Abstract
Human astroviruses (HAstVs) are a common etiological agent of infantile gastroenteritis. Recent studies revealed that novel astrovirus (AstV) strains of the MLB clade (MLB-AstVs) and VA clade (VA-AstVs), which are genetically distinct from the classic HAstVs, are circulating in the human population. In the present study, we quantified classic HAstVs as well as carried out a genetic analysis of classic and novel HAstVs in wastewater in Japan. The concentration of classic HAstVs in the influent water samples ranged from 104 to 105 copies per liter, and the amount removed by wastewater treatment was determined to be 2.4 ± 0.3 log10. Four types of classic HAstV strains (HAstV types 1, 2, 5, and 4/8) as well as novel AstV strains belonging to the MLB-2, VA-1, and VA-2 clades were identified using reverse transcription-PCR (RT-PCR) assays, including assays newly developed for the detection of strains of the MLB and VA clades, followed by cloning and nucleotide sequencing. Our results suggest that genetically diverse AstV strains are circulating among the human population in Japan. The newly developed (semi)nested RT-PCR assays for these novel AstV clades are useful to identify and characterize the novel AstVs in environmental waters.
INTRODUCTION
Human astroviruses (HAstVs) are considered one of the most important causes of infantile gastroenteritis worldwide (1). The infections typically occur sporadically, and the cause of 2 to 10% of total viral gastroenteritis cases is attributed to HAstVs (1–6). Astroviruses (AstVs) possess nonenveloped, icosahedrally shaped virions containing approximately 6,800 nucleotides (nt) of single-stranded positive-sense RNA (7). It has been reported that the diameter of AstV particles shed in fecal specimens ranges from 28 to 31 nm, while the diameter of those grown in cell culture is 41 nm (8). Their viral genome consists of three open reading frames (ORFs), namely, ORF1a, ORF1b, and ORF2, encoding the serine protease, RNA-dependent RNA polymerase, and capsid proteins, respectively (7). HAstVs are antigenically or genetically classified into eight different types (HAstV type 1 [HAstV-1] to HAstV-8). Previous studies revealed that HAstV-1 is the most prevalent type among infected individuals and in the environment, followed by HAstV-2, -3, -4, and -5 (3, 5, 9–13).
Because HAstV can be excreted in the feces of infected individuals at a high concentration (up to 1015 particles/g) (14), examination of municipal wastewater samples could be an effective approach to understand the actual prevalence and epidemiology of these viruses (15, 16). In spite of their importance as enteric pathogens, the occurrence and other characteristics of HAstVs in water compared to those of other enteric viruses, such as noroviruses, rotaviruses, and adenoviruses, have not been well documented (17, 18). Due to the scarcity of knowledge, the fate of HAstVs in water environments and wastewater treatment processes is not well understood.
Recent studies based on viral metagenomic analysis have identified some novel AstVs, namely, clade MLB AstVs (MLB-AstVs) and clade VA (HMO) AstVs (VA-AstVs), in human feces (19, 20). The novel AstV strains are genetically distinct from the classic HAstV strains (20–22); in fact, VA-AstVs are genetically more closely related to mink/ovine AstVs than to classic HAstVs (20, 22). To date, MLB-AstVs have been divided into 3 genotypes (MLB-AstV genotype 1 [MLB-AstV-1] to MLB-AstV-3) and VA-AstVs have been divided into 4 genotypes (VA-AstV genotype 1 [VA-AstV-1] to VA-AstV-4) (7, 20, 23, 24). It has been suggested that VA-AstV-1 and MLB-AstV-1 are associated with gastroenteritis outbreaks (23, 25), but the prevalence and etiological role of the novel AstV strains in humans remain unclear (26, 27). Some recent studies reported on the frequent detection of the novel AstVs in fecal samples (24, 26). This prompted us to investigate the occurrence of HAstVs in wastewater, which contains viruses shed from all populations regardless of symptoms.
On the basis of the background presented above, we performed the present study to investigate the occurrence and genetic diversity of classic HAstVs as well as novel AstVs in wastewater in Japan. Classic HAstV genomes were quantified by real-time quantitative PCR (qPCR) assays to assess the abundance of classic HAstVs at a wastewater treatment plant (WWTP) and the level of reduction of classic HAstVs by the wastewater treatment process. In addition, we employed nested and seminested PCR assays, including assays newly designed for the detection of novel AstVs, to amplify the viral genomes. The strains were further characterized on the basis of the sequence of the 3′ end of ORF1b or the sequence of the 5′ end of ORF2.
MATERIALS AND METHODS
Collection and concentration of wastewater samples.
Wastewater samples were collected monthly from October 2007 to March 2008 at a WWTP in an urban area in Japan as described in our previous study (28). The characteristics of the WWTP are described in Table S1 in the supplemental material. This WWTP employs chlorination and sand filtration after the secondary treatment. During the 6-month study period, a total of 24 samples were collected from four locations in the treatment train: influent, after secondary treatment, after chlorination, and after sand filtration (effluent). The samples (100 ml for the influent samples and 1,000 ml for each of the other samples) were concentrated using an electronegative membrane (type HA; diameter, 90 mm; pore size, 0.45 μm; Millipore, Tokyo, Japan) and a centrifugal ultrafiltration device (Centriprep YM-50; Millipore) to obtain a final volume of approximately 0.7 ml, as previously described (29). As a process control, 2.2 × 107 genome copies of murine norovirus (MNV) were inoculated into 140 μl of the concentrate.
Viral RNA extraction and RT.
Viral RNA was extracted from 140 μl of the concentrated sample inoculated with MNV using a QIAamp viral RNA minikit (Qiagen, Hilden, Germany) to obtain a final volume of 60 μl, according to the manufacturer's protocol. The reverse transcription (RT) reaction was performed using a high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA). Briefly, 10 μl of extracted RNA was added to 10 μl of an RT mixture containing 2 μl of 10× reverse transcription buffer, 0.8 μl of deoxynucleoside triphosphates (dNTPs), 2 μl of 10× random hexamers, 50 U of MultiScribe reverse transcriptase, and 20 U of RNase inhibitor. The RT reaction mixture was incubated at 25°C for 10 min, followed by 37°C for 120 min and, finally, 85°C for 5 min to inactivate the enzyme.
