Abstract
Glioblastoma multiforme (GBM) is the deadliest and most common form of malignant primary brain tumor in humans. However, until now, little is known about the glioma genesis and progression at the molecular level. Here we report that overexpression of sine oculis homeobox homolog 1 (Six1), a developmental transcription factor implicated in tumor onset and progression, can significantly promote glioblastoma cell proliferation and invasion by upregulating connective tissue growth factor (CTGF). Our results revealed that expression of Six1 mRNA was increased and small hairpin RNAi silencing of Six1 could dramatically inhibit cell proliferation and invasion in GBM. Moreover, it was found that CTGF gene could be transcriptionally regulated by Six1. Its overexpression induced CTGF up-regulation in GBM at both the mRNA and protein level, and significantly enhanced the activity of CTGF promoter in these tumor cells, while decreasing CTGF expression impeded Six1-induced cell proliferation and invasion, revealing that CTGF is required for Six1-mediated GBM growth and metastasis. Collectively, these findings suggest that Six1 overexpression may contribute to cell proliferation and invasion via upregulation of CTGF in GBM. Our study provides new insights into the important roles of Six1 and CTGF in tumor regulation, suggesting that Six1 might be a potential therapeutic target for preventing proliferation and metastasis of GBM.
Keywords: Six1, CTGF, glioblastoma, cell proliferation, invasion
Introduction
Glioblastoma multiforme (GBM) is the most common and most aggressive malignant primary brain tumor in adults, with a median survival of 15 months post-diagnosis [1]. Despite advances in surgery, chemotherapy and radiation, treatment of GBM still remains a big challenge in clinical oncology [2]. One of the most challenging problems in therapy of GBM is its extremely complex and heterogeneous molecular biology [3]. However, very little is known about the processes by which it develops until now. Therefore, the identification of new molecular pathways involved in tumor biology and invasiveness, as well as novel therapeutic targets for GBM is desperately needed.
Six1 belongs to sine oculis homeobox (SIX) protein family and is highly conserved from flatworms to humans [4,5]. It plays pivotal roles in the development of various organs including head, retina, brain, ear, nose, and kidney [6]. Also, Six1 has been implicated in the tumor onset and progression [4]. It is aberrantly expressed in a variety of human cancers, including breast cancer [7,8], ovarian cancer [9], pancreatic cancer [10] etc. where it leads to increased cell proliferation, survival and metastasis. Recently, it was reported that Six1 and its cofactors, EYA1 and DACH2, were dysregulated in glioma tumor progenitor cells [11]. These findings suggest that Six1 may play a critical role in glioma genesis or progression.
In this study, we detected a high level of Six1 in GBM, and Six1 overexpression promoted glioblastoma cell proliferation and invasion. In addition, Six1 directly regulated the expression of connective tissue growth factor (CTGF) in the tumor cells. Our findings suggest that Six1 may promote glioblastoma cell proliferation and invasion through upregulation of CTGF.
Materials and methods
Cell lines and tissue samples
Human glioblastoma cell lines U-118 (HTB-15) and U-87 (HTB-14) were obtained from American Type Culture Collection ATCC (Rockville, MD, USA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT, USA), 100 units/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen, California, USA) in 5% CO2 atmosphere at 37°C. Fresh GBM tissue samples (n = 55) and adjacent normal tissues (n = 35) were collected at the Yichang Central People’s Hospital and The First Affiliated Hospital of China Three Gorges University between 2013 and 2014. These tissue samples were frozen in liquid nitrogen and stored at -80°C immediately after surgery until RNA extraction. The collection of tissue samples was approved by the Research Ethics Committee of Three Gorges University. Written informed consents were obtained from all patients.
Stable transfection of glioblastoma cells
The human Six1 cDNA was subcloned into plasmid pcDNA4/TO according to previous report [10]. U-118 and U-87 cells were transfected with pcDNA4/TO-Six1 or control vector for stable transfection. Stably transfected cell lines were isolated by 100 μg/ml Zeocin selection.
