Abstract
Glycosylation plays a central role in plant defense against xenobiotics, including mycotoxins. Glucoconjugates of Fusarium toxins, such as deoxynivalenol-3-O-β-d-glucoside (DON-3G), often cooccur with their parental toxins in cereal-based food and feed. To date, only limited information exists on the occurrence of glucosylated mycotoxins and their toxicological relevance. Due to a lack of analytical standards and the requirement of high-end analytical instrumentation for their direct determination, hydrolytic cleavage of β-glucosides followed by analysis of the released parental toxins has been proposed as an indirect determination approach. This study compares the abilities of several fungal and recombinant bacterial β-glucosidases to hydrolyze the model analyte DON-3G. Furthermore, substrate specificities of two fungal and two bacterial (Lactobacillus brevis and Bifidobacterium adolescentis) glycoside hydrolase family 3 β-glucosidases were evaluated on a broader range of substrates. The purified recombinant enzyme from B. adolescentis (BaBgl) displayed high flexibility in substrate specificity and exerted the highest hydrolytic activity toward 3-O-β-d-glucosides of the trichothecenes deoxynivalenol (DON), nivalenol, and HT-2 toxin. A Km of 5.4 mM and a Vmax of 16 μmol min−1 mg−1 were determined with DON-3G. Due to low product inhibition (DON and glucose) and sufficient activity in several extracts of cereal matrices, this enzyme has the potential to be used for indirect analyses of trichothecene-β-glucosides in cereal samples.
INTRODUCTION
Infestation of cereals by phytopathogenic fungi is a global threat to the human food supply. In addition to economically devastating losses through yield and quality deterioration of agricultural products, contamination with mycotoxins poses a serious challenge for food safety (1). Among the most relevant mycotoxin producers worldwide are Fusarium species, causing Fusarium head blight disease of small-grain cereals (FHB; also known as Fusarium ear blight or scab). Trichothecene class mycotoxins inhibit eukaryotic protein synthesis and are important Fusarium virulence factors. Furthermore, they can cause apoptotic cell death and immunosuppression and trigger proinflammatory responses in humans and animals (1–3). The most important groups with regard to food safety are type A (T-2 toxin and HT-2 toxin [HT2]) and type B (nivalenol [NIV] and deoxynivalenol [DON]) trichothecenes (4). The type A trichothecene T-2 toxin possesses high acute toxicity and has caused fatal outbreaks of alimentary toxic aleukia in the last century (1). DON is the predominant trichothecene toxin produced by the Fusarium graminearum species complex and ranks among the most frequent contaminants of cereals. Although the acute toxicity of DON is lower than that of type A trichothecenes, its ubiquitous presence in Fusarium-infected cereals creates an important food safety issue (2). Acute symptoms of DON ingestion are gastroenteritis and emesis (hence, the colloquial term vomitoxin). Maximum levels for DON in cereal-based foodstuff are set in Commission Regulation 1881/2006 (5), and indicative levels are provided for the sum of T-2 toxin and HT-2 toxin in Commission Recommendation 2013/165/EU (6) in Europe to protect consumers.
The glycosylation of small molecules is a major route to inactivate endogenous and exogenous (xenobiotic) metabolites in plants (7–9). For example, formation of DON-3-O-β-d-glucopyranoside (DON-3G) is an important factor in plant defense against FHB and has been proposed to be the molecular basis of the still-unidentified FHB1 gene (Fusarium head blight resistance quantitative trait locus) in wheat (10). DON-3G has been detected in a wide range of cereal commodities, with concentrations of about 20% relative to that of DON (11, 12). However, high regional and seasonal variations have been reported, and in some cases, the content of DON-3G even exceeded that of DON (13, 14). Evidence for glucosylated metabolites of NIV and type A trichothecenes has been presented as well (15–17).
Such glucoconjugates of plant origin have been termed masked mycotoxins (13, 18), implying that they escape detection through routine analytical protocols, and reconstitution of the parental toxins through hydrolysis during food processing or in the digestive tract is possible. For example, most food-fermenting lactic acid bacteria possess β-glucosidase activities (19) and may release toxins from glucosylated precursors. DON-3G is known to be resistant to acidic hydrolysis, but it can be cleaved by several intestinal bacterial species, such as Lactobacillus spp. and Bifidobacterium spp. (20). Evidence that DON-3G is almost completely hydrolyzed in the digestive tract of rats, pigs, and humans has been presented (21–23).
Sensitive analytical methods for direct detection and quantitation of β-glucosides of trichothecene toxins are of high interest but require expensive and sophisticated analytical equipment. Further challenges are discrimination in sample preparation and analysis due to different polarities compared to the parental toxins and the present unavailability of commercial analytical standards, except for DON-3G. Therefore, indirect detection of glucosylated mycotoxins through hydrolysis prior to measurement, preferably through the aid of hydrolytic enzymes, has been suggested as an alternative approach (13, 24). Chemical hydrolysis by super acids has been proposed (25–27) but found unsuitable for this purpose (62). A β-1,3-glucosidase active toward DON-3G was reported recently (24), but this enzyme was not suited for direct application in cereal samples due to strong end product inhibition.
The aim of this study was to identify a β-glucosidase with the capacity to efficiently hydrolyze trichothecene glucosides such as DON-3G in cereal samples. The substrate specificities of several fungal and bacterial glycoside hydrolase family 3 (GH3) members were investigated. Their possible usefulness for indirect analysis of masked mycotoxins was evaluated, and a particularly promising enzyme from Bifidobacterium adolescentis was identified.
MATERIALS AND METHODS
Cloning, expression, and purification of β-glucosidases.
β-Glucosidase genes were amplified from genomic DNA of Lactobacillus brevis DSM 20054 (ATCC 14869) (LbBgl; GenBank accession number ERK40902.1, locus HMPREF0495_02581) and Bifidobacterium adolescentis DSM 20083 (ATCC 15703) (BaBgl; YP_910057.1; BAD_1194). The oligonucleotide primers used were LbBglF (5′-GATATACATATGGACATCGAACGAACGC-3′), LbBglR (5′-GTGGTGCTCGAGTTGACGTAATAAGGTGTTTGC-3′), BaBglF (5′-GATATACATATGAGCGAAAACACCTATC-3′), and BaBglR (5′-GTGGTGCTCGAGTTCGGCGGTTTCGG-3′). The restriction sites (NdeI and XhoI) used for cloning into the pET21a expression vector (C-terminal His6 tag; Novagen, Madison, WI) are underlined. Escherichia coli BL21 Star (DE3) (Invitrogen, Carlsbad, CA) was used as the expression host. Protein production was carried out in terrific broth supplemented with ampicillin (100 mg liter−1) by induction with isopropyl β-d-1-thiogalactopyranoside (IPTG; 0.5 mM), which was added at an optical density at 600 nm (OD600) of 0.5. Upon induction, incubation was continued for 16 h at 25°C with shaking (100 rpm).
Enzymes were purified from crude cell extracts on Ni2+-charged chelating Sepharose fast flow (15-ml total volume; column dimensions, 2 cm2 by 7.5 cm; GE Healthcare, Vienna, Austria) and eluted with imidazole according to the supplier's instructions. Anion exchange chromatography on Source 15Q (20-ml total volume; column dimensions, 2 cm2 by 10 cm; GE Healthcare) was performed as a second purification step. Proteins were bound to the column in 25 mM Tris-Cl (pH 7.0) and eluted with 1 M NaCl in the same buffer by applying a linear gradient of 10 column volumes. Desalting/buffer change between the purification steps was performed by size-exclusion chromatography on a HiPrep desalting column (5 cm2 by 10 cm; GE Healthcare). Enzyme activity of obtained fractions was assayed using the p-nitrophenol method as described below. The purified enzymes were stored in 25 mM Tris-Cl (pH 7) containing 150 mM NaCl at −80°C. All experiments reported in this study were conducted with the same batches of frozen enzyme(s).