Quantification of classic HAstV genomes by qPCR.
TaqMan-based qPCR for classic HAstVs was performed with a LightCycler 480 real-time PCR instrument II (Roche Diagnostics, Mannheim, Germany) as described previously (30). Briefly, 5 μl of cDNA was mixed with 20 μl of a reaction buffer containing 12.5 μl of a 2× TaqMan gene expression master mix (Applied Biosystems), 400 nM (each) sense and antisense primers, 100 nM TaqMan minor groove binder (MGB) probe, and nuclease-free water. Real-time PCR was performed under the following thermal cycling conditions: initial denaturation at 95°C for 15 min to activate the DNA polymerase, followed by 50 cycles of amplification with denaturation at 94°C for 15 s and annealing and extension at 62°C for 60 s. Fluorescence readings were collected and analyzed with LightCycler 480 software (version 1.5; Roche Diagnostics). The genome copy numbers of classic HAstVs were determined on the basis of a standard curve prepared with 10-fold serial dilutions of plasmid DNA containing an insert of the 5′ end of the ORF2 region of classic HAstV-1 (Oxford strain) at concentrations ranging from 105 to 101 genome copies per reaction mixture on the basis of the plasmid DNA concentration, determined by measuring the optical density at 260 nm by using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Kanagawa, Japan). Two PCR tubes were used for each sample, and the average cDNA copy numbers for the two tubes were used for subsequent calculations. Negative controls were included to avoid false-positive results due to cross-contamination. No false-positive qPCR signal was observed. The qPCR assay was designed to detect only classic HAstV strains (30). In fact, numerous mismatches between the nucleotide sequences of the novel AstVs and the primer and TaqMan probe sequences were observed (see Table S2 in the supplemental material), indicating that the assay does not detect the novel AstVs.
Determination of viral RNA extraction and RT-qPCR efficiency.
The inoculated MNV was recovered by RT-qPCR to determine the virus detection efficiency, as described previously (28). Briefly, the RNA extract was subjected to RT and qPCR as described above. A primer set and TaqMan probe designed by Kitajima et al. (31) were used for the reactions. Plasmid DNA containing an insert of the target sequence was used for generating a standard curve as described above. The virus detection efficiency was determined by comparing the observed genome copy number in a sample to that in a positive control prepared by inoculating MNV into Milli-Q water. If the ratio between the amount of MNV quantified from a sample and that quantified from the positive-control sample was less than 10%, the quantification was regarded as unreliable due to low RNA extraction and/or RT-qPCR efficiencies. Since MNV was spiked to determine the viral RNA extraction and RT-qPCR efficiencies, the classic HAstV concentration obtained was not corrected even if the quantification was determined to be unreliable.
Design of PCR primers for MLB-AstV and VA-AstV detection.
A set of seminested PCR primers for amplification of a 644-nt fragment in the 3′ end of ORF1b of MLB-AstVs was designed on the basis of the nucleotide sequence alignment of 11 MLB-AstV strains in the GenBank database (see Fig. S1A in the supplemental material). Similarly, a set of nested PCR primers for amplification of a 618-nt fragment in the 5′ end of ORF2 of VA-AstVs was designed on the basis of the nucleotide sequence alignment of 5 VA-AstV strains in the GenBank database (see Fig. S1B in the supplemental material). The accession numbers of the novel AstVs used for primer design were as follows: ET065575, ET065582, FJ222451, FJ227124, FJ227125, FJ227127, GQ502188, GQ502189, GQ502190, GQ502191, and GQ502192 for MLB-AstVs and FJ973620, GQ502193, GQ502194, GQ502195, and GQ502196 for VA-AstVs. The newly designed primers are shown in Table 1. A nucleotide BLAST search of each primer showed no significant homology to nontarget sequences (data not shown).
TABLE 1.
Primers and TaqMan probe used in this study
| Target | Assay | Primer or probe name | Function | Sequence (5′ → 3′)a | Locationc | Reference or source |
|---|---|---|---|---|---|---|
| Classic HAstV | qPCR | HuAstVf2240 | Sense primer | CAGGTAACTGTTGAGGTCA | 4349–4367 | 30 |
| qPCR | HuAstVf2140 | Sense primer | GCAAGTYACTGTTGAGGTCA | 4348–4367 | 30 | |
| qPCR | HuAstVf2239T4 | Sense primer | GAAGTCACTGTGGAGGTC | 4349–4366 | 30 | |
| qPCR | HuAstVr | Antisense primer | GTTTWGGTCCTGTGACACC | 4544–4562 | 30 | |
| qPCR | HuAstV/1-8/TP | TaqMan probe | FAM-TTACGGACACGYTGA-MGB-NFQb | 4501–4515 | 30 | |
| Nested PCR (first round) | AHAstVF1 | Sense primer | AATCACTCCATGGGAAGCTCCT | 4139–4160 | 33 | |
| Nested PCR (first round) | AHAstVR1 | Antisense primer | CCTARCGCYTGCACDGG | 4697–4713 | 33 | |
| Nested PCR (second round) | AHAstVF2 | Sense primer | CAGAAGAGCAACTCCATCGCAT | 4280–4301 | 33 | |
| Nested PCR (second round) | AHAstVR2 | Antisense primer | GTRCTYCCWGTAGCRTCCTTAAC | 4664–4686 | 33 | |
| MLB-AstV | Seminested PCR (first round) | SF0073 | Sense primer | GAYTGGACWCGATTTGATGGTAC | 3110–3132 | 25 |
| Nested PCR (first round) | AHMLBR1 | Antisense primer | CAGGYTTAGGCCCAGTTGTA | 4016–4035 | This study | |
| Seminested PCR (second round) | AHMLBR2 | Antisense primer | CGAGTGAAGCGCCTTGGTAAG | 3778–3798 | This study | |
| VA-AstV | Nested PCR (first round) | AHVAF1 | Sense primer | TATGGGAARCTCCTTTGCTAYCGC | 4025–4048 | This study |
| Nested PCR (first round) | AHVAR1 | Antisense primer | ARTTTCTTGACAAACCACCAWCC | 5211–5233 | This study | |
| Nested PCR (second round) | AHVAF2 | Sense primer | ATGCTGGATAGRCTTTGGAGGG | 4172–4193 | This study | |
| Nested PCR (second round) | AHVAR2 | Antisense primer | SCTCCCTCTTCATTKGTRTCTGT | 4812–4834 | This study |
The mixed bases in degenerate primer and probe are as follows: Y, C or T; W, for A or T; R, A or G; D, A, G, or T; S, G or C; K, T or G.
