Abstract
In response to cardiac damage, a mesothelial tissue layer enveloping the heart called the epicardium is activated to proliferate and accumulate at the injury site. Recent studies have implicated the epicardium in multiple aspects of cardiac repair: a source of paracrine signals for cardiomyocyte survival or proliferation; a supply of perivascular cells and possibly other cell types like cardiomyocytes; and, a mediator of inflammation1-9. Yet, the biology and dynamism of the adult epicardium is poorly understood. Here, we created a transgenic line to ablate this cell population in adult zebrafish. We find that genetic depletion of epicardium after myocardial loss inhibits cardiomyocyte proliferation and delays muscle regeneration. The epicardium vigorously regenerates after its ablation, through proliferation and migration of spared epicardial cells as a sheet to cover the exposed ventricular surface in a wave from the chamber base toward its apex. By reconstituting epicardial regeneration ex vivo, we show that extirpation of the bulbous arteriosus (BA), a distinct, smooth muscle-rich tissue structure that distributes outflow from the ventricle, prevents epicardial regeneration. Conversely, experimental repositioning of the BA by tissue recombination initiates epicardial regeneration and can govern its direction. Hedgehog (Hh) ligand is expressed in the BA, and treatment with Hh signaling antagonist arrests epicardial regeneration and blunts the epicardial response to muscle injury. Transplantation of Shh-soaked beads at the ventricular base stimulates epicardial regeneration after BA removal, indicating that Hh signaling can substitute for the BA influence. Thus, the ventricular epicardium has pronounced regenerative capacity, regulated by the neighboring cardiac outflow tract and Hh signaling. These findings extend our understanding of tissue interactions during regeneration and have implications for mobilizing epicardial cells for therapeutic heart repair.
Keywords: heart regeneration, epicardium, ventricle, outflow tract, zebrafish, Hedgehog
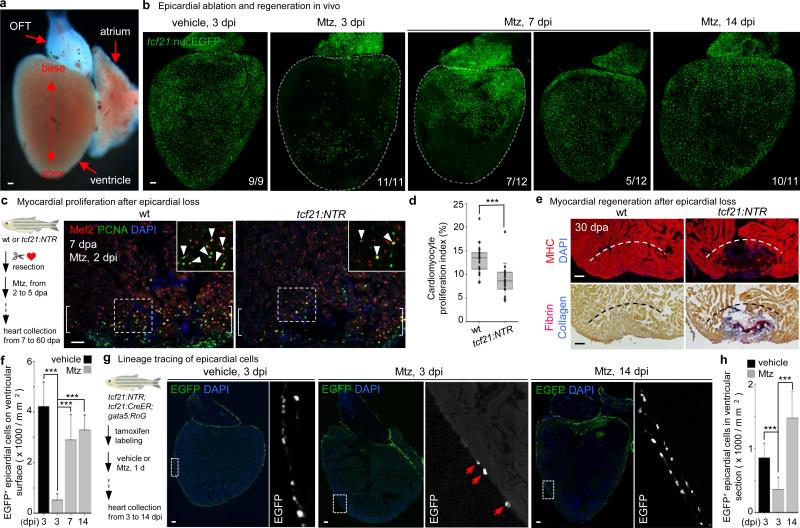
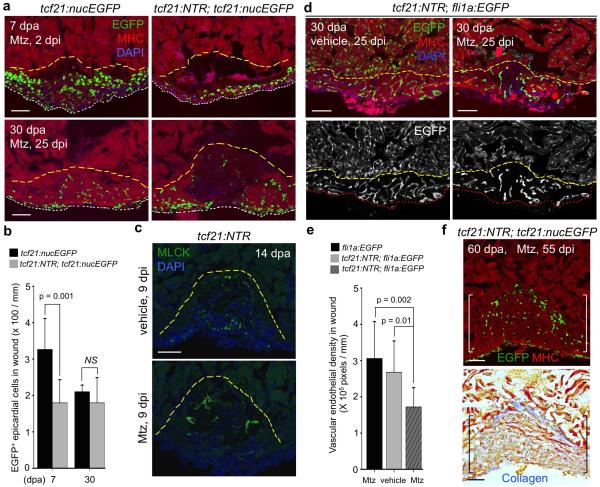
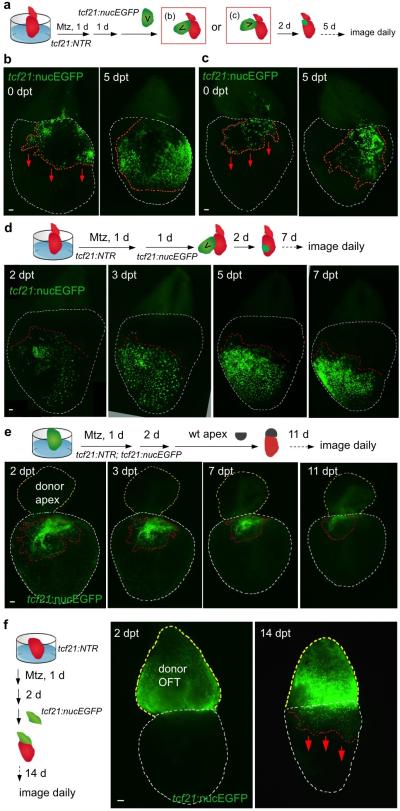
To assess homeostatic properties of the epicardium, we employed an inducible cell ablation system in adult zebrafish. Targeted expression of bacterial Nitroreductase (NTR) depletes specific cell types via conversion of a non-toxic substrate metronidazole (Mtz) to a cytotoxin10-12. We used tcf21 regulatory sequences, which in zebrafish drive the most widespread epicardial expression of known DNA elements2, to create an NTR transgenic line for lesioning this tissue without direct myocardial damage. After treatment of adult tcf21:NTR; tcf21:nucEGFP animals with Mtz, ~90% of EGFP+ epicardial nuclei on average were ablated from the ventricular surface in large patches (Fig. 1a, b, f).
Figure 1. Epicardial ablation and regeneration.
a, Adult zebrafish heart. OFT, outflow tract. b, tcf21:NTR; tcf21:nucEGFP adults were incubated with Mtz or vehicle, and hearts collected by random sampling at 3, 7, or 14 days post-incubation (dpi). Proportion of total animals with indicated phenotype is in lower right corner. All 3 dpi ventricles showed major ablation, averaging ~90% loss. c, (Left) Schematic for tests of epicardial ablation on muscle regeneration. (Right) Ventricular cardiomyocyte proliferation at 7 dpa. Brackets, injury site. Arrowheads, proliferating cardiomyocytes. d, Quantified PCNA+ cardiomyocyte indices in injury sites in experiments from (c). ***P < 0.001; Mann-Whitney Rank Sum test; n = 18 (wt) and 19 (tcf21:NTR) animals from two experiments. e, Section images of ventricles at 30 dpa, assessed for muscle recovery (MHC) and scar indicators (fibrin, collagen). One of 11 tcf21:nucEGFP and 8 of 12 tcf21:NTR; tcf21:nucEGFP ventricles showed myocardial gaps. Dashed line, approximate resection plane. **P < 0.01; Fisher Irwin exact test. f, Quantified EGFP+ nuclei from experiments in (b). ***P < 0.001; Student's two-tailed t-test. g, (Left) CreER-based strategy for permanent labeling of tcf21+ progeny. (Right) Section images of lineage-labeled EGFP+ epicardial progeny through 14 dpi, indicating derivation from pre-existing epicardium. Arrows at 3 dpi, EGFP+ cells spared by epicardial ablation. h, Quantified EGFP+ cells from experiments in (g). ***P < 0.001; Student's two-tailed t-test; n = 10 (vehicle, 3 dpi), 13 (Mtz, 3 dpi), and 15 (Mtz, 14 dpi). White dashed lines in (b), ventricle. Insets in (c, g), high magnifications of boxed areas. Scale bars, 50 μm. Error bars, s.d.