SDS-PAGE with Coomassie blue staining was performed using the Mini-Protean system with precast gels (4 to 20%) from Bio-Rad (Vienna, Austria). The molecular mass marker used was a high-precision, dual-color marker (10- to 250-kDa range; Bio-Rad).
Enzyme assays.
Commercial β-glucosidase preparations were obtained from Megazyme (Wicklow, Ireland) and Sigma-Aldrich (Vienna, Austria); a full description of the enzymes used is provided in Table 1. Serial enzyme dilutions were prepared with 25 mM Tris-Cl buffer, pH 7.0, supplemented with bovine serum albumin at 0.1 mg ml−1.
TABLE 1.
Hydrolysis of DON-3G by fungal and bacterial β-glucosidasesd
| Preparation/source | Catalog no. | GH family | Protein in assay (mg ml−1) | DON-3G hydrolysis (%) | Hydrolysis measured as: |
|
|---|---|---|---|---|---|---|
| μmol h−1 mg−1 | μmol h−1 ml−1 | |||||
| Aspergillus nigera (AnBgl) | E-BGLUC | 3 | 0.19 | 24 | 6.9 × 10−3 | 5.3 × 10−3 |
| Agrobacterium sp.a | E-BGOSAG | 1 | 0.48 | 1.1 | 1.3 × 10−4 | 2.4 × 10−4 |
| Thermotoga maritimaa | E-BGOSTM | 1 | 0.78 | 21 | 1.5 × 10−3 | 4.6 × 10−3 |
| Phanerochaete chrysosporiuma (PcBgl) | E-BGOSPC | 3 | 1.1 | 88 | 4.3 × 10−3 | 1.9 × 10−2 |
| Glucosidase from almondsb,c | G0395 | 5 | Not detectable | |||
| Glucosidase from A. nigerb,c | 49291 | 5 | 3.3 | 3.6 × 10−5 | 7.3 × 10−4 | |
| Novozyme 188b | C6105 | Not specified | 38 | 8.4 × 10−3 | ||
| Lactobacillus brevis (LbBgl) | 3 | 1 | 100 | >5.12 × 10−3 | >2.2 × 10−2 | |
From Megazyme International (Wicklow, Ireland); concentrations in assays result from the supplied solution.
From Sigma-Aldrich (Vienna, Austria).
Supplied in solid form.
Hydrolysis of DON-3G was performed with a 4-h reaction time, pH 7 (100 mM Tris-Cl), at 37°C.
Photometric enzyme assays were performed with chromogenic p-nitrophenyl (pNP) glycosides obtained from Sigma-Aldrich (Table 2). Standard assay conditions with these substrates were 10 mM substrate concentration, 100 mM Tris-Cl, pH 7.0, 37°C, 5-min reaction time. The assays were stopped by adding a 2-fold volumetric excess of 0.5 M Na2CO3. The absorption of released p-nitrophenol was measured at 400 nm on a Beckman Coulter DU800 spectrophotometer using a molar extinction coefficient of 18,300 M−1 cm−1. pH dependence was determined with Britton-Robinson buffers ranging from pH 3.0 to 9.5 (28).
TABLE 2.
Specific activities of GH3 glycosidases LbBgl, BaBgl, AnBgl, and PcBgl for synthetic and natural substratesa
| Substrate | Concn (mM) | Sp act (μmol min−1 mg−1) |
|||
|---|---|---|---|---|---|
| LbBgl | BaBgl | AnBgl | PcBgl | ||
| pNP-β-d-glucopyranoside | 10 | 42 ± 2 | 49 ± 1 | 4.2 ± 0.1 | 22 ± 2 |
| pNP-β-d-xylopyranoside | 10 | 5.3 ± 0.1 | 29 ± 0 | ND | ND |
| pNP-α-l-arabinofuranoside | 10 | 0.34 ± 0.01 | 0.97 ± 0.01 | ND | ND |
| pNP-β-d-galactopyranoside | 10 | NDc | 0.73 ± 0.02 | ND | ND |
| pNP-β-d-mannopyranoside | 10 | ND | ND | ND | ND |
| Cellobiose | 10 | 0.71 ± 0.02 | 0.064 ± 0.001 | 17 ± 0 | 0.53 ± 0.01 |
| Salicin | 10 | 36 ± 1 | 43 ± 0 | 2.3 ± 0.2 | 1.3 ± 0.0 |
| Quercetin-3-O-β-d-glucopyranosideb | 0.7 | ND | 0.075 ± 0.005 | ND | ND |
| n-Octyl-β-d-glucopyranoside | 10 | 10 ± 0 | 28 ± 0 | 4.3 ± 0.0 | 8.4 ± 0.6 |
| Deoxynivalenol-3-O-β-d-glucopyranoside | 10 | 0.082 ± 0.003 | 11 ± 1 | 0.039 ± 0.002 | 0.026 ± 0.001 |
| Nivalenol-3-O-β-d-glucopyranoside | 1 | 0.0036 ± 0.0001 | 0.18 ± 0.02 | 0.0012 ± 0.0002 | ND |
| HT-2-toxin-3-O-β-d-glucopyranoside | 2 | 0.017 ± 0.000 | 3.5 ± 0.1 | 0.040 ± 0.000 | <0.001 |
Specific activities were determined at 37°C, pH 7.0 (100 mM Tris-Cl). All values represent the mean values from triplicate determinations ± standard deviations.
Dissolved in 80% ethanol (20% in assay).
ND, not detectable.
Assays with cellobiose, salicin, quercetin-3-β-d-glucoside, and n-octyl-β-d-glucoside were performed as described above but stopped by heat inactivation (90°C, 5 min). Enzyme activity toward these substrates was determined by quantifying released glucose through high-performance liquid chromatography (HPLC). Equipment and conditions were a HPLC Summit Dionex with a P680 pump and ASI-100 autosampler (all from Dionex, Sunnyvale, CA). Separation was performed on an Aminex HPX87-K column coupled to a Micro-Guard cation H cartridge (both from Bio-Rad) at a temperature of 80°C. The mobile phase was H2O with an isocratic flow of 0.5 ml min−1 with a 20-μl injection volume. Analytes were monitored with a Shodex RI-100 detector (Showa Denko, Tokyo, Japan). Calibration curves with cellobiose and/or glucose standards were prepared in the range from 0.05 to 5 g liter−1.
DON-3G, nivalenol-3-O-β-d-glucopyranoside (NIV-3G), and HT-2-toxin-3-O-β-d-glucopyranoside (HT2-3G) were enzymatically prepared with a recombinant family 1 UDP-glucosyltransferase (29), purified by preparative HPLC, and structurally confirmed by 1H- and 13C-nuclear magnetic resonance (NMR) methods (H. Michlmayr, A. Malachova, E. Varga, M. Lemmens, S. Newmister, I. Rayment, F. Berthiller, and G. Adam, unpublished data). Assays with these substrates were performed under the same conditions as those described above (100 mM Tris-Cl, pH 7.0, 37°C) but stopped by transferring 15 μl reaction mix to 135 μl methanol. The samples were centrifuged for 5 min at 20,000 × g, further diluted with deionized water, and transferred to HPLC vials. Cereal samples were finely ground in a coffee mill and extracted 1:4 (wt/vol) with 125 mM Tris, pH 7. Beer was degassed and the pH adjusted to 7.0 with 0.1 M KOH. Beer and cereal extracts were spiked with DON-3G at 12.5 mg liter−1, and enzyme assays were done with 80 μl of spiked extracts and 20 μl enzyme solution (final concentration, 10 mg liter−1 DON-3G). The assays were stopped and prepared for analysis as described above.
Analysis of glucosides and released toxins by LC-MS.
The screening of different enzymes for their hydrolytic activity against DON-3G was performed on an 1100 series HPLC system (Agilent Technologies, Waldbronn, Germany) coupled to a QTrap liquid chromatography-tandem mass spectrometry (LC-MS/MS) system (Applied Biosystems, Foster City, CA) with atmospheric pressure chemical ionization. The method was based on reference 30 with slight modifications concerning the LC conditions using an Zorbax eclipse XDB-C8 column (150 by 4.6 mm, 5 μm; Agilent Technologies).
