E07-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman W.C., Sillibourne J., Rosa J., and Doxsey S.J.. 2004. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 15:3642–3657. 10.1091/mbc.E03-11-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.