Abstract
Photosystem I comprises 13 subunits in Chlamydomonas reinhardtii, four of which—the major reaction center I subunits PsaA and PsaB, PsaC and PsaJ—are chloroplast genome-encoded. We demonstrate that PSI biogenesis involves an assembly-governed regulation of synthesis of the major chloroplast-encoded subunits where the presence of PsaB is required to observe significant rates of PsaA synthesis and the presence of PsaA is required to observe significant rates of PsaC synthesis. Using chimeric genes expressed in the chloroplast, we show that these regulatory processes correspond to autoregulation of translation for PsaA and PsaC. The downregulation of translation occurs at some early stage since it arises from the interaction between unassembled PsaA and PsaC polypeptides and 5′ untranslated regions of psaA and psaC mRNAs, respectively. These assembly-dependent autoregulations of translation represent two new instances of a control by epistasy of synthesis process that turns out to be a general feature of protein expression in the chloroplast of C. reinhardtii.
Keywords: CES process, Chlamydomonas reinhardtii, chloroplast, photosystem I assembly, regulation of translation
Introduction
Many enzymatic functions are carried out by hetero-oligomeric proteins. The assembly of their constitutive subunits in an appropriate stoichiometry can be regarded as spontaneous, being a thermodynamically favoured process. Still, biological systems have optimised the rate of protein production in order to avoid wasteful accumulation of unassembled subunits. For instance, in prokaryotes, the operonal organisation of many genes allows a fine-tuning of the coupled transcription/translation rates of subunits from the same protein complex. In many other instances, a proteolytic disposal of unassembled subunits operates as a backup system in biogenesis pathways. The biogenesis of oligomeric protein complexes in the energy transducing membranes of organelles has unique features of particular complexity. Most of these protein complexes comprise more than 10 distinct subunits, some of which are nucleus-encoded, whereas the others are encoded by the organellar genome (Fox, 1996; Wollman et al, 1999). Because the gene copy numbers for nucleus- versus organelle-encoded subunits from a same protein complex can differ by as much as four orders of magnitude, organelle-based regulation processes, as well as crosstalks between the nucleo-cytosolic and organelle compartments, should be at work to regulate the level of expression of the various subunits of these oligomeric proteins.
Studies of respiratory mutants from yeast or of photosynthetic mutants from the unicellular green alga Chlamydomonas reinhardtii have been instrumental in this major issue in cell biology (Fox, 1996; Wollman et al, 1999). These studies showed that the accumulation of the various subunits of these oligomeric proteins is a concerted process: most mutant strains deficient for the expression of a major protein subunit present a pleiotropic loss of the whole set of subunits of the complex. Two main mechanisms are responsible for this concerted accumulation in the chloroplast of C. reinhardtii (reviewed in Wollman et al, 1999; Choquet and Vallon, 2000). Many unassembled subunits undergo a rapid proteolytic degradation but others show an assembly-dependant regulation of their rates of synthesis. We have defined this assembly-dependent regulatory process as a ‘control by epistasy of synthesis' or CES process (Wollman et al, 1999; Choquet and Vallon, 2000). In the case of cytochrome f, encoded by the chloroplast petA gene, we were able to characterise the molecular mechanism underlying the CES process as an autoregulation of petA mRNA translation (Choquet et al, 1998) that involves negative feedback from the C-terminal domain of the unassembled polypeptide (Choquet et al, 2003).
In the present study, we provide evidence that assembly-dependent autoregulation of translation initiation may be a central mechanism in the biogenesis of chloroplast oligomeric proteins in C. reinhardtii. Photosystem I (PSI) is a thylakoid-embedded pigment–protein complex that performs light-induced charge separation and drives electron transfer from plastocyanin to ferredoxin. In C. reinhardtii, PSI is made of 13 subunits: PsaA, B, C and J are chloroplast-encoded, whereas PsaD, E, F, G, H, K, L, N and O are nucleus-encoded (for a review, see Webber and Bingham, 1998). The core components of PSI are two large transmembrane subunits, PsaA and PsaB, which share strong sequence similarity and may have arisen from gene duplication (reviewed in Baymann et al, 2001). Both subunits comprise 11 transmembrane helices. The N-terminus of each subunit faces the chloroplast stroma, whereas their C-termini protrude on the lumenal side of the thylakoid (Sun et al, 1997). PsaA and PsaB assemble at an early step of PSI biogenesis, forming the chlorophyll a–protein complex I (CPI) that binds most of the pigments and redox cofactors of PSI. CPI is the template for assembly of the extrinsic PsaC subunit on the stromal side of the membranes. PsaC, a 9 kDa polypeptide, coordinates the Fe–S clusters FA and FB through two cysteine-rich domains. The stromal subunits PsaD and PsaE then assemble coordinately around PsaC (Yu et al, 1995).
PSI mutants lacking either PsaA, PsaB or PsaC display the same severe drop in the accumulation of all PSI subunits, demonstrating that accumulation of these proteins is a concerted process. Strains deleted for psaC show wild-type rates of synthesis of PsaB and PsaA, which are then rapidly degraded (Takahashi et al, 1991). In contrast, psaA mutants show wild-type rates of PsaB synthesis (Girard-Bascou et al, 1987; Goldschmidt-Clermont et al, 1990) but reduced rates of PsaC synthesis (Takahashi et al, 1991). Finally, mutants lacking expression of PsaB show no detectable PsaA synthesis, whether the strains contain mutations in the chloroplast psaB gene itself (Girard-Bascou et al, 1987), or in the nuclear TAB1 gene, which is required for psaB mRNA translation initiation (Stampacchia et al, 1997). A chloroplast mutation in the psaB 5′ untranslated region (UTR) suppresses the effect of this nuclear defect and restores translation of both PsaB and PsaA subunits, arguing for a role of PsaB availability in PsaA translation (Stampacchia et al, 1997).
In the present work, we used chimeric genes expressing reporter proteins translated under the control of psaA or psaC 5′ UTRs to provide evidence that the biogenesis of PSI involves a cascade of autoregulation of translation, most likely at the level of initiation, mediated by the unassembled CES subunits PsaA or PsaC.