Lentivirus-mediated shRNA knockdown of Six1 or CTGF expression
Lentiviral production, titration, and infection were performed as previously described [12]. Briefly, lentiviral plasmids expressing Six1 shRNAs or scrambled shRNA were co-transfected with pHelper plasmids in 293T cells. Lentiviral particles were harvested from the media 48 hours after transfection, and purified with ultracentrifugation. Cells were infected with lentiviruses encoding Six1 shRNAs or a scrambled shRNA as control. Cells were harvested at 72 hours after infection and the knockdown efficiency was evaluated by quantitative PCR and western blot analysis. Similar to Six1 knockdown, lentiviral constructs expressing CTGF shRNA (RHS3979-962913, Open Biosystems) was used for knockdown of CTGF and subsequently used for further experiments.
RNA extraction and real-time PCR
Total RNA was isolated using the RNeasy mini kit (Qiagen, Germany) and the cDNA was prepared using the SuperScript® III First-Strand Synthesis System (Invitrogen, California, USA). Quantitative PCR was performed using SYBR Green dye on an Applied Biosystems 7300 Real-time PCR system (Applied Biosystems, Foster City, CA). Primer sets were used as previously reported.
Analysis of microarray data
Oncomine Cancer Microarray database (http://www.oncomine.org) [13] was used to study gene expression of Six1 in glioblastoma samples as we previously described. Gene expression data were also obtained from NCBI Gene Expression Omnibus (GEO) database (accession numbers: GSE4290, GSE7696 and GSE4536) [14-16]. Expression data for Six1 were log-transformed, median centered per array, and the standard deviation was normalized to one per array.
Western blot analysis
Western blot analysis was performed as we previously described [17]. Briefly, cells were lysed in cold lysis buffer, proteins (20-30 μg) were resolved on SDS-PAGE, transferred onto PVDF membranes, and probed with antibodies for Six1 (sc-9709, santa cruz), CTGF (sc-14939, santa cruz), and GAPDH (sc-32233, santa cruz) at 4°C overnight. Detection was performed with the SuperSignal West Femto Maximum Sensitivity Substrate Trial Kit (Pierce, Rockford, IL, USA). The band images were digitally captured and quantified with a FluorChem FC2 imaging system (Alpha Innotech, San Leandro, CA, USA).
MTT assay
Cells plated in 96-well plates were incubated for different periods of time and then added 20 μL of MTT (tetrazolium bromide, 5 mg/mL, GE Healthcare) into each well. After incubation for 4 h, 150 μL of DMSO was added to solubilize the crystals for 20 min at room temperature and the absorbance at 570 nm was read by an ELISA plate reader (Model 680, Bio-Rad, CA).
Colony formation assay
Cells were seeded into 6-well plates. Two weeks later, cells were stained by 0.5% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) in methanol for 10 min. Colonies (more than 50μm diameter) were counted directly on the plate.
Cell invasion assays
The Boyden chamber and polycarbonate membrane precoated with matrigel (BD Biosciences) was used to evaluate invasion ability of glioblastoma cells. DMEM with 10% fetal bovine serum was added to the lower compartment as a chemoattractant. Cells suspended in 50 μL DMEM with 0.5% BSA was loaded onto the upper compartment of each chamber. After an incubation of 24 hours at 37°C, cells migrated through the chamber were stained by hematoxylin and eosin (H & E) and subsequently counted under the microscope.
Gene reporter assays
Gene reporter assays were performed as previously reported. Cells were transfected with expression vectors (empty vector or Six1), the human CTGF promoter pGL3-CTGF (-2000 bp nucleotides) and renilla reniformis luciferase. After 24 hours, luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA), according to the manufacturer’s instructions.
Statistical analysis
All data were expressed as mean ± standard error of the mean (SEM). Between groups and among groups comparisons were conducted with Student t test and ANOVA, respectively. Mann-Whitney U test was used for nonparametric variables. Statistical analysis was performed using GraphPad Prism software version 4.0 (PRISM4) (GraphPad Software Inc, LaJolla, CA), and P < 0.05 was considered significant.
Results
Six1 was overexpressed in glioblastoma cells
To determine the relative expression of Six1 in human normal brain and GBM tissues, we used semi-quantitative PCR specific Six1 primers. As shown in Figure 1A, the relative expression of Six1 mRNA in GBM samples (n = 55) was significantly higher than in adjacent non-tumor brain tissues (n = 35) after normalization using GAPDH (P = 0.0004). Furthermore, we analyzed the expression of Six1 by querying the ONCOMINE database. In three microarray expression studies (GSE4290, GSE7696 and GSE4536), the expression of Six1 mRNA was dramatically elevated in GBM tissues than in normal brain tissue; the mRNA levels increase between 1.95 to 8.70 fold (P < 0.05; Figure 1B-D).