FAM, 6-carboxyfluorescein; MGB, minor groove binder; NFQ, nonfluorescent quencher.
(Semi)nested PCR.
For amplification of each AstV target, a nested PCR assay for classic HAstVs and the newly designed (semi)nested PCR assays for novel AstVs (Table 1) were performed separately using KOD plus (version 2; Toyobo, Osaka, Japan), a high-fidelity PCR polymerase. Briefly, the first round of PCR amplification was performed in 50 μl of a reaction mixture containing 5 μl of cDNA, 1.0 U of KOD plus polymerase, 5 μl of 10× buffer for KOD plus polymerase (version 2), 200 nM dNTPs, 1.5 μl of 25 mM MgSO4, 400 nM (each) sense and antisense primers, and nuclease-free water. Amplifications of all three target genes were performed on a Verti 96-well thermal cycler (Applied Biosystems) under the following thermal cycling conditions: initial denaturation at 94°C for 2 min, followed by 40 cycles of amplification with denaturation at 98°C for 10 s, primer annealing at 54°C for 30 s, and an extension reaction at 68°C for 1 min and then a final extension at 68°C for 7 min. The second round of PCR was performed in a 50-μl reaction mixture containing 2 μl of the product of the first round of PCR amplification, 1.0 U of KOD plus polymerase, 5 μl of 10× buffer for KOD plus polymerase (version 2), 200 nM dNTPs, 1.5 μl of 25 mM MgSO4, 400 nM (each) sense and antisense primers, and nuclease-free water. PCR amplification was performed under the following thermal cycling conditions: initial denaturation at 94°C for 2 min, followed by 30 cycles of amplification with denaturation at 98°C for 10 s, primer annealing at 54°C for 30 s, and extension reaction at 68°C for 1 min and then a final extension at 68°C for 7 min.
Cloning, sequencing, and phylogenetic analysis.
The products from the second PCR amplification were separated by electrophoresis in a 1.5% agarose gel and visualized under a UV lamp after ethidium bromide staining. PCR products of the expected size (i.e., approximately 362 bp, 645 bp, and 618 bp for classic HAstVs, MLB-AstVs, and VA-AstVs, respectively) were excised from the gel and purified with a QIAquick gel extraction kit (Qiagen). The purified products were cloned into a Zero Blunt TOPO pCR 2.1 vector (Invitrogen, Carlsbad, CA), and the plasmid constructs were then transformed into Escherichia coli One Shot TOP10 chemically competent cells (Invitrogen). The transformants were incubated at 37°C on an LB agar plate containing 20 μg/ml of kanamycin. Six to eight colonies were selected, and insertion sizes were checked by direct colony PCR amplification using KOD plus polymerase (version 2) and an M13 forward (5′-GTAAAACGACGGCCAG-3′) and reverse (5′-CAGGAAACAGCTATGAC-3′) primer set. The PCR products were purified with a PCR purification kit (Qiagen), and both strands were sequenced with a BigDye cycle sequencing kit (version 3.1; Applied Biosystems) and a 3130 genetic analyzer (Applied Biosystems). Nucleotide sequences were assembled using the program Sequencher (version 4.2.2; Gene Codes Corporation, Ann Arbor, MI) and aligned by use of the Clustal W program (version 1.83; http://clustalw.ddbj.nig.ac.jp/top-e.html). The distances were calculated using Kimura's two-parameter method (32), and phylogenetic dendrograms from a bootstrap analysis with 1,000 replicates were generated by the neighbor-joining method.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study were deposited in GenBank under accession numbers LC009636 to LC009664 (classic HAstVs), LC009665 to LC009677 (MLB-AstVs), and LC009678 to LC009684 (VA-AstVs).
RESULTS
Quantification of classic HAstV genomes in wastewater by RT-qPCR.
The virus detection efficiencies obtained using inoculated MNV are summarized in Table S2 in the supplemental material. All samples showed at least 30% RNA extraction and RT-qPCR efficiencies, and these values are acceptably high, indicating that AstVs were detected at sufficient efficiencies.
All influent samples (n = 6) were positive for classic HAstVs, with concentrations ranging from 1.9 × 104 (October 2007) to 7.5 × 105 (December 2007) copies per liter (Fig. 1). The partially or fully treated wastewater samples were also positive for classic HAstVs, except for the samples collected in October 2007, when the influent samples showed the lowest concentration. The concentrations of classic HAstV genomes in secondary treated and chlorinated wastewater samples ranged from 102 to 103 copies per liter, and those in effluent samples ranged from 101 to 102 copies per liter, except for the samples negative for classic HAstVs. Among the samples positive for classic HAstVs, the geometric mean reduction of classic HAstV genomes brought about by the whole wastewater treatment process was determined to be 2.4 ± 0.3 log10.
FIG 1.