Results
Assessment of regulation of translation initiation in the CES behaviour of PsaA, using the aadA reporter gene
One of the two major reaction centre (RCI) subunits of PSI, PsaA, is a CES subunit since it displays a reduced rate of synthesis in the absence of the other RCI subunit, PsaB. This may result from regulation of translation initiation, controlled by the 5′ UTR of the psaA gene. To test that hypothesis, we constructed a chimeric gene bearing the psaA 5′ UTR fused immediately upstream of the bacterial aadA gene coding sequence, which confers resistance to the antibiotics spectinomycin and streptomycin (Goldschmidt-Clermont, 1991). By biolistic transformation, this chimeric gene was inserted downstream of the chloroplast petA gene in the wild type and in mutant strains unable to accumulate PsaB (Figure 1A). We chose the tab1-F15 nuclear mutant strain, which is deficient for translation of the psaB messenger (Stampacchia et al, 1997), and the chloroplast mutant strain C3 (Girard-Bascou et al, 1987), which expresses a truncated version of PsaB that is rapidly degraded (see Supplementary Figure I). Transformants were recovered from each recipient strain on spectinomycin (100 μg ml−1)–Tris-acetate-phosphate (TAP) plates. The level of antibiotic resistance conferred by the chimeric gene was determined by plating the transformants on TAP medium supplemented with increasing concentrations of antibiotics, as illustrated in Figure 1B (see also Supplementary Figure II and Supplementary Table I). While the chimeric gene allowed the growth of strains derived from the wild type on antibiotic concentrations higher than 1000 μg ml−1 of spectinomycin plus 100 μg ml−1 of streptomycin, transformants devoid of PsaB (tab1-F15 and C3 in Figure 1B) no longer grew when the concentration of antibiotics reached 500+50 μg ml−1 of spectinomycin+streptomycin. Since the chimeric 5′psaA-aadA mRNAs accumulated to the same level in all transformed strains (Figure 1C), these results point to a specific downregulation of translation of the psaA 5′ UTR-driven aadA gene when expressed in the absence of PsaB.
Figure 1.
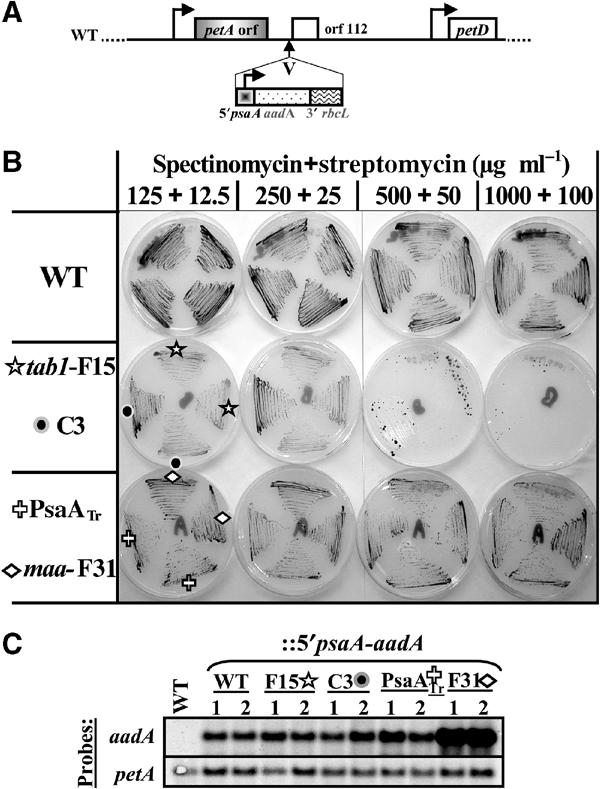
The psaA 5′ UTR confers a PsaB-dependent expression to the reporter gene aadA. (A) Schematic map of the petA-petD chloroplast region where the 5′psaA-aadA chimeric gene has been inserted, in direct orientation with respect to the petA gene, at the neutral EcoRV site (V). ↱ indicates transcription start sites. (B) Growth of independent transformants, derived from the recipient strains listed in the left, in the presence of increasing concentrations of antibiotics. The PsaATr and maa-F31 strains are devoid of PsaA expression (see later). (C) Accumulation of the chimeric aadA (and petA, as a loading control) mRNAs in the wild-type strain and in some of the transformants presented in panel B.
We could exclude that the reduced resistance to antibiotics observed in strains lacking PsaB was a mere consequence of PSI deficiency by transforming mutant strains lacking expression of PsaA with the same chimeric gene: the transformed strains that were recovered, although deficient for PSI accumulation, presented the same growth properties on TAP–antibiotics plates as those derived from the wild-type recipient strain (PsaATr and maa-F31 in Figure 1B and C).
The psaA 5′ UTR is sufficient to confer a PsaB-dependent rate of synthesis to the cytochrome f reporter protein
In the above experiments however, we monitored only indirectly expression of the chimeric 5′psaA-aadA gene through resistance of the transformed strains to antibiotics. As an alternative, we constructed another chimeric gene, 5′psaA-petA, allowing cytochrome f to be translated under the control of the psaA 5′ UTR (Figure 2A). We introduced the chimera in the chloroplast genome of the ΔpetA strain (Table I). Recovery of phototrophic transformants aAf indicated that the psaA 5′ UTR allowed high enough expression of cytochrome f to sustain photoautotrophic growth.
Figure 2.
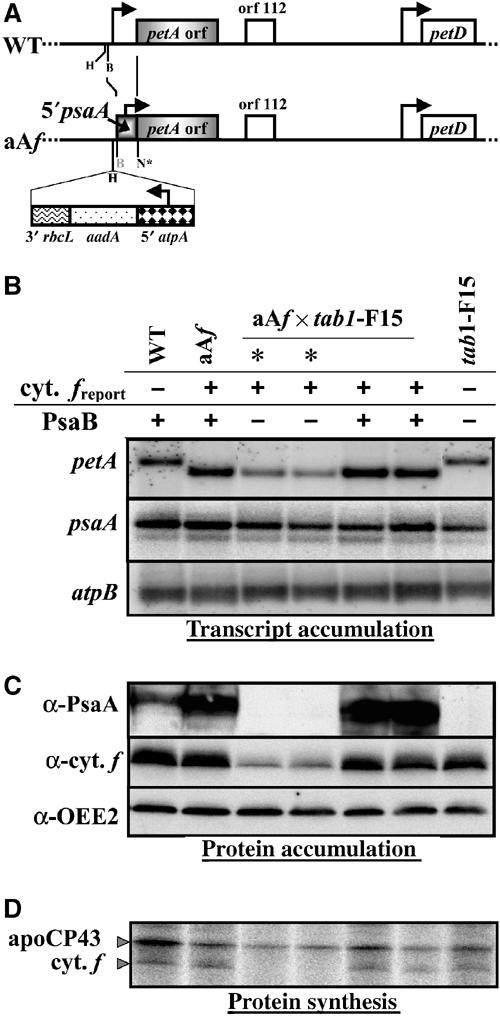
The CES behaviour for the PsaA subunit corresponds to a translational regulation mediated by the psaA 5′ UTR. (A) Map of the chloroplast petA gene in wild-type and aAf strains. Relevant restriction sites are indicated: B, BglII; N*, an NcoI site introduced by site-directed mutagenesis around the petA initiation codon for cloning purposes; H, HincII. (B) Accumulation of petA, psaA and atpB (as a loading control) transcripts in a representative tetrad progeny (out of seven) of the cross aAf × tab1-F15 and from parental and wild-type strains, revealed by hybridisation to petA, psaA (exon 3) and atpB-specific probes. (C) Accumulation of cytochrome f, PsaA and OEE2 (as a loading control), detected with specific antibodies on whole-cell proteins extracted from those strains. Lack of PsaA signs the tab1 progeny (marked by *). (D) Synthesis of cytochrome f, determined by 5 min pulse labelling with [14C]acetate in the presence of 8 μg ml−1 cycloheximide preventing cytosolic synthesis, in the same strains. Positions of cytochrome f and CP43 (providing an incorporation and loading control) are marked.