To determine whether epicardial depletion impacts the well-documented capacity of the zebrafish heart to regenerate13, we transiently incubated tcf21:NTR zebrafish with Mtz after resection of the ventricular apex. Mtz treatment reduced epicardial cell number in the 7 days post-amputation (dpa) injury site by ~45%, while reducing cardiomyocyte proliferation indices by ~33% (Fig. 1c, d, Extended Data Figs. 1a, b and 3c). Myofibroblasts were represented similarly in vehicle- and Mtz-treated clutchmates by 14 dpa (Extended Data Fig. 1c). Injured ventricles of Mtz-treated animals displayed reduced vascularization and muscularization by 30 dpa (Fig. 1e and Extended Data Fig. 1d, e), associated with fibrin and collagen retention (Fig. 1e). By 60 dpa, ventricles from Mtz-treated animals consistently showed normal muscularization and a large complement of tcf21-positive cells, along with minor collagen deposits (Extended Data Fig. 1f). Thus, depletion of epicardial tissue inhibits cardiomyocyte proliferation and vascularization after resection injury, reducing the efficacy of heart regeneration.
These experiments suggested a high capacity of epicardial cells to regenerate after major depletion. To test this directly, we examined otherwise uninjured hearts at different times after epicardial ablation. Ventricular epicardial cells typically have a low proliferation index (Extended Data Fig. 2a). However, within 3 days of Mtz treatment (3 dpi), many spared epicardial cells entered the cell cycle (Extended Data Fig. 2b, c). At 7 dpi, ventricles displayed quantifiable epicardial recovery that was more prominent at the chamber base (Fig. 1b). By 14 dpi, and as early as 7 dpi, ventricles were fully covered to their apices with tcf21:nucEGFP+ epicardial cells (Fig. 1b, f). The temporal variation in recovery likely reflects variation in location/pattern of epicardial cells spared by ablation among clutchmates, or in chamber size (Extended Data Fig. 3a). To examine origins of regenerated epicardium, we employed inducible Cre-based genetic fate-mapping to permanently label tcf21-expressing cells and their progeny prior to injury2. Labeling and subsequent fate-mapping experiments indicated that pre-existing epicardial cells, and not a tcf21-negative precursor, are a primary source for regeneration (Fig. 1g, h). In total, these experiments reveal that adult epicardium regenerates after substantial genetic ablation, through a mechanism of expansion by spared epicardial cells.
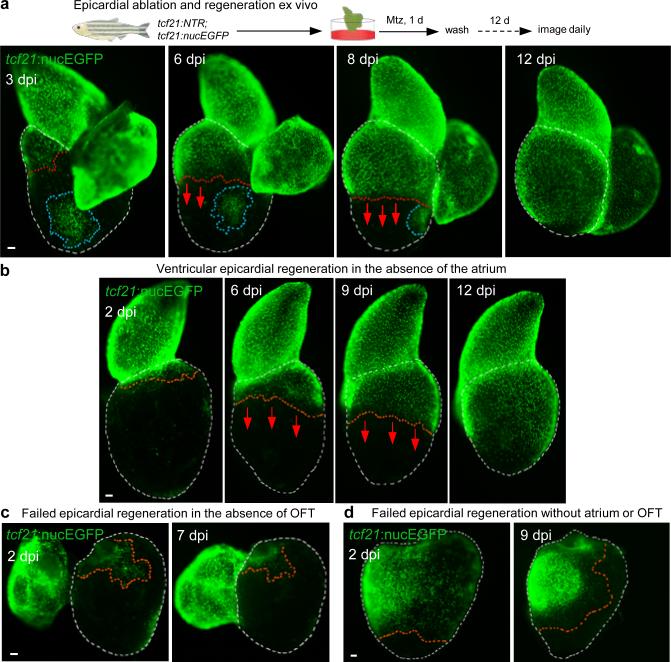
To expand our range of experimental manipulations, we refined protocols such that freshly dissected hearts contracted for several weeks ex vivo (Supplementary Video 1)14,15. When Mtz was added transiently to culture medium for one day, ventricular epicardial cells were potently ablated. Epicardial layers of the atrium and the BA (alternatively referred to as outflow tract) were less effectively depleted (Fig. 2a), likely due to differential expression of the NTR transgene among cardiac chambers (Extended Data Fig. 3b). Daily imaging of these hearts confirmed observations from in vivo experiments, demonstrating regeneration of the epicardium from base to apex that is typically completed in 2 weeks (Fig. 2a). Hearts from animals given partial ventricular resections injuries in vivo showed a similar pattern of epicardial regeneration after ex vivo ablation (Extended Data Fig. 4a). Cardiac muscle regeneration was ineffective in explanted hearts in our experiments. Increases in cell number occurred concomitantly with movement across the myocardial surface during epicardial regeneration, with spared epicardial cell patches away from the leading edge eventually incorporated into the sheet (Fig. 2a).
Figure 2. Cardiac outflow tract is required for regeneration of adjacent ventricular epicardium.
a, (Top) Schematic for epicardial ablation and regeneration in hearts cultured ex vivo. (Bottom) Regeneration occurs in a base-to-apex direction (arrows). Isolated patches (circled by blue dashed lines) do not participate in regeneration until contacted by the leading edge. b, Ventricular epicardium regenerates in the absence of the atrium (n = 19; behavior seen in all samples). Arrows, direction of regeneration. c, d, Ventricular epicardium fails to regenerate in the absence of the BA. Ventricular epicardium showed defective regeneration in these experiments with (c) (n = 6; all samples) or without atrium (d) (n = 14; all samples), even when, in rare cases, many basal epicardial cells were spared (d). Red dashed lines, epicardium or epicardial leading edge. White dashed lines, ventricle. Scale bars, 50 μm.
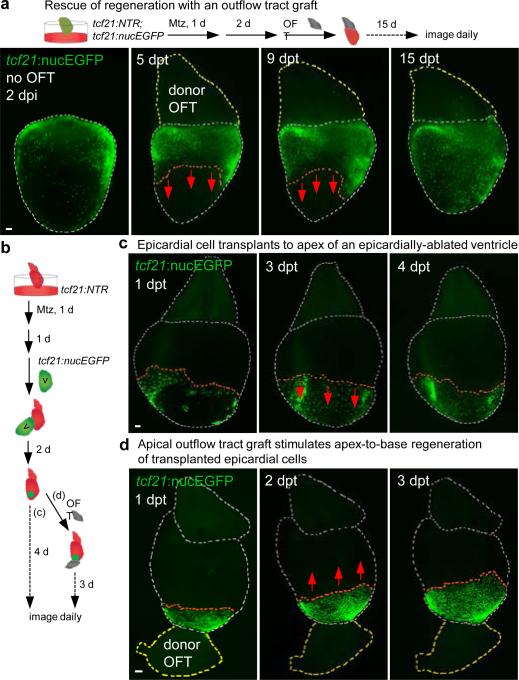
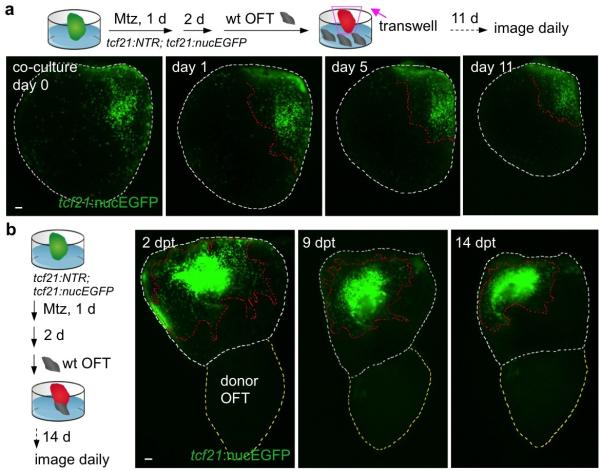
To identify possible intrinsic differences in epicardial cells from different ventricular regions, we examined behaviors of basal or apical epicardial tissue patches transplanted to ablated ventricles. In these experiments, transplanted cells of either origin consistently repopulated the ventricular surface in a base-to-apex direction after transplantation (Extended Data Fig. 5a-d), revealing no proliferative bias in ventricular epicardial cells that could explain the directional flow of regeneration. To assess potential extrinsic influences on epicardial regeneration, we removed the atrium or BA from its attachment at the ventricular base prior to epicardial ablation. Atrial extirpation did not noticeably affect regeneration of ventricular epicardium (Fig. 2b and Supplementary Video 2). By contrast, removal of outflow tissue blocked epicardial cell recovery, an arrest that persisted for at least two weeks (Fig. 2c, d and data not shown). To test whether this arrest was solely a consequence of mechanical tissue disruption, we ablated the epicardium after host BA removal, before grafting a non-transgenic BA to the ventricular base 2 days later. In most of these tissue recombination procedures (13 of 21), host tcf21:nucEGFP+ epicardium regenerated to cover the ventricle (Fig. 3a). This effect was not observed when a portion of donor ventricular apex was inverted and transplanted to the host ventricular base (Extended Data Fig. 5e). Complementary grafting experiments indicated that BA could contribute epicardial cells to the ventricular surface, as a potential supplement to expansion of the ventricular epicardial cell pool (Extended Data Fig. 5f). Thus, our experiments indicate that outflow tissue provides an essential interaction for regeneration from existing ventricular epicardial cells.
