Abstract
The most common cancer in children is acute lymphoblastic leukemia (ALL) and it had high cure rate, especially for B-precursor ALL. However, relapse due to drug resistance and overdose treatment reach the limitations in patient managements. In this study, integration of gene expression microarray data, logistic regression, analysis of microarray (SAM) method, and gene set analysis were performed to discover treatment response associated pathway-based signatures in the original cohort. Results showed that 3772 probes were significantly associated with treatment response. After pathway analysis, only apoptosis pathway had significant association with treatment response. Apoptosis pathway signature (APS) derived from 15 significantly expressed genes had 88% accuracy for treatment response prediction. The APS was further validated in two independent cohorts. Results also showed that APS was significantly associated with induction failure time (adjusted hazard ratio [HR] = 1.60, 95% confidence interval [CI] = [1.13, 2.27]) in the first cohort and significantly associated with event-free survival (adjusted HR = 1.56, 95% CI = [1.13, 2.16]) or overall survival in the second cohort (adjusted HR = 1.74, 95% CI = [1.24, 2.45]). APS not only can predict clinical outcome, but also provide molecular guidance of patient management.
Keywords: Acute lymphoblastic leukemia, apoptosis, gene signature, prediction and clinical outcome
Introduction
Acute lymphoblastic leukemia (ALL) is the most common cancer in the children [1]. Because suitable treatment strategies of children ALL were established, overall cure rate was approximately 80% [2], especially for B-precursor ALL [3]. Despite higher cure rate, treatment resistance, toxicity, and reduction of adverse effects of treatment need to be addressed [2,4].
Risk classifications for ALL are based on genetic abnormalities (hypodiploidy, hyperdiploidy, trisomy, and chromosome translocation resulted in gene fusions), clinical characteristics (age, WBC count, sex, and central nervous system involvement), and minimum residual disease [5]. However, some patients still have poor response to the serious treatments and some receive overdose treatment [2,6]. High-throughput technologies such as microarrays could detect expression levels of thousands of genes at once. Gene expressions in different experimental conditions can be used to explore the underlying molecular biology mechanism [6]. Global gene expression profiling could also reveal heterogeneity of cancer cells and provide predictions of drug resistance and clinical outcome of pediatric ALL [6-9]. Until now, several studies demonstrated that gene expression microarrays revealed subtypes of pediatric ALL [10,11] and associations with drug resistance or clinical outcome [12-14], especially for childhood high-risk (HR) B-precursor ALL [3,7,15].
Current studies used single gene approach to identify major differential expressed genes in different treatment response status or clinical outcomes of the diseases [3,7,13,16]. However, because some subtle differential genes expressions might be interactive to result in diverse physiological states [17], it would be better to explore a group of co-expressed genes in the specific biological pathway [18]. Hence, gene set enrichment analysis incorporated biological pathway information could not only increase power and reduce the dimensions of the underlying statistics problem, but also understand the functional mechanism in a cell [19].
In this study, gene set analysis approach was used to identify genes in the same biological pathway associated with early treatment response of HR B-precursor ALL from a published microarray data [15]. Identified pathway-based signature had high accuracy for prediction of treatment response and clinical outcome. The results were validated in two independent cohorts. The findings of this study may have potential for high risk patients selection and improve the clinical managements of HR B-precursor ALL.
Materials and methods
Study population and gene expression microarray data
Published clinical and microarray data including 99 patients with National Cancer Institute-defined HR B-precursor ALL (age ≥ 10 years and/or WBC ≥ 50,000/μl) were used in this study [15]. These patients were treated uniformly with the Children’s Oncology Group (COG) 1961 protocol and received four-drug induction. Treatment response was assessed by minimal residual disease (MRD) testing on day 7 and classified into slow early responses (SER) or rapid early responders (RER) with 25% blasts in the marrow as a threshold. Long-term clinical outcome was evaluated whether achieved complete continuous remission (CCR) for at least 4 years after initial diagnosis. Clinical characteristics of all subjects were briefly summarized in the Table 1.
Table 1.