Table 1.
Transformation experiments
| Transformed strains | Recipient strainsa | Plasmid used | Selection |
|---|---|---|---|
| WT::aAK | WT (SpS) | pfaAK | Spec. resistance |
| F15::aAK | tab1-F15 (SpS), (1) | pfaAK | Spec. resistance |
| C3::aAK | C3 (SpS), (2) | pfaAK | Spec. resistance |
| aAf | ΔpetA (SpR), (3) | pKaAf | Phototrophy |
| aACf | ΔpetA (SpR), (3) | pKaACf | Phototrophy |
| {C3, aAf} | C3 (SpS), (2) | pKaAf | Spec. resistance |
| PsaATrb1 | WT (SpS) | pKrPsaATr | Spec. resistance |
| {PsaATr, aAf} | psaATr (SpS)b2 | pKaAf | Spec. resistance |
| aCf | WT (SpS) | pKaCf | Spec. resistance |
| {ΔpsaC, aCf} |
ΔpsaC (SpS) (4) |
pKaCf |
Spec. resistance |
| aAll recipient strains were mt+, and either resistant (SpR) or sensitive (SpS) to spectinomycin. (1) Stampacchia et al (1997), (2) Girard-Bascou et al (1987), (3) Kuras and Wollman (1994), (4) Takahashi et al (1991). | |||
| b1The PsaATr strain was initially selected for spectinomycin resistance due to the presence of the recycling aadA cassette. b2After excision of the cassette according to Fischer et al (1996), the strain became SpS and was used as a recipient strain for a second round of transformation with plasmid pKaAf, based on selection for spectinomycin resistance. | |||
Strain aAf was then crossed to the tab1-F15 nuclear mutant strain that lacks PsaB. Each member of the resulting tetrads carried the chloroplast chimeric gene, uniparentally transmitted from the mt+ parent aAf. Half of them (indicated by * in Figure 2B–D) had fluorescence transients typical of impaired PSI activity (Chua et al, 1975) (data not shown) and harboured the nuclear tab1 mutation, which shows Mendelian segregation; the other half inherited a wild-type nuclear genome. Therefore, analysis of cytochrome f expression among tetrad progeny by RNA hybridisation, protein pulse labelling and immunodetection with specific antibodies allowed us to compare the expression of the chimeric gene in the presence or absence of the PsaB subunit. We observed a 2:2 segregation in cytochrome f expression, in agreement with the above experiments using aadA as a reporter: the tab1 progeny exhibited a drastic decrease in the accumulation of the reporter protein, reaching only 10–15% of that observed in the parental strain aAf or in the progeny with a wild-type nuclear genome (a preliminary report of this experiment has been presented at the 673rd meeting of the Biochemical Society; Choquet et al, 2001) (Figure 2C). This resulted from a decrease in translation of the 5′psaA-petA reporter gene (Figure 2D). However, the chimeric messenger, which migrates faster than wild-type petA mRNA, was less accumulated in the tab1 offspring from aAf × tab1-F15 crosses (Figure 2B). In contrast, the psaA mRNA was similarly accumulated in all progeny, even those lacking detectable PsaA product (tab1 progeny).
Since RNA stability determinants can be found in coding sequences (Singh et al, 2001), we reproduced the same set of experiments with a third chimeric gene, 5′psaAC-petA, which contained an extension of 60 nucleotides (nt), corresponding to the first 20 residues of the PsaA protein fused in-frame with the petA coding sequence, in addition to the promoter and 5′ UTR of psaA. After transformation of the ΔpetA strain, phototrophic transformants aACf were recovered and crossed to the tab1-F15 mutant strain. The expression of cytochrome f was similar in strains aACf and aAf and showed the same decrease in expression in the tab1 progeny of the crosses. With this construct, the drop in the rate of expression of the reporter protein in the absence of PsaB was not accompanied by any change in the mRNA level (see Supplementary Figure III).
Thus we conclude that both the aadA gene product and cytochrome f reporters, expressed under control of the 5′psaA UTR, showed a drop in translation in the absence of PsaB. In the subsequent experiments, we chose to use the chimeric gene 5′psaA-petA, because it contains only sequences from the psaA 5′ UTR.
In order to rule out the possibility that the decrease in expression of the chimeric gene in the absence of PsaB resulted from a pleiotropic effect of the tab1 mutation on translation of both psaA and psaB messengers, we used the chloroplast mutant strain C3, which expresses a truncated PsaB, but has a wild-type nuclear genome. Upon transformation with the chimeric gene 5′psaA-petA, the resulting strains, hereafter named {C3, aAf}, were used in pulse-labelling experiments. We observed a low rate of cytochrome f translation in all {C3, aAf} transformants tested when compared to the aAf strain (Figure 3C), similar to that observed in the aAf, tab1 progeny indicated by * in Figure 2D.
Figure 3.