For testing the specific activities and obtaining the kinetic constants, the method was transferred to a QTrap 4000 LC-MS/MS system (AB Sciex, Foster City, CA) to gain sensitivity. Furthermore, the following analytes were included in the method: NIV, NIV-3G, HT2, and HT2-3G. Chromatographic separation was achieved on a Gemini C18 column (150 by 4.6 mm, 5 μm; Phenomenex, Aschaffenburg, Germany) at 25°C with a flow rate of 0.8 ml min−1. The following water-methanol gradient (eluent A, 80:20 [vol/vol]; eluent B, 3:97 [vol/vol]; both containing 5 mM ammonium acetate) was used. Initial conditions at 0% B were a hold for 1 min, followed by a linear increase to 50% B within 5 min and an increase to 100% B within another 3 min. After holding with 100% B for 2.5 min, a fast switch to the initial conditions was performed, followed by column equilibration until 14 min. For the first 6.5 min the mass spectrometer was operated in negative electrospray ionization mode, whereas in the last 7.5 min the positive electrospray ionization mode was used. The following source settings were used: temperature, 550°C; ion spray voltage, 4 kV (positive mode) and −4 kV (negative mode); curtain gas, 30 lb/in2 (207 kPa of >99% nitrogen); source gas one and two, both 50 lb/in2 (345 kPa of zero-grade air); and collision gas (nitrogen) set to high. For quantitation, two selected reaction-monitoring transitions per compound were acquired with a dwell time of 25 ms. In the first period, the acetate adducts of the analytes (m/z 355.1 for DON, m/z 371.1 for NIV, m/z 517.3 for DON-3G, and m/z 533.1 for NIV-3G) were chosen as precursors, and the declustering potential (DP) was −40 V for DON and NIV, −50 V for DON-3G, and −60 V for NIV-3G. The following product ions were chosen as quantifier and qualifier, respectively: for DON, m/z 59.2 (collision energy [CE] of −40 V) and m/z 265.2 (CE of −22 V); for NIV, m/z 59.1 and 281.1 (CE of −38 V for both); for DON-3G, m/z 427.1 (CE of −30 V) and m/z 59.1 (CE of −85 V); and for NIV-3G, m/z 263.0 (CE of −30 V) and m/z 443.0 (CE of −26 V). In the second period, the ammonium adducts of HT2 (m/z 442.2; DP, 70 V) and HT2-3-G (m/z 604.4; DP, 51 V) were chosen as precursors, and the following product ions were selected: for HT2, m/z 215.1 (CE of 19 V) and m/z 197.1 (CE of 25 V); for HT2-3G, m/z 263.3 (CE of 27 V) and m/z 215.1 (CE of 25 V). A sample chromatogram of standards of DON/DON-3G, NIV/NIV-3G, and HT2/HT2-3G is shown in Fig. S1 in the supplemental material.
RESULTS
Substrate specificity and selectivity.
Several commercially available β-glucosidase preparations and a previously described GH3 enzyme from Lactobacillus brevis (31, 32), here designated LbBgl, were assayed for their capacity to hydrolyze DON-3G at concentrations of 10 mg liter−1 (22 μM) within 4 h. Almond β-glucosidase (Sigma-Aldrich) apparently was inactive toward DON-3G. The highest conversion rates were observed with the β-glucosidases from Aspergillus niger, Phanerochaete chrysosporium, and LbBgl. The latter completely hydrolyzed DON-3G under these conditions.
It was previously reported that B. adolescentis is able to hydrolyze DON-3G in vitro (20). The genome of B. adolescentis ATCC 15703 (GenBank accession number NC_008618.1) contains six genes encoding putative GH3 hydrolases. BAD_1194 is the enzyme/gene with the highest amino acid sequence similarity to LbBgl (58% similarity and 41% identity by BLASTp [33]) and was selected for biochemical characterization. This enzyme subsequently is referred to as BaBgl.
The substrate specificities of recombinant LbBgl and BaBgl (purified to apparent homogeneity) (Fig. 1) for several synthetic and natural substrates, including DON-3G and two other trichothecene-toxin β-glucosides available in our laboratory, were determined. The two fungal GH3 enzymes from A. niger (AnBgl) and P. chrysosporium (PcBgl) that were included in the DON-3G hydrolysis assays described above (Table 1) were characterized analogously to obtain a comparative view on the substrate range of these glycosidases. All assays were performed under conditions (37°C, 100 mM Tris-Cl, pH 7) that do not necessarily reflect the optimal reaction conditions of the individual catalysts. One reason for choosing these conditions was to compromise on a pH value that agrees with most of the commonly used inorganic and organic buffers, which is also relevant in practical terms. Second, all enzyme assays were performed under identical conditions to obtain a comparative view of the catalytic potential of different catalysts.
FIG 1.
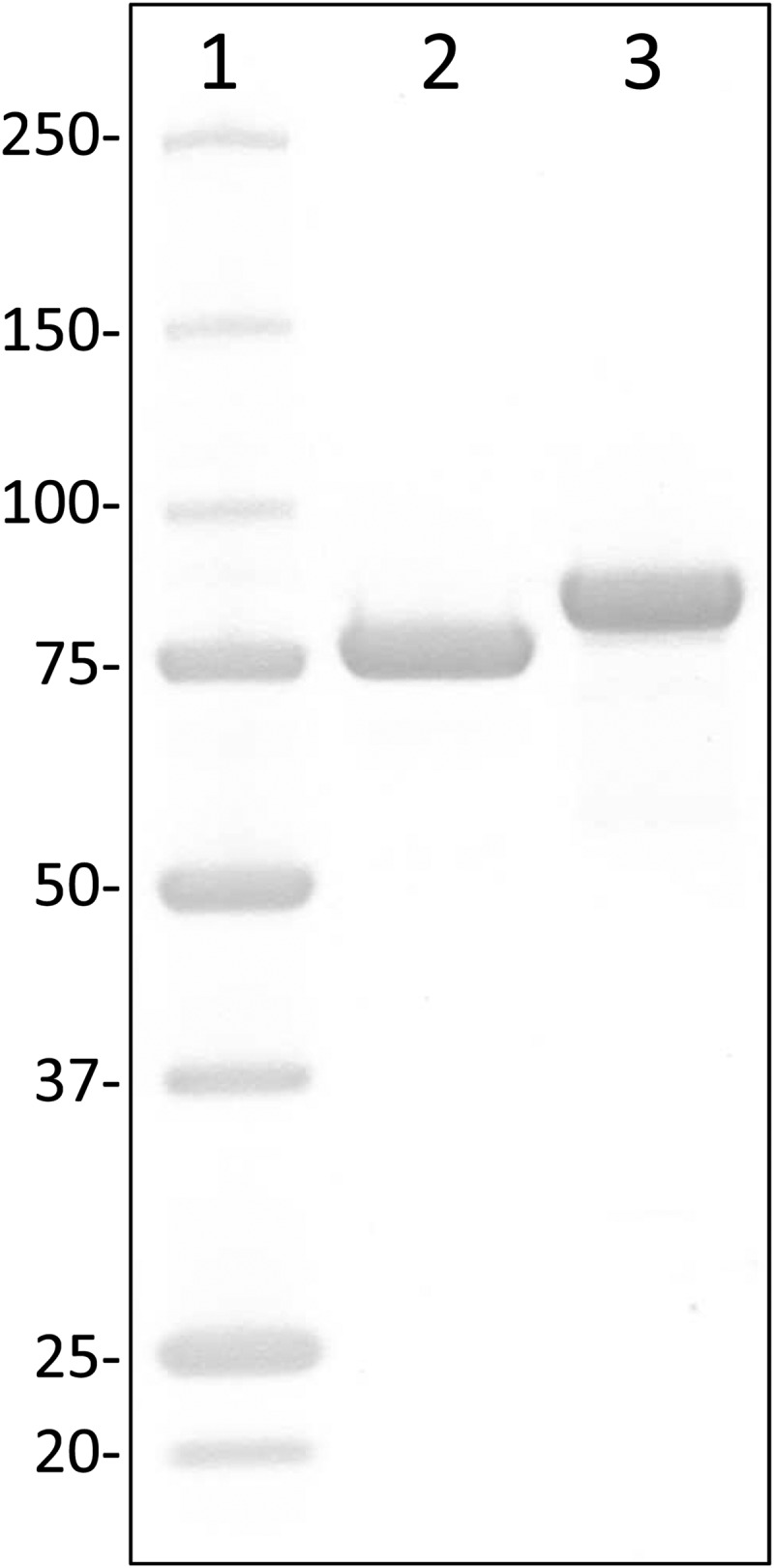
SDS-PAGE of two-step-purified His6-tagged LbBgl and BaBgl with theoretical molecular masses of 83.6 and 82.2 kDa, respectively. Lane 1, Precision Plus protein standard (Bio-Rad), molecular mass in kilodaltons; lane 2, LbBgl (4 μg); lane 3, BaBgl (4 μg).