Clinical characteristics of 99 patients in the children’s Oncology Group (COG) 1961
| Variable | RER* | SER* | p-value |
|---|---|---|---|
| N | 59 | 40 | |
| Age (months) | 132.47 ± 59.19 | 107.60 ± 63.48 | 0.05§ |
| WBC (×109/L) | 88391 ± 119156 | 121085 ± 123159 | 0.19§ |
| Gender | 0.84£ | ||
| Female | 22 (37.29) | 16 (40.00) | |
| Male | 37 (62.71) | 24 (60.00) | |
| Translocation | 0.01£ | ||
| t (12;21) | 4 (6.78) | 4 (10.00) | |
| t (1:19) | 9 (15.25) | 0 (0.00) | |
| t (4,11) | 3 (5.08) | 1 (2.50) | |
| t (9,22) | 1 (1.69) | 5 (12.50) | |
| non | 42 (71.19) | 30 (75.00) | |
| MRD | |||
| M1 | 42 (71.19) | 0 (0.00) | |
| M2 | 17 (28.81) | 0 (0.00) | |
| M3 | 0 (0.00) | 40 (100.00) | |
| Outcome | 0.42£ | ||
| CCR | 19 (32.20) | 9 (22.50) | |
| relapse | 17 (28.81) | 14 (35.00) | |
| missing | 23 (38.98) | 17 (42.50) |
RER, rapid early response; SER, slowly early response;
p-value of t test;
p-value of Fisher’s exact test.
Affymetrix HG-U133Plus2.0 microarray raw data (cel files) were downloaded from the National Center for Biotech-nology Information Gene Expression Omnibus (GEO) (series accession number GSE7440) [15]. Microarray raw data were analyzed with Affymetrix Microarray Suite (MAS 5.0) for further analysis.
Two independent microarray data sets download from public databases were used for validation of the outcome signatures. The first validation cohort was 220 patients with pediatric B-precursor ALL treated on Pediatric Oncology Group (POG) 9006 phase III clinical trial, and the microarray expression data was downloaded from the National Cancer Institute Cancer Array Informatics website (https://catissuesuite.ecmc.ed.ac.uk/caarray/home.action, Experiment ID 1015897590271440) [10]. The second validation cohort was 207 patients with high-risk B-precursor ALL enrolled in Children’s Oncology Group (COG) Clinical Trial P9906, and the microarray expression data was downloaded from the Gene Expression Omnibus (GEO) database of the National Center for Biotechno-logy Information (series accession number GSE11877) [3,7]. Microarray platforms of two datasets were Affymetrix U95Av2 and U133-Plus2.0, respectively.
Statistical analysis
To reduce variation among microarrays, the intensity values of each sample were normalized by quantile-normalized method [20]. Finally, each intensity value was taken logarithm transformation with base 2. For determining whether the expression level of genes from microarray data associated with treatment response of HR B-precursor ALL, multivariate logistic regression with clinical covariates (age, sex, WBC, and chromosome translocation status) was performed to select potential candidate genes. If the p-value of regression coefficient of a gene was less than 0.05, this gene was selected as candidate gene. Next, significantly analysis of microarray (SAM) method was used to control false discovery rate (< 0.05) [21].
Efron-Tibshirani’s GSA maxmean statistics for gene set analysis [22] was employed to identify the treatment response associated biological pathways in HR B-precursor ALL. The Hotelling T2-test [23] was used to identify the significantly different expression profiles in the specified pathway between different treatment response groups. Besides, MetaCore (Thomson Reuters, city, state) was applied to determine the disease related genes.
In order to investigate the relevant pathway impact on treatment in HR B-precursor ALL, the pathway-based risk score method was calculated in each subject. It was a linear combination of expression values of significantly differential genes in the associated pathway and weighted by coefficients of multivariate logistic regression of genes. The pathway-based risk score was compared between RER and SER by t test. Receiver operating characteristic (ROC) curve with Youden index [24] for cut off point seleciton was used to evaluate the predictive accuracy of the pathway-based signatures.
For validating signatures in the specific pathway as pathway-base signatures of HR B-precursor ALL, pathway-based risk scores calculation, ROC curve with Youden index for cut-point selection as high- or low-risk group grouping were also applied in the 2 independent cohorts. The Kaplan-Meier method was used to estimate overall survival (OS), event-free survival (EFS), or induction failure time. The log-rank test was used to test the survival difference between two groups. Multivariate Cox proportional hazards regression analysis with clinical covariates such as age, sex, WBC count, and chromosome translocation was carried out to evaluate the independent prognostic factor for pathway-based risk scores. All statistical analyses were computed by SAS (SAS Institute Inc. Cary, NC), R (http://www.R-project.org), and BRB tools (http://linus.nci.nih.gov/BRB-ArrayTools.html). All tests were two-tailed. P values < 0.05 were considered significant.
