Abstract
Accurate optical reporting of electrical activity in genetically defined neuronal populations is a long-standing goal in neuroscience. Here we describe Accelerated Sensor of Action Potentials 1 (ASAP1), a novel voltage sensor design in which a circularly permuted green fluorescent protein is inserted within an extracellular loop of a voltage-sensing domain, rendering fluorescence responsive to membrane potential. ASAP1 demonstrates on- and off- kinetics of 2.1 and 2.0 ms, reliably detects single action potentials and subthreshold potential changes, and tracks trains of action potential waveforms up to 200 Hz in single trials. With a favorable combination of brightness, dynamic range, and speed, ASAP1 enables continuous monitoring of membrane potential in neurons at KHz frame rates using standard epifluorescence microscopy.
Understanding how information is processed in the brain would benefit from precise spatio-temporal recording of electrical activity in individual neurons and larger neuronal circuits. Genetically encoded fluorescent indicators are promising tools for optical reporting of brain activity, as they allow monitoring of genetically-defined neuronal circuits, and do not require chemical access. An ideal genetically-encoded indicator would produce large fluorescence responses, facilitating detection of action potentials (APs), or spikes, that represent neuronal outputs, as well as the monitoring of the amplitude and spatio-temporal distributions of inputs that are received and integrated by neurons1,2. An ideal indicator would also have millisecond-timescale kinetics, enabling interrogation of synchrony and temporal coding in individual neurons and across large neuronal populations3. In particular, tracking high-frequency firing would be useful for visualizing how bursts of neurotransmission are decoded by postsynaptic neurons4 and for understanding how the 50–200 Hz firing of fast-spiking interneurons regulates information processing in the brain5,6 or is affected in disease7.
A genetically encoded sensor with these capabilities had not previously been developed. After intense engineering efforts, fluorescent protein-based calcium reporters can now detect single action potentials8. However, they do not provide a direct readout of membrane potential changes. Furthermore, because calcium transients can persist in neurons for hundreds of milliseconds, calcium responses cannot track high-frequency AP trains9. For example, GCaMP6f, the fastest variant in the latest iteration of calcium sensors, has mean half decay times (τ1/2) of 142 ± 11 ms (1 AP, mouse V1 cortex), 400 ± 41 ms (10 APs, dissociated neuronal cultures), and ~650 ms (zebrafish tectum)8. Accurate reporting of neuronal activity would thus benefit from sensors that more directly report membrane potential.
Existing fluorescent voltage sensors are constructed from one of two types of voltage-sensing proteins: seven-helix microbial rhodopsins, and four-helix voltage-sensing domains (VSDs) from voltage sensitive phosphatases or ion channels. Rhodopsin-based sensors typically show larger fluorescence dynamic range, but their use can be complicated by the dependence of voltage sensitivity on illumination intensity and wavelength, and nonlinear increase in fluorescence with increasing illumination intensity10. Additionally, they are still slow compared to the typical 2-ms duration of action potentials in pyramidal neurons, and are not sufficiently bright to report neuronal activity over background fluorescence in brain slices or in vivo11,12. Voltage sensors using four-helix VSDs are brighter than rhodopsin-based sensors, but produce suboptimal fluorescence responses to neuronal activity13–19, exhibit inactivation kinetics that are too slow for following fast trains of action potentials13,15,16,19,20, and/or require excitation at wavelengths below 450 nm where phototoxicity and autofluorescence are more problematic17,18. The recently developed ArcLight sensor produces the largest fluorescence response to APs among previously reported VSD-based sensors and is excited at ~488 nm, but slow kinetics limit its ability to resolve spikes separated by less than ~50 ms15,21. Thus no existing genetically encoded activity sensor possesses all the characteristics needed for accurate optical reporting of neuronal activity in vivo. We therefore sought to develop a voltage sensor with sufficient brightness, dynamic range and kinetics for detection of neuronal activity ranging from subthreshold potentials to rapid trains of action potentials.
RESULTS
The extracellular loop between the third (S3) and fourth (S4) transmembrane segments of VSDs is thought to undergo substantial conformational changes upon depolarization22. In particular, crystal structures of a VSD isolated from the seasquirt Ciona intestinalis voltage sensitive phosphatase (VSP) suggests that repolarization causes upward reorientation of S3-S4 loop residues, partial unwinding of S3 at its extracellular end, and disappearance of a short helix in the middle of the S3-S4 loop23. We reasoned that insertion into this location of a green fluorescent protein (GFP) might allow voltage-induced movements to perturb GFP fluorescence. We also hypothesized that circular permutation of GFP, so that its termini are located near the chromophore, would enhance conformational coupling between the VSD and fluorescent protein domains. We chose a VSD from the chicken Gallus gallus as an initial candidate VSD because it has a shorter S3-S4 loop than Ciona intestinalis VSD (Supplementary Figure 1a), which we hypothesized would increase coupling between voltage-induced movements and GFP barrel distortions. We thus constructed and tested fusions of the circularly permuted GFP (cpGFP) from GCaMP324 to the S3-S4 loop of the Gallus gallus voltage-sensing domain (GgVSD). We included a R153Q mutation previously shown to shift the voltage response of Ciona intestinalis VSPs to a less negative range of potentials 25.
We obtained several protein constructs comprising cpGFP inserted into GgVSD that were well expressed at the plasma membrane of HEK293A human embryonic kidney cells (Supplementary Fig. 1b) and showed a fluorescence decrease in response to membrane depolarization (Supplementary Fig. 1c). Conveniently, because these construct were in their higher fluorescence state at resting membrane potentials, no additional fluorescent protein marker was required to detect transfected cells. Beginning with the brightest variant, where cpGFP was inserted between residues 147 and 148 of GgVSD, we tested substitutions of various fluorescent proteins (Supplementary Fig. 1d), and found that the OPT variant of circularly permuted superfolder GFP26 (cpsfGFP-OPT) improved both brightness and dynamic range while maintaining efficient expression at the membrane. We named this protein, whose basic design is depicted in Fig. 1a, Accelerated Sensor of Action Potentials 1 (ASAP1).