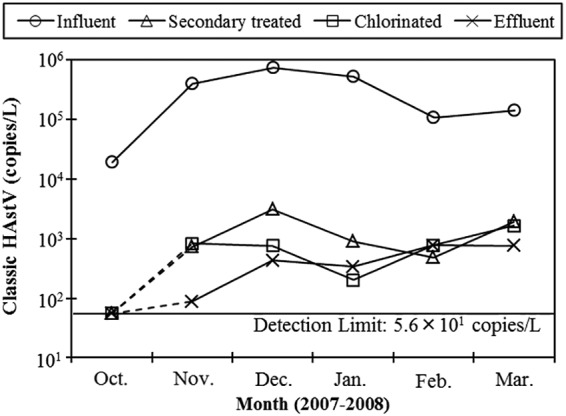
Concentrations of classic HAstVs in wastewater samples determined by RT-qPCR. The results for samples negative for classic HAstVs (secondary treated, chlorinated, and effluent samples collected in October 2007) were plotted on a line indicating the detection limit (5.6 × 101 copies per liter) and are connected by dashed lines.
Genetic diversity of classic HAstVs and MLB- and VA-AstVs in wastewater.
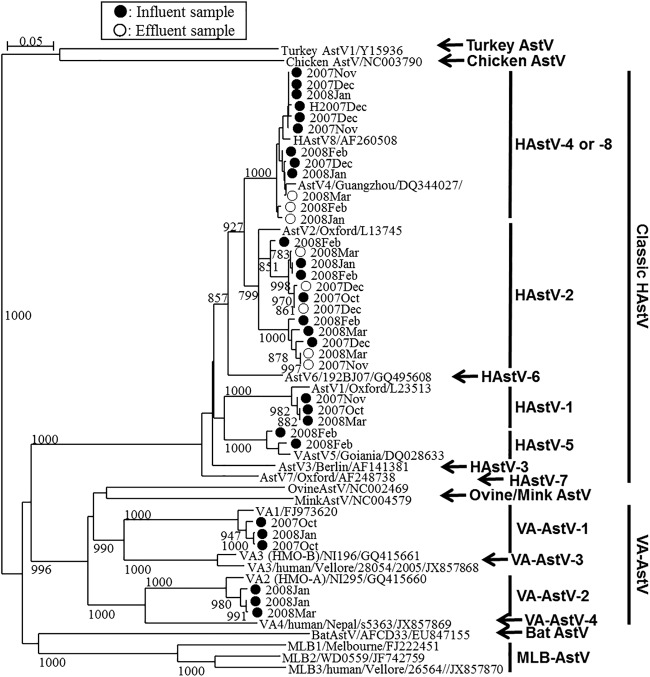
Among the influent and effluent samples tested (one sample of each type per month), classic HAstV-positive PCR products were obtained from all the influent samples and 4 effluent samples by nested RT-PCR; no PCR products were obtained from the effluent samples in October and November 2007, and of these samples, the October 2007 sample tested negative by qPCR as well, but the November 2007 sample was qPCR positive (Fig. 1). On the basis of the results of nucleotide sequencing analysis, a total of 29 classic HAstV strains were identified in the wastewater samples (Fig. 2). Type 2 and type 4/8 (types 4 and 8 could not be distinguished [33]) were found in both influent and effluent wastewater samples at relatively high frequencies. Type 1 and type 5 were found in 2 and 1 influent wastewater samples, respectively, but these types were not found in effluent wastewater samples (Fig. 2).
FIG 2.
Phylogenetic tree of classic HAstV and VA-AstV strains obtained using an approximately 360-nt fragment at the 5′ end of ORF2. Strains detected from influent samples (marked ●) and strains detected from effluent samples (marked ○), along with corresponding historical AstV strains, including MLB-AstVs and other animal AstVs strains, are included. Bootstrap values greater than 700 among 1,000 replicates are shown on each branch.
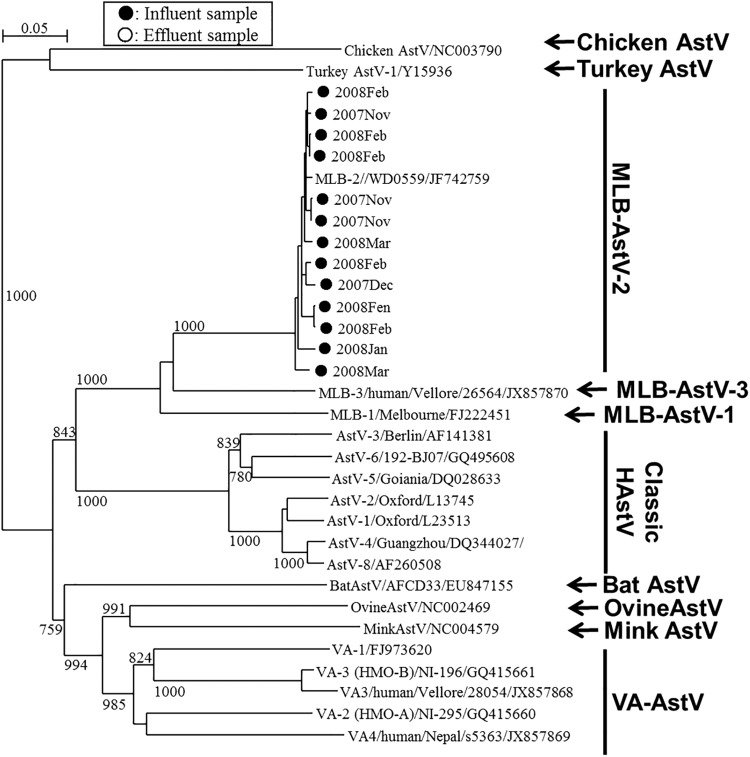
MLB- and VA-AstVs were identified in 5 and 3 influent samples, respectively, but they were not detected in any of the effluent samples (Fig. 2 and 3 and Table 2). Nucleotide sequence analysis of the 3′ end of ORF1b (MLB-AstVs) and the 5′ end of ORF2 (VA-AstVs) revealed that all the MLB-AstVs identified in the present study (13 strains) were closely related to type 2 and the VA-AstVs (6 strains) were classified as type 1 or type 2 (Fig. 2 and 3). An influent sample collected in January 2008 contained both type 1 and type 2 VA-AstVs.
FIG 3.