Results
Differentially expressed genes between responder and non-responder
Multivariate logistic regressions were used to evaluate the associations between genes and treatment response status. The results showed that expression values of 3798 probes out of 54675 probes were significantly associated with early treatment response of pediatric HR B-precursor ALL. Comparing to SAM analysis, 3772 probes were still significant under false discovery rate < 0.05. These significant genes were most pertinent to leukemia disease (Figure 1). In order to incorporate biological knowledge into the analysis, we applied gene set analysis using maxmean statistics. Apo-ptosis pathway was the only highly significant pathway in the KEGG database under 200 permutations (p-value < 0.005). Total fifteen differential expression genes were in this apoptosis pathway, four were protected genes (odds ratio (OR) < 1) and eleven were risk genes (OR > 1) (Table 2). Significantly differential expression profile of fifteen genes in the apoptosis pathway between treatment response groups was observed (Hotelling T2-test, p-value < 0.001).
Figure 1.
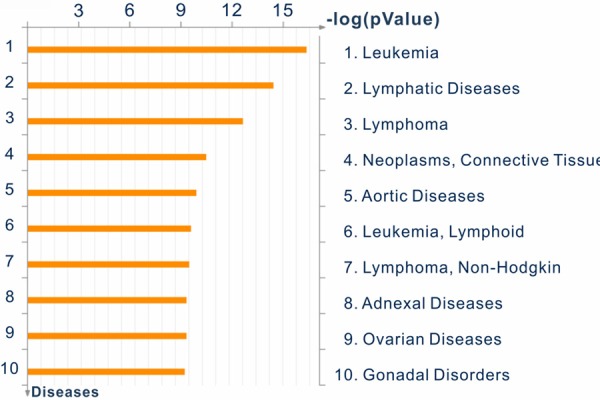
Significant differential expressed genes enriched in specific disease categories.
Table 2.
Differential expressed genes (n = 15) in the apoptosis pathway associated treatment response for childhood HR B-precursor ALL
| Gene Symbol | Crude p-value | Adjusted p-valueb | Adjusted OR (95% CI)b | Probe sets |
|---|---|---|---|---|
| IL1RAPa | 0.0012 | 0.0025 | 1.98 (1.27, 3.10) | 219489_s_at, 210233_at |
| IRAK3a | 0.0002 | 0.0005 | 2.60 (1.52, 4.46) | 213817_at, 1568830_at, 220034_at |
| IRAK2 | 0.0150 | 0.0267 | 1.48 (1.05, 2.08) | 1553740_a_at |
| PPP3CA | 0.0167 | 0.0210 | 1.87 (1.10, 3.18) | 202429_s_at |
| PRKAR2B | 0.0037 | 0.0008 | 1.85 (1.29, 2.64) | 203680_at |
| CHP | 0.0139 | 0.0115 | 0.59 (0.39, 0.89) | 207993_s_at |
| TNFRSF10B | 0.0224 | 0.0291 | 1.81 (1.06, 3.09) | 209295_at |
| IKBKG | 0.0145 | 0.0262 | 2.98 (1.14, 7.80) | 209929_s_at |
| CFLAR | 0.1264 | 0.0355 | 0.53 (0.30, 0.96) | 209939_x_at |
| PIK3R1 | 0.0616 | 0.0468 | 0.55 (0.31, 0.99) | 212240_s_at |
| FAS | 0.0102 | 0.0281 | 1.40 (1.04, 1.89) | 215719_x_at |
| XIAP | 0.0008 | 0.0011 | 4.73 (1.86, 12.02) | 225858_s_at |
| PIK3R5 | 0.0002 | 0.0010 | 2.17 (1.36, 3.44) | 227645_at |
| CYCS | 0.0051 | 0.0231 | 0.40 (0.18, 0.88) | 229415_at |
| BCL2a | 0.0006 | 0.0006 | 3.16 (1.64, 6.12) | 207005_s_at, 207004_at, 203685_at |
Note: p-value and adjusted ORs were calculated from logistic regression.
Gene expression level was the average of multiple probes.
Estimates was adjusted for covariates (age, presenting white blood cell count, gender, chromosome translocation status).
Abbreviations: OR, odds ratio; 95% CI, 95% confidential interval.
Apoptosis-pathway signature (APS) for treatment response and clinical outcome
Gene expression profiles of fifteen genes were used to construct apoptosis pathway-based risk score. Pathway-based risk score was significant difference between RER and SER groups (p-value < 0.0001) (Figure 2A). The ROC curve also showed that the accuracy was 0.8809 (p-value < 0.0001) (Figure 2C). Positive predicted value (PPV) and Negative predicted value (NPV) were 84.85% and 81.82%, respectively. Because of positive correlations between treatment response and clinical outcome for pediatric ALL, we used this pathway-based signature to predict clinical outcome of B-precursor ALL. Pathway-based risk score between CCR and relapse groups was significant difference (p-value < 0.0001) (Figure 2B). The ROC curve analysis also showed that accuracy was still around 80% (p-value < 0.0001) (Figure 2D).