Figure 3. Outflow tract tissue is sufficient to initiate and redirect epicardial regeneration.
a, (Top) A non-transgenic BA (donor OFT) was transplanted to a transgenic ventricular base after epicardial ablation ex vivo. (Bottom) Base-to-apex epicardial regeneration (arrows) was observed from host tissue in 13 of 21 ventricles. dpt, days post-transplantation. b, (Top) Experimental design in (c, d). c, tcf21:nucEGFP+ epicardial cells transplanted to an epicardially ablated ventricular apex were static or regenerated (arrows) toward the apex (n = 12, all samples), but not toward the base. d, tcf21:nucEGFP+ epicardial cells were transplanted to the apex of an epicardially-ablated ventricle, followed by apical grafting of a donor BA. tcf21:nucEGFP+ cells regenerated in a reversed apex-to-base direction (arrows) in 9 of 14 ventricles. Twelve of 18 host ventricles with the host BA removed before donor BA grafting also showed apex-to-base regeneration. Red dashed lines in (a, c, d), epicardial leading edge. White dashed lines in (a, c, d), ventricle and host BA. Yellow dashed lines in (a, d), donor BA. Scale bars, 50 μm.
To test if outflow tract tissue is sufficient to stimulate epicardial regeneration, we ectopically positioned experimentally manipulated cardiac structures. Co-culture of several BAs in a transwell assay with an epicardially ablated ventricle did not restore regeneration in the absence of host BA (Extended Data Fig. 6a). Similarly, a BA graft placed at the ventricular apex showed no evidence of directing regeneration of basally located host epicardial cells toward the apex (Extended Data Fig. 6b). Thus, we could not detect BA effects requiring long-range diffusion through tissue or culture medium. Next, we transplanted an tcf21:nucEGFP+ epicardial cell patch to the apex of an ablated host ventricle, after which we grafted a wild-type BA to the apex (Fig. 3b). Remarkably, the apical BA was capable of stimulating apex-to-base regeneration from the nearby epicardial patch in a high proportion (21 of 32) of experiments, effectively reversing the stereotypic direction of recovery (Fig. 3c, d). Together, these experiments indicate that the cardiac outflow tract is necessary and sufficient for epicardial regeneration, and that this neighboring tissue provides a short-range influence(s) that directs regeneration from base to apex.
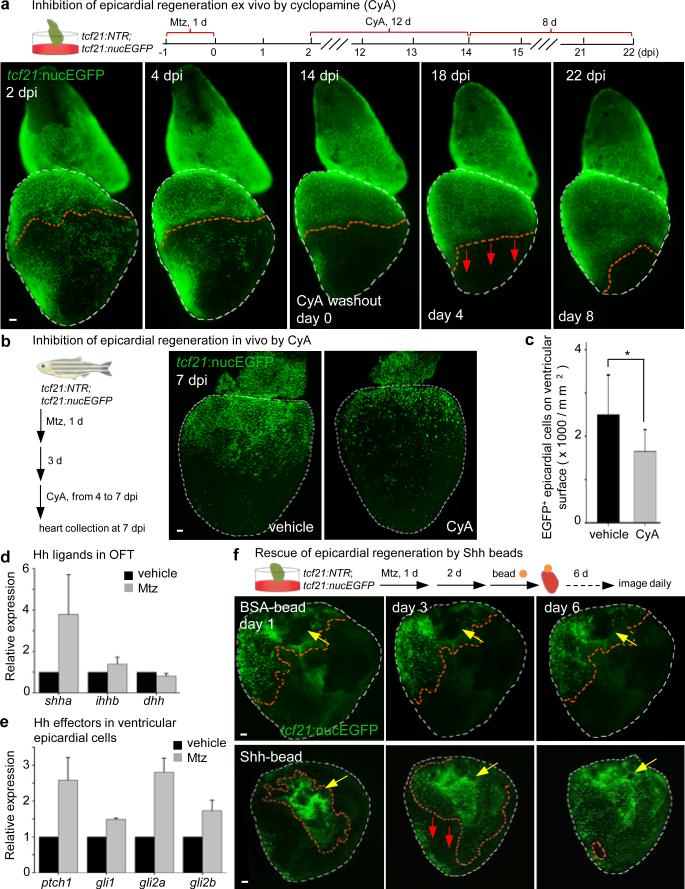
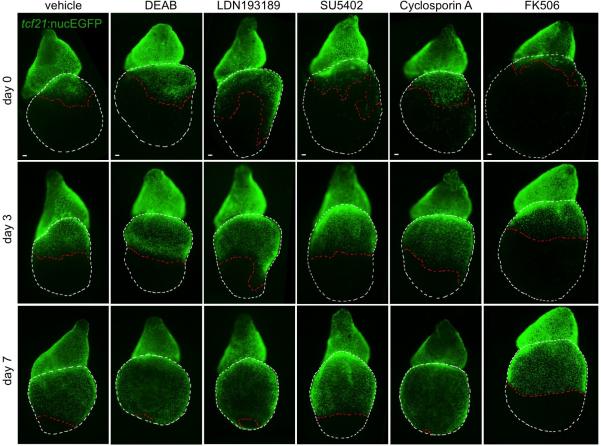
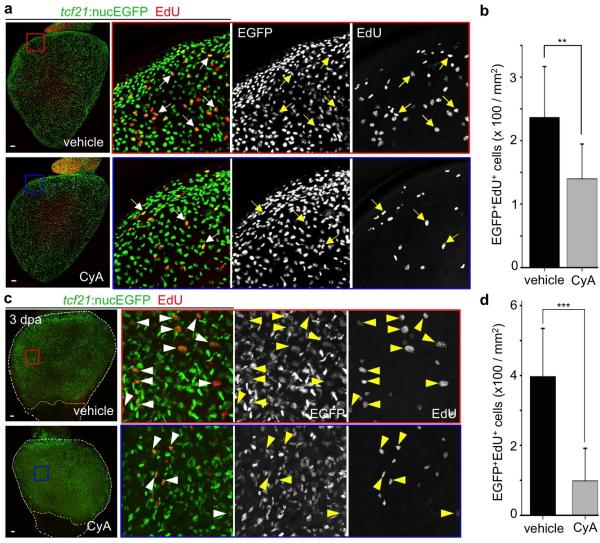
What is the molecular nature of interactions between outflow tract and ventricular epicardium? To address this, we applied a small panel of signaling pathway effectors to epicardially ablated hearts cultured ex vivo. Among several compounds (Extended Data Fig. 7), the Smoothened (Smo) antagonist cyclopamine (CyA) blocked regeneration; yet, regeneration initiated normally after drug washout (Fig. 4a). CyA treatment reduced spontaneous epicardial cell EdU incorporation occurring in the first 2 days of explant culture, suggesting that intact Hh signaling promotes epicardial proliferation (Extended Data Fig. 8a, b). CyA also disrupted in vivo epicardial regeneration, not only in adults but in larvae, an additional developmental setting in which we identified base-to-apex regeneration (Fig. 4b, c and Extended Data Fig. 9a-c). CyA treatment from 2 to 4 dpf also reduced the initial epicardial occupancy of the larval ventricle (Extended Data Fig. 9d-f), indicating that epicardial regeneration recapitulates at least one pathway influential in morphogenesis. Finally, we observed inhibitory effects of CyA on the epicardial proliferative response to muscle resection in vivo, and in coverage of these injuries ex vivo (Extended Data Fig. 4b and Extended Data Fig. 8c, d).