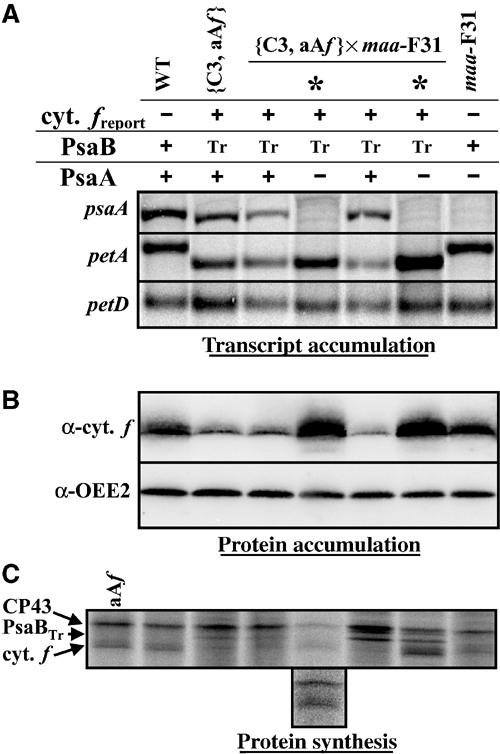
The expression of the 5′psaA-petA reporter gene is no longer repressed in the absence of PsaB when the endogenous PsaA subunit is lacking. (A) Accumulation of petA, psaA and petD (as a loading control) transcripts in progeny from the cross {C3, aAf} × maa-F31 and in wild-type and parental strains, detected with probes specific for petA, psaA (exon 1) and petD. Absence of mature psaA mRNA signs the maa progeny, designated by *. (B) Accumulation and (C) translation of cytochrome f in the same strains. OEE2 accumulation provides a loading control. In (C), the translation of the 5′psaA-petA chimeric gene in the presence of PsaB (lane aAf) is shown for comparison. (Inset) Longer exposure of the gel for the second progeny of the tetrad, which incorporated poorly radiolabelled 14C. Positions of neosynthesised CP43, truncated PsaB (Tr) and cytochrome f are indicated. In the two progeny with a wild-type genome, 14C incorporation is lower in cytochrome f than in CP43, while it is higher in the maa progeny.
Together, these experiments proved that the absence of PsaB causes decreased expression of not only psaA but also of chimeric genes translated under the control of the psaA 5′ UTR. Therefore, the CES regulation in psaA expression occurs before translation elongation or before cotranslational degradation of the nascent PsaA polypeptide, which would both depend on the coding sequence. Rather, some early step in the initiation process is regulated, either an activation step prior to initiation, translation initiation itself or a transition step between initiation and elongation of translation.
Downregulation of PsaA expression in the absence of PsaB is due to autoregulation of translation
In the absence of the PsaB protein, downregulation of psaA translation could occur either because PsaB would act as an activator for PsaA translation (‘transactivation' hypothesis; Figure 4, right panel) or because unassembled PsaA, which may accumulate to some extent, would expose a translational repressor domain (autoregulation hypothesis; Figure 4, left panel). Because the psaA 5′ UTR was sufficient to confer the PsaA CES behaviour to chimeric genes, these two hypotheses could be discriminated by looking at the expression of the 5′psaA-driven cytochrome f reporter in the absence of both the PsaA and PsaB subunits (Figure 4, bottom). In the case of a transactivation hypothesis, the absence of the positive regulator brought along with the PsaB subunit should result in poor expression of the chimeric gene, either in the presence or in the absence of PsaA. By contrast, in the autoregulation hypothesis, a strain lacking the repressor domain carried by the unassembled CES protein, PsaA, should show a high expression of the chimera, even in the absence of the assembly partner PsaB.
Figure 4.
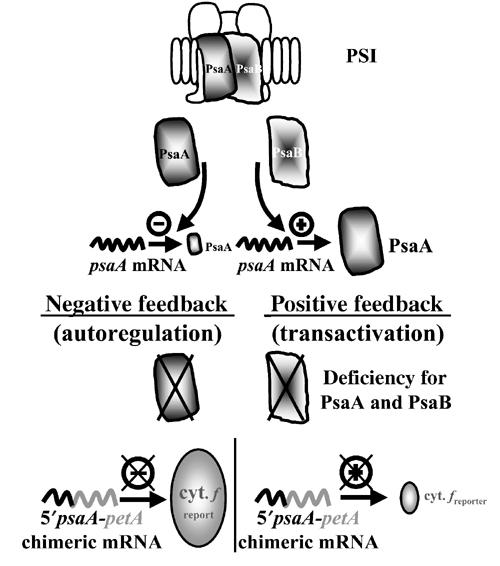
The two models for the repression of psaA mRNA translation in the absence of the dominant protein PsaB can be discriminated by looking at the expression of a 5′psaA-driven reporter gene in the absence of both PsaA and PsaB.
We thus crossed the {C3, aAf} strain, which expresses the 5′psaA-petA reporter gene, but does not accumulate PsaB, because it expresses only a truncated and unstable polypeptide, with the maa-F31 strain, defective for PsaA expression. This mutant is impaired in trans-splicing of precursor transcripts carrying exon 1 and exon 2 of psaA. Therefore, it lacks full-length psaA mRNA but accumulates the exon 1 precursor RNA (Goldschmidt-Clermont et al, 1990). As a preliminary control, we first checked that the maa mutation did not prevent expression of the chimeric gene by crossing the aAf strain with the maa-F31 strain. The rates of synthesis and accumulation of the reporter cytochrome f were identical in all tetrad progeny (see Supplementary Figure IV). Thus cytochrome f expressed from the chimeric 5′psaA-petA gene remains insensitive to the maa nuclear mutation.
Analysis of cytochrome f expression in one out of five tetrads from the cross {C3, aAf} × maa-F31 is shown in Figure 3A–C. Each tetrad progeny harboured both the chimeric gene, as demonstrated by the higher mobility of petA mRNAs (Figure 3A), and the psaB mutation, as revealed by the synthesis of a truncated PsaB polypeptide (Figure 3C). Only half of the progeny, identified by the absence of mature psaA mRNA (Figure 3A), inherited the maa nuclear mutation and are indicated by * in Figure 3. The two progeny with a wild-type nuclear genome were similar to the parental strain {C3, aAf}: they showed low synthesis and accumulation of the psaA-driven cytochrome f (Figure 3B and C) due to the absence of PsaB. In contrast, the maa progeny, which express neither full-length PsaB nor PsaA, recovered a high expression of the reporter gene, similar to that observed in the aAf strain.
Therefore, translation of the 5′psaA-driven cytochrome f is no longer decreased in the absence of the PsaB subunit, when the CES protein PsaA cannot accumulate in the thylakoid membranes. These experiments strongly support an autoregulation of psaA translation rather than a transactivation hypothesis.