Phylogenetic tree of MLB-AstVs strains obtained using a 645-nt fragment at the 3′ end of ORF1b. Strains detected from influent samples (marked ●) along with corresponding historical AstV strains, including VA-AstVs and other animal AstVs strains, are included. Bootstrap values greater than 700 among 1,000 replicates are shown on each branch.
TABLE 2.
Astrovirus genotypes identified in wastewater samples
| Mo and yr of sample collection | Genotype(s) identified in: |
|||||
|---|---|---|---|---|---|---|
| Influent |
Effluent |
|||||
| Classic HAstV | MLB-AstV | VA-AstV | Classic HAstV | MLB-AstV | VA-AstV | |
| Oct. 2007 | 2, 4/8a | NDb | 1 | ND | ND | ND |
| Nov. 2007 | 1, 4/8 | 2 | ND | ND | ND | ND |
| Dec. 2007 | 2, 4/8 | 2 | ND | 2 | ND | ND |
| Jan. 2008 | 4/8 | 2 | 1, 2 | 2, 4/8 | ND | ND |
| Feb. 2008 | 2, 4/8, 5 | 2 | ND | 4/8 | ND | ND |
| Mar. 2008 | 1, 2 | 2 | 2 | 2, 4/8 | ND | ND |
| Positive ratioc | 6/6 | 5/6 | 3/6 | 4/6 | 0/6 | 0/6 |
Type 4 or 8.
ND, not detected by RT-qPCR or RT-(semi)nested PCR.
The data represent the number of samples in which the indicated genotype(s) was detected/total number of samples tested.
DISCUSSION
Classic HAstVs are responsible for 2 to 10% of cases of infantile viral gastroenteritis and are recognized to be important enteric pathogens (1). The goal of the present study was to reveal the prevalence and genetic diversity of human AstVs, including AstVs of emerging clades MLB and VA, in municipal wastewater. Since cell culture assays for classic HAstVs are time-consuming, less sensitive than RT-PCR, and not specific, RT-PCR-based assays are more effective for determination of the occurrences of classic HAstVs. In general, the amplification efficiency/sensitivity of PCR for environmental virus detection can be affected by several factors, such as the primer and probe sequences used and the sample matrix (34, 35). Because environmental samples contain multiple virus strains, selection of a broadly reactive RT-PCR assay is important to obtain representative results.
We first investigated the occurrence of classic HAstVs in wastewater by using qPCR assays. We reported the concentration of classic HAstVs in wastewater samples determined by a TaqMan-based qPCR assay for classic HAstVs (developed by Le Cann et al. [36]) in our previous study (28). This assay has been widely used for the detection and quantification of classic HAstV genomes in clinical as well as environmental samples (37–39); however, we found that the primer and probe sequences of the assay (targeting the 3′ end of ORF2) have considerable mismatches with the sequences of classic HAstV strains, based on the sequence alignment of a total of 281 classic HAstV nucleotide sequences (33). This prompted us to consider another TaqMan-based assay reported by Yokoi and Kitahashi (30), which targets the 5′ end of ORF2 and shows substantially fewer mismatches, for the more accurate detection/quantification of genetically diverse classic HAstVs (see Fig. S2 in the supplemental material). Using this assay, we examined the same wastewater sample set used in the previous study and obtained a positive ratio (21 positive samples out of 24 samples tested) higher than that obtained in our previous study (11 positive samples out of 24 samples tested) using the less broadly reactive assay (36). Furthermore, the concentration observed in the present study was 0.89 ± 0.30 log10 higher than that observed in the previous study for each sample (see Fig. S3 in the supplemental material). The assay targeting the 5′ end of ORF2 (30) showed improved detection and quantification compared to those of the other assay targeting the 3′ end of ORF2 (36). The genetic sequences on the 5′ end of the ORF2 region are more conserved than those on the 3′ end of the ORF2 region among classic HAstV strains (40). Thus, the concentration of classic HAstVs determined in our previous study using the assay targeting the 3′ end of ORF2 was underestimated. The concentrations of classic HAstVs in the influent samples (1.9 × 104 to 7.5 × 105 copies per liter, as determined by the assay targeting the 5′ end of ORF2) were similar to those of genotype II (GII) noroviruses (8.9 × 104 to 9.5 × 105 copies per liter), the leading cause of viral gastroenteritis, according to the findings of our previous study which analyzed the same sample set analyzed in the present study (28). This suggests that classic HAstVs are widespread in humans in Japan, at least during the autumn and winter seasons. Classic HAstV infections are observed sporadically in general (1), while GII noroviruses frequently cause gastroenteritis outbreaks in winter (41). This indicates that there were considerable numbers of underreported cases that caused mild or asymptomatic infections in the sewage service area. The improved detection/quantification also implies that the risk of classic HAstV infection via water may be higher than was previously thought. It is possible that the risk of infection with HAstVs is comparable to that with other widespread viruses causing gastroenteritis, such as norovirus, rotavirus, and adenovirus.
The reduction of classic HAstVs by the conventional activated sludge process (2.4 ± 0.3 log10) was comparable to that of many of other enteric viruses tested in our previous study (28). A difference in the stability of different AstVs in the environment has been suggested. For example, Morsy El-Senousy et al. (42) reported that genogroup B classic HAstVs (types 6 and 7) are more resistant to wastewater treatments than other classic HAstV genogroups. In our current study, however, this genogroup was not detected in wastewater (Fig. 2), which agrees with the findings of previous clinical studies reporting that this genogroup is not prevalent among gastroenteritis patients in Japan (10, 43). Future studies should investigate using type/clade-specific qPCR assays to detect differences in the levels of reduction of different AstV genotypes/groups, including the novel clades, attained with treatment.