In assays with pNP-glycosides (Table 2), both fungal enzymes solely hydrolyzed pNP-β-d-glucopyranoside. The bacterial enzymes displayed additional β-d-xylopyranosidase and lower α-l-arabinofuranosidase side activities, which is consistent with the known possible functionalities of GH3 hydrolases (34). Concerning aglycon specificities, AnBgl clearly preferred cellobiose as the substrate and PcBgl displayed its maximum activities with the synthetic substrates pNP-β-d-glucopyranoside and n-octyl-β-d-glucopyranoside. Both bacterial enzymes demonstrated poor hydrolytic activity toward cellobiose and differed considerably in their substrate specificities, especially with regard to the trichothecene-β-glucosides. Among the tested enzymes, BaBgl showed the highest specific activities for DON-3G, NIV-3G, HT2-3G, salicin, quercetin-3-β-d-glucoside, and n-octyl-β-d-glucoside (Table 2). Kinetic constants obtained with selected substrates (Table 3) clearly confirm that both bacterial enzymes are highly inefficient with regard to cellobiose hydrolysis. Comparison of the kcat/Km values obtained with pNP-β-glucoside, pNP-β-xyloside, and DON-3G are a further indication that BaBgl is less selective than LbBgl. Although the affinities of LbBgl and BaBgl for DON-3G appear to be in a similar range as judged from apparent Km values (2.8 and 5.4 mM, respectively) (Table 3), the catalytic efficiency (kcat/Km) of BaBgl exceeded that of LbBgl 80-fold.
TABLE 3.
Kinetic constants of the GH3 glycosidases from LbBgl and BaBgla
| Enzyme and substrate | Kinetic constant |
|||
|---|---|---|---|---|
| Km (mM) | Vmax (μmol min−1 mg−1) | kcatb (s−1) | kcat/Km (s−1 mM−1) | |
| LbBgl | ||||
| pNP-β-d-glucopyranoside | 0.63 ± 0.09 | 47 ± 1 | 66 | 104 |
| pNP-β-d-xylopyranoside | 2.6 ± 0.2 | 6.6 ± 0.2 | 9.3 | 3.5 |
| Deoxynivalenol-3-O-β-d-glucopyranoside | 2.8 ± 0.4 | 0.11 ± 0.00 | 0.15 | 0.053 |
| Cellobiose | 63 ± 4 | 5.3 ± 0.2 | 7.4 | 0.12 |
| BaBgl | ||||
| pNP-β-d-glucopyranoside | 1.1 ± 0.1 | 68 ± 2 | 94 | 87 |
| pNP-β-d-xylopyranoside | 4.2 ± 0.3 | 39 ± 1 | 53 | 13 |
| Deoxynivalenol-3-O-β-d-glucopyranoside | 5.4 ± 0.5 | 16 ± 1 | 22 | 4.0 |
| Cellobiose | 73 ± 12 | 0.50 ± 0.05 | 0.69 | 0.0094 |
Kinetic constants from LbBgl and BaBgl were determined at 37°C, pH 7.0 (100 mM Tris-Cl).
Calculations are based on the theoretical molecular mass of the His6-tagged proteins: LbBgl, 83.6 kDa; BaBgl, 82.2 kDa.
Product inhibition and physicochemical characterization.
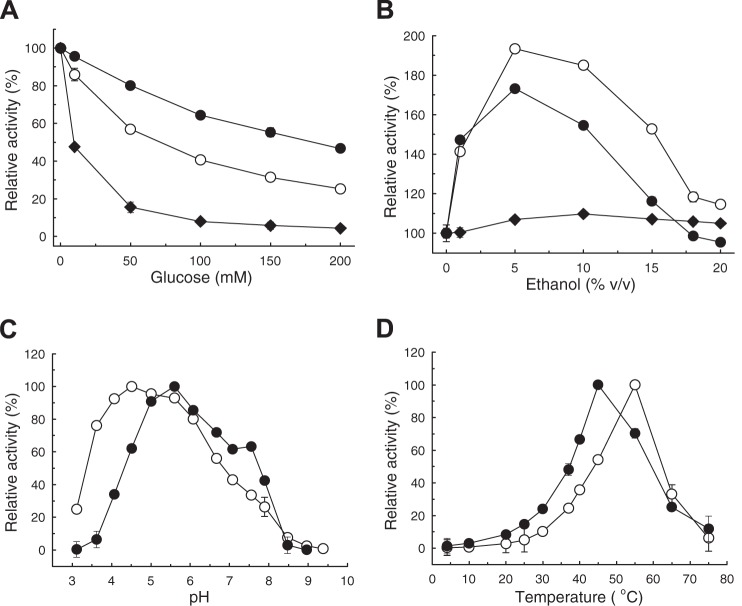
Compared to the Aspergillus cellobiase AnBgl, the bacterial enzymes LbBgl and BaBgl were moderately inhibited by glucose (Fig. 2A): 50% activity reduction was estimated at 11 mM, 67 mM, and 180 mM glucose for AnBgl, LbBgl, and BaBgl, respectively. At 12 mM DON (3.5 g liter−1), a concentration high above the DON levels that can be expected in contaminated cereal samples (milligram per liter range), DON did not appear to exert an inhibitory effect on pNP-β-d-glucoside hydrolysis by LbBgl and BaBgl (see Table S1 in the supplemental material). Increased activity in the presence of ethanol (<20% vol/vol) was observed with LbBgl and BaBgl but not with AnBgl (Fig. 2B).
FIG 2.
Influence of glucose (A) and ethanol (B) on activities of LbBgl (○), BaBgl (•), and AnBgl (◆) in the standard pNP-β-glucopyranoside assay (37°C, pH 7). pH (C) and temperature (D) dependence of LbBgl (○) and BaBgl (•). (C) Assays (37°C) performed with Britton-Robinson buffers (28) in the range of pH 3.0 to 9.5. (D) Standard assay (100 mM Tris, pH 7) performed at 4 to 75°C. All data represent the averages from triplicate determinations, and error bars indicate standard deviations. One hundred percent relative activity refers to standard assay conditions without additive (A and B) or maximum activity (C and D).
The pH (Fig. 2C) and temperature (Fig. 2D) profiles of LbBgl and BaBgl indicate maximum hydrolytic activities in the range of pH 5 to 6 and 50 to 60°C. Inclusion of EDTA and salts of several mono- and divalent metal ions implied that LbBgl and BaBgl do not depend on metal ions for hydrolysis (see Table S1 in the supplemental material). Both enzymes were active in several commonly used organic and inorganic buffers (Table 4). At pH 7, highest activities were determined in citrate-phosphate buffer prepared according to McIlvaine (35), and the lowest activities were recorded in Tris-Cl buffer, which was used as the standard assay buffer in this study.
TABLE 4.