Figure 1.
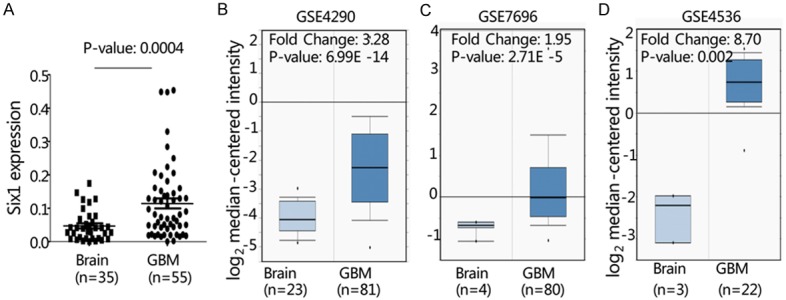
Expression of Six1 in GBM. A. The relative expression of Six1 mRNA was determined by quantitative PCR in primary GBM (n = 55) and adjacent non-tumor brain tissue samples (n = 35). B. ONCOMINE database was used to analyze previously published microarray data (GSE4290, GSE7696 and GSE4536). All results, including P-values, were calculated using ONCOMINE data.
Six1 promoted glioblastoma cell proliferation and invasion
To determine the functional role of Six1 in GBM, we examined the effect of Six1 overexpression on the cell proliferation of U-87 and U-118 cells. These cells were stably transfected with either pcDNA4/TO-Six1 or control vector plasmids. Cell proliferation was assessed by MTT assay and colony formation assay. As shown in Figure 2A, MTT assays showed that cell growth rate of U-87 cells with overexpressed Six1 was significantly higher than that of the control cells at 5 and 6 days after plating (*, P < 0.05). In the colony formation assay, stable overexpression of Six1 in U-87 and U-118 cells caused a significant increase in anchorage dependent growth as evidenced by an increase in the number of colonies compared with control cells. In colony formation assay for U-87 cells, the numbers of colonies formed for vector control and pcDNA4/TO-Six1 group were 45.0 ± 8.5 and 88.5 ± 18.6, respectively (*, P < 0.05, Figure 2B). Similar results of colony formation assay were also obtained in U-118 cells (*, P < 0.05, Figure 2B). These results suggest that overexpression of Six1 increases the anchorage dependent growth of human glioblastoma cells. To determine whether Six1 is required for the glioblatoma cell growth, we silenced Six1 in U-118 cells using Small hairpin RNAi (shRNA) silencing method. We found that cell proliferation was significantly reduced after knockdown of Six1 in U-118 cells (Figure 2C). We next evaluated the effect of Six1 on the cell invasion using matrigel-coated transwell assay. As shown in Figure 2D, overexperssion of Six1 significantly enhanced cell invasion ability of U-118 cells. Invaded cells of control and Six1 group were 22.1 ± 2.6 and 31.1 ± 3.6, respectively. Conversely, suppression of endogenous Six1 attenuated the cell invasion ability of U-118 cells. Invaded cells of control shRNA, Six1 shRNA-1 and Six1 shRNA-2 were 23.2 ± 2.5, 14.1 ± 1.6 and 12.5 ± 4.6, respectively (*, P < 0.05, Figure 2D). Collectively, these data suggest that Six1 may be involved in promoting glioblastoma cell proliferation and invasion.
Figure 2.