In our previous study, we designed nested PCR primers targeting the conserved 5′ end of the ORF2 region (33). The assay showed sensitivity comparable to that of the broadly reactive RT-qPCR assay in this study (Fig. 2 and Table 2). Classic HAstV-1 has been the type that has been the most frequently found in clinical samples worldwide (10, 44, 45), as well as in urban wastewater and surface water samples (9, 46). In the present study, however, classic HAstV-1 was detected in only 2 influent samples, while classic HAstV-2 and classic HAstV-4/8 were more frequently detected in both influent and effluent samples. These results also suggest that classic HAstV infections, which may be caused by classic HAstV-2 and -4/8 in the study area, are underreported.
We developed seminested PCR primers targeting the 3′ end of ORF1b and nested PCR primers targeting the 5′ end of ORF2 of MLB- and VA-AstV, respectively, for the specific detection of each clade. We found that each region was conserved among genetically diverse MLB- and VA-AstV genotypes (see Fig. S1 in the supplemental material). We identified type 2 MLB-AstVs and type 1 and 2 VA-AstV strains, showing the considerable genetic diversity of AstVs in the wastewater samples (Fig. 2 and 3). Even though MLB-AstVs were detected in the influent samples at a relatively high positive ratio (5 positive samples of 6 samples tested), they were not detected in any of the effluent samples. On the contrary, classic HAstVs showed a high positive ratio in both influent samples (6 positive samples of 6 samples tested) and effluent samples (4 or 5 positive samples of 6 samples tested). This implies that MLB-AstVs are present at lower concentration in the influent samples and/or that MLB-AstVs are more readily reduced by the wastewater treatments. For further investigation of the occurrence of the novel AstVs and their levels of reduction with wastewater treatment, the application of quantitative assays which have not yet been developed to a number of samples would be indispensable.
The presence of these novel AstVs has been documented in several countries, including Australia, China, India, Nepal, the Netherlands, Nigeria, Pakistan, and the United States, but not in Japan (19, 20, 22, 23, 47, 48). We identified MLB-AstVs in all influent samples collected between November 2007 and March 2008. This trend was similar to that for the classic HAstVs, which showed the lowest concentration in October 2007, indicating that the epidemiological trends for MLB-AstVs and classic HAstVs are similar. In contrast, VA-AstVs were less frequently detected in the influent samples, suggesting that VA-AstVs are not as prevalent as classic HAstVs or MLB-AstVs in humans. Similarly, clinical studies investigating infantile diarrheal samples in India, the United States, Egypt, and China reported the more frequent occurrence of MLB-AstVs than VA-AstVs (22, 49, 50). A previous clinical study reported the less frequent detection of MLB-AstV, but the study investigated samples from adults with diarrhea and children without diarrhea (20). These observations suggest that MLB-AstVs may be more prevalent than VA-AstVs among children with diarrheal disease. All MLB-AstV strains found in this study were classified as type 2 strains, which have been found in nasopharyngeal swab and plasma samples obtained from febrile children and have been hypothesized to infect extraenteric tissues (7, 50, 51). Our results suggest that the prevalence of nonenteric MLB-AstVs as well as enteric viruses can be assumed by investigating wastewater samples.
The occurrence of the novel AstVs in wastewater samples strongly implies their circulation in Japan and potential waterborne transmission. A previous study suggested an association of VA-AstVs with gastroenteritis outbreaks, although the pathogenicity of the novel AstVs has not been well documented to date (23). Finkbeiner et al. (22) pointed out that the novel AstVs could be the cause of gastroenteritis diseases whose etiologies are undetermined. Further studies are needed for characterization of the novel AstVs with respect to their pathogenicity, epidemiology, and fate in the water environment.
In summary, we identified genetically diverse AstV strains, including emerging MLB- and VA-AstV strains, in wastewater in Japan. To our knowledge, this is the first study describing the occurrence and genetic diversity of MLB- and VA-AstVs in wastewater. MLB-AstVs seemed to be prevalent during the autumn and winter seasons, while the seasonal prevalence of VA-AstVs remains unclear. Most AstV strains identified in the wastewater samples were clinically undetermined types, suggesting that these strains may be causing underreported (mild or asymptomatic) infections and that AstV strains which were found to be circulating in the study area are more genetically diverse than was previously appreciated.
Our results demonstrate the importance of investigation of water samples to explore the actual prevalence of AstVs among human populations. Future studies, such as a yearlong surveillance and quantitative detection of the novel AstVs, are required to obtain a better understanding of the epidemiology of AstVs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Core Research for Evolutional Science and Technology (CREST).
We thank Jason Torrey at The University of Tokyo for critically reviewing the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00563-15.