Figure 2.
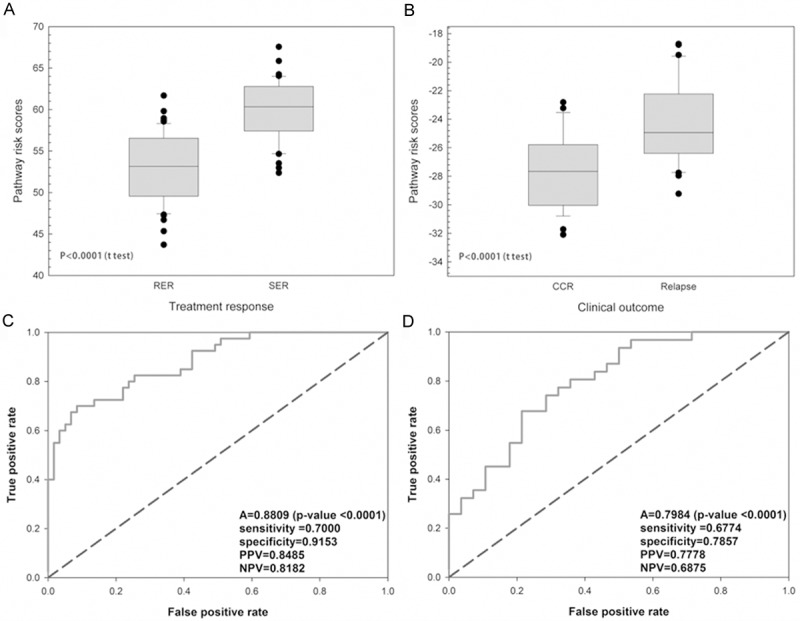
Risk score distribution and prediction ability of APS in the different outcomes. The APS-based risk score incorporated fifteen differential expressed genes. Distributions of score were significantly different between (A) treatment response and (B) clinical outcome, respectively. Receiver operating characteristics (ROC) curves of the predicted score of (C) treatment response reached the 88.09% accuracy and (D) clinical outcome reached the about 80% accuracy.
Validation of APS in the two independent cohorts
Two large independent cohorts (sample size = 220 and 207) were used to validate APS. Patients were classified into the high or low risk group based on APS. The cut-off point was calculated according the Youden index of the ROC curve.
The median follow-up for 220 patients with B-precursor ALL in the POG 9006 phase III clinical trial was about 33 months. We selected 135 patients as HR B-precursor ALL according to NCI definition such as age greater than 10 or WBC greater than 50000 per microliter. The median follow up was about 28 months. After risk classification, the induction failure time of the high-risk group in HR B-precursor ALL was shorter than low-risk group (p-value = 0.0019) (Figure 3A). The adjusted hazard ratio (HR) for APS was 1.60 (95% confidence interval [CI] = 1.13 to 2.27, p value = 0.0080) (Table 3). Results of survival analysis in the B-precursor ALL were similar to those in the HR B-precursor ALL. The high-risk group had a shorter induction failure time than in low-risk group (p-value = 0.0002) (Figure 3B). The multivariate Cox regression analysis with clinical covariates showed that APS was significantly associated with induction failure of B-precursor ALL (adjusted HR = 1.54, 95% CI = 1.13 to 2.10, p-value = 0.0065) (Table 3). APS was also associated with induction failure of B-precursor ALL, not only for HR B-precursor ALL.
Figure 3.