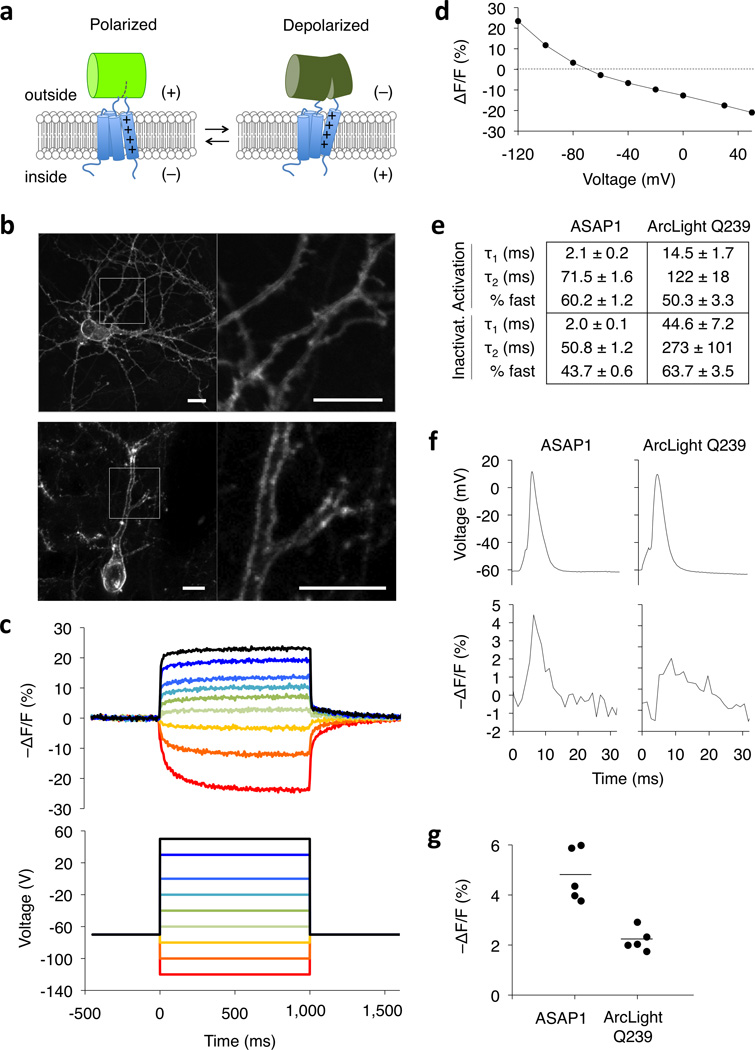
Figure 1.
ASAP1 design and voltage response characteristics. (a) ASAP1 is a circularly permuted GFP inserted into the extracellular S3-S4 loop of a voltage-sensing domain. Depolarization leads to decreased fluorescence. (b) ASAP1 was localized to the plasma membrane in a 12-day-in-vitro dissociated rat hippocampal neuron imaged by confocal microscopy (top) and in a fixed brain slice from an 8-week-old mouse transfected in utero and imaged by two-photon microscopy (bottom). Right panels show details from the left panels. Scale bar, 10 μm. Quantification of membrane localization in 22 neurons is in Supplementary Fig. 5. (c) ASAP1 responses in a representative HEK293A cell (top) to voltage steps from −120 to 50 mV (bottom). Responses were measured at 5-ms intervals and were normalized to fluorescence at the −70 mV holding potential. (d) Mean ASAP1 response to transmembrane voltage in HEK293A cells (n = 10 cells). Error bars are standard error of the mean (SEM). (e) Comparison of activation and inactivation kinetics of ASAP1 (n = 4 cells) and ArcLight Q239 (n = 6) in HEK293A cells. Numbers are mean ± standard error of the mean. (f) Comparison of ASAP1 and ArcLight Q239 responses to representative single trial recordings of action potentials (APs) induced by current injection in cultured hippocampal neurons. AP full widths at half-maximum (FWHM) of the voltage traces (top) were 3.3 and 3.6 ms for ASAP1- and ArcLight Q239-expressing neurons, respectively. The corresponding FWHM of the fluorescence responses (bottom) were 3.7 ms and 6.5 ms for ASAP1 and ArcLight Q239, respectively. (g) ASAP1 produces larger responses to current-triggered APs in cultured hippocampal neurons than ArcLight Q239 (p = 0.001, n = 5 neurons from 3 litters for each sensor). Each data point is the average response of an individual neuron over 12–25 APs per neuron (91 APs total for ASAP1 and 87 APs total for ArcLight Q239). For each sensor, the mean response over all tested neurons is depicted using a horizontal bar.
We created ASAP1 variants to test whether our initial choice of GgVSD with an R153Q mutation was optimal. Substituting VSDs from Danio rerio or Xenopus laevis in place of GgVSD in ASAP1 produced voltage sensors with comparable membrane expression but lowered dynamic range (Supplementary Fig. 2a,b). Sensors containing the VSD from Ciona intestinalis did not localize well to the plasma membrane (Supplementary Fig. 2a). As fusions with fluorescent protein domains at either or both intracellular termini are well localized to the membrane13,18,25, this localization sensitivity of Ciona intestinalis VSD appears specific to the insertion of a fluorescent protein in the S3-S4 loop. Reversion of the R153Q mutation did not affect membrane expression of ASAP1 but lowered the fluorescence response to hyperpolarizing signals (Supplementary Fig. 2a,c).
We next investigated structural determinants necessary for the voltage sensitivity of ASAP1. Dimming upon depolarization suggests that VSD movement disrupts hydrogen-bonding interactions between the beta-barrel and the chromophore that stabilize the anionic form of the chromophore. Modifying the circular permutation breakpoint in GFP reduced the response amplitude, although interestingly some variants had inverted responses (Supplementary Fig. 3a-c). Removing the first residue of cpsfGFP-OPT (immediately following the junction, Phe-145 in the original GFP numbering) severely reduced response. In contrast, removing other amino acids at the GgVSD/cpsfGFP-OPT junction, such as the last residue of cpsfGFP-OPT, had modest effects on dynamic range (Supplementary Fig. 3d,e). Certain conservative mutations of Phe-145 also reduced dynamic range (Supplementary Fig. 4). Notably, mutation of Phe-145 to Tyr markedly reduced dynamic range, consistent with the smaller response amplitude of our initial sensors using cpGFP from GCaMP3, which contains Tyr-145. These results suggest that the modulation of cpsfGFP-OPT by voltage is sensitive to the structure near the breakpoint, as previously seen with calcium sensors based on circularly permuted fluorescent proteins27,28. In particular, they suggest that cpsfGFP-OPT responds to a conformational change on the S3 side of the linker, consistent with models of VSD structural transitions23.
We next tested ASAP1 expression in neurons. ASAP1 produced excellent membrane localization in living dissociated hippocampal neurons and fixed brain slices, both at the cell body and in individual dendrites (Fig. 1b and Supplementary Fig. 5). To determine if ASAP1 expression altered membrane capacitance, we transfected cultured neurons with 1 to 4 times the amount of ASAP1 plasmid used in typical fluorescence imaging experiments. Under these conditions, we observed no change in membrane capacitance upon ASAP1 expression (Supplementary Fig. 6).
We next quantified ASAP1’s dynamic range and kinetics. When expressed in HEK293A cells, ASAP1 responded to voltage steps from −120 to 50 mV with a total fluorescence change (ΔF/F) of −44.4 ± 2.4% (mean ± standard error of the mean (SEM), Fig. 1c,d). The response to a 100-mV depolarization from a holding potential of −70 mV was −17.5 ± 1.0 % ΔF/F (Fig. 1d), an order of magnitude larger than previous cpGFP-based voltage sensors such as ElectrikPk14. ASAP1 also produced large changes to hyperpolarizing signals, with ΔF/F of 11.7 ± 0.6 % and 23.4 ± 0.6 % in response to 30 and 50 mV hyperpolarizations, respectively. In HEK293A cells at room temperature, ASAP1 had rapid biexponential kinetics, with activation and inactivation characterized by a fast component with time constants of 2.1 ± 0.2 ms and 2.0 ± 0.1 ms, respectively (Fig.1e). The fast component accounted for 60.2 ± 1.2 % and 43.7 ± 0.6 % of the full activation and deactivation kinetics, respectively. For comparison, we characterized ArcLight Q239, the voltage sensor with the largest fluorescence response to APs from of all previous VSD-based sensors. The fast components of the ASAP1 response for activation and inactivation were 7.0 (p = 0.00063) and 22.5 (p = 0.002) times as fast as those of ArcLight Q239, respectively, while the slow components were 1.7 (p = 0.034) and 5.4 (p = 0.0095, two-tail Mann-Whitney U test) times as fast.