REFERENCES
- 1.Guix S, Bosch A, Pintó RM. 2005. Human astrovirus diagnosis and typing: current and future prospects. Lett Appl Microbiol 41:103–105. doi: 10.1111/j.1472-765X.2005.01759.x. [DOI] [PubMed] [Google Scholar]
- 2.Andreasi MSA, Cardoso DDDP, Fernandes SM, Tozetti IA, Borges AMT, Fiaccadori FS, Santos RAT, Souza M. 2008. Adenovirus, calicivirus and astrovirus detection in fecal samples of hospitalized children with acute gastroenteritis from Campo Grande, MS, Brazil. Mem Inst Oswaldo Cruz 103:741–744. doi: 10.1590/S0074-02762008000700020. [DOI] [PubMed] [Google Scholar]
- 3.Dai Y-C, Xu Q-H, Wu X-B, Hu G-F, Tang Y-L, Li J-D, Chen Q, Nie J. 2010. Development of real-time and nested RT-PCR to detect astrovirus and one-year survey of astrovirus in Jiangmen City, China. Arch Virol 155:977–982. doi: 10.1007/s00705-010-0664-6. [DOI] [PubMed] [Google Scholar]
- 4.Khamrin P, Okame M, Thongprachum A, Nantachit N, Nishimura S, Okitsu S, Maneekarn N, Ushijima H. 2011. A single-tube multiplex PCR for rapid detection in feces of 10 viruses causing diarrhea. J Virol Methods 173:390–393. doi: 10.1016/j.jviromet.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 5.Mustafa H, Palombo EA, Bishop RF. 2000. Epidemiology of astrovirus infection in young children hospitalized with acute gastroenteritis in Melbourne, Australia, over a period of four consecutive years, 1995 to 1998. J Clin Microbiol 38:1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tran A, Talmud D, Lejeune B, Jovenin N, Renois F, Payan C, Leveque N, Andreoletti L. 2010. Prevalence of rotavirus, adenovirus, norovirus, and astrovirus infections and coinfections among hospitalized children in northern France. J Clin Microbiol 48:1943–1946. doi: 10.1128/JCM.02181-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch A, Pintó RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Risco C, Carrascosa JL, Pedregosa AM, Humphrey CD, Sánchez-Fauquier A. 1995. Ultrastructure of human astrovirus serotype 2. J Gen Virol 76:2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 9.Aw TG, Gin KY. 2011. Prevalence and genetic diversity of waterborne pathogenic viruses in surface waters of tropical urban catchments. J Appl Microbiol 110:903–914. doi: 10.1111/j.1365-2672.2011.04947.x. [DOI] [PubMed] [Google Scholar]
- 10.Chan-it W, Thongprachum A, Okitsu S, Mizuguchi M, Ushijima H. 2010. Epidemiology and molecular characterization of sapovirus and astrovirus in Japan, 2008-2009. Jpn J Infect Dis 63:302–303. [PubMed] [Google Scholar]
- 11.Rodríguez-Díaz J, Querales L, Caraballo L, Vizzi E, Liprandi F, Takiff H, Betancourt WQ. 2009. Detection and characterization of waterborne gastroenteritis viruses in urban sewage and sewage-polluted river waters in Caracas, Venezuela. Appl Environ Microbiol 75:387–394. doi: 10.1128/AEM.02045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silva PA, Cardoso DD, Schreier E. 2006. Molecular characterization of human astroviruses isolated in Brazil, including the complete sequences of astrovirus genotypes 4 and 5. Arch Virol 151:1405–1417. doi: 10.1007/s00705-005-0704-9. [DOI] [PubMed] [Google Scholar]
- 13.Yokoi H, Kitahashi T, Tanaka T, Utagawa E. 2001. Detection of astrovirus RNA from sewage works, seawater and native oysters samples in Chiba City, Japan using reverse transcription-polymerase chain reaction. Kansenshogaku Zasshi 75:263–269. (In Japanese.) doi: 10.11150/kansenshogakuzasshi1970.75.263. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Mitchell DK, Afflerbach C, Jakab F, Walter J, Zhang YJ, Staat MA, Azimi P, Matson DO. 2006. Quantitation of human astrovirus by real-time reverse-transcription-polymerase chain reaction to examine correlation with clinical illness. J Virol Methods 134:190–196. doi: 10.1016/j.jviromet.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 15.Gofti-Laroche L, Gratacap-Cavallier B, Demanse D, Genoulaz O, Seigneurin JM, Zmirou D. 2003. Are waterborne astrovirus implicated in acute digestive morbidity (E.MI.R.A. study)? J Clin Virol 27:74–82. doi: 10.1016/S1386-6532(02)00130-0. [DOI] [PubMed] [Google Scholar]
- 16.Scarcella C, Carasi S, Cadoria F, Macchi L, Pavan A, Salamana M, Alborali GL, Losio MM, Boni P, Lavazza A, Seyler T. 2009. An outbreak of viral gastroenteritis linked to municipal water supply, Lombardy, Italy, June 2009. Euro Surveill 14(29):pii=19274 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19274. [DOI] [PubMed] [Google Scholar]
- 17.Bertrand I, Schijven JF, Sánchez G, Wyn-Jones P, Ottoson J, Morin T, Muscillo M, Verani M, Nasser A, de Roda Husman AM, Myrmel M, Sellwood J, Cook N, Gantzer C. 2012. The impact of temperature on the inactivation of enteric viruses in food and water: a review. J Appl Microbiol 112:1059–1074. doi: 10.1111/j.1365-2672.2012.05267.x. [DOI] [PubMed] [Google Scholar]
- 18.Yang BY, Liu XL, Wei YM, Wang JQ, He XQ, Jin Y, Wang ZJ. 2014. Rapid and sensitive detection of human astrovirus in water samples by loop-mediated isothermal amplification with hydroxynaphthol blue dye. BMC Microbiol 14:38. doi: 10.1186/1471-2180-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, Zaidi SZ, Delwart E. 2009. Multiple novel astrovirus species in human stool. J Gen Virol 90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkbeiner SR, Kirkwood CD, Wang D. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J 5:117. doi: 10.1186/1743-422X-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, Kang G, Wang D. 2009. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol J 6:161. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, Virgin HW, Anderson LJ, Vinjé J, Wang D, Tong S. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol 83:10836–10839. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang H, Holtz LR, Bauer I, Franz CJ, Zhao G, Bodhidatta L, Shrestha SK, Kang G, Wang D. 2013. Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes. Virology 436:8–14. doi: 10.1016/j.virol.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkbeiner SR, Le BM, Holtz LR, Storch GA, Wang D. 2009. Detection of newly described astrovirus MLB1 in stool samples from children. Emerg Infect Dis 15:441–444. doi: 10.3201/eid1503.081213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holtz LR, Bauer IK, Rajendran P, Kang G, Wang D. 2011. Astrovirus MLB1 is not associated with diarrhea in a cohort of Indian children. PLoS One 6:e28647. doi: 10.1371/journal.pone.0028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto T, Wangchuk S, Tshering K, Yahiro T, Zangmo S, Dorji T, Tshering K, Mitui MT, Nishizono A, Ahmed K. 2013. Complete genome sequences of two astrovirus MLB1 strains from Bhutanese children with diarrhea. Genome Announc 1(4):e00485-13. doi: 10.1128/genomeA.00485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hata A, Kitajima M, Katayama H. 2013. Occurrence and reduction of human viruses, F-specific RNA coliphage genogroups and microbial indicators at a full-scale wastewater treatment plant in Japan. J Appl Microbiol 114:545–554. doi: 10.1111/jam.12051. [DOI] [PubMed] [Google Scholar]