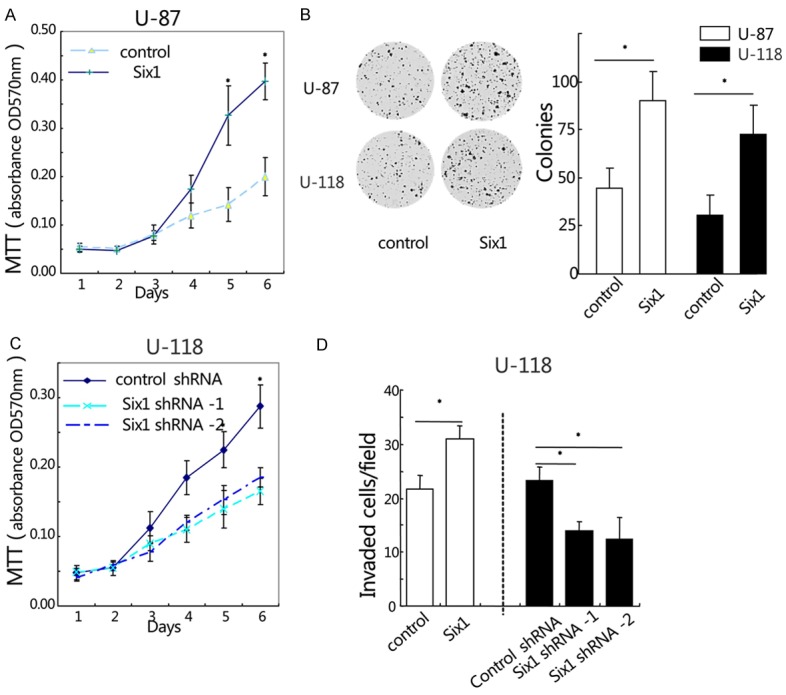
Six1 promotes glioblastoma cell growth and invasion. (A and B) The effect of Six1 overexpression on glioblastoma cell proliferation was measured by MTT assay (A) and colony formation assay (B). (C) MTT assay was performed to determine the effect of Six1 knockdown on cell growth rate in U-118 cells. (D) Cell invasion was determined by matrigel-coated transwell assay. Cells crossed the Matrigel-coated filter were fixed, stained and counted. *P < 0.05. Data represent the mean ± SEM of three independent experiments.
Six1 upregulated CTGF expression in glioblastoma cells
Previous reports showed that CTGF played a key role in the development and tumor progression of GBM [18-20]. Here we found that Six1 was coordinately expressed with CTGF in GBM samples as shown in Figure 3A (r = 0.325, P = 0.020). We also found that forced expression of Six1 in U-87 cells significantly increased the expression of CTGF protein and mRNA (Figure 3B and 3C). Conversely, inhibition of Six1 expression dramatically decreased the expression of CTGF protein and mRNA in U-118 cells (*, P < 0.05, Figure 3B and 3C). Since Six1 is a transcription factor, we next asked whether Six1 could directly regulate the activity of CTGF promoter. U-87 cells were transfected with the human CTGF promoter reporter in the presence of increasing concentrations of human Six1 expression plasmid. It showed that Six1 significantly enhanced the luciferase activity of CTGF promoter reporter in a dose-dependent manner (*, P < 0.05, Figure 3D). These results indicate that Six1 could upregulate CTGF expression at both the mRNA and protein level in GBM.
Figure 3.
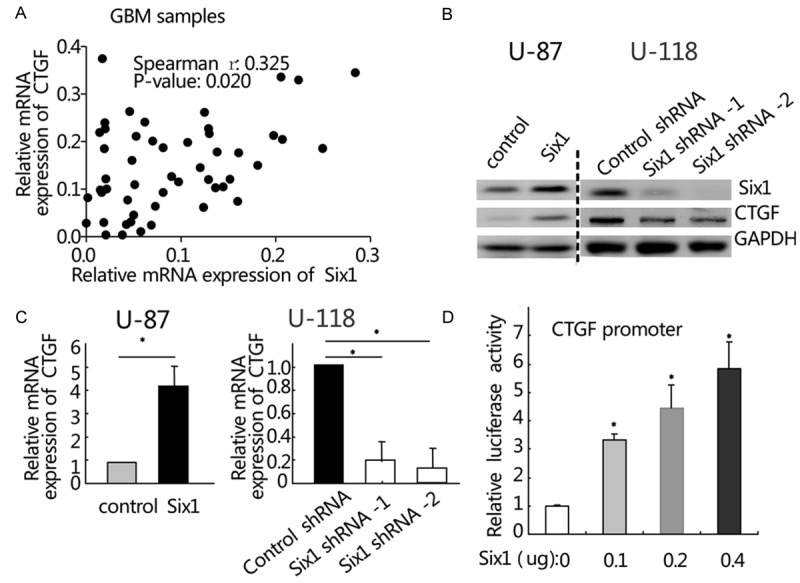
Six1 upregulates CTGF expression in glioblastoma cells. (A) Six1 correlates with CTGF in GBM tissue samples as determined by quantitative PCR (r = 0.325, P = 0.020). (B and C). The effect of Six1 on the expression of CTGF protein (B) and mRNA (C) was examined in U-87 and U-118 cells. GAPDH was used as an internal control. (D). CTGF promoter activity is shown to be responsive to increased amounts of a Six1 expression vector, using a luciferase reporter. *P < 0.05. Data represent the mean ± SEM of three independent experiments.