We predicted that ASAP1’s faster kinetics compared to ArcLight Q239 would increase the fidelity by which fluorescence tracks transmembrane voltage, and would enhance responses to fast events such as APs. Indeed, while ArcLight Q239 produced a broad fluorescence trace in response to single APs in cultured hippocampal neurons, as previously reported15, the fluorescence response of ASAP1 to single APs more closely resembled the corresponding voltage response (Fig. 1f). Averaging over the entire neuron, ASAP1 produced a fluorescence response of −4.8 ± 0.5 %, 2.2 times larger than the −2.2 ± 0.2 % response we observed with Arclight Q239 (Fig. 1g). The larger response to single action potentials with ASAP1 is consistent with its ~7-fold faster on-kinetics, which are better matched to the kinetics of action potentials.
We next tested the ability of ASAP1 to track various membrane potential waveforms. In voltage-clamped HEK293A cells, ASAP1 was able to track trains of up to 200 Hz while clearly discerning individual peaks in single trials without filtering. In contrast, ArcLight Q239 traces at 100 Hz appeared flattened with an elevated baseline and poor peak discrimination (Fig. 2a, b). In cultured hippocampal pyramidal neurons, ASAP1 was able to detect subthreshold depolarizations in the form of simulated excitatory postsynaptic potentials (EPSPs) and inhibitory postsynaptic potentials (IPSPs) of 5–20 mV amplitude, comparing favorably to ArcLight Q239 (Fig. 3a, b). Importantly, ASAP1 was able to resolve spikes superimposed on a large EPSP, whereas ArcLight Q239 was not (Fig. 3c and Supplementary Fig. 7). ASAP1 also detected spontaneous spikes, spontaneous 10-Hz bursting, and subthreshold potential changes between spikes in neurons (Fig. 4a,b and Supplementary Fig. 8). As observed previously under voltage-clamp (Fig. 3b), ArcLight Q239 failed to detect or produced minimal responses to spontaneous APs superposed on large EPSPs (Fig. 4a and Supplementary Fig. 8). ASAP1 responses to slow low-amplitude changes were disproportionally larger than to the fast component of APs (Fig. 4a); this is expected from the relative steepness of the fluorescence response near the resting potential (Fig.1d) and a slow component in the ASAP1 response (Fig. 1e).
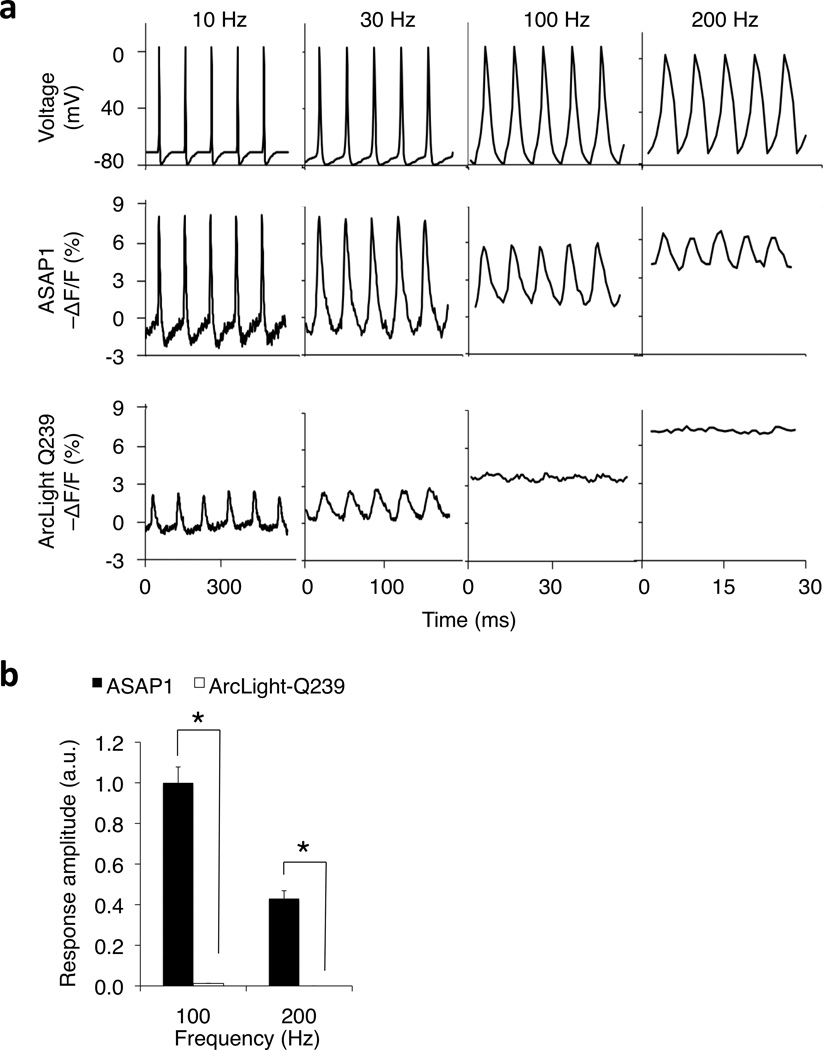
Figure 2.
Monitoring simulated AP trains in voltage-clamped HEK293A cells. (a) ASAP1 followed 200-Hz trains of AP waveforms, while ArcLight Q239 followed trains of 30 Hz but not 100 Hz or 200 Hz. For each frequency, simulated trains of APs (2.0-ms FWHM, 75-mV peak amplitude) were applied for 1 second. Traces shown are the fluorescence response to 5 AP waveforms at 500 ms from the start of each train. (b) We quantified the frequency response of ASAP1 and ArcLight-Q239 to 100 Hz and 200 Hz simulated spike trains described above. We first estimated the power spectra density of the response using fast Fourier transforms. For 100 Hz trains, we quantified the amplitude of the power density peak at 100 Hz; correspondingly, we quantified the amplitude of the 200 Hz peak for 200 Hz spike trains. Consistent with subpanel (a) (bottom row) ArcLight Q239 produced little or no response at these frequencies, with mean peak amplitudes of 0.012 ± 0.001 (100 Hz) and < 0.001 (200 Hz). ASAP1 showed greater mean response amplitudes to both 100 Hz and 200 Hz simulated AP trains (100 Hz, p = 0.021; 200 Hz, p = 0.031). Differences are statistically significant following Holm-Bonferroni correction for multiple comparisons. For each train, n = 5 HEK293A cells per construct. Error bars, SEM.
Figure 3.