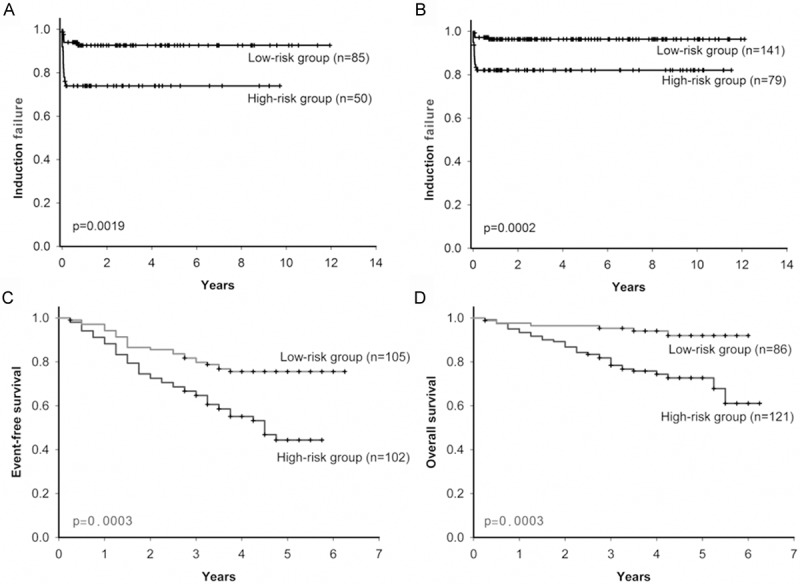
Kaplan-Meier estimates of clinical outcome based on APS in the two independent cohorts. In the POG 9006 phase III clinical trial, (A) high-risk group had significantly shorter induction failure time than low-risk group in HR Precursor-B ALL (n = 135); (B) in all precursor-B ALL (n = 220), high-risk group also had significantly shorter induction failure time than low-risk group. In the COG clinical trial P9906, (C) high-risk group both had shorter median event-free survival and (D) median overall survival than low-risk group.
Table 3.
Validations of APS in two independent cohorts
| Independents cohorts | Prediction outcome | Crude | Adjusted with clinical variables | Adjusted with cytogenetic abnormality | Adjusted with clinical variables and cytogenetic abnormality | |||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||
| HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | |||
| POG 9006 phase III clinical trial | ||||||||||
| B-precursor ALL | Treatment failure | 1.72 (1.32-2.25) | < 0.0001 | 1.58 (1.20-2.08)b | 0.0013b | 1.56 (1.16-2.10)c | 0.0034c | 1.54 (1.13-2.10)d | 0.0065d | |
| HR B-precursor ALLa | Treatment failure | 1.75 (1.30-2.34) | 0.0002 | 1.68 (1.24-2.28)b | 0.0008b | 1.56 (1.13-2.17)c | 0.0069c | 1.60 (1.13-2.27)d | 0.0080d | |
| COG Clinical Trial P9906 | ||||||||||
| HR B-precursor ALL | Death or relapse | 1.56 (1.14-2.13) | 0.0057 | 1.56 (1.13-2.17)e | 0.0072e | 1.56 (1.14-2.13)f | 0.0051f | 1.56 (1.13-2.16)g | 0.0071g | |
| HR B-precursor ALL | Death | 1.81 (1.32-2.48) | 0.0002 | 1.71 (1.24-2.37)e | 0.0011e | 1.88 (1.34-2.64)f | 0.0003f | 1.74 (1.24-2.45)g | 0.0013g | |
According to High-risk B-precursor ALL (HR B-precursor ALL, age ≥ 10 years and/or presenting WBC ≥ 50,000/μL) definition by the National Cancer Institute, the subset from B-precursor ALL in the POG 9006 phase III clinical trial were selected.
Adjusted for age, gender, presenting white blood cell count, and race.
Adjusted for t(12;21), t(1;19), t(9;22), t(4;11), hyperdiploid, and hypodiploid.
Adjusted for age, gender, presenting white blood cell count, race, t(12;21), t(1;19), t(9;22), t(4;11), hyperdiploid, and hypodiploid.
Adjusted for age, gender, presenting white blood cell count, race, and central nervous system status at diagnosis.
Adjusted for E2A-PBX1 translocations, and MLL translocations.
Adjusted for age, gender, presenting white blood cell count, race, and central nervous system status at diagnosis, E2A-PBX1 translocations, and MLL translocations.
Abbreviations: HR B-precursor ALL, High-risk B-precursor ALL; POG, Pediatric Oncology Group; COG, Children’s Oncology Group; HR, hazard ratio; 95% CI, 95% confidential interval.
We also validated this APS in another independent cohort with 207 HR B-precursor ALL patients enrolled in Children’s Oncology Group (COG) Clinical Trial P9906. Patients who were grouped into the high-risk group had shorter event-free survival than in the low-risk group (p-value = 0.0003) (Figure 3C). The similar results were also found in the overall survival (p-value = 0.0003) (Figure 3D). Results of multivariate Cox regression analyses showed that the APS was significantly associated with relapse or death (adjusted HR = 1.56, 95% CI = 1.13 to 2.16, p-value = 0.0071) (Table 3) or overall survival (adjusted HR = 1.74, 95% CI = 1.24 to 2.45, p-value = 0.0013) (Table 3).