- 29.Katayama H, Shimasaki A, Ohgaki S. 2002. Development of a virus concentration method and its application to detection of enterovirus and Norwalk virus from coastal seawater. Appl Environ Microbiol 68:1033–1039. doi: 10.1128/AEM.68.3.1033-1039.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yokoi H, Kitahashi T. 2009. Astrovirus RNA detection using real-time reverse transcription-polymerase chain reaction. Kansenshogaku Zasshi 83:120–126. (In Japanese.) [DOI] [PubMed] [Google Scholar]
- 31.Kitajima M, Tohya Y, Matsubara Y, Haramoto E, Utagawa E, Katayama H, Ohgaki S. 2008. Use of murine norovirus as a novel surrogate to evaluate resistance of human norovirus to free chlorine disinfection in drinking water supply system. Environ Eng Res 45:361–370. (In Japanese.) [Google Scholar]
- 32.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 33.Hata A, Kitajima M, Tajiri-Utagawa E, Katayama H. 2014. Development of a high resolution melting analysis for detection and differentiation of human astroviruses. J Virol Methods 200:29–34. doi: 10.1016/j.jviromet.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 34.Gentry J, Vinjé J, Lipp EK. 2009. A rapid and efficient method for quantitation of genogroups I and II norovirus from oysters and application in other complex environmental samples. J Virol Methods 156:59–65. doi: 10.1016/j.jviromet.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Kitajima M, Oka T, Haramoto E, Katayama H, Takeda N, Katayama K, Ohgaki S. 2010. Detection and genetic analysis of human sapoviruses in river water in Japan. Appl Environ Microbiol 76:2461–2467. doi: 10.1128/AEM.02739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Cann P, Ranarijaona S, Monpoeho S, Le Guyader F, Ferré V. 2004. Quantification of human astroviruses in sewage using real-time RT-PCR. Res Microbiol 155:11–15. doi: 10.1016/j.resmic.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Ferreira MS, Xavier MDPTP, Tinga ACDC, Rose TL, Fumian TM, Fialho AM, de Assis RM, Carvalho Costa FA, de Oliveira SA, Leite JPG, Miagostovich MP. 2012. Assessment of gastroenteric viruses frequency in a children's day care center in Rio De Janeiro, Brazil: a fifteen year study (1994-2008). PLoS One 7:e33754. doi: 10.1371/journal.pone.0033754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rezaeinejad S, Vergara GG, Woo CH, Lim TT, Sobsey MD, Gin KW. 2014. Surveillance of enteric viruses and coliphages in a tropical urban catchment. Water Res 58:122–131. doi: 10.1016/j.watres.2014.03.051. [DOI] [PubMed] [Google Scholar]
- 39.Victoria M, Guimarães F, Fumian T, Ferreira F, Vieira C, Leite JP, Miagostovich M. 2009. Evaluation of an adsorption-elution method for detection of astrovirus and norovirus in environmental waters. J Virol Methods 156:73–76. doi: 10.1016/j.jviromet.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Méndez-Toss M, Romero-Guido P, Munguía ME, Méndez E, Arias CF. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J Gen Virol 81(Pt 12):2891–2897. [DOI] [PubMed] [Google Scholar]
- 41.Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. 2014. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morsy El-Senousy W, Guix S, Abid I, Pintó RM, Bosch A. 2007. Removal of astrovirus from water and sewage treatment plants, evaluated by a competitive reverse transcription-PCR. Appl Environ Microbiol 73:164–167. doi: 10.1128/AEM.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dey SK, Phan GT, Nishimura S, Mizuguchi M, Okitsu S, Ushijima H. 2010. Molecular and epidemiological trend of sapovirus, and astrovirus infection in Japan. J Trop Pediatr 56:205–207. doi: 10.1093/tropej/fmp082. [DOI] [PubMed] [Google Scholar]
- 44.Clark B, McKendrick M. 2004. A review of viral gastroenteritis. Curr Opin Infect Dis 17:461–469. doi: 10.1097/00001432-200410000-00011. [DOI] [PubMed] [Google Scholar]
- 45.de Grazia S, Martella SV, Chironna M, Bonura F, Tummolo F, Calderaro A, Moschidou P, Giammanco GM, Medici MC. 2013. Nationwide surveillance study of human astrovirus infections in an Italian paediatric population. Epidemiol Infect 141:524–528. doi: 10.1017/S0950268812000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nadan S, Walter JE, Grabow WO, Mitchell DK, Taylor MB. 2003. Molecular characterization of astroviruses by reverse transcriptase PCR and sequence analysis: comparison of clinical and environmental isolates from South Africa. Appl Environ Microbiol 69:747–753. doi: 10.1128/AEM.69.2.747-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chu DK, Chin AW, Smith GJ, Chan KH, Guan Y, Peiris JS, Poon LL. 2010. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J Gen Virol 91(Pt 10):2457–2462. doi: 10.1099/vir.0.022764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smits SL, van Leeuwen M, van der Eijk AA, Fraaij PL, Escher JC, Simon JH, Osterhaus AD. 2010. Human astrovirus infection in a patient with new-onset celiac disease. J Clin Microbiol 48:3416–3418. doi: 10.1128/JCM.01164-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed SF, Sebeny PJ, Klena JD, Pimentel G, Mansour A, Naguib AM, Bruton J, Young SY, Holtz LR, Wang D. 2011. Novel astroviruses in children, Egypt. Emerg Infect Dis 17:2391–2393. doi: 10.3201/eid1712.110909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Li Y, Jin Y, Li DD, Li X, Duan ZJ. 2013. Recently identified novel human astroviruses in children with diarrhea, China. Emerg Infect Dis 19:1333–1335. doi: 10.3201/eid1908.121863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wylie KM, Mihindukulasuriya KA, Sodergren E, Weinstock GM, Storch GA. 2012. Sequence analysis of the human virome in febrile and afebrile children. PLoS One 7:e27735. doi: 10.1371/journal.pone.0027735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.