A truncated PsaA polypeptide escapes the psaA CES control
We noted that accumulation of chimeric 5′psaA-petA transcripts was higher in the maa nuclear background (Figure 3A). This has been observed previously with other 5′psaA-containing transcripts in various psaA trans-splicing mutants, for example with the psaA exon 1 precursor itself (Choquet et al, 1988). The high expression of the 5′psaA-driven cytochrome f in the maa progeny from the cross {C3, aAf}, observed in the above experiment, may be due to the overaccumulation of the chimeric messenger. In addition, some nucleus-encoded factors, required for the translation of psaA mRNA, may not be able to bind to the precursor transcript of psaA exon 1 and thus become fully available for translation of the sole chimeric 5′psaA-petA mRNA. For a critical assessment of these alternative hypotheses, we caused a premature termination of translation 155 residues after the initiation methionine by introducing a frameshift in the third exon of psaA, thereby preventing accumulation of full-length PsaA (Figure 5A). In this case, the putative nuclear regulatory factors participate normally in the translation of a truncated psaA product, which should be rapidly degraded because of its inability to assemble into a PSI complex.
Figure 5.
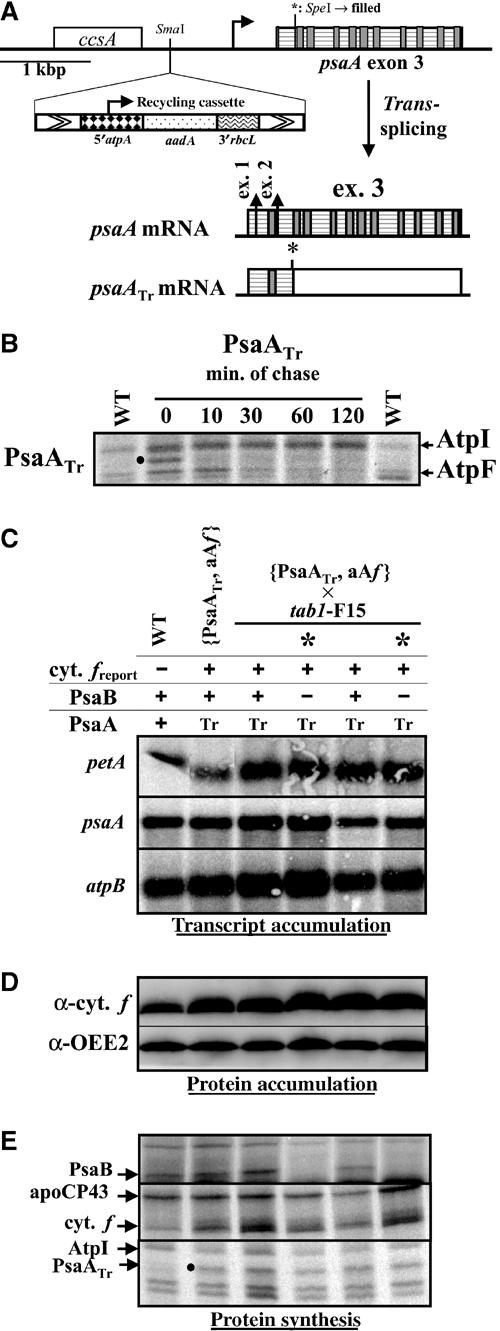
Expression of the 5′psaA-petA reporter gene in strains expressing a truncated PsaA. (A) Strategy used to introduce a mutation (*) in the third exon of psaA. Relevant restriction sites are shown, as well as transmembrane helices coding regions (grey boxes). Coding sequences are indicated by horizontal hatches. Due to the mutation *, most of the psaA mRNA (after trans-splicing) is not translated in the mutant strain (white rectangle). (B) Pulse labelling of the PsaATr strain (time 0) followed by a chase for the indicated times in the presence of an excess of nonradioactive acetate and 200 μg ml−1 chloramphenicol. The position of truncated PsaA is marked with a dot. As already described (Delepelaire, 1983), the AtpF protein is short-lived. (C) Accumulation of petA, psaA and atpB (as a loading control) transcripts in a representative tetrad from the cross {PsaATr, aAf} × tab1-F15 and in wild-type and {PsaATr, aAf} strains, detected using probes specific for petA, psaA (exon 1) and atpB. (D) Accumulation of cytochrome f (and OEE2 as a loading control) in the strains. (E) Chloroplast translates in the same strains. Lack of PsaB synthesis signs the tab1 tetrad progeny, designated by *.
A two-step procedure was used to introduce both the psaA frameshift and the 5′psaA-petA reporter into the same strain. We first associated the psaA mutation with a ‘recycling' spectinomycin resistance cassette that allowed us to select transformants on spectinomycin-supplemented TAP medium before losing specifically the cassette—but not the mutant psaA allele—once the selective pressure is released (Fischer et al, 1996). The resulting strains, PsaATr, were screened for PSI deficiency by fluorescence kinetics and showed normal accumulation of the mutated psaA messenger (data not shown). A truncated PsaA polypeptide of about 16 kDa was detected by a pulse-labelling experiment, between two CF0 subunits of the ATP synthase (Lemaire and Wollman, 1989). Its rate of synthesis—as quantified by a PhosphorImager scan of the 14C labelling of the gel and corrected for the number of carbon atoms—is similar to that of the PsaB and PsaA polypeptides in a wild-type strain. In a pulse-chase experiment (Figure 5B), the truncated form of PsaA proved to be very unstable, with a half-life of less than 10 min. Therefore, it does not accumulate to any significant extent in thylakoid membranes.
Next, we allowed spontaneous excision of the recycling cassette from the chloroplast genome of PsaATr transformants, and used them as recipient strains for a second round of transformation with the 5′psaA-petA reporter gene, associated with a new spectinomycin resistance cassette. Transformants {PsaATr, aAf}, selected on antibiotic-supplemented TAP medium, were crossed with the nuclear mutant strain tab1-F15 to compare expression of the reporter gene in the absence of PsaA alone or in the absence of both PsaA and PsaB. All tetrad progeny from that cross inherited the psaA 5′ UTR-driven cytochrome f (Figure 5C) and the truncated psaA allele carried by the chloroplast genome, because of the uniparental inheritance from the mt+ parent (Figure 5E). Half of the progeny inherited the tab1 nuclear mutation and were identified in pulse-labelling experiments by their lack of PsaB synthesis (Figure 5E), while the other half had a wild-type nuclear genome and translated PsaB normally. The rate of synthesis of the truncated PsaA polypeptide was identical in the four progeny (PsaATr, indicated by a dot in Figure 5E). Therefore, the translation of this truncated and unstable PsaA was no longer dependent on the presence of PsaB. Similarly, the 5′psaA-driven petA reporter gene was expressed at the same high level in all progeny of the cross {PsaATr, aAf} × tab1-F15 (Figure 5D and E), in contrast to what was observed among the progeny of the cross aAf × tab1-F15 (where translation was repressed in the tab1 members of the tetrad; Figure 2D). Therefore, when PsaA cannot accumulate in thylakoid membranes, translation of the 5′psaA-petA transcript remains high, even in the absence of the assembly partner, PsaB. That the presence of PsaB does not stimulate any 5′psaA-driven translation fully excludes the transactivation hypothesis. Our data point to an autoregulation of translation where unassembled PsaA exerts a negative feedback on the translation of psaA mRNA.