Monitoring simulated hyperpolarizations and subthreshold potentials in voltage-clamped neurons. (a) ASAP1 can detect subthreshold potential and hyperpolarization waveforms in cultured hippocampal neurons. Subthreshold depolarizations and hyperpolarizations have peak amplitudes of 5, 10, 15, and 20 mV, and peak full width at half maximum of 17 ms (depolarizations) and 38 ms (hyperpolarizations). (b) Quantification of the fluorescence responses to subthreshold depolarizations (top) and hyperpolarizations (bottom). Asterisks identify statistically significant differences from pairwise two-tailed t-tests adjusted for multiple comparisons using the Holm-Bonferroni method (p = 0.006, −5 mV waveform; p = 0.0005, 0.008, 0.007 and 0.003 for the −5, −10, −15 and −20mV waveforms, respectively). Multiple comparisons adjustments were performed separately for depolarizations and hyperpolarizations. n = 6 (ASAP1) and 4 (ArcLight Q239) neurons from the same litter. Error bars, SEM. (c) ASAP1’s faster kinetics allow improved resolution of a 100-Hz, three-AP waveform sequence in cultured hippocampal neurons (ASAP1, n = 8 neurons; ArcLight Q239, n = 6 neurons; all cells from same litter). Command voltage spike FWHM is 1.8 ms. Additional examples are in Supplementary Fig. 7.
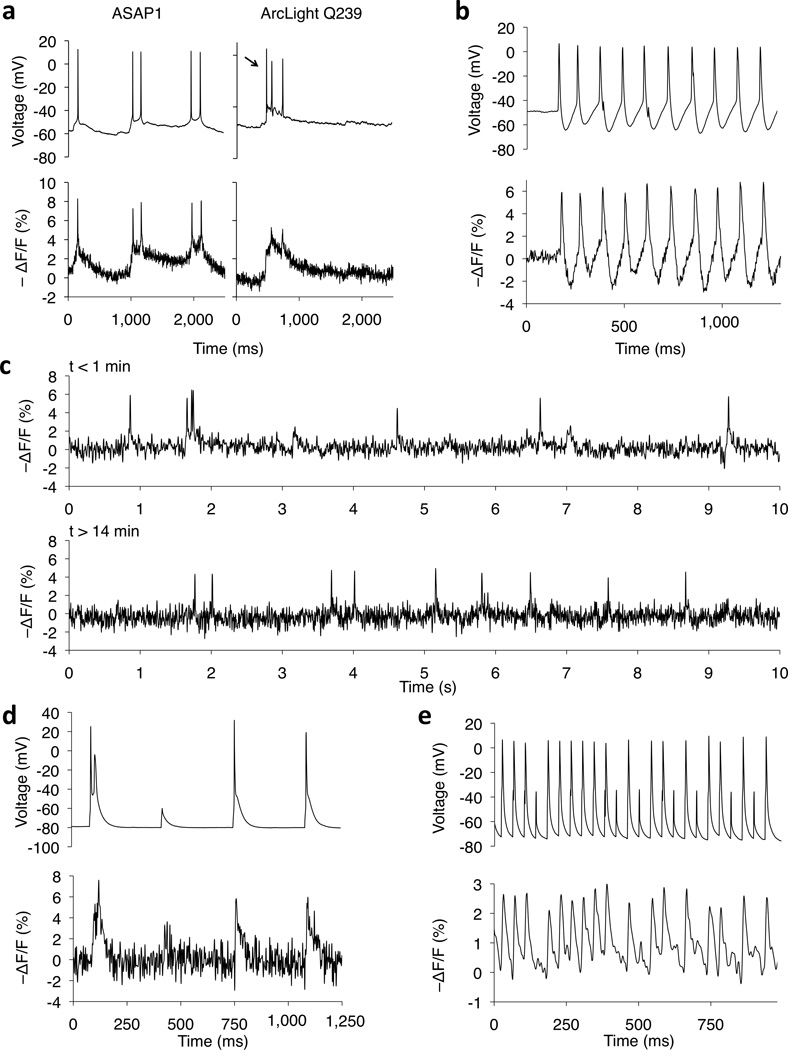
Figure 4.
Imaging neural activity in current-clamp from cortical slices and dissociated hippocampal cultures. (a) Fluorescence responses of ASAP1 (left) and ArcLight Q239 (right) to spontaneous subthreshold potentials and APs in cultured hippocampal neurons. From cell to cell, ASAP1 mean fluorescence responses ranged from −4.8 to −8.1 %, averaging −6.3 ± 0.6 % (n = 6 neurons from 5 litters, ≥ 10 APs per neuron). Arrow, AP not detected by ArcLight Q239. Additional examples are in Supplementary Figure 8. (b) ASAP1 followed a spontaneous AP train in a cultured hippocampal neuron (ΔF/F = −6.2 ± 0.5% mean ± SEM, n = 10 APs). Spontaneous bursts are rare events in cultured neurons and we made this observation only a single time in all our recordings. (c) ASAP1 responses to spontaneous activity in a cultured hippocampal neuron at the beginning (top) and end (bottom) of 15 min of continuous illumination (0.036 mW/mm2). Similar observations were made in 4 neurons from 3 litters; additional examples are shown in Supplementary Fig. 10. (d) In an acute cortical slice from a mouse brain transfected in utero, ASAP1 produced large responses to individual current-induced APs in a Layer-5 pyramidal cell (ΔF/F = −6.2 ± 0.2% mean ± sem, n = 10 spikes; single observation). (e) ASAP1 tracked APs and subthreshold depolarizations in a Layer-2/3 neuron injected with current pulses at 25 Hz. ΔF/F = −1.5 to −3.2 %, across 98 APs total from 4 neurons, each from a different slice from the same animal. All traces are from single trials, without filtering (a,b,d), with LOWESS smoothing (c), or with a 100-Hz 4th-order low-pass Butterworth filter (e).
To investigate ASAP1’s suitability for imaging neuronal activity over longer durations, we first determined its photobleaching time constant in neurons under continuous illumination (0.12 mW/mm2 of 470- to 490-nm light) as 35.0 ± 2.5 min (Supplementary Fig. 9). For comparison with probes we developed previously, we estimated the time over which the signal/noise ratio (SNR) for AP detection decays from 4 to 2 due to photobleaching at a fixed excitation intensity to be 49.1 ± 9.8 min (mean ± sem, n = 4 neurons), longer than FRET-based voltage sensors16. As this calculation was based on extrapolations from short recordings, we also directly tested ASAP1’s ability to monitor spontaneous neuronal activity over longer durations. We easily detected APs in cultured neurons over 15 minutes of constant illumination (Fig. 4c and Supplementary Fig. 10). Peak SNR decreased modestly from 14.6 ± 2.3 to 9.4 ± 1.3 (n = 4 neurons from 3 litters; paired t-test, p = 0.047), due in part to a 19.9 ± 5.1 % reduction in the AP response (paired t-test, p = 0.014). Baseline noise also increased 23.3 ± 20.5 %, but this was not statistically significant (paired t-test, p = 0.34). These results suggest that ASAP1 has sufficient dynamic range, brightness, and photostability for experiments requiring long durations. However, the magnitude of photobleaching and photodamage will likely depend on experimental conditions, such as illumination power, ASAP1 expression levels, neuron type, and specimen preparation.
Finally, we investigated ASAP1 performance in acute cortical slices from mouse brains transfected in utero. ASAP1 reported current-induced APs in Layer-5 pyramidal neurons in single trials without filtering or averaging (Fig. 4d) and was able to distinguish between subthreshold depolarizations and APs. ASAP1 was also to reliably report current-induced 25-Hz AP trains in Layer-2/3 neurons (Fig. 4e), detecting all APs and most subthreshold depolarizations. Thus, ASAP1 is able to discriminate between subthreshold voltage changes, single action potentials, and closely spaced action potentials in neurons in culture and in brain slices. While we provide evidence that ASAP1 can report neuronal activity in intact circuits, we are currently conducting experiments to more fully evaluate the performance of ASAP1 across a broader set of conditions in brain slices, and in living animals.
