Abstract
Glycosylation is one of the most common protein modifications. Each glycoprotein can be glycosylated at multiple glycosites, and each glycosites can be modified by different glycans. Due to this heterogeneity of glycosylation, it has proven difficult to study the structure–function relationship of specific glycans and their affected glycoproteins. Here, we report a novel method for rapid and quantitative identification of glycoproteins containing specific glycans. Lectin affinity isolations are followed by chemical immobilization of the captured glycopeptides, allowing the identification of glycoproteins containing specific glycans by subsequent mass spectrometry. The application of the method should be useful to facilitate our understanding of how changes in glycan associate with diseases, and to discover novel glycoproteins with certain glycans that could serve as biomarkers or therapeutic targets.
Glycosylation of proteins is known to play an important role in various biological processes, and aberrant glycosylation also plays an important role in a large number of human diseases.1–4 Since glycoproteins typically contain variable glycan structures at each glycosylation site (glycosite),5,6 the glycans present at each glycosite on the glycoproteins have therefore attracted great interest, and elucidation of their precise nature would greatly advance our understanding of their role in health and disease. However, there are currently no methods available that are sufficiently specific and sensitive for the analysis of glycans at each glycosite in complex samples.
Recently, two popular glycoprotein enrichment methods have been established: affinity capture based on glycan-lectin interaction and solid-phase extraction of glycopeptides (SPEG). For example, Kaji et al. isolated N-linked glycoproteins and glycopeptides using a lectin affinity capture method, which was followed by tandem mass spectrometry (MS/MS).7 Several studies reported the use of lectins for the characterization of glycoproteins in serum and clinical samples.8–12 However, the capture specificity of glycopeptides, which is defined as the percentage of identified peptides containing consensus N-linked glycosylation motif (NXT/S sequence, while X is any amino acids except P), is insufficient. Presumably, the low specificity of this method is caused mostly by binding of nonglycopeptides within proteins to lectins due to the low affinity of the lectin–glycan interaction.
Chemical immobilization methods, such as solid-phase extraction of glycosite-containing peptides (SPEG), allows for specific isolation and identification of glycopeptides.13,14 Glycans of the cis-diol groups are oxidized and covalently coupled onto a solid surface using hydrazide chemistry. The nonglycopeptides that are not coupled are removed, and the glycopeptides are subsequently released using endoglycosidase PNGase F prior to analysis using MS/MS. It has been demonstrated that by using the SPEG method, the glycopeptides could be captured with high specificity, and then identified in a rapid, sensitive, and reproducible manner.15–19 However, the SPEG method cannot provide any structural information about glycans, since the glycans are removed from the peptides before MS detection.
Here, we developed a novel method that allows for identification of N-linked glycosites using tandem mass spectrometry using lectin affinity capture followed by SPEG (LecSPEG). For proof-of-principle, we applied this method to compare the glycosite-containing peptides in the serum sample. We showed that this method was able to identify and quantify those glycopeptides from glycoproteins interacting with specific lectins with high specificity and sensitivity.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Aleuria aurantia lectin (AAL)-, Sambucus nigra lectin (SNA)-, and wheat germ agglutinin (WGA)-conjugated agarose beads were purchased from Vector Laboratories (Burlingame, CA). Tris (2-carboxythyl) phosphine (TCEP) was purchased from Pierce (Rockford, IL). Sequencing-grade trypsin was from Promega (Madison, WI). Peptide-N-glycosidase F (PNGase) was from ProZyme (San Leandro, CA). Sodium periodate, hydrazide resin, and P6 desalting spin columns were from Bio-Rad (Hercules, CA). C18 and MCX desalting columns were from Waters (Milford, MS). The high-performance liquid chromatography–mass spectrometry (HPLC–MS) platform used in this study includes an Eksigent 2D nanoLC system (Dublin, CA) and a Thermo Scientific Orbitrap Velos mass spectrometer (Waltham, MA). Magic C-18 packing material was from Michrom Bioresources (Auburn, CA), and all other chemicals were purchased from Sigma (St. Louis, MO).
Serum Samples
The serum specimens and relevant clinical information were obtained with informed consent and with the approval of the Institutional Review Board of the Johns Hopkins University. To establish the method, pooled serum samples from healthy males were used.