Synthesis of the PsaC CES subunit is controlled at the level of translation initiation
As indicated in Introduction, PSI contains another CES protein, PsaC, whose synthesis is decreased in the absence of PsaA. To determine whether translation initiation of PsaC was indeed regulated by the availability of PsaA, we constructed a 5′psaC-petA gene (including the first 30 nt of PsaC coding sequence), associated with a spectinomycin resistance cassette (Figure 6A). Transformants containing this reporter, named aCf, were selected for growth on spectinomycin–TAP plates and were found to grow phototrophically despite accumulating only ∼10% of the wild-type level of cytochrome f (see Figure 6C, lane aCf). This low expression of cytochrome f when driven by the psaC 5′ UTR prevented its detection in pulse-labelling experiments.
Figure 6.
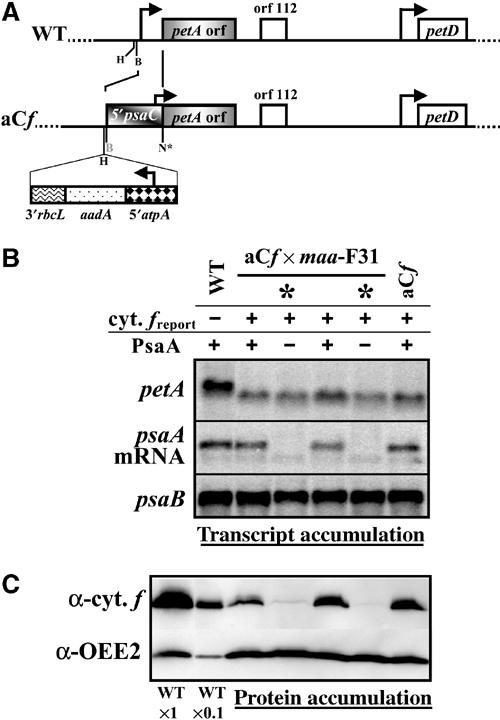
The CES behavior for the RCI subunit PsaC corresponds to a translational regulation mediated by the psaC 5′ UTR. (A) Maps of the petA gene in wild-type and aCf strains. Same conventions as in Figure 2A. (B) petA, psaA and psaB (as a loading control) transcripts accumulation in a tetrad of the cross aCf × maa-F31 and in wild-type and parental strains, detected using probes specific for petA, psaA (exon 3) and psaB. Progeny lacking the mature psaA transcript (indicated by *) bear the nuclear maa mutation. (C) Cytochrome f (and OEE2, as a loading control) accumulations in the same strains.
To study the expression of the chimeric 5′psaC-petA gene in the absence of PsaA, we crossed the aCf strain with the maa-F31 strain. The whole progeny carried the chloroplast chimeric gene, uniparentally transmitted, as demonstrated by the faster migration of its transcript upon electrophoresis (Figure 6B). Half of the progeny, identified by the lack of mature psaA mRNA (Figure 6B), inherited the maa nuclear mutation (indicated by * in Figure 6). Cytochrome f accumulation was nearly undetectable in those progeny (Figure 6C), whereas offspring harbouring a wild-type nuclear genome displayed the same accumulation of cytochrome f as did the parental strain aCf. Since RNA hybridisation analysis showed no correlation in the tetrad progeny between changes in chimeric mRNA content and changes in cytochrome f accumulation (Figure 6B and C), we conclude that translation of the chimeric 5′psaC-petA gene was decreased in the absence of the PsaA subunit. Thus, the psaC 5′ UTR is able to confer CES behaviour of the PsaC subunit on a petA reporter gene. PsaC is a bona fide CES subunit, whose synthesis is regulated at some early step of the translation process.
The decreased PsaC synthesis is due to autoregulation of translation mediated by the unassembled PsaC subunit
To test the mechanism of PsaC translational downregulation in the absence of the PsaA protein, we followed the same rationale as we did for PsaA. Expression of the 5′psaC-petA gene was examined in the absence of both PsaA, here the assembly partner, and PsaC, the CES subunit.
We introduced the chimeric 5′psaC-petA gene, associated with the spectinomycin resistance cassette, in the chloroplast genome of a ΔpsaC deletion strain (Takahashi et al, 1991), in the place of the endogenous petA gene. Transformants {ΔpsaC, aCf} were selected for spectinomycin resistance and crossed to the maa-F31 nuclear mutant. All progeny that lacked PsaC, but had either a wild-type nuclear genome or a maa mutant background (indicated by *, see Figure 7A), displayed the same rates of cytochrome f synthesis and accumulation (Figure 7B and C) as the parental strain {ΔpsaC, aCf}. It amounted up to about 50% of the wild-type rate. This is a much higher rate than that observed in transformants expressing the chimeric gene in the presence of the endogenous PsaC protein (about 10% of the wild-type level; Figure 7D).
Figure 7.