Discussion
APS for HR B-precursor ALL was derived from gene expression microarray and gene set analysis enriched in biological pathways. APS was explored by detecting the differential expressed genes in the specific pathway associated with treatment response. Then treatment-related signature APS was applied to predict treatment response and clinical outcome. The results showed that APS had high accuracy (80%) for predictions of treatment response and clinical outcome. Furthermore, APS were validated in two independent cohorts. Results not only showed that patients with high risk signature had shorter event-free or overall survival, but also had shorter induction failure time in the HR B-precursor ALL and all B-precursor ALL.
The findings of this study were different with previous study [15] which used the same microarray and clinical data of HR B-precursor ALL. Bhojwani et al. found that 24 significant probe sets had prediction power for treatment response. Interestingly, the 23 out of 24 probe sets were also obtained in our significant probe set list (3772 probes) but not enriched in the apoptosis pathway. Only gene BCL2 was in our apoptosis-gene-signature. The different findings between Bhojwani et al. and this study may due to different analysis strategies and statistical methods. In particular, treatment response associated specific apoptosis-gene-signature was found in this study.
HR precursor-B ALL patients with the same clinical characteristics and cytogenetic features showed different treatment and clinical outcomes and introduced relapse or the treatment toxicity [2]. It indicated it is the heterogeneous disease. Hence, current clinical features and cytogenetic markers based risk stratification strategy for children ALL treatment may reach its limitations. Gene expression profiling using microarray technologies could explore subtype of ALL and provide important insight for the drug response and clinical outcome in childhood ALL [13,25,26]. In advance, the gene set analysis strategy incorporated biological pathways information and gene expression profiling to explore outcome associated important pathway and had higher prediction power for treatment response or clinical outcome [27].
The majority of B-cell ALL are found to be associated with chromosome aberration such as chromosome translocation and hyperdiploid [28]. Genomic structure variation can be used to do risk classification and decision of treatment strategy in B-precursor ALL [29]. Through developments of microarray and next generation sequencing, combinations of genomic mutations and chromosome structure variations will be useful tool for subtypes classification of ALL [29]. However, subtype grouping is almost based on genomic information at this time. Gene expression level might add more clues to predict patients’ prognosis or treatment response [30,31]. In this study, independent of genomic structure variation and clinical characteristics, APS combining pathway and gene expression information could predict clinical outcome of precursor B-ALL. This would give more clues for pathogenesis exploring and add value for patients’ risk stratification for treatment strategy.
Some studies showed that alterations in the expression of apoptosis associated genes may involve in the progress of B-cell leukemia [32] and affect treatment sensitivity [33]. The vast majority of associated genes were upregulated in patients with relapse [34]. In our study, results also show that eleven of fifteen differential expressed genes had ORs greater than 1. Patients with higher expressed values of these risk genes would tend to have higher risk of worse treatment response. Identification of important apoptosis genes could not only understand the role of apoptosis involved in progression mechanism but also may provide novel tools for diagnosis and targets for drug development in pediatric ALL [32]. Recent studies demonstrated that targeting therapies for pediatric ALL could improve cure rate and possibly minimize toxicity [35]. In this study, three out of 15 genes (BCL2, XIAP, and TNFRSF10B) in the apoptosis pathway have been reported as potential therapeutic targets in pediatric ALL and investigated in clinical trials [35].
APS was significantly associated with treatment response and clinical outcome of HR B-precursor ALL. This signature not only could be used for risk stratification but provided potential molecular targets for drug development.
Acknowledgements
Supported by grants from the Institute of Statistical Science of the Academia Sinica, AS-100-TP-AB1, AS-100-TP-AB2, NSC 98-3112-B-001-034, NSC 99-2314-B-001-003-MY3, NSC 100-2325-B-001-027, NSC 101-2325-B-002-071, NSC 102-2325-B-002-078, NSC 101-2319-B-002-002, NSC 102-2319-B-002-002, NSC 102-2911-I-002-303, NSC 101-2911-I-002-303, and NSC 102-2911-I-002-303, DOH101-TD-B-111-001, and 102R7557.
Disclosure of conflict of interest
None.