Glycoprotein Extraction Using Affinity Capture by Lectin-Conjugated Agarose Beads (Lec Meothd)
Lectin-conjugated agarose beads (250 μL) were prewashed for three times with 1 mL Tris-HCl buffer (0.2 M Tris-HCl, pH 7.5). The beads were mixed with Tris-HCl buffer for 5–10 min, and the buffer was removed following centrifugation at 6000g for 5 min. Aliquot of serum pooled (25 μL) was diluted 1:10 in Tris-HCl buffer then incubated with the beads at 4 °C overnight with gentle shaking. The unbound nonglycoproteins were removed by washing the beads with Tris-HCl buffer for three times. Beads were then incubated with 100 μL lectin-specific elution buffers (100 mM fucose in Tris-HCl buffer for AAL, 0.5 mM lactose in Tris-HCl buffer for SNA, and 0.8 M N-acetyl-D-glucosamine in 0.2 M acetic acid for WGA) for 30 min with gentle shaking to elute glycoproteins. The supernatant was collected and desalted on a P6 column. Glycoproteins were denatured, reduced, and digested with trypsin as previously described13,14 Trypsin was then deactivated by incubating the sample at 80 °C for 15 min and 0.5 μL of PNGase F was used to remove N-linked glycans from the peptides. The peptides were desalted using a C-18 column, and the eluate was dried using a SpeedVac and resuspended in 10 μL of 0.4% acetic acid per sample.
Glycopeptide Extraction Using Affinity Capture by Lectin-Conjugated Agarose Beads (Lec+Lec Method)
Glycoproteins were extracted from a 25 μL aliquot of serum pooled from healthy men using the Lec method as described above. The eluted glycoproteins were digested with trypsin. The peptides were then desalted with a C-18 column and incubated with 100 μL of prewashed lectin-conjugated agarose beads for glycopeptide enrichment using the same procedures for glycoprotein extraction described above. The eluted glycopeptides were desalted using a C-18 column, and the glycopeptides were treated with 0.5 μL of PNGase F to release N-linked glycans from the peptides. The peptides were desalted again and dried using a SpeedVac, before being resuspended in 10 μL of 0.4% acetic acid per sample.
Extraction of N-Linked Glycosite-Containing Peptides Using the Combination of Lectin Affinity Capture and SPEG Method (LecSPEG Method)
The peptides enriched by lectins were oxidized with 10 mM sodium periodate at 4 °C for 1 h in the dark. The oxidized peptides were purified using C18 desalting columns to remove the oxidant and conjugated to hydrazide beads at room temperature for 4 h in 80% acetonitrile. Nonconjugated peptides were removed by washing the resin for three times with 800 μL washing buffer (1.5 M NaCl, H2O, and 100 mM of NH4HCO3). The N-linked glycosite-containing peptides were released from the resin with 0.5 μL of PNGase F in 100 mM of NH4HCO3 and incubated at 37 °C overnight. After the final purification using a MCX desalting column, the peptides were dried by SpeedVac and resuspended in 10 μL of 0.4% acetic acid solution for subsequent mass spectrometry analysis.
Analysis of N-Linked Glycosite-Containing Peptides Using a HPLC-Orbitrap-MS Platform
Peptide identification by liquid chromatography tandem mass spectrometry (LC–MS/MS) analysis was performed using an Orbitrap MS/MS (Thermo Fisher Scientific) interfaced with a nanoLC system (Eksigent). A 5 μL sample of isolated peptides was loaded onto the HPLC-Orbitrap-MS platform. The HPLC mobile phase A were composed of 0.2% formic acid in HPLC-grade water, while phase B were composed of 0.2% formic acid in HPLC-grade acetonitrile. The mobile phase B was increased linearly from 5% to 40% over 90 min at 300 nL/min. A C18 column (75 μm ID × 10 cm, Magic C18 material, 5 μm, 100 Å) was used for peptide separation. Mass analysis was performed using a data-dependent analysis of the top ten precursors and a dynamic exclusion of 30 s. The data was acquired at 30000 resolution for the precursor scans and 7500 for MS/MS. Target values of 1e6 for MS and 1e5 for MS/MS were set with maximum injection times of 100 and 300 ms, respectively. Data was acquired used the monoisotopic precursor selection (MIPS) method, with predictive AGC enabled.
The acquired data was searched against the Homo sapiens taxonomy of the RefSeq database (version 40, 04/16/2010) using the Mascot (Matrix Science version 2.2.0) search algorithm within Proteome Discoverer (Thermo Scientific version 1.1). The data was searched with 2 missed cleavages allowed and a tolerance of 15 ppm on the precursors and 0.02 Da on the fragment ions. Modifications allowed included carbamidomethyl cysteines set to static and deamidation of asparagines and oxidation of methionines set to variable. Data were searched against a decoy database and filtered to a 1% false discovery rate (FDR). The data set was further filtered to ensure that all the identified peptides had Mascot ion scores greater than 50.