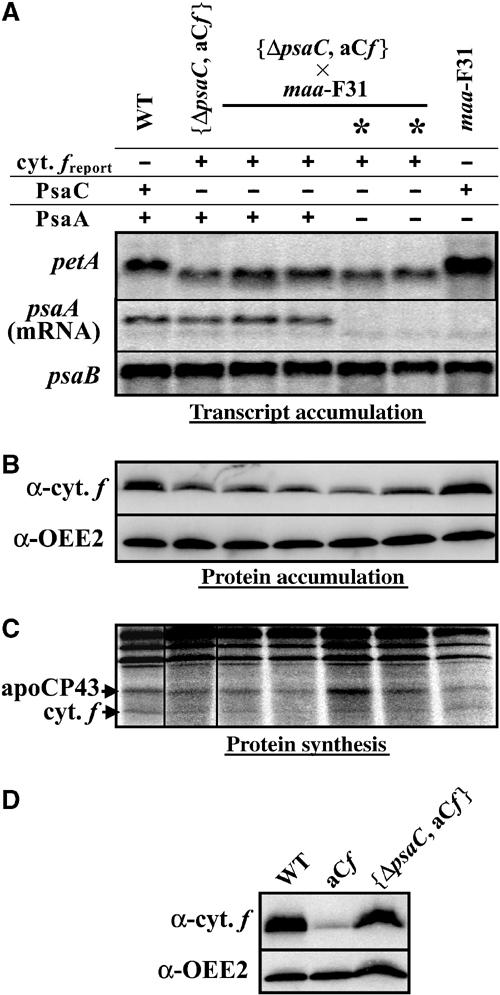
: Cytochrome f translated under the control of the psaC 5′ UTR is no longer repressed in the absence of the PsaA subunit, when PsaC is lacking. (A) Accumulation of petA, psaA and psaB (as a loading control) transcripts in a representative tetrad (out of six) from the cross {ΔpsaC, aCf} × maa-F31 and in the wild-type and parental strains. * designates the maa members of the tetrad, lacking psaA messenger. (B) Cytochrome f and OEE2 (as a loading control) accumulation in the same strains. (C) Rate of translation of the chimeric gene 5′psaC-petA in these strains. The positions of the neosynthesised cytochrome f and CP43 (as a loading and incorporation control) are indicated. The lane {ΔpsaC, aCf} is boxed because it originates from another region from the same gel. (D) Cytochrome f expressed from the 5′psaC-petA chimeric gene (normalised to that of the loading control OEE2) accumulates to 9 and 54% of the wild-type level, respectively, in strains expressing (aCf) or lacking PsaC ({ΔpsaC, aCf}).
The expression of the chimeric 5′psaC-petA gene is thus increased in strains lacking PsaC, independent of the assembly partner PsaA. This excludes the transactivation hypothesis, where the PsaA subunit would activate psaC translation. Rather, PsaC translation is autoregulated by its ability to assemble with PsaA.
Discussion
Critical assessment of the reporter constructs used to study the CES process
Our attempt to characterise the molecular mechanism of the CES processes at work in PSI biogenesis critically depends on the expression of reporter proteins, translated under the control of the 5′ UTR of putative CES genes, in C. reinhardtii chloroplasts. The expression of these 5′ UTR CES-driven reporters should faithfully reflect the behaviour of the original subunits. We used two constructs, one expressing the aminoglycoside adenine transferase (aadA), a heterologous, soluble protein from Escherichia coli, and the other cytochrome f, a thylakoid-bound protein from C. reinhardtii. The level of aadA translation can be measured only indirectly through the level of resistance to antibiotics. Indeed, changes in the level of expression of a resident CES protein can be mimicked with aadA when driven by the corresponding CES 5′ UTR (this study and Choquet et al, 1998). In order to measure directly changes in the rates of protein synthesis, we also used cytochrome f, which is particularly well suited for use as a reporter protein: (i) At variance with other reporters, such as aadA gene product, GUS or GFP, its rate of translation can be accurately determined in vivo by pulse-labelling experiments and its accumulation can be measured by immunodetection with specific antibodies. (ii) Whether assembled or not into the cytochrome b6f complex, cytochrome f is a very stable protein (Kuras and Wollman, 1994; Choquet et al, 1998). Its accumulation is therefore a faithful measure of its rate of translation, even when the latter is too low to be measured directly. (iii) Finally, cytochrome f, synthesised as a precursor protein with a lumen-targeting sequence of 31 amino acids, can accommodate, at least, 35 extra amino-acid residues (Y Choquet and K Wostrikoff, unpublished observations). As the presequence is cleaved after membrane insertion, these extra residues are not part of mature cytochrome f and do not alter its stability.
The fact that cytochrome f is itself a CES protein (Choquet et al, 1998) does not interfere with the study of other CES processes since it is conferred only by the 5′ UTR of the petA gene, which is no longer present in the reporter constructs. Indeed, the two reporter genes used in this study (petA and aadA) gave similar results.
CES processes in the biogenesis of PSI are born by the 5′ UTR of PsaA and PsaC
Previous work clearly established that PsaA was a CES protein, which requires the presence of the PsaB protein to be synthesised at a wild-type level (Girard-Bascou et al, 1987; Stampacchia et al, 1997). Here we show that the psaA 5′ UTR is sufficient to confer a PsaB-dependent rate of synthesis to the aadA reporter gene. In a previous study, Stampacchia et al (1997) failed to observe a decrease in expression of the chimeric 5′psaA-aadA gene, in the absence of PsaB. However, the antibiotic concentration they used (1000 μg ml−1 of spectinomycin alone) was too low to observe the regulation. We confirmed our finding using psaA-driven petA genes. In preliminary experiments, we observed that indeed the loading of psaA mRNA on polysomes was drastically reduced (at least four-fold) in the C3 strain compared to the wild type. We observed a similar reduction in mRNA loading on polysomes in a ΔpsaB deletion strain (data not shown). This appears conflicting with the observation of Dauvillée et al (2003) that psaA mRNA was only slightly less (70%) associated with polysomes in the tab2-F14 nuclear mutant strain, specifically deficient for the translation of the psaB mRNA. However, these results may be difficult to compare since different protocols were used for the isolation and analysis of polysome fractions in the two studies.
Any significant contribution of pretranslational steps to the regulation of psaA expression can be excluded because the accumulation of the endogenous psaA mRNA is not decreased in strains lacking PsaB, such as C3 or tab1 strains, that show no detectable expression of the PsaA protein and because we found no decrease in the accumulation of the 5′psaA-aadA or 5′psaAC-petA messengers in the absence of PsaB, despite their decreased expression. A correlation between accumulation and translation of the chimeric messenger was only observed with the 5′psaA-petA reporter gene. The 5′psaA-petA messenger could be less stable than psaA or petA transcripts and protected from degradation by translating ribosomes. The decreased accumulation of the 5′psaA-petA transcript in strains lacking PsaB would thus be an effect, rather than a cause, of the decreased synthesis of the psaA-driven cytochrome f.
We also demonstrated that the psaC 5′ UTR (together with the first 30 nt downstream of the psaC initiation codon) is sufficient to confer a PsaA-dependent regulation to the reporter protein cytochrome f. The 5′psaC-petA chimeric gene is less translated in the absence of PsaA than in its presence, with no changes in the mRNA level, as was confirmed by the drastic decrease of psaC mRNA loading on polysomes (data not shown). These experiments account for the observation by Takahashi et al (1991) that PsaC synthesis is decreased in mutants lacking PsaA. Since PsaC interacts with residues from both PsaA and PsaB in assembled PSI (Jordan et al, 2001), CPI, rather than PsaA alone, is likely to be the actual protein partner whose assembly with PsaC controls its rate of translation initiation.