Figure 4. Hedgehog signaling controls epicardial regeneration.
a, (Top) Experimental schematic. (Bottom) CyA arrested regeneration while present (n = 7; all samples), and regeneration initiated (arrows) after CyA removal (6 of 7 ventricles). b, tcf21:NTR; tcf21:nucEGFP adults were incubated with Mtz and randomly separated into two groups for treatment with CyA or vehicle. c, Quantified tcf21:nucEGFP+ epicardial cells from experiments in (b). *P < 0.05; Student's two-tailed t-test; n = 13 (vehicle) and 12 (CyA). d, Quantitative RT-PCR detecting shha and ihhb expression in outflow tract after epicardial ablation (3 dpi), from 3 separate experiments on pooled tissues using 90 total zebrafish. e, Quantitative RT-PCR of FACS-purified ventricular epicardial cells showed increased ptch1 and gli2a expression after epicardial ablation (7 dpi), from 2 separate experiments using 209 zebrafish. f, (Top) BSA- or recombinant Shh-soaked beads were grafted to the base of epicardially ablated ventricles 2 days after 24 hours Mtz treatment. (Bottom) Shh-beads stimulated one-half or greater coverage (red arrows) in 9 of 32 ventricles, whereas BSA-beads (middle) did not (0 of 27). Yellow arrows, transplanted beads. Red dashed lines in (a, f), epicardium or epicardial leading edge. White dashed lines in (a, f), ventricle. Scale bars, 50 μm. Error bars, s.d.
Smo is an effector for several Hh family ligands, which have potent short-range effects in multiple contexts of embryonic development16-21. Quantitative PCR revealed shha, ihhb and dhh ligand transcripts in adult atrium, ventricle, and BA, where in situ hybridization detected shha and dhh transcript signals in smooth muscle tissue (Extended Data Fig. 10a-d). Epicardial ablation injury boosted BA and ventricular shha levels, as well as levels of ptch1 and gli2a in purified epicardial cells (Fig. 4d, e). Moreover, a shha:EGFP reporter strain visualized shha regulatory sequence-driven fluorescence in smooth muscle and epicardial tissues of the BA (Extended Data Fig. 10e). No additional in situ hybridization or shha:EGFP fluorescence patterns were detectable after epicardial ablation; however, apical resection injury induced fluorescence in ventricular epicardial tissue by 2 dpa (Extended Data Fig. 10d, f). To test whether local Hh ligand delivery is sufficient to substitute for the BA, we removed atrium and BA from cardiac explants, ablated epicardial cells, and applied beads soaked with Shh protein to the exposed ventricular base. Shh-soaked beads stimulated epicardial regeneration (one-half or greater coverage) in 9 of 32 ventricles, whereas this level of recovery never occurred after transplantation of BSA-soaked beads ((0 of 27) ventricles; Fig. 4f). We speculate that these effects of Hh on the epicardial sheet might involve cytoplasmic extensions or a factor transport system22,23. Together, our findings support a model in which Hh ligand from outflow tract, and possibly additional tissues, guides the base-to-apex regeneration of ventricular epicardium.
In conclusion, we have identified a requirement for the mesothelial covering of the zebrafish heart in the proficiency of muscle regeneration. Moreover, we show that the ventricular epicardium itself has high endogenous renewal capacity, vigorously regenerating as a sheet from the base of the chamber to its apex after genetic depletion. Our results point to the outflow tract as an unexpected signaling center and source of Hh, and possibly additional influences, that can promote epicardial regeneration. It is likely that tissue regeneration is similarly regulated in trans in other contexts; for example, to maintain the mesothelium lining abdominal organs. As a mediator of epicardial regeneration, Hh signaling can be integrated into new strategies to modulate repair of the damaged heart.
METHODS
Zebrafish maintenance and procedures
Adult zebrafish of the Ekkwill and Ekkwill/AB strains were maintained as described and resection injuries were performed as described24,13. Animals between 4 and 12 months of both sexes were used. Transgenic lines used in this study were Tg(tcf21:mCherry-NTR)pd108 (tcf21:NTR, described below), Tg(tcf21:nucEGFP)pd41 24, Tg(tcf21:CreER)pd42 2, Tg(gata5:loxpmCherry-loxp-nucEGFP)pd40 25, Tg(fli1a:EGFP)y1 26 and Tg(shha:EGFP)sb15 27. All transgenic strains were analyzed as hemizygotes. For epicardial ablation experiments in adults, animals were bathed for 24 hours in 10 mM Mtz (Sigma) as described and returned to water12. If ablation was performed after ventricular resection, we used a protocol of daily changes of 1 mM Mtz solution for 3 days that had similar ablation effects (Extended Data Fig. 3c). This period corresponds with the early epicardial proliferative response to resection injury (ref #7 and data not shown), and was intended to extend the ablation window and improve animal survival. For larval epicardial ablation, 6 hpf embryos were bathed for 48 hours in 10 mM Mtz before washout. For ex vivo epicardial ablation, dissected hearts were bathed for 24 hours in 1 mM Mtz before washout. Cyclopamine (CyA, Selleckchem) was dissolved in ethanol to a final concentration of 20 mM. CyA was used at 10 μM for in vivo treatment of adult animals and 5 μM for ex vivo culture and embryo treatments. For EdU incorporation experiments that followed epicardial ablation, animals were injected with 10 mM EdU 4 hours prior to collection. Experiments with uninjured animals used 3 daily 10 mM EdU injections. For lineage tracing, strains carrying tcf21:NTR; tcf21:CreER, and gata5:loxp-mCherry-loxp-nucEGFP transgenes were placed in a small beaker of aquarium water containing 5 μM tamoxifen. Fish were maintained for 24 hours, rinsed with fresh aquarium water, and returned to a recirculating aquatic system for 24 hours, before repeating this incubation twice. After 3 days of rinsing, Mtz was added for an additional 24 hours. As is common when using Cre-based tools, we could not genetically label all epicardial cells in these experiments or rule out minor contributions by tcf21-negative cells. Animal procedures were performed in accordance with Duke University guidelines.