Relative activities of LbBgl and BaBgl in organic and inorganic buffers at pH 7d
| Buffer | Relative enzyme activity (%) |
|
|---|---|---|
| LbBgl | BaBgl | |
| Tris (100 mM) | 100 ± 0 | 100 ± 2 |
| McIlvainea | 158 ± 2 | 174 ± 1 |
| Phosphate buffer (100 mM) | 127 ± 2 | 143 ± 1 |
| MOPSb (100 mM) | 163 ± 2 | 158 ± 2 |
| HEPES (100 mM) | 145 ± 0 | 132 ± 1 |
| Succinic acid (100 mM) | 124 ± 2 | 135 ± 2 |
| Britton-Robinson bufferc | 136 ± 1 | 141 ± 1 |
Citrate-phosphate buffer according to reference 35.
Morpholinepropanesulfonic acid.
Described in reference 28.
Relative activities of LbBgl and BaBgl in organic and inorganic buffers at pH 7 were determined with pNP-β-d-glucopyranoside at 37°C. All values represent the means from triplicate determinations ± standard deviations.
DON-3G hydrolysis in cereal samples.
Judged from the catalytic efficiency with DON-3G and low product inhibition, the results described above indicated that BaBgl is an interesting candidate for the hydrolysis of DON-3G and possibly structurally similar trichothecene-glucosides in cereal samples. As a proof of concept, extracts of several cereal samples and degassed beer were spiked with DON-3G (10 mg liter−1) to test the performance of BaBgl in realistic sample matrices. A naturally contaminated barley sample containing both DON and DON-3G was included as well (Table 5). BaBgl efficiently hydrolyzed DON-3G in all sample matrices (Table 5), as judged from the release of DON and the fact that DON-3G was not detectable after 15 min in all samples. While in beer DON-3G still was detectable, its levels were already below the limit of quantification (Table 5). Furthermore, NIV-3G and HT2-3G (both at 10 mg liter−1) were completely hydrolyzed by BaBgl within 15 min in spiked wheat extract.
TABLE 5.
Hydrolysis of DON-3G in aqueous cereal sample extracts
| Time (min) | Hydrolysis (mM) in extract ofa: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Malt |
Wheat |
Rice |
Beerb |
Barleyc |
||||||
| DON-3G | DON | DON-3G | DON | DON-3G | DON | DON-3G | DON | DON-3G | DON | |
| 0 | 23 | ND | 22 | ND | 22 | ND | 23 | ND | 15 | 39 |
| 5 | ND | 21 | ND | 21 | <2.2 | 18 | 7.8 | 13 | ND | 50 |
| 10 | ND | 21 | ND | 20 | ND | 19 | <2.2 | 19 | ND | 50 |
| 15 | ND | 23 | ND | 21 | ND | 20 | <2.2 | 19 | ND | 49 |
Hydrolysis of DON-3G by BaBgl (0.8 mg ml−1) to DON in aqueous (100 mM Tris-Cl, pH 7) cereal sample extracts (1:5, wt/vol) spiked with 10 mg liter−1 (22 μM) DON-3G. All values represent the means from triplicate determination, and concentrations are displayed in millimolars. ND, not detectable (signal-to-noise ratio below 3:1).
Degassed and adjusted to pH 7 with 0.1 M KOH.
Naturally contaminated sample. Concentrations refer to dry sample (concentration in assay, 1:5).
Sequence comparison of GH3 hydrolases.
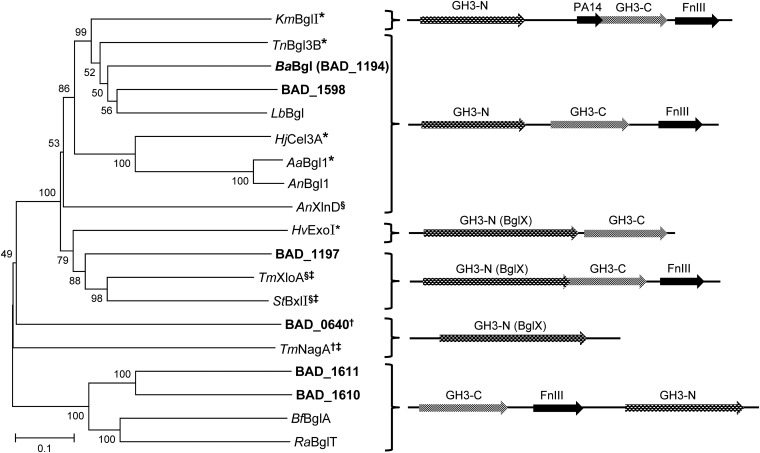
GH3 is a large and divergent enzyme family (at the time of writing, >6,000 sequences are in CAZy [34]) with considerable differences in known domain structures and arrangements (36). Figure 3 illustrates the relationships between the six putative GH3 enzymes encoded by the B. adolescentis genome relative to other GH3 hydrolases with known function and resolved crystal structure. BAD_0640 is a putative N-acetyl-β-d-glucosaminidase, and BAD_1197 could be related to GH3 xylosidases. BAD_1610 and BAD_1611 seem to belong to a different GH3 subclass with reversed domain arrangement. Together with BAD_1598 and LbBgl, BaBgl (BAD_1194) appears similar to β-glucosidase TnBgl3B from Thermotoga neapolitana (Fig. 3). TnBgl3B (37) consists of three domains, and overall sequence similarities suggest that LbBgl (36% identity, 97% sequence coverage) and BaBgl (37% identity, 99% coverage) are structured in a similar fashion. The architecture of TnBgl3 comprises an N-terminal TIM barrel-like (α/β)8 fold (domain 1, GH3 N-terminal domain; pfam00933) followed by a α/β sandwich fold (domain 2, GH3 C-terminal domain; pfam01915). Both domain structures are conserved within GH3, and the substrate binding pocket of TnBgl3B is located at their interface. Domain one contains the catalytic nucleophile (D242 in TnBgl3), and several conserved residues that have been identified as pivotal for glycon (β-d-glucopyranosyl-) binding and orientation. The acid/base catalytic residue (E458) is located on a less conserved region on domain two (37) (see Fig. S1 in the supplemental material). C-terminal domain three of TnBgl3B is a fibronectin type III-like fold (FnIII; pfam14310). This domain often is present in GH3 hydrolases (Fig. 3), and it also has been identified in eukaryotic GH3 β-glucosidases (38–40), but its function is unknown to date.
FIG 3.
Neighbor-joining tree of putative and experimentally confirmed GH3 β-glucosidases and related enzymes. §, β-xylosidase; †, N-acetyl-β-d-glucosaminidase; *, three-dimensional structure is available from the references listed below; ‡, crystal structure not yet published (www.cazy.org). Information on domain organization as schematically depicted on the right is derived from the available crystal structures and by sequence alignments: GH3-N, conserved N-terminal GH3 domain (pfam00933); GH3-C, C-terminal GH3 domain (pfam01915); FnIII, fibronectin type III-like domain (pfam14310); PA14 domain (pfam07691); and BglX, glucosidase related hydrolases (COG1472). Sequences of Bifidobacterium adolescentis (NC_008618.1, loci BAD_0640, BAD_1194 [BaBgl], BAD_1197, BAD_1598, BAD_1610, and BAD_1611) are highlighted in boldface. KmBglI (Kluyveromyces marxianus, PDB entry 3AC0, reference 39), TnBgl3B (Thermotoga neapolitana, PDB entry 2X40, reference 37), LbBglI (Lactobacillus brevis, ERK40902.1), HjCel3A (Hypocrea jecorina, anamorph of Trichoderma reesei, PDB entry 3ZYZ, reference 40), AaBglI (Aspergillus aculeatus, PDB entry 4IIB, reference 38), AnBglI (Aspergillus niger, CAB75696.1, reference 56), AnXlnD (A. niger, CAB06417.1 reference 57), HvExoI (Hordeum vulgare, PDB entry 1EX1, reference 58), TmXloA (Thermotoga maritima, AAD35170.1), StBxlI (Streptomyces thermoviolaceus, BAD02389.1, reference 59), TmNagA (T. maritima, AAD35891.1), BfBglA (Butyrivibrio fibrisolvens, P16084.1, references 36 and 60), and RaBglT (Ruminococcus albus, CAA33461.1, references 36 and 61). Amino acid sequence alignment and phylogenetic analysis was performed with Mega 6. The Muscle algorithm was used for alignment, and the tree was constructed with the p-distance model and 1,000 bootstrap iterations.