RESULTS
Detection of glycosite-containing peptides with special glycan motifs using lectin affinity capture and chemical immobilization.
To compare the LecSPEG method with the two lectin enrichment methods, glycoprotein enrichment (Lec method) and affinity enrichment of glycoproteins followed by enrichment of glycopeptides from the enriched glycoproteins using lectins (Lec + Lec method), we applied these three methods to aliquots from the same pooled sample of human sera to determine the number of identified peptides (Figure 1), the number of identified glycopeptides, and the specificity of identified glycopeptides to total peptides.
Figure 1.
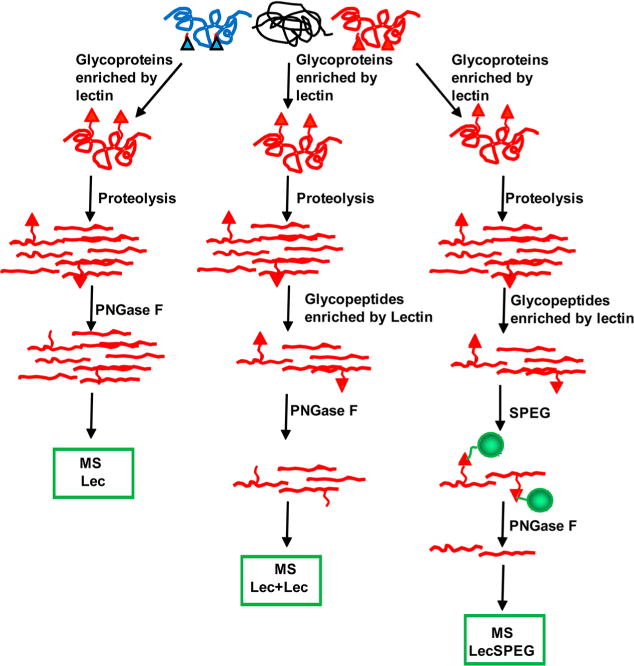
Schematic diagram of the analytical procedure of glycosite detection using a lectin affinity capture method combined with chemical immobilization.
First, we employed the lectin SNA-conjugated agarose beads to capture the glycoproteins by using the Lec method. A total number of 85 peptides were identified from the serum sample, with only 2 peptides containing N-linked glycosylation sites with NXT/S motif. The low capture specificity (2%) of glycosite-containing peptide identified by this method suggests that it is not suitable for glycosite analysis (Table 1 of the Supporting Information).
Next, lectin SNA-conjugated agarose beads were first used to capture glycoproteins from the serum sample, and the isolated glycoproteins were digested to peptides. The glycopeptides were then further isolated by lectin SNA-conjugated agarose beads and analyzed by LC–MS/MS. Using this tandem lectin affinity capture method, 14 glycosite-containing peptides were identified from the same serum sample with the percentage of specificity or identification of glycosite-containing peptides at 36%. Thus, the Lec + Lec procedure increased the number of identified glycosite-containing peptides and capture specificity when compared with the Lec method (Table 1 of the Supporting Information).
Finally, LecSPEG was used to analyze glycoproteins from the same serum sample, after isolation of glycopeptides by Lec + Lec method. They were captured by SPEG method and analyzed by LC–MS/MS. We were able to identify 46 unique glycosite-containing peptides from the serum sample with the capture specificity of 82% (Table 1 of the Supporting Information). Clearly the lecSPEG method has the power to identify the most glycosites and has the highest specificity for the capture in crude serum among the three methods tested.
To determine if the LecSPEG method possesses a principal advantage for glycopeptide detection with specific glycan motifs, we compared the three methods using SNA, AAL, and WGA-conjugated beads for glycopeptide identification. SNA binds to glycans containing α-2,6-linked sialic acid; AAL binds to glycans containing fucose; and WGA binds to glycans containing terminal β-N-acetylglucosamine or sialic acid.20 In each case, more glycopeptides were identified with higher capture specificity using the LecSPEG method (Table 1 of the Supporting Information).