DISCUSSION
Despite decades of development29, previous voltage-sensing proteins have limitations hindering their use for high-fidelity recording of neuronal activity. Early sensors consisting of GFP fused to ion channels produce slow or small responses in oocytes29,30 and no responses in neurons due to failure of membrane trafficking31. More recent sensors, with fluorescent proteins fused to intracellular termini of VSDs from voltage sensitive phosphatases, show improved membrane localization and can detect voltage changes in neurons. These sensors can be divided into three main classes. In the first, a large change (> 10 %) in fluorescence resonance energy transfer between two fluorescent proteins occurs in response to depolarization, but kinetics are slow (τ > 10 ms)13,16. In a second class, represented by ArcLight, depolarization produces a large decrease in fluorescence of a pH-sensitive GFP fused to the VSD C-terminus, but also with slow kinetics15. In a third class of sensor, the fluorescence of single fluorescent proteins (with or without circular permutation) attached to the VSD C-terminus can be perturbed by voltage with rapid kinetics (τ < 5 ms) but with very small dynamic range (< 5 %)14,32.
We designed ASAP1 with a different architecture from previous sensors. Instead of attaching fluorescent proteins to intracellular locations in VSDs, we inserted cpGFP in the extracellular S3-S4 loop of a VSD. This location enables direct coupling between conformational changes in the S3-S4 linker and β-strand 7 of cpGFP, which interacts with the chromophore to stabilize its deprotonated state. Indeed, the large and rapid responses of ASAP1 indicate that movement of the S4 helix in response to depolarization is able to rapidly modulate protonation of the chromophore.
In conclusion, ASAP1 is distinct from earlier sensors in combining efficient membrane localization, high brightness, large dynamic range, and fast kinetics, allowing for sensitive detection discrimination of a wide repertoire of membrane voltage responses in neurons with standard optical equipment. In particular, its fast responses to single action potentials and ability to track high-frequency trains make ASAP1 an excellent sensor for action potential detection and counting, either in unitary events or in rapid trains, while its large fluorescence changes near the resting membrane potential make it a sensitive reporter for subthreshold excitatory and inhibitory potentials. ASAP1 should thus facilitate efforts to map the activity of genetically defined neurons in the brain and decode neuronal communication in time and space33.
METHODS
Plasmid construction
Plasmids were constructed by standard molecular biology methods and verified by sequencing of all cloned fragments. ASAP1 expression plasmid and its complete sequence can be obtained via Addgene (http://www.addgene.org, plasmid 52519). ArcLight Q23915 and ASAP1 variants were cloned between the NheI-HindIII sites of pcDNA3.1/Puro-CAG16. All variants contain identical Kozak sequences. To evaluate sensor brightness or membrane localization, we coexpressed sensors with a fusion of mCherry with a farnesylation motif (CAAX) from the CAG promoter by using the encephalomyocarditis virus internal ribosome entry site (IRES); a schematic of the resulting plasmids is shown in Supplementary Figure 5a.
We constructed or obtained four circularly permuted fluorescent proteins for use in voltage sensor variants (Supplementary Figure 1b,d). First, we constructed a circularly permuted GFP from GCaMP3 by inserting amino acids 145–148 (‘YNSH’) of EGFP to the N terminus of cpGFP149-144 from plasmid pEGFP-N1-GCaMP324 (Addgene 22692). Second, we designed a circularly permuted Clover16 GFP by alignment to cpGFP from GCaMP3, creating new termini at original amino acid positions 145 and 144, and linking its original N- and C- termini using the peptide ‘GGTGGS’ between the C-terminal lysine (K239) and the original starting methionine. Third, we obtained cpsfGFP-OPT145-144 (abbreviated as cpsfGFP-OPT) from Luke Oltrogge (Steve Boxer lab, Stanford). cpsfGFP-OPT is a circular permutant of a superfolder GFP variant; it was evolved for use in a split GFP system, and contains seven mutations (“GFP1-10 OPT” mutations) not present in the original superfolder GFP34. The full sequence of cpsfGFP-OPT is provided as part of ASAP1. Finally, we designed circularly permuted superecliptic pHluorinA227D15 (cpsepHluorinA227D) by alignment to cpsfGFP-OPT, creating new termini at original amino acid positions 145 and 144, and linking its original N- and C- termini using the peptide ‘GTGGSAS’ between the C-terminal lysine (K239) and a lysine originally present near its N-terminus (K4).
ASAP1 variants with alternate voltage-sensing domains (VSD) were constructed by inserting cpsfGFP-OPT immediately after amino acids G146 (Xenopus laevis VSD), amino acid G147 (Danio rerio VSD), or amino acids L203 to G214.
HEK293A cell culture and transfection
HEK293A cells (Life Tech) were maintained in high-glucose Dulbecco’s Modified Eagle Medium (DMEM, HyClone) supplemented with 5% fetal bovine serum (FBS, Life Tech) and 2 mM glutamine (Sigma) at 37 °C in air with 5% CO2. Cells were plated onto glass-bottom 24-well plates (In Vitro Scientific) for standard imaging, or onto uncoated No. 0 12-mm coverslips (Glaswarenfabrik Karl Hecht GmbH & Co) for patch clamping experiments. Transfections were carried out using FuGene HD (Promega) according to manufacturer instructions, except that cells were transfected at ~50% confluence with lower amounts of DNA (200 ng) and transfection reagent (0.6 µl) to reduce cell toxicity. Cells were cultured for ~48 h before experiments were performed.
HEK293A imaging without patch clamping
We used an IX81 microscope with a 60× 1.42-numerical aperture (NA) PlanApo oil-immersion objective (Olympus). Fluorescence excitation was delivered using a 120W Mercury vapor short arc lamp (X-Cite 120PC, Exfo) through a 485/22-nm (GFP) or 545/30-nm (RFP) filter. Fluorescence emission was passed through a 540/40-nm (GFP) or 605/50-nm (RFP) filter, and recorded using an Orca ER CCD (Hamamatsu) with Micro-Manager35 software. Fluorescent images shown in Supplementary Figures are representative images selected out of multiple fields of view.