Figure S2 in the supplemental material (residue numbering refers to that of the TnBgl3B sequence) highlights the conserved amino acid residues identified in GH3 β-glucosidases (37, 40) compared to three GH3 β-xylosidase sequences. Notable differences are that D58 and W243 appear not to be conserved in GH3 xylosidases. Both residues have been shown to be crucial for glucose accommodation in the glycon binding subsite of TnBgl3B.
DISCUSSION
In addition to the primarily analytical objective of this study, the substrate specificities of two bacterial and two fungal GH3 hydrolases were of interest. While the two fungal GH3 enzymes (A. niger and P. chrysosporium) appear to be specific β-glucosidases, LbBgl and BaBgl possess considerable β-d-xylosidase and low α-l-arabinofuranosidase side activities. Glycon recognition motifs usually are highly conserved within GH families, yet the mechanistic details determining β-glucosidase or β-xylosidase activity in GH3 so far have not been clarified sufficiently. Sequence alignment of LbBgl and BaBgl (see Fig. S2 in the supplemental material) does not indicate differences in conserved glucose binding residues compared to other GH3 β-glucosidases. However, it was reported that aglycon binding also can have an effect on glycon orientation (41), and the side activities of LbBgl and BaBgl could be related to their broad aglycon specificities.
In contrast to the conserved glycon recognition motifs, the aglycon binding sites of glycosidases usually are much less defined. Aglycon accommodation at the active site is determined mainly by hydrophobic interactions, which allows for additional freedom in substrate positioning (42, 43). While this accounts for flexibility, it currently is not possible to predict aglycon specificities solely based on sequence analyses. Understanding the biochemical functionality of such β-glucosidases requires evaluation of a broad range of possible substrates which often is limited by unavailability or poor solubility of analytes of interest. It is further possible that some enzyme classes have been evolutionarily tailored to be versatile. Therefore, the quest to identify the true substrate (i.e., thermodynamically ideal substrate in terms of affinity or catalytic efficiency) may not always yield the best results, especially with regard to a possible physiological function. Thus, the true substrate of BaBgl most likely has not been identified in this study. DON-3G hydrolysis by BaBgl appears to be caused by its unselective nature rather than by a particular preference for this structure, which is also reflected in the relatively high Km value (5.4 mM for DON-3G). This stands in contrast to a 1,3-β-glucanase (catalog no. L9259 [product discontinued]; Sigma-Aldrich) recently reported to possess surprisingly high affinity (Km of 4.5 μM) for DON-3G (24). However, this enzyme was almost completely inhibited by glucose at low concentrations (10 μM). Consequently, inconvenient sample cleanup was necessary to eliminate glucose originally present in the sample prior to enzymatic treatment.
BaBgl exerted adequate hydrolytic activity with DON-3G, NIV-3G, and HT2-3G and was able to hydrolyze these compounds in cereal samples as well. Its unspecific nature suggests that BaBgl or functionally related glycosidases are able to hydrolyze an even wider spectrum of masked mycotoxins. Although DON is the most frequently occurring trichothecene toxin worldwide (44), regional and seasonal variations of F. graminearum chemotypes and of other Fusarium species (45, 46) imply that occurrences of different trichothecenes and their (putative) β-glucosides underlie high geographic and seasonal fluctuations. For example, nivalenol is more prevalent in Asian countries (47), and high incidences of T-2 toxin and HT-2 toxin have been reported recently in northern European countries (48); F. graminearum strains isolated in the United States can produce a previously unknown type A trichothecene (NX-3) (49).
Practical applications of BaBgl may involve hydrolysis of glucosylated trichothecene toxins in aqueous cereal extracts prior to analysis. In principle, this would be of interest for immunological detection methods, such as enzyme-linked immunosorbent assay (ELISA) kits that are widely used for rapid estimation of DON levels in cereals. However, the utility of this approach is limited by the high cross-reactivities of DON-specific antibodies with DON-3G, which have been reported to range from 52 to 157% (50, 51). For example, assuming a cross-reactivity of 50%, the increase of the signal intensity due to the enzymatic cleavage of DON-3G to DON would be visible only if more than 20% DON-3G compared to the level of DON on a molar basis is present in the sample. The reason for this is the repeatability of ELISA methods (typically 10 to 20% relative standard deviation [RSD]), which would make it impossible to distinguish between the DON content before and after enzymatic hydrolysis. A more suitable application could be the hydrolysis of glucoconjugated trichothecenes prior to analysis by conventional HPLC and gas chromatography methods, which show better repeatabilities (often 5 to 10% RSD). This should reveal an increase in the parental toxin content equivalent to the molar concentration of the masked compound.
As a final remark, it is worthwhile to note that previous studies investigating glycoside hydrolase activities of bifidobacteria were concerned mainly with their positive implications. Examples include the metabolism of prebiotics (52) and a considerable number of publications on the release of isoflavones (phytoestrogens) and other potentially health-beneficial plant metabolites from their β-glucoside precursors (for examples, see references 53–55). One aim of this study was to point out the possible functional and structural diversity of (bacterial) GH3 β-glucosidases. By considering this and the high number of putative GH3 genes reported in individual Bifidobacterium genomes (52), it appears that a wide range of β-glucosidase functionalities in these bacteria can be expected. Therefore, it is crucial to consider that such intestinal species possess the capability to increase the bioavailability not only of health-beneficial plant metabolites but also of masked dietary toxins.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the support of the Vienna Science and Technology Fund (WWTF; grant LS12-021). Furthermore, financial support by the Austrian Federal Ministry of Science, Research and Economy, the National Foundation of Research, Technology and Development, and the Lower Austrian Government is acknowledged.
This work is dedicated to the memory of Klaus D. Kulbe.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01061-15.