Furthermore, we tested the reproducibility of the LecSPEG method for glycopeptide analysis. The glycopeptides were isolated in quadruplicates from 25 μL serum samples by LecSPEG method using SNA-conjugated agarose beads. Using this hybrid method, 42 glycopeptides were identified in all four replicate samples (Figure 2; Table 2 of the Supporting Information). The CVs of the peak intensities of the 42 glycopeptides ranged from 2.1% to 39.1%. Only four glycopeptides displayed CVs greater than 30%, and the majority of glycopeptides (38 of 42, 90.5%) had CVs lower than 30%, which indicates that this multistep method was able to generate reproducible results.
Figure 2.
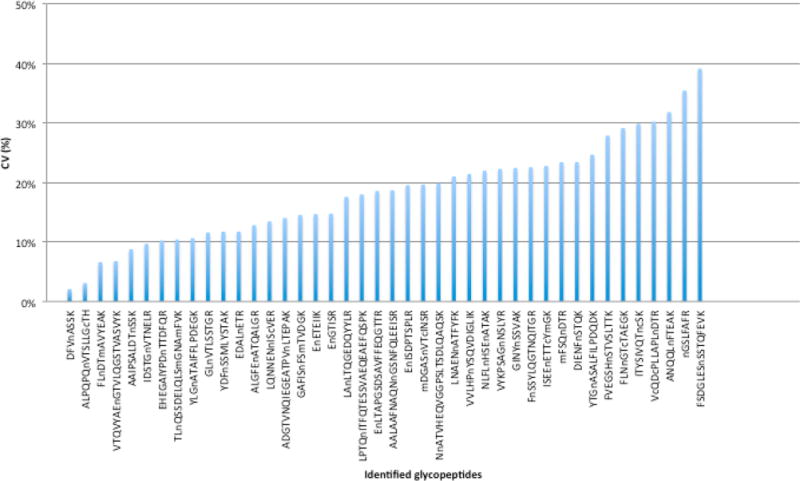
Reproducibility of glycopeptides captured by LecSPEG method using SNA conjugated agarose beads analyzed in quadruplicate.
DISCUSSION
The interactions between lectins and glycans are relatively weak, and a large proportion of peptides captured by lectins are nonglycopeptides. When we sequentially combined lectin affinity capture with the SPEG method, the capture specificity of glycopeptides was significantly improved from 2% (Lec) to 36% (Lec+Lec), to further 82% (LecSPEG), while the number of identified unique glycosites increased from 2 (Lec) to 14 (Lec+Lec), to 46 (LecSPEG) using SNA-conjugated agarose beads. Similar results were also observed when using more lectins for glycopeptide capture, including AAL and WGA (Table 1 of the Supporting Information).
In addition to its high specificity and sensitivity, the reproducibility of the method for glycopeptide analysis is important, especially for clinical research, which usually requires sample comparison. For the tandem SNA lectin and SPEG capture methods, the CVs of most identified glycopeptides were lower than 30%, which is comparable to those of tryptic peptides analyzed by LC–MS.19,21,22 Thus, our data indicate that our system is feasible for quantification analysis of clinical samples.
Moreover, the glycan motif information can be obtained for each glycosite-containing peptide using LecSPEG detection. The glycosite-containing peptides identified by SNA-LecSPEG method contained sialic acids (α2–α6) on glycans. The glycopeptides identified by AAL-LecSPEG and WGA-LecSPEG contained fucose and terminal β-N-acetylglucosamine or sialic acids, respectively. When the serum samples were profiled using the LecSPEG method, the glycan motif information and quantification data based on the LC–MS peak intensity clearly showed the heterogeneity of glycans at each glycosite, that individual glycoproteins could contain multiple glycosites and each glycosite has its own glycan motif patterns (Table 3 of the Supporting Information). For example, the glycosite DFVnASSK of Heparin cofactor II protein contains glycan motifs of fucose and terminal β-N-acetylglucosamine that bound to lectins AAL and WGA, respectively (Table 3 of the Supporting Information). We can determine the relative quantities of the specific glycan motif at each glycopeptides from individual samples by directly comparing the LC–MS data between different samples.
In summary, we developed a lectin affinity followed by SPEG capture method for glycopeptide analysis and tested the analytical performance of the LecSPEG in this study. We demonstrated that our method can be used to profile glycopeptides with specific glycan motifs. The newly developed LecSPEG method will be a useful tool for subsequent glycoproteomic studies in both biological and clinical studies.