References
- 1.Belson M, Kingsley B, Holmes A. Risk factors for acute leukemia in children: a review. Environ Health Perspect. 2007;115:138–145. doi: 10.1289/ehp.9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 3.Harvey RC, Mullighan CG, Wang X, Dobbin KK, Davidson GS, Bedrick EJ, Chen IM, Atlas SR, Kang H, Ar K, Wilson CS, Wharton W, Murphy M, Devidas M, Carroll AJ, Borowitz MJ, Bowman WP, Downing JR, Relling M, Yang J, Bhojwani D, Carroll WL, Camitta B, Reaman GH, Smith M, Hunger SP, Willman CL. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia with gene expression profiling: correlation with genome-wide DNA copy number alterations, clinical characteristics and outcome. Blood. 2010;116:4874–4884. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaynon PS, Angiolillo AL, Carroll WL, Nachman JB, Trigg ME, Sather HN, Hunger SP, Devidas M. Long-term results of the children’s cancer group studies for childhood acute lymphoblastic leukemia 1983-2002: a Children’s Oncology Group Report. Leukemia. 2010;24:285–297. doi: 10.1038/leu.2009.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schultz KR, Pullen DJ, Sather HN, Shuster JJ, Devidas M, Borowitz MJ, Carroll AJ, Heerema NA, Rubnitz JE, Loh ML, Raetz EA, Winick NJ, Hunger SP, Carroll WL, Gaynon PS, Camitta BM. Risk- and response-based classification of childhood B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG) Blood. 2007;109:926–935. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Szczepanek J, Styczynski J, Haus O, Tretyn A, Wysocki M. Relapse of acute lymphoblastic leukemia in children in the context of microarray analyses. Arch Immunol Ther Exp (Warsz) 2011;59:61–68. doi: 10.1007/s00005-010-0110-1. [DOI] [PubMed] [Google Scholar]
- 7.Kang H, Chen IM, Wilson CS, Bedrick EJ, Harvey RC, Atlas SR, Devidas M, Mullighan CG, Wang X, Murphy M, Ar K, Wharton W, Borowitz MJ, Bowman WP, Bhojwani D, Carroll WL, Camitta BM, Reaman GH, Smith MA, Downing JR, Hunger SP, Willman CL. Gene expression classifiers for relapse-free survival and minimal residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115:1394–1405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhojwani D, Moskowitz N, Raetz EA, Carroll WL. Potential of gene expression profiling in the management of childhood acute lymphoblastic leukemia. Paediatr Drugs. 2007;9:149–156. doi: 10.2165/00148581-200709030-00003. [DOI] [PubMed] [Google Scholar]
- 9.Flotho C, Coustan-Smith E, Pei DQ, Cheng C, Song GC, Pui CH, Downing JR, Campana D. A set of genes that regulate cell proliferation predicts treatment outcome in childhood acute lymphoblastic leukemia. Blood. 2007;110:1271–1277. doi: 10.1182/blood-2007-01-068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin SB, Mosquera-Caro MP, Potter JW, Davidson GS, Andries E, Kang H, Helman P, Veroff RL, Atlas SR, Murphy M, Wang X, Ar K, Xu Y, Chen IM, Schultz FA, Wilson CS, Harvey R, Bedrick E, Shuster J, Carroll AJ, Camitta B, Willman CL. Gene expression overlap affects karyotype prediction in pediatric acute lymphoblastic leukemia. Leukemia. 2007;21:1341–1344. doi: 10.1038/sj.leu.2404640. [DOI] [PubMed] [Google Scholar]
- 11.Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, Olofsson T, Rade J, Fontes M, Porwit-Macdonald A, Behrendtz M, Hoglund M, Johansson B, Fioretos T. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 12.Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, Condos G, Hunger SP, Raetz E, Saffery R, Relling MV, Bhojwani D, Morrison DJ, Carroll WL. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118:5218–5226. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holleman A, Cheok MH, den Boer ML, Yang W, Veerman AJ, Kazemier KM, Pei D, Cheng C, Pui CH, Relling MV, Janka-Schaub GE, Pieters R, Evans WE. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351:533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 14.Yang YL, Lin SR, Chen JS, Lin SW, Yu SL, Chen HY, Yen CT, Lin CY, Lin JF, Lin KH, Jou ST, Hu CY, Chang SK, Lu MY, Chang HH, Chang WH, Lin KS, Lin DT. Expression and prognostic significance of the apoptotic genes BCL2L13, Livin, and CASP8AP2 in childhood acute lymphoblastic leukemia. Leuk Res. 2010;34:18–23. doi: 10.1016/j.leukres.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Bhojwani D, Kang H, Menezes RX, Yang W, Sather H, Moskowitz NP, Min DJ, Potter JW, Harvey R, Hunger SP, Seibel N, Raetz EA, Pieters R, Horstmann MA, Relling MV, den Boer ML, Willman CL, Carroll WL. Gene expression signatures predictive of early response and outcome in high-risk childhood acute lymphoblastic leukemia: A Children’s Oncology Group Study [corrected] . J. Clin. Oncol. 2008;26:4376–4384. doi: 10.1200/JCO.2007.14.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zembutsu H, Yanada M, Hishida A, Katagiri T, Tsuruo T, Sugiura I, Takeuchi J, Usui N, Naoe T, Nakamura Y, Ohno R. Prediction of risk of disease recurrence by genome-wide cDNA microarray analysis in patients with Philadelphia chromosome-positive acute lymphoblastic leukemia treated with imatinib-combined chemotherapy. Int J Oncol. 2007;31:313–322. [PubMed] [Google Scholar]