Construction of tcf21:NTR zebrafish
The translational start codon of tcf21 in the BAC clone DKEYP-79F12 was replaced with the mCherry-NTR cassette by Red/ET recombineering technology (Gene Bridges)12. The 5’ and 3’ homologous arms for recombination were a 50-base pair (bp) fragment upstream and downstream of the start codon, and were included in PCR primers to flank the mCherry-NTR cassette. To avoid aberrant recombination between the mCherry-NTR cassette and the endogenous loxp site in the BAC vector, we replaced the vector-derived loxp site with an I-SceI site using the same technology. The final BAC was purified with Nucleobond BAC 100 kit (Clontech) and co-injected with I-SceI into one-cell-stage zebrafish embryos.
Ex vivo cardiac explants
Adult hearts were rinsed several times in PBS after collection and cultured in dishes with DMEM medium plus 10% fetal bovine serum, 1% non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-Mercaptoethanol, while rotating at 150 rpm. Primocin™ (InvivoGen) was added to prohibit microbial contaminants during the first 3 days primary culture. For transplantation experiments, outflow tract or ventricular tissues were grafted by mounting in 1% low-melting point agarose with ablated hearts in culture dishes, and covering with medium. After 2 days of culturing, attached tissues were released from the agarose; if transplanting an epicardial patch, the ventricular donor tissue was removed carefully using forceps. Fluorescent transgenes in these cardiac explants were monitored using a Leica MZ05FA stereofluorescence microscope.
Recombinant mouse Sonic Hedgehog (C25II), N-terminus protein (R & D Systems) was reconstituted at 100 μg/ml in phosphate buffered saline (PBS) containing 0.1% bovine serum albumin (BSA). Affi-Gel Blue beads (Bio-Rad) were prepared by thoroughly washing the beads in PBS, then incubating them in the Shh solution for 2 hours at room temperature. A solution with the same concentration of BSA protein was used as the control. The beads were then applied to the base of ventricular explants that were settled in low-melting point agarose in serum-free DMEM supplemented as above. After 24 hours, the ventricles were released from the agarose with the attached beads and cultured in supplemented, serum-free DMEM. Under these ex vivo culture conditions we observed no increase in cardiomyocyte proliferation upon resection injury.
Preparation of outflow tract and ventricular epicardial cells and RT-qPCR
Adult hearts were dissected from tcf21:NTR or tcf21:NTR; tcf21:nucEGFP animals 3 and 7 days post-treatment with vehicle or Mtz. Outflow tracts were frozen in liquid nitrogen. Ventricular nucEGFP+ epicardial cells were isolated as described previously28 with modifications. Briefly, ventricles were collected on ice and washed several times to remove blood cells. Ventricles were digested in an Eppendorf tube with 0.5 ml HBSS plus 0.13 U/ml Liberase DH (Roche) and 1% sheep serum at 37°C, while stirring gently with a Spinbar® magnetic stirring bar (Bel-Art Products). Supernatants were collected every 5 min and neutralized with sheep serum. Dissociated cells were spun down and sorted using a BD FACSVantage SE sorter for EGFP-positive cells. Total RNA was extracted using an RNAqueous®-Micro isolation kit (Ambion) according to the manufacturer's instructions. Reverse transcription was carried out using a SuperScript III First-Strand synthesis kit (Life Technologies). qPCR was carried out in triplicate using an Roche LightCycler® 480 II system with the LightCycler® 480 Probes Master Mix and the Universal Probe Library (UPL) (Roche). rpl13a served as the control. Primers and the UPL# used in this study were:
shha-f aagcccacattcattgctct and shha-r ccttctgtcctccgtcctg, UPL #54;
shhb-f gcaagtatgggatgctatccag and shhb-r tcctgatttagcagccactg, UPL #16;
ihha-f tgggtctactatgagtccaaagc and ihha-r ggttttagcagccacagagtg, UPL #86;
ihhb-f tcttgttatgctgcggtgaa and ihhb-r aagcgtagaggtgcaaaagc, UPL #140;
dhh-f cgtgcactgctctgtcaaa and dhh-r aaacatgacatgggcttttgt, UPL #156;
ptch1-f tggcttaagggcagctaatc and ptch1-r gccgtgtgtacttgagttcct, UPL #87;
ptch2-f ccatgacatcaactggaatga and ptch2-r gaatgctcccatgaacaacc, UPL #6;
gli1-f ctgcagcaaagagttcgaca and gli1-r ctccgtggatgtgctcatta, UPL #68;
gli2a-f cctcacccaccacagcat and gli2a-r cgatcgggattggtgtgt, UPL #39;
gli2b-f cctgccagaatacttcacatca and gli2b-r ctcaacctgggcgtcatac, UPL #48;
rpl13a-f gcggaccgattcaataagg and rpl13a-r gaaagacgaccgaggagatg, UPL #147.
Histology
Analyses of cardiomyocyte proliferation were performed as previously described by counting Mef2+ and PCNA+ nuclei in wound sites14. To quantify vascular endothelial cells in the wound site by 30 dpa with fli1a:EGFP or tcf21NTR; fli1a:EGFP animals, three medial, longitudinal sections were selected from each heart. Images of single optical slices of green fluorescence in the wound site were acquired using a 20× objective (1024×1024 pixels). EGFP+ areas were quantified in pixels by ImageJ software, and the ratio of EGFP+ area versus the length of the outlined apical wound was calculated for each heart. To quantify tcf21+ epicardial cells in the wound site at 7 and 30 dpa with tcf21:nucEGFP or tcf21:NTR; tcf21:nucEGFP animals, three medial, longitudinal sections were selected from each heart. EGFP+ cells were counted in the wound area, and the ratio of EGFP+ cells versus the length of the outlined apical wound was calculated for each heart. Acid Fuchsin-Orange G and immunostaining were performed as described13. Primary antibodies used in this study were anti-Myosin heavy chain (MHC; F59, mouse; Developmental Studies Hybridoma Bank), anti-GFP (rabbit; Invitrogen), anti-Mef2 (rabbit; Santa Cruz Biotechnology), anti-MLCK (mouse, K36; Sigma), and anti-PCNA (mouse; Sigma). Secondary antibodies (Invitrogen) used in this study were: Alexa Fluor 488 goat anti-rabbit; Alexa Fluor 594 goat anti-rabbit, goat anti-rat and goat anti-mouse; and Alexa 633 goat anti-mouse. EdU was detected through a click reaction as described previously29 with fluorescent azide (Alexa Fluor® 594 or 647, Invitrogen). Whole-mounted and sectioned ventricular tissues were imaged used a Zeiss 700 confocal microscope.
Data collection and statistics
Clutchmates (or hearts collected from clutchmates) were randomized into different treatment groups for each experiment. No animal or sample was excluded from the analysis unless the animal died during the procedure. All experiments were performed with at least 2 biological replicates, using appropriate numbers of samples for each replicate. Sample sizes were chosen based on previous publications and experiment types, and are indicated in each figure legend. For expression patterns, at least 6 fish were examined. For assessment of epicardial ablation and consequences on muscle regeneration, at least 9 fish were examined. At least 12 hearts of each group were pooled for RNA purification and subsequent RT-qPCR. For ex vivo epicardial ablation experiments, at least 6 hearts were used for each treatment. An exception was the small compound screen, where at least 4 hearts were used for each drug. Quantification of cell proliferation and calculation of statistical outcomes were assessed by a person blinded to the treatments. All statistical values are displayed as Mean +/- Standard Deviation. Sample sizes, statistical tests, and P values are indicated in the figures or the legends. Student's t-tests (two-tailed) were applied when normality and equal variance tests were passed. The Mann-Whitney Rank Sum test was used when these failed. Fisher Irwin exact tests or Chi-square tests were used where appropriate.