Simultaneous patch clamping and imaging of HEK293A cells
We performed patch-clamp experiments at room temperature (22°C), using an Axopatch 700B amplifier (Axon Instruments) and borosilicate glass electrodes with resistances of 2.5–5 MΩ. Cells were superfused in a chamber mounted on the stage of an Axiovert 100M inverted microscope with a 40× 1.3-N.A. oil-immersion objective (Zeiss). Fluorescence excitation was delivered using a high-power blue light-emitting diode (LED, UHP-MIC-LED-460, Pryzmatix) through a 472/30-nm filter at a power density of 0.4–5.2 mW/mm2 at the sample plane. Fluorescence emission was passed through a 525/50-nm filter, and recorded using either an ORCA-Flash4.0 V2 C11440-22CU (Hamamatsu) scientific CMOS camera (experiments for Fig. 1c,d), or an iXon 860 electron-multiplied charge-coupled device camera (Andor) cooled to −80 °C (all other experiments). Unless otherwise indicated, step voltage depolarizations were applied to change the membrane potential from a holding voltage of −70 mV to voltages ranging from −120 mV to 50 mV for 0.5–1.0 s. For these voltage steps experiments, we captured images at 200 Hz without binning, and the fluorescence response was measured from pixels at the perimeter of the cell (plasma membrane). For experiments with trains of artificial AP waveforms, we captured images at 827 Hz with 4×4 binning, and fluorescence response was measured using pixels from the entire cell. The action potential waveform, recorded from a hippocampal neuron, has a full width at half-maximum (FWHM) of 2.0 ms and peak amplitude of 75 mV. To measure the kinetics of ASAP1 and ArcLight Q239, emission was passed through a 520/40-nm band-pass filter to a PMM02 photomultiplier tube (Thorlabs) and sampled at 5–10 kHz. Double-exponential models were applied to the rising and falling portions of the imaging trace during command step voltages. For all experiments, fluorescence traces were corrected for photobleaching; electrophysiological data was recorded with Clampex (Molecular Devices) while fluorescence images were acquired with Solis (Andor) or Micro-Manager35.
Neuronal cell culture and transfection
Animal experiments were performed in accordance with the rules of the Stanford University Administrative Panel on Laboratory Animal Care. Primary hippocampal neurons were dissected from Sprague-Dawley rats on embryonic day 18 and digested with 10–20 U/mL papain (Worthington) in Hank’s Balanced Salt Solution (HBSS) with 2 U/µL DNaseI for 25 min at 37° C. Neurons were then dissociated by gentle trituration in Neurobasal (Life Tech) with 10% FBS. Neurons were plated at 4 × 105 cells/cm2 on 12-mm No. 0 coverslips (Glaswarenfabrik Karl Hecht GmbH & Co) precoated for 24 h with > 300-kDa poly-D-lysine (Sigma) in water. Neurons were cultured overnight at 37° C in air with 5% CO2 in Neurobasal with 1× B27 (Life Tech), 2 mM GlutaMAX (Life Tech), and 10% FBS. The following day, 90% of the media was replaced with identical media without FBS. Cytosine β-D-arabinofuranoside (Sigma) was added to 2 μM at 5 days in vitro (DIV). Neurons were transfected at 7–10 DIV using calcium phosphate36. For each well, we transfected 400 ng of sensor DNA and 1.1 µg of empty pcDNA3.1/Puro-CAG vector.
Confocal imaging of live dissociated neurons
Live neurons were imaged 2 days post-transfection in HBSS supplemented with 10 mM HEPES pH 7.4, 1× B27, 2 mM GlutaMAX, and 1 mM sodium pyruvate on an IX81 microscope with a FluoView FV1000 laser-scanning confocal unit operated using the FV10-ASW v3.01 software (Olympus). Fluorescence excitation was delivered using a 488-nm laser through a 40× 1.3-NA oil-immersion objective (Olympus). Emission was passed through a 530/40-nm emission filter. A single slice close to the bottom of the neuron was imaged using a 1-Airy pinhole setting. Images shown in Fig. 1b are representative images selected out of multiple fields of view.
Antibody staining and confocal imaging of fixed dissociated neurons
DIV10 neurons were transfected with a plasmid vector expressing both ASAP1 and mCherry-CAAX (see Plasmid construction section & Supplementary Fig. 5a) as described above, except that a range of plasmid concentrations was used (0.2–1.6 µg of sensor DNA, supplemented to 1.6µg total DNA with empty pcDNA3.1/Puro-CAG vector). Four days post-transfection, neurons were washed in HBSS, fixed for 15 min at room temperature with 4% paraformaldehyde, and washed three times with HBSS. For preparation of intact (non-permeabilized neurons), we blocked non-specific staining by incubating neurons for 30 min in blocking buffer: 5% goat serum (Life Tech) in phosphate-buffered saline (PBS, HyClone/Thermo Scientific); neurons were then washed three times with PBS. Primary antibody staining was performed with a rabbit anti-GFP polyclonal antibody (ab6556, Abcam) at 1:2000 dilution for 2 h, followed by three washes with PBS. Secondary antibody antibody staining was performed with a goat anti-rabbit antibody conjugated with Alexa Fluor 633 (AF633) dye (A21071, Life Tech) for 2 h in the dark. Multiple published studies have used ab655637,38 and A2107139,40 for similar applications. Cells were washed once with PBS, and twice with PBS + 0.1% Triton X100 (Amresco), for 20 min in each wash. All steps were performed at room temperature. Permeabilized neurons were prepared as above, except that Triton X100 was added to the blocking and staining solutions to a final concentration of 0.1%; washes were performed identically. Coverslips were mounted using Vectashield HardSet mounting medium (H-1400, Vector Laboratories) supplemented with 0.01% sodium azide. Neurons were imaged using a 40× 1.3-NA oil-immersion objective on an IX81 microscope with a FluoView FV1000 laser-scanning confocal unit operated using the FV10-ASW v3.01 software (Olympus). mCherry-CAAX was imaged with a 559-nm laser and a 603/35-nm emission filter. ASAP1-associated Alexa Fluor 633 was imaged with a 635-nm laser and a 705/100-nm emission filter. Images were acquired using a 1-Airy pinhole setting and filtered with a two-pass Kalman filter. All neurons with visible membrane-localized mCherry-CAAX were selected for analysis. For each neuron and wavelength, a Z-stack from −3 to +3 µm relative to the mid-cell position was acquired, with 0.5-µm vertical spacing between slices. The slice showing the best mCherry-CAAX membrane localization was used for quantifying ASAP1 (AF633) membrane localization; this slice was typically at or close to the mid-cell position. For Supplementary Fig. 5e, membrane and cytosol regions of interest were hand-drawn using the corresponding mCherry-CAAX image to identify region boundaries.
Simultaneous patch clamping and imaging of dissociated neurons
Two days post-transfection, cells were patch-clamped at room temperature (22° C) using borosilicate glass electrodes with resistances of 3–5 MOhm attached to an Axopatch 700B amplifier (Axon Instruments). Cells were superfused with extracellular solution containing 110 mM NaCl, 26 mM sucrose, 23 mM glucose, 5 mM HEPES-Na, 5 mM KCl, 2.5 mM CaCl2, and 1.3 mM MgSO4, adjusted to pH 7.4. The intracellular (pipette) solution contained 115 mM K-gluconate, 10 mM HEPES-Na, 10 mM EGTA, 10 mM glucose, 8 mM KCl, 5 mM MgCl2, and 1 mM CaCl2, adjusted to pH 7.4. Cells were imaged using the same microscope, camera, filters, and illumination as HEK293A cells (see “Simultaneous patch clamping and imaging of HEK293A cells”). Images were captured at 827 Hz (current-induced APs) or 417 Hz (spontaneous APs) with 4×4 binning, and fluorescence response was in the entire cell. Fluorescence traces were acquired while cells were voltage-clamped or current-clamped in whole-cell mode. For all experiments, fluorescence traces were corrected for photobleaching. To generate action potentials, ~800 pA of current was injected for 2 ms. We discarded from future analysis neurons with resting membrane potential greater than −50 mV or membrane resistance lower than 100 MOhm. Electrode voltages and currents were recorded using pClamp (Axon Instruments) and analyzed using custom software written in MATLAB. Voltage traces were corrected for the junction potential post hoc. The neuron resting membrane potential was measured at I = 0 in current clamp immediately after membrane rupture. Membrane capacitance was determined by pClamp v10 (Molecular Devices) preset routines using the current response to 5-mV square pulses.