REFERENCES
- 1.Bhat R, Rai RV, Karim AA. 2010. Mycotoxins in food and feed: present status and future concerns. Compr Rev Food Sci Food Saf 9:57–81. doi: 10.1111/j.1541-4337.2009.00094.x. [DOI] [PubMed] [Google Scholar]
- 2.Rotter BA, Prelusky DB, Pestka JJ. 1996. Toxicology of deoxynivalenol (vomitoxin). J Toxicol Environ Health 48:1–34. [DOI] [PubMed] [Google Scholar]
- 3.Pestka JJ. 2010. Deoxynivalenol: mechanisms of action, human exposure, and toxicological relevance. Arch Toxicol 84:663–679. doi: 10.1007/s00204-010-0579-8. [DOI] [PubMed] [Google Scholar]
- 4.McCormick SP, Stanley AM, Stover NA, Alexander NJ. 2011. Trichothecenes: from simple to complex mycotoxins. Toxins 3:802–814. doi: 10.3390/toxins3070802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.European Commission. 2006. Commission Regulation (EC) no 1881/2006 of 19 December 2006 setting maximum levels for certain contaminants in foodstuffs. Off J Eur Union L364:5–24. [Google Scholar]
- 6.European Commission. 2013. Commission recommendation of 27 March 2013 on the presence of T-2 and HT-2 toxin in cereals and cereal products. Off J Eur Union L91:12–15. [Google Scholar]
- 7.Coleman JOD, Blake-Kalff MMA, Davies TGE. 1997. Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2:144–151. doi: 10.1016/S1360-1385(97)01019-4. [DOI] [Google Scholar]
- 8.Bowles D, Isayenkova J, Lim EK, Poppenberger B. 2005. Glycosyltransferases: managers of small molecules. Curr Opin Plant Biol 8:254–263. doi: 10.1016/j.pbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Gachon CMM, Langlois-Meurinne M, Saindrenan P. 2005. Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci 10:542–549. doi: 10.1016/j.tplants.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Lemmens M, Scholz U, Berthiller F, Dall'Asta C, Koutnik A, Schuhmacher R, Adam G, Buerstmayr H, Mesterházy A, Krska R, Ruckenbauer P. 2005. The ability to detoxify the mycotoxin deoxynivalenol colocalizes with a major quantitative trait locus for Fusarium head blight resistance in wheat. Mol Plant Microbe Interact 18:1318–1324. doi: 10.1094/MPMI-18-1318. [DOI] [PubMed] [Google Scholar]
- 11.Berthiller F, Dall'Asta C, Corradini R, Marchelli R, Sulyok M, Krska R, Adam G, Schuhmacher R. 2009. Occurrence of deoxynivalenol and its 3-β-d-glucoside in wheat and maize. Food Addit Contam 26:507–511. doi: 10.1080/02652030802555668. [DOI] [PubMed] [Google Scholar]
- 12.Desmarchelier A, Seefelder W. 2011. Survey of deoxynivalenol and deoxynivalenol-3-glucoside in cereal-based products by liquid chromatography electrospray ionization tandem mass spectrometry. World Mycotoxin J 4:29–35. doi: 10.3920/WMJ2010.1236. [DOI] [Google Scholar]
- 13.Berthiller F, Crews C, Dall'Asta C, Saeger SD, Haesaert G, Karlovsky P, Oswald IP, Seefelder W, Speijers G, Stroka J. 2013. Masked mycotoxins: a review. Mol Nutr Food Res 57:165–186. doi: 10.1002/mnfr.201100764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostelanska M, Hajslova J, Zachariasova M, Malachova A, Kalachova K, Poustka J, Fiala J, Scott PM, Berthiller F, Krska R. 2009. Occurrence of deoxynivalenol and its major conjugate, deoxynivalenol-3-glucoside, in beer and some brewing intermediates. J Agric Food Chem 57:3187–3194. doi: 10.1021/jf803749u. [DOI] [PubMed] [Google Scholar]
- 15.Busman M, Poling SM, Maragos CM. 2011. Observation of T-2 toxin and HT-2 toxin glucosides from Fusarium sporotrichioides by liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS). Toxins 3:1554–1568. doi: 10.3390/toxins3121554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakagawa H, Ohmichi K, Sakamoto S, Sago Y, Kushiro M, Nagashima H, Yoshida M, Nakajima T. 2011. Detection of a new Fusarium masked mycotoxin in wheat grain by high-resolution LC-Orbitrap MS. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 28:1447–1456. doi: 10.1080/19440049.2011.597434. [DOI] [PubMed] [Google Scholar]
- 17.Nakagawa H, Sakamoto S, Sago Y, Nagashima H. 2013. Detection of type A trichothecene di-glucosides produced in corn by high-resolution liquid chromatography-Orbitrap mass spectrometry. Toxins 5:590–604. doi: 10.3390/toxins5030590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rychlik M, Humpf H-U, Marko D, Dänicke S, Mally A, Berthiller F, Klaffke H, Lorenz N. 2014. Proposal of a comprehensive definition of modified and other forms of mycotoxins including “masked” mycotoxins. Mycotoxin Res 30:197–205. doi: 10.1007/s12550-014-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michlmayr H, Kneifel W. 2014. β-Glucosidase activities of lactic acid bacteria: mechanisms, impact on fermented food and human health. FEMS Microbiol Lett 352:1–10. doi: 10.1111/1574-6968.12348. [DOI] [PubMed] [Google Scholar]
- 20.Berthiller F, Krska R, Domig KJ, Kneifel W, Juge N, Schuhmacher R, Adam G. 2011. Hydrolytic fate of deoxynivalenol-3-glucoside during digestion. Toxicol Lett 206:264–267. doi: 10.1016/j.toxlet.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagl V, Schwartz H, Krska R, Moll W-D, Knasmüller S, Ritzmann M, Adam G, Berthiller F. 2012. Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in rats. Toxicol Lett 213:367–373. doi: 10.1016/j.toxlet.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagl V, Woechtl B, Schwartz-Zimmermann HE, Hennig-Pauka I, Moll W-D, Adam G, Berthiller F. 2014. Metabolism of the masked mycotoxin deoxynivalenol-3-glucoside in pigs. Toxicol Lett 229:190–197. doi: 10.1016/j.toxlet.2014.06.032. [DOI] [PubMed] [Google Scholar]
- 23.Dall'Erta A, Cirlini M, Dall'Asta M, Del Rio D, Galaverna G, Dall'Asta C. 2013. Masked mycotoxins are efficiently hydrolyzed by human colonic microbiota releasing their aglycones. Chem Res Toxicol 26:305–312. doi: 10.1021/tx300438c. [DOI] [PubMed] [Google Scholar]
- 24.Nielen M, Weijers C, Peters J, Weignerová L, Zuilhof H, Franssen M. 2014. Rapid enzymatic hydrolysis of masked deoxynivalenol and zearalenone prior to liquid chromatography mass spectrometry or immunoassay analysis. World Mycotoxin J 7:107–113. doi: 10.3920/WMJ2013.1662. [DOI] [Google Scholar]
- 25.Liu Y, Walker F, Hoeglinger B, Buchenauer H. 2005. Solvolysis procedures for the determination of bound residues of the mycotoxin deoxynivalenol in Fusarium species infected grain of two winter wheat cultivars preinfected with barley yellow dwarf virus. J Agric Food Chem 53:6864–6869. doi: 10.1021/jf050831u. [DOI] [PubMed] [Google Scholar]
- 26.Zhou B, Li Y, Gillespie J, He G-Q, Horsley R, Schwarz P. 2007. Doehlert matrix design for optimization of the determination of bound deoxynivalenol in barley grain with trifluoroacetic acid (TFA). J Agric Food Chem 55:10141–10149. doi: 10.1021/jf0722957. [DOI] [PubMed] [Google Scholar]
- 27.Tran ST, Smith TK. 2011. Determination of optimal conditions for hydrolysis of conjugated deoxynivalenol in corn and wheat with trifluoromethanesulfonic acid. Anim Feed Sci Technol 163:84–92. doi: 10.1016/j.anifeedsci.2010.10.008. [DOI] [Google Scholar]
- 28.Britton HTS, Robinson RA. 1931. CXCVIII. Universal buffer solutions and the dissociation constant of veronal. J Chem Soc 1931:1456–1462. [Google Scholar]
- 29.Schweiger W, Pasquet JC, Nussbaumer T, Kovalsky Paris MP, Wiesenberger G, Macadré C, Ametz C, Berthiller F, Lemmens M, Saindrenan P, Mewes HW, Mayer KFX, Dufresne M, Adam G. 2013. Functional characterization of two clusters of Brachypodium distachyon UDP-glycosyltransferases encoding putative deoxynivalenol detoxification genes. Mol Plant Microbe Interact 26:781–792. doi: 10.1094/MPMI-08-12-0205-R. [DOI] [PubMed] [Google Scholar]
- 30.Berthiller F, Dall'Asta C, Schuhmacher R, Lemmens M, Adam G, Krska R. 2005. Masked mycotoxins: determination of a deoxynivalenol glucoside in artificially and naturally contaminated wheat by liquid chromatography-tandem mass spectrometry. J Agric Food Chem 53:3421–3425. doi: 10.1021/jf047798g. [DOI] [PubMed] [Google Scholar]