Supplementary Material
Acknowledgments
This work was supported from Early Detection Research Network (NIH/NCI/EDRN) Grant U01CA152813 (H.Z.) and U24CA115102 (D.W.C), the NHLBI proteomic contract NHLBI-HV-10-05 (J.V.E), the Patrick C. Walsh Prostate Cancer Research Fund (H.Z.), the United States Department of Defense Grant PC081386 (Y.L.), the National Natural Science of China, Grant 31270909 (Y.L.), and the Outstanding Technical Talent Award of Chinese Academy of Sciences (Y.L.). We would like to thank Xiaer Sun and Helen Fedor for technical assistance, Dr Robert Cole, Robert O’Meally, and Tatiana Boronina for assistance in mass spectrometry and data analyses, and Torsten Juellch for critical reading of the manuscript. Serum samples were obtained from The Brady Urological Research Institute in Johns Hopkins University, which is funded by NIH/NCI Prostate SPORE (P50CA58236).
Footnotes
Supporting Information
N-linked glycosite identifications by Lec, Lec+Lec, and LecSPEG method, reproducibility of glycopeptides captured by LecSPEG method using SNA conjugated agarose beads, and distribution of glycan forms of individual glycopeptides with different lectin binding specificity. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Spiro RG. Glycobiology. 2002;12:43R–56R. doi: 10.1093/glycob/12.4.43r. [DOI] [PubMed] [Google Scholar]
- 2.Drake PM, Cho W, Li B, Prakobphol A, Johansen E, Anderson NL, Regnier FE, Gibson BW, Fisher SJ. Clin Chem. 2010;56:223–236. doi: 10.1373/clinchem.2009.136333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dennis JW, Granovsky M, Warren CE. Bioessays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Durand G, Seta N. Clin Chem. 2000;46:795–805. [PubMed] [Google Scholar]
- 5.Schachter H. Glycobiology. 1991;1:453–461. doi: 10.1093/glycob/1.5.453. [DOI] [PubMed] [Google Scholar]
- 6.Neumann U, Brandizzi F, Hawes C. Ann Bot (London, UK) 2003;92:167–180. doi: 10.1093/aob/mcg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K, Takahashi N, Isobe T. Nat Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 8.Drake RR, Schwegler EE, Malik G, Diaz J, Block T, Mehta A, Semmes OJ. Mol Cell Proteomics. 2006;5:1957–1967. doi: 10.1074/mcp.M600176-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Calvano CD, Zambonin CG, Jensen ON. J Proteomics. 2008;71:304–317. doi: 10.1016/j.jprot.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 10.McDonald CA, Yang JY, Marathe V, Yen TY, Macher BA. Mol Cell Proteomics. 2009;8:287–301. doi: 10.1074/mcp.M800272-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Narimatsu H, Sawaki H, Kuno A, Kaji H, Ito H, Ikehara Y. FEBS J. 2010;277:95–105. doi: 10.1111/j.1742-4658.2009.07430.x. [DOI] [PubMed] [Google Scholar]
- 12.Ito S, Hayama K, Hirabayashi J. Methods Mol Biol. 2009;534:195–203. doi: 10.1007/978-1-59745-022-5_14. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Li XJ, Martin DB, Aebersold R. Nat Biotechnol. 2003;21:660–666. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 14.Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. Nat Protoc. 2007;2:334–339. doi: 10.1038/nprot.2007.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan S, Zhang H, Rush J, Eng J, Zhang N, Patterson D, Comb MJ, Aebersold R. Mol Cell Proteomics. 2005;4:182–190. doi: 10.1074/mcp.M400161-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Yi EC, Li XJ, Mallick P, Kelly-Spratt KS, Masselon CD, Camp DG, Smith RD, Kemp CJ, Aebersold R. Mol Cell Proteomics. 2005;4:144–155. doi: 10.1074/mcp.M400090-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Y, Aebersold R, Zhang H. Anal Chem. 2007;79:5826–5837. doi: 10.1021/ac0623181. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H, Liu AY, Loriaux P, Wollscheid B, Zhou Y, Watts JD, Aebersold R. Mol Cell Proteomics. 2007;6:64–71. doi: 10.1074/mcp.M600160-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Sokoll LJ, Rush J, Meany D, Zou N, Chan DW, Zhang H. Proteomics: Clin Appl. 2009;3:597–608. [Google Scholar]
- 20.Tao SC, Li Y, Zhou J, Qian J, Schnaar RL, Zhang Y, Goldstein IJ, Zhu H, Schneck JP. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hawkridge AM, Wysocky RB, Petitte JN, Anderson KE, Mozdziak PE, Fletcher OJ, Horowitz JM, Muddiman DC. Anal Bioanal Chem. 2010;398:737–749. doi: 10.1007/s00216-010-3979-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagaraj N, Mann M. J Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