Extended Data
Extended Data Figure 1. Epicardial ablation and responses to resection injury.
a, tcf21:nucEGFP or tcf21:NTR; tcf21:nucEGFP clutchmates underwent resection injury and were treated for 3 days with 1 mM Mtz, before collection of ventricles at 7 (n = 8 animals per group) and 30 dpa (n = 11). tcf21:NTR; tcf21:nucEGFP wounds had less epicardial cells at 7 dpa and comparable occupancy by 30 dpa compared to tcf21:nucEGFP wounds. White dashed lines, wound edge. b, Quantification of EGFP+ epicardial cells at 7 and 30 dpa with respect to wound edge lengths from (a). NS, not significant; Mann-Whitney Rank Sum Test. c, tcf21:NTR clutchmates underwent resection injury and were treated for 3 days with 1 mM Mtz or vehicle with random separation into two groups for treatment. MLCK+ cells indicating myofibroblasts had comparable presence in both groups at 14 dpa (n = 7 for each group). d, tcf21:NTR; fli1a:EGFP clutchmates underwent resection injury and were treated for 3 days with 1 mM Mtz or vehicle with random separation into two groups for treatment. Epicardial ablation led to lower vascular density in 30 dpa wounds compared with controls. Red dashed lines, wound edge. e, Quantification of EGFP+ vascular endothelial pixel area in wounds of tcf21:NTR; fli1a:EGFP zebrafish treated with Mtz (n = 12) or vehicle (n = 6), or of fli1a:EGFP zebrafish treated with Mtz (n = 6), with respect to the wound edge lengths. Student's two-tailed t-test. f, By 60 dpa, 55 days after epicardial ablation protocols, muscularization (top) and wound collagen deposition (bottom) were grossly normal (n = 23). Brackets, area of regeneration. Yellow dashed lines in (a, c, d), approximate amputation plane. Scale bars, 50 μm. Error bars, s.d.
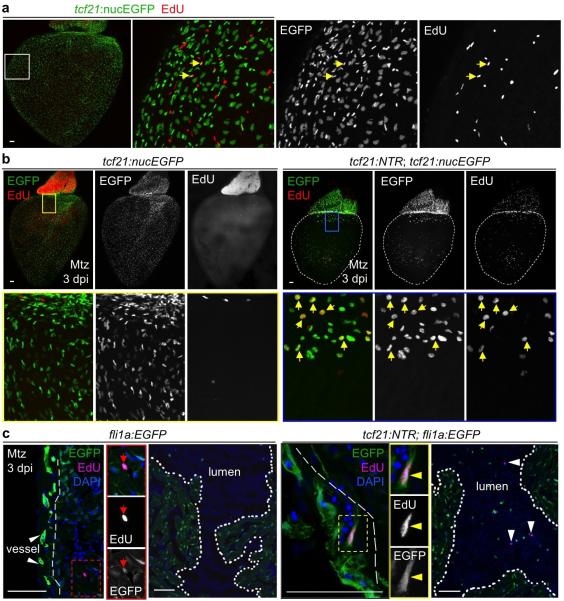
Extended Data Figure 2. Epicardial cell proliferation without injury and after epicardial ablation.
a, Limited epicardial cell proliferation on the ventricular surface. tcf21:nucEGFP fish were injected with 10 mM EdU once daily for 3 days and collected one day after the last injection. 105 ventricular nucEGFP+ cells were assessed for EdU reactivity in 15 animals, from which 608 cells were positive (a 0.6% rate for 4 days EdU labeling). Whole mount image is shown, and arrows in enlarged boxed area indicate EGFP+EdU+ nuclei. b, tcf21:nucEGFP or tcf21:NTR; tcf21:nucEGFP fish were injected with 10 mM EdU at 3 days post-Mtz treatment, and hearts were collected 4 hours later. Boxed areas in images of whole-mounted hearts show magnified views. Yellow arrows in (a, b), representative EGFP+ (Green) EdU+ (Red) nuclei. c. fli1a:EGFP or tcf21:NTR; fli1a:EGFP fish were injected with 10 mM EdU at 3 days post-Mtz treatment, and hearts were collected 4 hours later. Red arrows, representative EGFP+ (Green) EdU+ (Magenta) endocardial cell nuclei. Yellow arrowheads, representative EGFP+ (Green) EdU+ (Magenta) vascular endothelial cell nuclei. White arrowheads, EdU+ (Magenta) nuclei within the ventricular lumen, ostensibly erythrocyte nuclei. Scale bars, 50 μm.
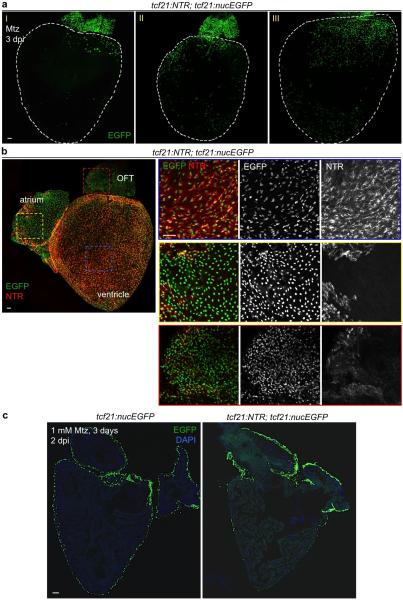
Extended Data Figure 3. Mosaic NTR expression and patterns of spared epicardial cells after ablation.
a, Whole-mounted examples of varied location/pattern of spared epicardial cells in ventricles from tcf21:NTR; tcf21:nucEGFP adult clutchmates 3 days after incubation with 10 mM Mtz. White dashed lines, ventricle. b, Differential expression of the NTR transgene among cardiac chambers. In adult tcf21:NTR; tcf21:nucEGFP hearts, EGFP expression is comparable in epicardial tissue covering the atrium, OFT, and ventricle. By contrast, NTR (red, indicated by mCherry) expression is patchy and/or weak in atrium and OFT compared with ventricular expression. c, Section images of ventricles from tcf21:nucEGFP (left) or tcf21:NTR; tcf21:nucEGFP zebrafish (right) treated with 1 mM Mtz (right) for 3 days, and collected 2 days later. Ventricular epicardium was ablated effectively in these experiments. Scale bars, 50 μm.
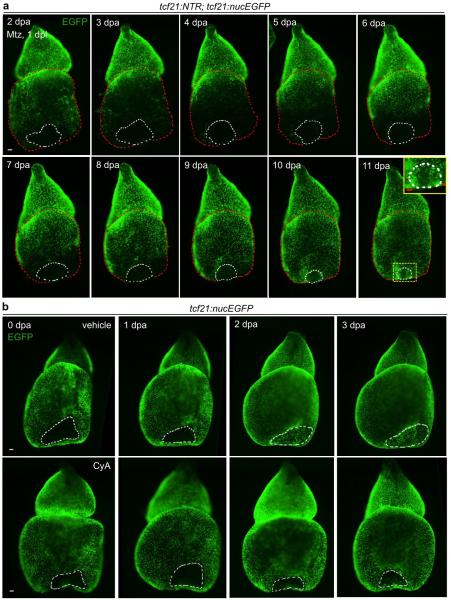
Extended Data Figure 4. Epicardial regeneration after ventricular resection.
a, Hearts were removed from tcf21:NTR; tcf21:nucEGFP fish immediately after ventricular resection injuries, followed by 24 hours of Mtz and a 24-hour washout ex vivo. A base-to-apex pattern of epicardial regeneration was observed, in this example covering the apical wound by 11 dpa (n = 18; behavior seen in all samples). Epicardial coverage of resection injuries in these ablation experiments is delayed compared to ventricles recovering with an intact epicardium (b, Top). Yellow boxed area, magnified view of the apical wound. b, Hearts were removed from tcf21:nucEGFP clutchmates immediately after apical resection injury and cultured ex vivo, before random separation two treatment groups. Epicardial cells covered the wound area by 3 dpa (n = 11; behavior seen in all samples), unless treated with CyA (n = 26; failed coverage in 20 of 26 ventricles). Red dashed lines in (a), ventricle. White dashed lines in (a, b), apical wounds. Scale bars, 50 μm.