- 31.Michlmayr H, Schümann C, Barreira Braz Da Silva N, Kulbe K, Del Hierro A. 2010. Isolation and basic characterization of a β-glucosidase from a strain of Lactobacillus brevis isolated from a malolactic starter culture. J Appl Microbiol 108:550–559. doi: 10.1111/j.1365-2672.2009.04461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michlmayr H, Schümann C, Wurbs P, da Silva NMBB, Rogl V, Kulbe KD, Andrés M. 2010. A β-glucosidase from Oenococcus oeni ATCC BAA-1163 with potential for aroma release in wine: cloning and expression in E. coli. World J Microbiol Biotechnol 26:1281–1289. doi: 10.1007/s11274-009-0299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McIlvaine TC. 1921. A buffer solution for colorimetric comparison. J Biol Chem 49:183–186. [Google Scholar]
- 36.Harvey AJ, Hrmova M, De Gori R, Varghese JN, Fincher GB. 2000. Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins 41:257–269. doi:. [DOI] [PubMed] [Google Scholar]
- 37.Pozzo T, Pasten JL, Karlsson EN, Logan DT. 2010. Structural and functional analyses of β-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J Mol Biol 397:724–739. doi: 10.1016/j.jmb.2010.01.072. [DOI] [PubMed] [Google Scholar]
- 38.Suzuki K, Sumitani JI, Nam YW, Nishimaki T, Tani S, Wakagi T, Kawaguchi T, Fushinobu S. 2013. Crystal structures of glycoside hydrolase family 3 β-glucosidase 1 from Aspergillus aculeatus. Biochem J 452:211–221. doi: 10.1042/BJ20130054. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida E, Hidaka M, Fushinobu S, Koyanagi T, Minami H, Tamaki H, Kitaoka M, Katayama T, Kumagai H. 2010. Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 beta-glucosidase from Kluyveromyces marxianus. Biochem J 431:39–49. doi: 10.1042/BJ20100351. [DOI] [PubMed] [Google Scholar]
- 40.Karkehabadi S, Helmich KE, Kaper T, Hansson H, Mikkelsen N-E, Gudmundsson M, Piens K, Fujdala M, Banerjee G, Scott-Craig JS. 2014. Biochemical characterization and crystal structures of a fungal family 3 β-glucosidase, Cel3A from Hypocrea jecorina. J Biol Chem 289:31624–31637. doi: 10.1074/jbc.M114.587766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ketudat Cairns JR, Esen A. 2010. β-Glucosidases. Cell Mol Life Sci 67:3389–3405. doi: 10.1007/s00018-010-0399-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grandits M, Michlmayr H, Sygmund C, Oostenbrink C. 2013. Calculation of substrate binding affinities for a bacterial GH78 rhamnosidase through molecular dynamics simulations. J Mol Catal B Enzym 92:34–43. doi: 10.1016/j.molcatb.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marana SR. 2006. Molecular basis of substrate specificity in family 1 glycoside hydrolases. IUBMB Life 58:63–73. doi: 10.1080/15216540600617156. [DOI] [PubMed] [Google Scholar]
- 44.Turner P. 2010. Deoxynivalenol and nivalenol occurrence and exposure assessment. World Mycotoxin J 3:315–321. doi: 10.3920/WMJ2010.1242. [DOI] [Google Scholar]
- 45.Goswami RS, Kistler HC. 2004. Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5:515–525. doi: 10.1111/j.1364-3703.2004.00252.x. [DOI] [PubMed] [Google Scholar]
- 46.Yli-Mattila T. 2010. Ecology and evolution of toxigenic Fusarium species in cereals in northern Europe and Asia. J Plant Pathol 92:7–18. [Google Scholar]
- 47.Yang L, Van der Lee T, Yang X, Yu D, Waalwijk C. 2008. Fusarium populations on Chinese barley show a dramatic gradient in mycotoxin profiles. Phytopathology 98:719–727. doi: 10.1094/PHYTO-98-6-0719. [DOI] [PubMed] [Google Scholar]
- 48.Nathanail AV, Helkama J, Malachova A, Jestoi M, Varga E, Michlmayr H, Adam G, Sieviläinen E, Berthiller F, Peltonen K. 3 May 2015. Simultaneous determination of major type A and B trichothecenes, zearalenone and their masked metabolites on Finnish cereal grains with novel validated liquid chromatography-tandem mass spectrometric method. Anal Bioanal Chem doi: 10.1007/s00216-015-8676-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varga E, Wiesenberger G, Hametner C, Ward TJ, Dong Y, Schöfbeck D, McCormick S, Broz K, Stückler R, Schuhmacher R, Krska R, Kistler HC, Berthiller F, Adam G. 18 November 2014. New tricks of an old enemy: isolates of Fusarium graminearum produce a type A trichothecene mycotoxin. Environ Microbiol doi: 10.1111/1462-2920.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ruprich J, Ostry V. 2008. Immunochemical methods in health risk assessment: cross reactivity of antibodies against mycotoxin deoxynivalenol with deoxynivalenol-3-glucoside. Cent Eur J Public Health 16:34. [DOI] [PubMed] [Google Scholar]
- 51.Tangni EK, Motte J-C, Callebaut A, Pussemier L. 2010. Cross-reactivity of antibodies in some commercial deoxynivalenol test kits against some fusariotoxins. J Agric Food Chem 58:12625–12633. doi: 10.1021/jf103025e. [DOI] [PubMed] [Google Scholar]
- 52.Van Den Broek LA, Voragen AG. 2008. Bifidobacterium glycoside hydrolases and (potential) prebiotics. Innovat Food Sci Emerg Technol 9:401–407. doi: 10.1016/j.ifset.2007.12.006. [DOI] [Google Scholar]
- 53.Ávila M, Hidalgo M, Sánchez-Moreno C, Pelaez C, Requena T, de Pascual-Teresa S. 2009. Bioconversion of anthocyanin glycosides by Bifidobacteria and Lactobacillus. Food Res Int 42:1453–1461. doi: 10.1016/j.foodres.2009.07.026. [DOI] [Google Scholar]
- 54.Raimondi S, Roncaglia L, De Lucia M, Amaretti A, Leonardi A, Pagnoni UM, Rossi M. 2009. Bioconversion of soy isoflavones daidzin and daidzein by Bifidobacterium strains. Appl Microbiol Biotechnol 81:943–950. doi: 10.1007/s00253-008-1719-4. [DOI] [PubMed] [Google Scholar]
- 55.Yang L, Akao T, Kobashi K, Hattori M. 1996. Purification and characterization of a novel sennoside-hydrolyzing beta-glucosidase from Bifidobacterium sp. strain SEN, a human intestinal anaerobe. Biol Pharm Bull 19:705–709. doi: 10.1248/bpb.19.705. [DOI] [PubMed] [Google Scholar]
- 56.Dan S, Marton I, Dekel M, Bravdo B-A, He S, Withers SG, Shoseyov O. 2000. Cloning, expression, characterization, and nucleophile identification of family 3, Aspergillus niger β-glucosidase. J Biol Chem 275:4973–4980. doi: 10.1074/jbc.275.7.4973. [DOI] [PubMed] [Google Scholar]
- 57.van Peij NN, Brinkmann J, Vršanská M, Visser J, de Graaff LH. 1997. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur J Biochem 245:164–173. doi: 10.1111/j.1432-1033.1997.00164.x. [DOI] [PubMed] [Google Scholar]
- 58.Varghese JN, Hrmova M, Fincher GB. 1999. Three-dimensional structure of a barley β-d-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7:179–190. doi: 10.1016/S0969-2126(99)80024-0. [DOI] [PubMed] [Google Scholar]
- 59.Tsujibo H, Takada C, Tsuji A, Kosaka M, Miyamoto K, Inamori Y. 2001. Cloning, sequencing, and expression of the gene encoding an intracellular β-d-xylosidase from Streptomyces thermoviolaceus OPC-520. Biosci Biotechnol Biochem 65:1824–1831. doi: 10.1271/bbb.65.1824. [DOI] [PubMed] [Google Scholar]
- 60.Lin L-L, Rumbak E, Zappe H, Thomson JA, Woods DR. 1990. Cloning, sequencing and analysis of expression of a Butyrivibrio fibrisolvens gene encoding a β-glucosidase. J Gen Microbiol 136:1567–1576. doi: 10.1099/00221287-136-8-1567. [DOI] [PubMed] [Google Scholar]
- 61.Ohmiya K, Takano M, Shimizu S. 1990. DNA sequence of a beta-glucosidase from Ruminococcus albus. Nucleic Acids Res 18:671. doi: 10.1093/nar/18.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Malachová A, Štočková L, Wakker A, Varga E, Krska R, Michlmayr H, Adam G, Berthiller F. 2015. Critical evaluation of indirect methods for the determination of deoxynivalenol and its conjugated forms in cereals. Anal Bioanal Chem doi: 10.1007/s00216-015-8793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.