CTGF was essential for Six1-induced glioblastoma cell proliferation and invasion
To address the role of CTGF in the Six1-induced glioblastoma cell proliferation and invasion, we used RNAi techniques to knock down the CTGF expression in U-87 and U-118 cells stably transduced with Six1. Both MTT and colony formation assay showed that shRNA-mediated silencing of CTGF gene significantly attenuated the growth advantage conferred by Six1 in U-87 cells (*, P < 0.05, Figure 4A and 4B). As shown in Figure 4C, Six1 overexpression enhanced the invasive ability of U-87 cells (control group and Six1 alone group, 19.2 ± 3.2 and 29.8 ± 3.5, respectively; *, P < 0.05). However, Six1-induced cell invasion was significantly inhibited after silencing of endogenous CTGF expression than Six1 alone group in U-87 cells (16.3 ± 4.2 versus 29.8 ± 3.5, *, P < 0.05, Figure 4C). Western blot was performed to determine the protein level of Six1 and CTGF in U-87 cells (Figure 4D). Taken together, these results suggest that CTGF is essential for Six1-induced glioblastoma cell proliferation and invasion.
Figure 4.
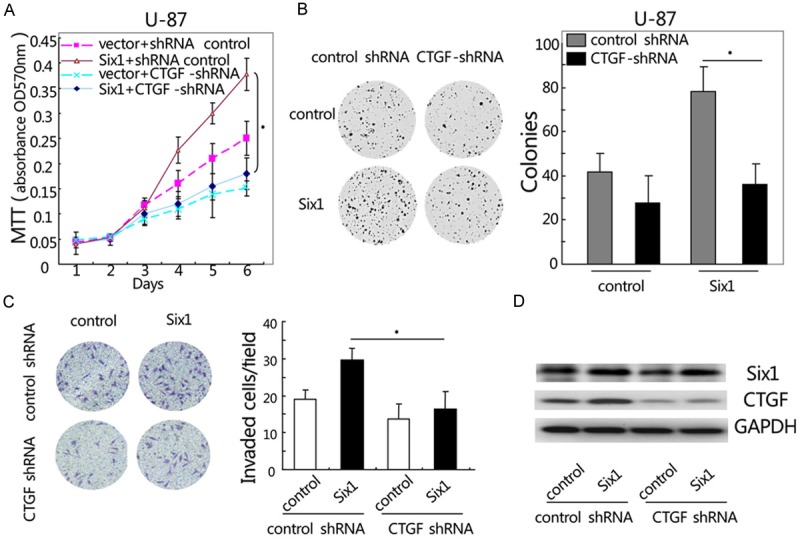
Knockdown of CTGF expression inhibits Six1-induced glioblastoma cell growth and invasion. (A) Suppression of CTGF decreases Six1-induced cell growth as determined by MTT (A) and colony formation assay (B) in U-87 cells. (C) Knockdown of CTGF inhibits Six1-induced U-87 cell invasion as determined by matrigel-coated transwell assay. (D) Western blot was performed to determine the protein level of Six1 and CTGF in U-87 cells. *P < 0.05. Data represent the mean ± SEM of three independent experiments.
Discussion
Six1 has been reported to be dysregulated in a variety of human tumors [4,5]; however, the expression of Six1 and its role in GBM development and progression are not well understood. In this report, we show that Six1 is overexpressed in GBM. Furthermore, by querying three independent datasets from ONCOMINE database, we confirmed the elevated expression of Six1 gene in GBM. However, the clinical significance of Six1 dysregulation and its prognostic value in GBM need to be further studied.