Extended Data Figure 5. Ex vivo grafts and epicardial regeneration.
a-c, (a) Schematic of the experimental design. Epicardial cells transplanted at the base of a an epicardially ablated host regenerated toward the apex regardless of basal (b) (n = 25, behavior seen in all samples) or apical (c) (n=27, all samples ) origin. d, (Top) Epicardial cells from the base of a transgenic donor ventricle were transplanted to the chamber midpoint of an epicardially ablated host ventricle and observed for regeneration. (Bottom) Transplanted cells eventually migrated toward the apex, not the base (n = 13; all samples). e, (Top) Following epicardial ablation, the host BA was replaced with a non-transgenic donor ventricular apex and observed for regeneration. (Bottom) Ventricular epicardium showed little or no regeneration in these experiments (n = 7; behavior seen in all samples). f, (Left) Following ex vivo epicardial ablation in a host tcf21:NTR ventricle, the host BA was replaced with a donor tcf21:nucEGFP BA. (Right) The host ventricular surface contained different amounts of EGFP+ nuclei in these ventricles (n = 3; behavior seen in all samples). Red dashed lines in (b-f), epicardium or epicardial leading edge. White dashed lines in (b-f), ventricle. Yellow dashed lines in (e, f), donor apex (d) or BA (f). Scale bars, 50 μm.
Extended Data Figure 6. Context-specific effects of outflow tract on epicardial regeneration.
a, (Top) Following ex vivo epicardial ablation and BA removal, ventricles were co-cultured with 10 outflow tracts in a transwell assay and observed for regeneration. (Bottom) No evidence for epicardial regeneration was observed in these experiments (n = 9; behavior seen in all samples). b, (Left) Following ex vivo epicardial ablation and BA removal, a non-transgenic BA (labeled as donor OFT) was transplanted to the apex and observed for regeneration. (Right) No evidence for regeneration of EGFP+ epicardium from apex to base was observed in these experiments (n = 10; behavior seen in all samples). Red dashed lines in (a, b), epicardium. White dashed lines in (a, b), ventricle. Yellow dashed lines in (b), donor outflow tract. Scale bars, 50 μm.
Extended Data Figure 7. Small-scale screen for compounds that inhibit epicardial regeneration.
Ex vivo ablation and regeneration of tcf21:NTR; tcf21:nucEGFP ventricles over 7 days. Mtz was added for 24 hours to freshly isolated hearts, washed out, and compounds were added after 2 days (Day 0). Hearts were treated with vehicle (n > 10), 10 μM DEAB (n = 5; Sigma-Aldrich), 100 nM LDN193189 (n = 4; Cayman Chemical), 10 μM SU5402 (n = 5; Santa Cruz Biotechnology), 1 μM CyclosporinA (n = 4; Sigma-Aldrich), or 0.1 μg/ml FK506 (n = 5; Sigma-Aldrich), in each case showing base-to-apex recovery (behavior seen in all samples). The dissected hearts were randomly separated into groups for drug treatment. Red dashed lines, epicardial leading edge. White dashed lines, ventricle. Scale bars, 50 μm.
Extended Data Figure 8. Epicardial proliferation is regulated by Hh signaling.
a, Freshly dissected tcf21:nucEGFP hearts were randomly separated into two groups and cultured for 47 hours with vehicle (n = 11) or 5 μM CyA (n = 8). Then, 25 μM EdU was added to the medium for one hour prior to collection at 48 hours. Cyclopamine (CyA) treatment decreases epicardial cell proliferation ex vivo. Arrows, representative EGFP+ (Green) EdU+ (Red) nuclei. b, Quantification of EGFP+EdU+ nuclei per mm2 on the ventricular surface, from hearts in (a). **P < 0.01; Student's two-tailed t-test. c, tcf21:nucEGFP adult fish were subjected to partial ventricular resection surgery, and randomly separated into two groups for treatment with vehicle (n = 8) or 10 μM cyclopamine (CyA) (n = 10) from 2 to 3 dpa. Then, 10 mM EdU was injected intraperitoneally 1 hour prior to collection. CyA treatment decreases epicardial cell proliferation in vivo. Arrowheads, representative EGFP+ (Green) EdU+ (Red) nuclei. d, Quantification of EGFP+EdU+ nuclei per mm2 on the ventricular surface, from hearts in (c). ***P < 0.001; Mann-Whitney Rank Sum Test. Yellow dashed lines in (c), resection plane. White dashed lines in (c), ventricle. Boxed areas in (a, c), magnified views. Scale bars, 50 μm. Error bars, s.d.
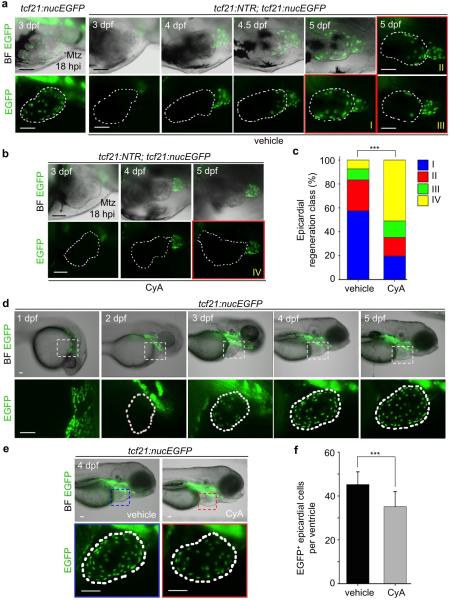
Extended Data Figure 9. Larval epicardial development and regeneration.
a, tcf21:nucEGFP or tcf21:NTR; tcf21:nucEGFP larval clutchmates were treated with 10 mM Mtz from 6 hpf to 54 hpf, and then imaged at different times from 3 to 5 dpf. tcf21:nucEGFP larvae show normal ventricular epicardial coverage at 3 dpf, while tcf21:NTR; tcf21:nucEGFP coverage is sparse. tcf21:NTR; tcf21:nucEGFP larvae with confirmed full ablation were imaged from 3 to 5 dpf, covering first the ventricular base and then the apex. Three different extents of regeneration at 5 dpf are shown: classes I, greater than two-thirds coverage; II, one-third to two-thirds coverage; and III, some cells but less than one-third coverage. b, A subset of tcf21:NTR; tcf21:nucEGFP larvae with confirmed full ablations were randomly separated and treated with vehicle or CyA, which limited regeneration in most cases (class IV, no ventricular epicardial cells). c, Quantification of extents of regeneration from experiments in (a) and (b). ***P < 0.001; Chi-square test; n = 54 embryos for vehicle, 51 for CyA. d, Epicardial morphogenesis visualized in tcf21:nucEGFP larvae. No epicardial cells are evident at or before 2 days post-fertilization (dpf). By 3 dpf, ventricles contained 17.6 +/- 6 epicardial cells on average (n = 23), whereas 4 dpf larvae contained 45.2 +/- 5.8 cells (n = 21). e, tcf21:nucEGFP larval clutchmates were randomly separated into two groups for treatment with vehicle or 5 μM CyA from 2 to 4 dpf. f, Quantification of ventricular EGFP+ epicardial cells from groups in (e). ***P < 0.001; Student's two-tailed t-test; n = 21 for each group. White dashed lines in (a, b, d, e), ventricle. Boxed areas in (d, e), magnified views. Scale bars, 50 μm. Error bars, s.d.