Photobleaching and long-term voltage sensing in dissociated neurons
Neurons were cultured and transfected as described above, but were left intact (unpatched). They were imaged with an ORCA-Flash4.0 V2 C11440-22CU (Hamamatsu) scientific CMOS camera as described above, except that we used a 480/20-nm excitation filter (Chroma HQ480/20x).
Estimating the concentration of membrane-localized ASAP1 molecules
To obtain a coarse estimate of the number of ASAP1 molecules per µm2 in dissociated neurons (see legend to Supplementary Figure 6f), we first purified and determined the concentration of a preparation of Clover GFP41 as previously described42. We injected dilutions of this preparation into a custom polydimethylsiloxane (PDMS) microfluidics device43. To obtain a correspondence between fluorescence and number of Clover molecules, we imaged channels of 10-µm depth using the microscopy system described above. We adjusted this relationship to measure ASAP1 molecules by factoring the difference in brightness between Clover and cpsfGFP-OPT44. We assumed that ASAP1 brightness is equivalent to cpsfGFP-OPT at the brightest point of the ΔF/F vs V curve (−150 mV, Fig. 1d), and then factored in the difference in ASAP1 brightness between −160 mV and the resting membrane potential of approximately −70 mV.
In utero electroporation, acute slice preparation and imaging
Embryos from pregnant CD-1 mice (Charles Rivers) were injected with 1–1.5 μL of 1–2 μg/μL ASAP1 plasmid DNA in PBS and electroporated using five 10-ms 50–55 V voltage pulses delivered at 1 Hz. At P15-P30, electroporated pups were anesthetized with 2% isofluorane, and perfused with a sucrose solution containing 234 mM Sucrose, 26 mM NaHCO3, 11 mM Glucose, 10 mM MgSO4, 2.5 mM KCl, 1.25 mM NaH2PO4, 0.5 mM CaCl2, and continuously bubbled with 95% O2 and 5% CO2. 300-µm slices were prepared and allowed to recover for 30 minutes in bubbled artificial cerebrospinal fluid (ACSF) containing 123 mM NaCl, 26 mM NaHCO3, 11 mM Glucose, 3 mM KCl, 2 mM CaCl2, 1 mM MgCl2. Slices were placed in an imaging chamber in a customized BX-51WI fluorescence microscope (Olympus), continuously perfused with ACSF, and visualized under a 20× 1.0-NA objective (Olympus). Cells were illuminated with a stable LED source (Heliophor, 89 North) at a power density of 8 mW/mm2, or a 488-nm diode laser (OBIS, Coherent) at a power density of 50 mW/mm2. Layer 5 cortical pyramidal cells expressing ASAP1 were patch clamped, held in current clamp mode and either monitored for spontaneous spiking or injected with 2- to 5-ms current pulses (600–1500 pA) to induce single spikes with minimal subthreshold contribution. Fluorescence traces were recorded at 400 Hz by an iXon DU-897 EMCCD camera (Andor). For all experiments, fluorescence traces were corrected for photobleaching.
Data analysis
Peak signal to noise ratio peak (SNR) was defined as the ratio of baseline-subtracted fluorescence intensity changes and the standard deviation of the baseline trace in a 500-ms windows near the AP, in regions with no apparent hyperpolarizations or subthreshold depolarizations. T4→2 is defined as the time a cell, illuminated so as to obtain an initial signal-to-noise ratio (SNR) of 4, can be imaged before the SNR drops to 2 as previously described16. Assuming a reciprocal relation between excitation intensity and photobleaching time, the time to decay from an AP SNR of θ to θ/2 is , where τoff is the decay time of the fluorescence transient induced by the spike. The signal-to-noise capacity (SNRC) is a dimensionless measure of a probe's ability to report activity and is defined as the instantaneous peak/noise ratio of a probe integrated over all time. For an single-channel probe, SNRC is defined as SNRC2 = (ΔF/F)2 N(t) τPB, with ΔF/F the fractional change in fluorescence to a spike, N(t) the rate of emission, and τPB the photobleaching time constant. We used fluorescence traces from stimulated APs in dissociated neurons to extract the SNRC and τoff (see “Simultaneous patch clamping and imaging of dissociated neurons”).
Statistical methods
Results presented in the form x ± y represent the mean ± standard error of the mean (SEM), unless indicated otherwise. Statistical comparisons of pre-identified measures of interest between two datasets were performed with the Student t-test unless otherwise indicated. Prior to performing such statistical comparisons, the Shapiro-Wilk method was used to test the null hypothesis that the data followed a Gaussian (normal) distribution. When this normality hypothesis could not be rejected, Student t-tests were performed; otherwise, the Mann-Whitney U non-parametric test was used. Prior to performing t-tests, we also tested the null hypothesis of equal variance between the two datasets, and employed Welch’s correction when the null hypothesis was rejected. Two-tailed tests were performed at significance level (α) of 0.05. Statistical tests were performed in Excel (Microsoft) and MATLAB (MathWorks). Except where otherwise indicated, neurons for a given experiment were from the same preparation of hippocampal neurons dissected from one rat litter. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed in the field. Data collection and analysis were not performed blind to the conditions of the experiments. However, as mentioned earlier, we chose cells for data collection and analysis using pre-defined selection criteria. Furthermore, data analysis was largely performed using automated Matlab and ImageJ routines.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following for providing rats or dissociated neurons: Hyokeun Park and Yang Geng (Lin lab); Maik Hintze and Subhashree Ganesan (Lu Chen lab, Stanford); Charu Ramakrishnan and Haley Swanson (Karl Deisseroth lab, Stanford). We also thank Xiaozhe Ding (Tsinghua University) for assistance with cloning ASAP1 variants; Yang Geng (Lin lab) for providing microfluidic chambers; Jun Chu (Lin lab) for a purified preparation of Clover GFP; Vincent Pieribone (The John B. Pierce Laboratory and Yale University) for the generous gift of GgVSD(153Q), DrVSD(R153Q), and XlVSD(R152Q); Luke Oltrogge (Steve Boxer lab, Stanford) for the generous gift of cpsfGFP-OPT; Jason Bant (Giocomo lab, Stanford) for advice on electrophysiological recordings; and members of the Lin lab for comments on the manuscript. This work was supported by National Science Foundation grant 1134416 (F.S.-P., M.Z.L.), a Walter V. and Idun Berry Postdoctoral Fellowship (Y.Y.), a Stanford University Bio-X Interdisciplinary Initiatives Project grant (M.Z.L., M.J.S.), the Stanford CNC Program (Y.G., J.D.M., M.J.S.), and the National Academy of Sciences Keck Futures Initiative (Y.G., J.D.M., M.J.S.). M.Z.L. is a Rita Allen Foundation Scholar.