Six1 belongs to the SIX protein family, which is a group of evolutionarily conserved transcription factors found in diverse organisms [5,6]. Six1 is expressed in various tissues including the nervous system during ontogenesis [21], and members of the SIX protein family play pivotal roles in the development of brain [22,23]. More recently, gene expression microarray of glioma tumor progenitor cells uncovered the critical and unrecognized role of Six1 signaling in glioma genesis or progression [11]. In this study, we demonstrate that forced overexpression of Six1 enhances glioblastoma cell proliferation and invasion, while inhibition of Six1 expression significantly reduces cancer cell proliferation and invasion. Our results suggest that Six1 might play an essential role in tumor growth and clinical progression of GBM. This is in agreement with previous report that SIX1-EYA1-DACH2 transcriptional complex plays a significant role in glial tumorigenesis [11].
CTGF belongs to the CCN (Cyr61/Cef10, connective tissue growth factor [CTGF], neuroblastoma overexpressed gene [Nov]) family [24]. It has been implicated in various biological processes, including stimulation of cell proliferation, invasion, angiogenesis and tumorigenesis [25,26]. Studies suggest that CTGF plays a key role in glioma progression by supporting tumor cells survival, invasion and drug resistance [20,27,28]. In this study, we find that Six1 promotes glioblastoma cell proliferation and invasion through upregulation of CTGF. Firstly, we show that Six1 induces CTGF expression and directly regulates CTGF promoter activity. Secondly, Six1 expression significantly correlates with CTGF in GBM samples. Lastly, knockdown of CTGF expression inhibits Six1-induced glioblastoma cell proliferation and invasion.
Taken together, these findings demonstrate that Six1 is overexpressed in GBM and its overexpression may promote glioblastoma cell proliferation and invasion by upregulation of CTGF. Our study provides new insights into the important roles of Six1 and CTGF in tumor regulation, suggesting that Six1 might be a potential therapeutic target for preventing proliferation and metastasis of GBM.
Acknowledgements
This study was supported by funds from the National Natural Science Foundation of China (81172118, 81301087 and 81402380), China Postdoctoral Science Foundation (2013M540574 and 2014T70689) and Youth Innovation Fund Project of The First Affiliated Hospital, Zhengzhou University (to Tian Tian).
Disclosure of conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Fine HA. New Strategies in Glioblastoma: Exploiting the New Biology. Clin Cancer Res. 2015;21:1984–1988. doi: 10.1158/1078-0432.CCR-14-1328. [DOI] [PubMed] [Google Scholar]
- 3.Kesari S. Understanding glioblastoma tumor biology: the potential to improve current diagnosis and treatments. Semin Oncol. 2011;38(Suppl 4):S2–10. doi: 10.1053/j.seminoncol.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Wu W, Ren Z, Li P, Yu D, Chen J, Huang R, Liu H. Six1: A critical transcription factor in tumorigenesis. Int J Cancer. 2015;136:1245–1253. doi: 10.1002/ijc.28755. [DOI] [PubMed] [Google Scholar]
- 5.Kumar JP. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009;66:565–583. doi: 10.1007/s00018-008-8335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen KL, Patrick AN, McCoy EL, Ford HL. The six family of homeobox genes in development and cancer. Adv Cancer Res. 2008;101:93–126. doi: 10.1016/S0065-230X(08)00405-3. [DOI] [PubMed] [Google Scholar]
- 7.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Müller-Tidow C, Golub TR, Kawakami K, Ford HL. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coletta RD, Christensen KL, Micalizzi DS, Jedlicka P, Varella-Garcia M, Ford HL. Six1 overexpression in mammary cells induces genomic instability and is sufficient for malignant transformation. Cancer Res. 2008;68:2204–2213. doi: 10.1158/0008-5472.CAN-07-3141. [DOI] [PubMed] [Google Scholar]
- 9.Behbakht K, Qamar L, Aldridge CS, Coletta RD, Davidson SA, Thorburn A, Ford HL. Six1 overexpression in ovarian carcinoma causes resistance to TRAIL-mediated apoptosis and is associated with poor survival. Cancer Res. 2007;67:3036–3042. doi: 10.1158/0008-5472.CAN-06-3755. [DOI] [PubMed] [Google Scholar]