- 17.Schadt EE. Molecular networks as sensors and drivers of common human diseases. Nature. 2009;461:218–223. doi: 10.1038/nature08454. [DOI] [PubMed] [Google Scholar]
- 18.Medina I, Montaner D, Bonifaci N, Pujana MA, Carbonell J, Tarraga J, Al-Shahrour F, Dopazo J. Gene set-based analysis of polymorphisms: finding pathways or biological processes associated to traits in genome-wide association studies. Nucleic Acids Res. 2009;37:W340–344. doi: 10.1093/nar/gkp481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackermann M, Strimmer K. A general modular framework for gene set enrichment analysis. BMC Bioinformatics. 2009;10:47. doi: 10.1186/1471-2105-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 21.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.B E, R T. On testing the significance of sets of genes. Annals of Applied Statistics. 2007;1:107–129. [Google Scholar]
- 23.Hotelling H. The generalization of Student’s ratio. Annals of Mathematical Statistics. 1931;2:360–378. [Google Scholar]
- 24.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Moos PJ, Raetz EA, Carlson MA, Szabo A, Smith FE, Willman C, Wei Q, Hunger SP, Carroll WL. Identification of gene expression profiles that segregate patients with childhood leukemia. Clin Cancer Res. 2002;8:3118–3130. [PubMed] [Google Scholar]
- 26.Yeoh EJ, Ross ME, Shurtleff SA, Williams WK, Patel D, Mahfouz R, Behm FG, Raimondi SC, Relling MV, Patel A, Cheng C, Campana D, Wilkins D, Zhou XD, Li JY, Liu HQ, Pui CH, Evans WE, Naeve C, Wong LS, Downing JR. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 27.Bild AH, Yao G, Chang JT, Wang QL, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Marks JR, Dressman HK, West M, Nevins JR. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J. Clin. Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 29.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122:3407–3415. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Den Boer ML, van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven LJ, Beverloo HB, Van der Spek PJ, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, Evans WE, Pieters R. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol. 2009;10:125–134. doi: 10.1016/S1470-2045(08)70339-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahgozar S, Moafi A, Abedi M, Entezar EGM, Moshtaghian J, Ghaedi K, Esmaeili A, Montazeri F. mRNA expression profile of multidrug-resistant genes in acute lymphoblastic leukemia of children, a prognostic value for ABCA3 and ABCA2. Cancer Biol Ther. 2014;15:35–41. doi: 10.4161/cbt.26603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullauer L, Gruber P, Sebinger D, Buch J, Wohlfart S, Chott A. Mutations in apoptosis genes: a pathogenetic factor for human disease. Mutat Res. 2001;488:211–231. doi: 10.1016/s1383-5742(01)00057-6. [DOI] [PubMed] [Google Scholar]
- 33.Mata JF, Silveira VS, Mateo EC, Cortez MA, Queiroz RG, Yunes JA, Lee ML, Toledo SR, Petrilli AS, Brandalise SR, Tone LG, Scrideli CA. Low mRNA Expression of the Apoptosis-Related Genes CASP3, CASP8, and FAS Is Associated With Low Induction Treatment Response in Childhood Acute Lymphoblastic Leukemia (ALL) Pediatr Blood Cancer. 2010;55:100–107. doi: 10.1002/pbc.22463. [DOI] [PubMed] [Google Scholar]
- 34.Bhojwani D, Kang H, Moskowitz NP, Min DJ, Lee H, Potter JW, Davidson G, Willman CL, Borowitz MJ, Belitskaya-Levy I, Hunger SP, Raetz EA, Carroll WL. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 2006;108:711–717. doi: 10.1182/blood-2006-02-002824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee-Sherick AB, Linger RM, Gore L, Keating AK, Graham DK. Targeting paediatric acute lymphoblastic leukaemia: novel therapies currently in development. Br J Haematol. 2010;151:295–311. doi: 10.1111/j.1365-2141.2010.08282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