Footnotes
ACCESSION CODES
ASAP1 nucleotide sequence: Genbank KJ598785. ASAP1 expression plasmid (pcDNA3.1/Puro-CAG-ASAP1): Addgene 52519.
AUTHOR CONTRIBUTIONS
M.Z.L. conceived the study. F.S.-P., Y.Y., J.D.M. and Y.G. designed and performed experiments, and analyzed data. M.Z.L. and M.J.S. provided ideas and advice. F.S.-P. and M.Z.L. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
A supplementary methods checklist is available
REFERENCES
- 1.Magee JC. Dendritic integration of excitatory synaptic input. Nat Rev Neurosci. 2000;1:181–190. doi: 10.1038/35044552. [DOI] [PubMed] [Google Scholar]
- 2.Zecevic D, et al. Imaging nervous system activity with voltage-sensitive dyes. Curr Protoc Neurosci. 2003 doi: 10.1002/0471142301.ns0617s23. Chapter 6, Unit 6.17. [DOI] [PubMed] [Google Scholar]
- 3.Castro-Alamancos MA. Cortical up and activated states: implications for sensory information processing. Neuroscientist. 2009;15:625–634. doi: 10.1177/1073858409333074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branco T, Hausser M. Synaptic integration gradients in single cortical pyramidal cell dendrites. Neuron. 2011;69:885–892. doi: 10.1016/j.neuron.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puig MV, Ushimaru M, Kawaguchi Y. Two distinct activity patterns of fast-spiking interneurons during neocortical UP states. Proc Natl Acad Sci U S A. 2008;105:8428–8433. doi: 10.1073/pnas.0712219105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Royer S, et al. Control of timing, rate and bursts of hippocampal place cells by dendritic and somatic inhibition. Nat Neurosci. 2012;15:769–775. doi: 10.1038/nn.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou FW, Roper SN. Altered firing rates and patterns in interneurons in experimental cortical dysplasia. Cereb Cortex. 2011;21:1645–1658. doi: 10.1093/cercor/bhq234. [DOI] [PubMed] [Google Scholar]
- 8.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murthy VN, Sejnowski TJ, Stevens CF. Dynamics of dendritic calcium transients evoked by quantal release at excitatory hippocampal synapses. Proc Natl Acad Sci U S A. 2000;97:901–906. doi: 10.1073/pnas.97.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maclaurin D, Venkatachalam V, Lee H, Cohen AE. Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin. Proc Natl Acad Sci U S A. 2013;110:5939–5944. doi: 10.1073/pnas.1215595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kralj JM, Douglass AD, Hochbaum DR, Maclaurin D, Cohen AE. Optical recording of action potentials in mammalian neurons using a microbial rhodopsin. Nat Methods. 2012;9:90–95. doi: 10.1038/nmeth.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gong Y, Li JZ, Schnitzer MJ. Enhanced Archaerhodopsin Fluorescent Protein Voltage Indicators. PLoS One. 2013;8:e66959. doi: 10.1371/journal.pone.0066959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akemann W, et al. Imaging neural circuit dynamics with a voltage-sensitive fluorescent protein. J Neurophysiol. 2012;108:2323–2337. doi: 10.1152/jn.00452.2012. [DOI] [PubMed] [Google Scholar]
- 14.Barnett L, Platisa J, Popovic M, Pieribone VA, Hughes T. A fluorescent, genetically-encoded voltage probe capable of resolving action potentials. PLoS One. 2012;7:e43454. doi: 10.1371/journal.pone.0043454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jin L, et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75:779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundby A, Mutoh H, Dimitrov D, Akemann W, Knopfel T. Engineering of a genetically encodable fluorescent voltage sensor exploiting fast Ci-VSP voltage-sensing movements. PLoS One. 2008;3:e2514. doi: 10.1371/journal.pone.0002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsutsui H, et al. Improved detection of electrical activity with a voltage probe based on a voltage-sensing phosphatase. J Physiol. 2013 doi: 10.1113/jphysiol.2013.257048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautam SG, Perron A, Mutoh H, Knopfel T. Exploration of fluorescent protein voltage probes based on circularly permuted fluorescent proteins. Front Neuroeng. 2009;2:14. doi: 10.3389/neuro.16.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staff NP, Jung HY, Thiagarajan T, Yao M, Spruston N. Resting and active properties of pyramidal neurons in subiculum and CA1 of rat hippocampus. J Neurophysiol. 2000;84:2398–2408. doi: 10.1152/jn.2000.84.5.2398. [DOI] [PubMed] [Google Scholar]
- 21.Cao G, et al. Genetically targeted optical electrophysiology in intact neural circuits. Cell. 2013;154:904–913. doi: 10.1016/j.cell.2013.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jensen MO, et al. Mechanism of voltage gating in potassium channels. Science. 2012;336:229–233. doi: 10.1126/science.1216533. [DOI] [PubMed] [Google Scholar]
- 23.Li Q, et al. Structural mechanism of voltage-dependent gating in an isolated voltage-sensing domain. Nat Struct Mol Biol. 2014 doi: 10.1038/nsmb.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrov D, et al. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 27.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel MS, Isacoff EY. A genetically encoded optical probe of membrane voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 30.Ataka K, Pieribone VA. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker BJ, et al. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods. 2007;161:32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Perron A, Mutoh H, Launey T, Knopfel T. Red-shifted voltage-sensitive fluorescent proteins. Chem Biol. 2009;16:1268–1277. doi: 10.1016/j.chembiol.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knopfel T. Genetically encoded optical indicators for the analysis of neuronal circuits. Nat Rev Neurosci. 2012;13:687–700. doi: 10.1038/nrn3293. [DOI] [PubMed] [Google Scholar]
Supplementary References
- 34.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 35.Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using microManager. Curr Protoc Mol Biol. 2010 doi: 10.1002/0471142727.mb1420s92. Chapter 14, Unit14.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 37.Watkins RJ, et al. A novel interaction between FRMD7 and CASK: evidence for a causal role in idiopathic infantile nystagmus. Hum Mol Genet. 2013;22:2105–2118. doi: 10.1093/hmg/ddt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teuber J, et al. The ubiquitin ligase Praja1 reduces NRAGE expression and inhibits neuronal differentiation of PC12 cells. PLoS One. 2013;8:e63067. doi: 10.1371/journal.pone.0063067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wouterlood FG, Boekel AJ, Kajiwara R, Belien JA. Counting contacts between neurons in 3D in confocal laser scanning images. J Neurosci Methods. 2008;171:296–308. doi: 10.1016/j.jneumeth.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Roepstorff K, et al. Stimulus-dependent regulation of the phagocyte NADPH oxidase by a VAV1, Rac1, and PAK1 signaling axis. J Biol Chem. 2008;283:7983–7993. doi: 10.1074/jbc.M708281200. [DOI] [PubMed] [Google Scholar]
- 41.Lin MZ, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shcherbo D, et al. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- 43.Butko MT, et al. Fluorescent and photo-oxidizing TimeSTAMP tags track protein fates in light and electron microscopy. Nat Neurosci. 2012;15:1742–1751. doi: 10.1038/nn.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang YM, Bystroff C. Complementation and reconstitution of fluorescence from circularly permuted and truncated green fluorescent protein. Biochemistry. 2009;48:929–940. doi: 10.1021/bi802027g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.