- 10.Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang L, Li X, Li L, Ma W, Wu J, Zhang M. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS One. 2013;8:e59203. doi: 10.1371/journal.pone.0059203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auvergne RM, Sim FJ, Wang S, Chandler-Militello D, Burch J, Al Fanek Y, Davis D, Benraiss A, Walter K, Achanta P, Johnson M, Quinones-Hinojosa A, Natesan S, Ford HL, Goldman SA. Transcriptional differences between normal and glioma-derived glial progenitor cells identify a core set of dysregulated genes. Cell Rep. 2013;3:2127–2141. doi: 10.1016/j.celrep.2013.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Tian T, Hu X, Zhang X, Li L, Nan F, Chang Y, Wang X, Sun Z, Lv F, Zhang M. Targeting Six1 by lentivirus-mediated RNA interference inhibits colorectal cancer cell growth and invasion. Int J Clin Exp Pathol. 2014;7:631–639. [PMC free article] [PubMed] [Google Scholar]
- 13.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J. Clin. Oncol. 2008;26:3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, Pastorino S, Purow BW, Christopher N, Zhang W, Park JK, Fine HA. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Tian T, Hu X, Zhang X, Nan F, Chang Y, Lv F, Zhang M. Six1 mediates resistance to paclitaxel in breast cancer cells. Biochem Biophys Res Commun. 2013;441:538–543. doi: 10.1016/j.bbrc.2013.10.131. [DOI] [PubMed] [Google Scholar]
- 18.Yin D, Chen W, O’Kelly J, Lu D, Ham M, Doan NB, Xie D, Wang C, Vadgama J, Said JW, Black KL, Koeffler HP. Connective tissue growth factor associated with oncogenic activities and drug resistance in glioblastoma multiforme. Int J Cancer. 2010;127:2257–2267. doi: 10.1002/ijc.25257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwards LA, Woolard K, Son MJ, Li A, Lee J, Ene C, Mantey SA, Maric D, Song H, Belova G, Jensen RT, Zhang W, Fine HA. Effect of brain- and tumor-derived connective tissue growth factor on glioma invasion. J Natl Cancer Inst. 2011;103:1162–1178. doi: 10.1093/jnci/djr224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox JL, Dews M, Minn AJ, Thomas-Tikhonenko A. Targeting of TGFβ signature and its essential component CTGF by miR-18 correlates with improved survival in glioblastoma. RNA. 2013;19:177–190. doi: 10.1261/rna.036467.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeong Y, Oh S. Identification of an evolutionarily conserved, functional noncoding element regulated by Six1 homeoprotein. Genes Genet Syst. 2010;85:233–240. doi: 10.1266/ggs.85.233. [DOI] [PubMed] [Google Scholar]
- 22.Appolloni I, Calzolari F, Corte G, Perris R, Malatesta P. Six3 controls the neural progenitor status in the murine CNS. Cereb Cortex. 2008;18:553–562. [Google Scholar]
- 23.Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, Russell HR, McKinnon PJ, Solnica-Krezel L, Oliver G. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu CY, Chang CC, Prakash E, Kuo ML. Connective tissue growth factor (CTGF) and cancer progression. J Biomed Sci. 2008;15:675–685. doi: 10.1007/s11373-008-9264-9. [DOI] [PubMed] [Google Scholar]
- 25.Dhar A, Ray A. The CCN family proteins in carcinogenesis. Exp Oncol. 2010;32:2–9. [PubMed] [Google Scholar]
- 26.Kubota S, Takigawa M. Cellular and molecular actions of CCN2/CTGF and its role under physiological and pathological conditions. Clin Sci (Lond) 2015;128:181–196. doi: 10.1042/CS20140264. [DOI] [PubMed] [Google Scholar]
- 27.Han N, Shahveranov A, Cheng Y, Qin K, Yu SY, Zhang MX. Effects of connective tissue growth factor (CTGF) gene silencing on the radiosensitivity of glioblastoma. Int J Clin Exp Med. 2014;7:2557–2563. [PMC free article] [PubMed] [Google Scholar]
- 28.Romão LF, Mendes FA, Feitosa NM, Faria JC, Coelho-Aguiar JM, de Souza JM, Moura Neto V, Abreu JG. Connective tissue growth factor (CTGF/CCN2) is negatively regulated during neuron-glioblastoma interaction. PLoS One. 2013;8:e55605. doi: 10.1371/journal.pone.0055605. [DOI] [PMC free article] [PubMed] [Google Scholar]