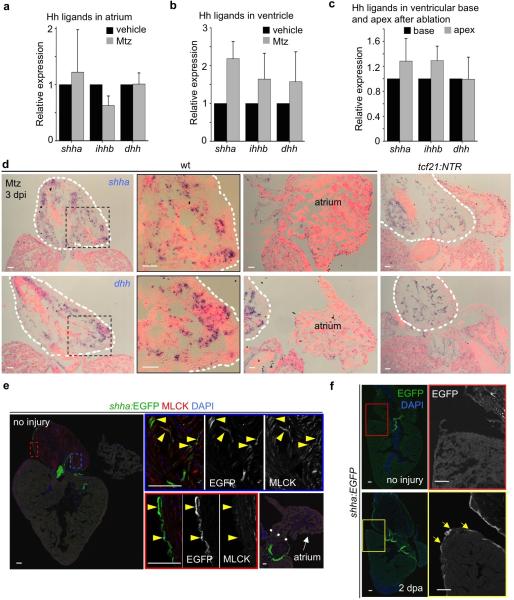
Extended Data Figure 10. Hedgehog ligand expression.
a-c, Quantitative RT-PCR revealing shha, ihhb and dhh expression in atrium (a) or ventricle (b) in uninjured hearts and 3 days post-ablation, or in separated ventricular basal (the basal third of the chamber) and apical (the apical third) tissue after ablation (c). Three separate quantitative RT-PCR experiments on pooled tissues were performed, using a total of 90 zebrafish for (a) and (b) experiments, and another 90 fish for (c). shhb and ihha were not detected in these tissues. d, In situ hybridization (ISH) for shha or dhh in wild-type (wt) or tcf21:NTR clutchmate hearts at 3 days post Mtz treatment, indicating expression in outflow tract (OFT) but not ventricle or atrium. OFT of uninjured and epicardially ablated hearts showed comparable shha and dhh signals by ISH, a qualitative/semi-quantitative assay. e, Section of adult shha:EGFP heart, indicating fluorescence in outflow tract tissues. Smooth muscle cells (MLCK, red) and epicardial cells (outer layer) in outflow tract showed clear EGFP signals, while there is no obvious EGFP fluorescence in ventricle and atrium. Valve mesenchyme also displays EGFP fluorescence. Arrowheads, EGFP signals in smooth muscle cells and epicardium. f, Ventricular resection induces shha:EGFP fluorescence in the basal ventricular epicardium at 2 dpa. Arrows, ventricular epicardial fluorescence. White dashed line in (d, e), outflow tract (d) or atrioventricular junction (e). Boxed areas in (d-f), magnified views. Scale bars, 50 μm. Error bars, s.d.
Supplementary Material
Acknowledgments
We thank J. Burris, N. Lee, A. Dunlap, and S. Davies for fish care, and M. Bagnat, B. Hogan, J. Kang, and R. Karra for comments on the manuscript. This work was funded by postdoctoral fellowships from the American Heart Association to J.W. and J.C., and grants from NIH (HL081674) and American Federation for Aging Research to K.D.P.
Footnotes
Author Contributions. J.W., J.C., and K.D.P. designed experimental strategy, analyzed data, and prepared the manuscript. J.W. generated the transgenic system for epicardial cell ablation and performed in vivo regeneration experiments and analysis. J.C. developed the ex vivo culture assay and performed ex vivo regeneration experiments and analysis. A.D. performed histology and data analysis. All authors commented on the manuscript.
Author Information. The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Huang GN, et al. C/EBP transcription factors mediate epicardial activation during heart development and injury. Science. 2012;338:1599–1603. doi: 10.1126/science.1229765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kikuchi K, et al. tcf21+ epicardial cells adopt non-myocardial fates during zebrafish heart development and regeneration. Development. 2011;138:2895–2902. doi: 10.1242/dev.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smart N, et al. De novo cardiomyocytes from within the activated adult heart after injury. Nature. 2011;474:640–644. doi: 10.1038/nature10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou B, et al. Adult mouse epicardium modulates myocardial injury by secreting paracrine factors. J. Clin. Invest. 2011;121:1894–1904. doi: 10.1172/JCI45529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang J, Karra R, Dickson AL, Poss KD. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013;382:427–435. doi: 10.1016/j.ydbio.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smart N, et al. Thymosin beta4 induces adult epicardial progenitor mobilization and neovascularization. Nature. 2007;445:177–182. doi: 10.1038/nature05383. [DOI] [PubMed] [Google Scholar]
- 7.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 8.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pisharath H, Rhee JM, Swanson MA, Leach SD, Parsons MJ. Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech. Dev. 2007;124:218–229. doi: 10.1016/j.mod.2006.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curado S, et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn. 2007;236:1025–1035. doi: 10.1002/dvdy.21100. [DOI] [PubMed] [Google Scholar]
- 12.Singh SP, Holdway JE, Poss KD. Regeneration of amputated zebrafish fin rays from de novo osteoblasts. Dev. Cell. 2012;22:879–886. doi: 10.1016/j.devcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 14.Kikuchi K, et al. Retinoic acid production by endocardium and epicardium is an injury response essential for zebrafish heart regeneration. Dev. Cell. 2011;20:397–404. doi: 10.1016/j.devcel.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J, Rubin N, Huang Y, Tuan TL, Lien CL. In vitro culture of epicardial cells from adult zebrafish heart on a fibrin matrix. Nat. Protoc. 2012;7:247–255. doi: 10.1038/nprot.2011.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75:1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 17.Roelink H, et al. Floor plate and motor neuron induction by different concentrations of the amino-terminal cleavage product of sonic hedgehog autoproteolysis. Cell. 1995;81:445–455. doi: 10.1016/0092-8674(95)90397-6. [DOI] [PubMed] [Google Scholar]
- 18.Fan CM, Tessier-Lavigne M. Patterning of mammalian somites by surface ectoderm and notochord: evidence for sclerotome induction by a hedgehog homolog. Cell. 1994;79:1175–1186. doi: 10.1016/0092-8674(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 19.Roelink H, et al. Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell. 1994;76:761–775. doi: 10.1016/0092-8674(94)90514-2. [DOI] [PubMed] [Google Scholar]
- 20.Echelard Y, et al. Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell. 1993;75:1417–1430. doi: 10.1016/0092-8674(93)90627-3. [DOI] [PubMed] [Google Scholar]
- 21.Johnson RL, Laufer E, Riddle RD, Tabin C. Ectopic expression of Sonic hedgehog alters dorsal-ventral patterning of somites. Cell. 1994;79:1165–1173. doi: 10.1016/0092-8674(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 22.Guerrero I, Kornberg TB. Hedgehog and its circuitous journey from producing to target cells. Sem. Cell Dev. Biol. 2014;33:52–62. doi: 10.1016/j.semcdb.2014.06.016. [DOI] [PubMed] [Google Scholar]
- 23.Sanders TA, Llagostera E, Barna M. Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature. 2013;497:628–632. doi: 10.1038/nature12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430. doi: 10.1242/dev.068601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kikuchi K, et al. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawson ND, Weinstein BM. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 2002;248:307–318. doi: 10.1006/dbio.2002.0711. [DOI] [PubMed] [Google Scholar]
- 27.Shkumatava A, Fischer S, Muller F, Strahle U, Neumann CJ. Sonic hedgehog, secreted by amacrine cells, acts as a short-range signal to direct differentiation and lamination in the zebrafish retina. Development. 2004;131:3849–3858. doi: 10.1242/dev.01247. [DOI] [PubMed] [Google Scholar]
- 28.Zhou B, Pu WT. Isolation and characterization of embryonic and adult epicardium and epicardium-derived cells. Meth. Mol. Biol. 2012;843:155–168. doi: 10.1007/978-1-61779-523-7_15. [DOI] [PubMed] [Google Scholar]
- 29.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.