CES processes in PSI biogenesis are due to autoregulation of translation mediated by unassembled PsaA and PsaC subunits
The PsaA and PsaB subunits are in close contact through their last five transmembrane helices in the PSI (Jordan et al, 2001). The downregulation of psaA translation in the absence of PsaB could be due to a negative feedback from a protein domain exposed by unassembled PsaA or it could result from the absence of a positive feedback by the PsaB subunit. The latter possibility can be excluded since the expression of the 5′psaA-petA reporter gene was high in strains lacking expression of both PsaA and PsaB, as was that of the truncated PsaA in strains lacking the PsaB protein. Translation of PsaA is therefore autoregulated by its unassembled form. Similarly, unassembled PsaC autoregulates its own synthesis since expression of the 5′psaC-driven petA reporter gene was stimulated—and not repressed—in the absence of both PsaC and PsaA subunits.
A critical point in these regulation mechanisms is the extent of accumulation that can be reached by the non-assembled CES proteins in the absence of their assembly partners. We definitely lack direct information of the concentrations of unassembled PsaA and PsaC subunits in strains lacking PsaB and PsaA, respectively. It proved difficult to assess the rate of PsaA synthesis in the absence of PsaB, since it migrates as a diffuse band in gels. Strains lacking expression of PsaB were found to accumulate less than 0.5% of wild-type levels of PsaA (Dauvillée et al, 2003). In a wild-type chloroplast, the accumulation of PSI (or PsaA) is in large excess over that of the psaA messenger (106 proteins versus 103 copies of the mRNA) (Rapp et al, 1992). Thus, the concentration of PsaA in strains devoid of PsaB, even reduced below detection level, remains probably high enough to prevent translation of psaA messenger. Unassembled PsaC could not be detected in the absence of PsaB (Boudreau et al, 1997), but the antibody failed to detect PsaC in a 20% dilution of wild-type cells. In the tab1 mutant, which lacks PsaB, the two peripheral subunits PsaD and PsaE, which assemble coordinately with PsaC, still accumulate to about 20% (Boudreau et al, 1997), whereas they are completely absent in a ΔpsaC deletion strain (Fischer et al, 1999). Therefore, strains lacking the PsaA/PsaB heterodimer probably accumulate some unassembled PsaC that would be associated with PsaD and PsaE in a protease-resistant form.
As the syntheses of the CES subunits PsaA and PsaC are autoregulated, we might expect an increased expression of the reporter genes in strains lacking the corresponding CES proteins, that is, either PsaA or PsaC, compared to a wild-type context. These strains should indeed be fully devoid of the repressor domain counteracting expression of the reporter gene, whereas in wild-type chloroplasts one may expect the presence of a subset of unassembled subunits, as was observed for the CES control of cytochrome b6f biogenesis (Choquet et al, 1998). This was indeed observed for psaC, because the 5′psaC-petA reporter gene is more highly expressed in ΔpsaC deletion strains than in a wild-type genetic context. However, in the case of PsaA, expression of the 5′psaA-petA reporter gene was similar in strains expressing or lacking PsaA, but dropped in the absence of PsaB. We conclude that the concentration of unassembled PsaA, available to repress its own translation, must be kept very low in the wild type. This subunit may bind up to 40 chlorophyll molecules, an efficient source of triplet chlorophyll, hence of damaging radicals, in the absence of energy transfer to the primary PSI donor P700.
The residues that form the PsaA or PsaC repressor domains have not been identified. For PsaA, they are likely located in the C-terminal part of the protein, which interacts with the corresponding region of PsaB. PsaA and PsaC are highly conserved from cyanobacteria to higher plants, especially in the C-terminal region of PsaA (Baymann et al, 2001). Since there is no evidence for a CES process in cyanobacteria (Choquet et al, 2003), it seems unlikely that these two proteins have evolved a protein domain capable of sequence-specific interactions with the 5′ UTR of their mRNA in C. reinhardtii. We note that an active RNA-binding domain has recently been proposed for another conserved CES protein of the photosynthetic apparatus, the large subunit of RuBisCo. However, its RNA-binding property was not sequence specific (Yosef et al, 2004), a requirement if it is the cause of a CES behaviour. Therefore, we would favour a model based on an indirect interaction involving a ternary effector, capable of competitive binding to both the 5′ UTR of the transcript and the unassembled subunit, as we have previously proposed for cytochrome b6f biogenesis (Wostrikoff et al, 2001; Choquet et al, 2003). Depending on its binding to the unassembled CES subunit, the ternary factor would modulate translation of the corresponding transcripts, thereby behaving as a translational activator. This class of nucleus-encoded factors is a major feature of chloroplast gene expression in C. reinhardti (Zerges, 2000). Recently, we identified a nuclear mutant specifically deficient for translation of psaA mRNA (unpublished results), the molecular characterisation of which is now in progress.
In the mitochondrion of the yeast Saccharomyces cerevisiae, it is likely that Cox1p is a CES subunit, because its apparent rate of synthesis decreases in mutants lacking Cox2p or Cox7p, with no decrease in the stability of the newly synthesised polypeptide (Calder and McEwen, 1991). Most interestingly, it was recently shown (Perez-Martinez et al, 2003) that one of the translational activators required for the expression of Cox1p, Mss51p, is also able to interact physically with newly synthesised Cox1p. The authors conclude that ‘Mss51p could tightly coordinate Cox1p synthesis with downstream events leading to cytochrome oxidase assembly'. These are the very properties one would expect for a CES ternary effector.
A CES cascade follows photosystem I assembly steps
The present study defines a ‘CES cascade' among the chloroplast-encoded subunits of PSI: PsaB is required for significant PsaA synthesis, which, in turn, is required for PsaC translation (Figure 8). Whether the other chloroplast-encoded PSI subunit, PsaJ, is also a CES protein is unknown. This CES hierarchy (PsaB>PsaA>PsaC) is reminiscent of the sequence of polypeptide assembly during PSI biogenesis. Since the CES process for PSI subunits relies on autoregulation mediated by the unassembled CES subunits, the translation of a given subunit can reach significant levels only when the substrate for its assembly is present, that is, when the previous steps in the sequential assembly of PSI have been completed.
Figure 8.
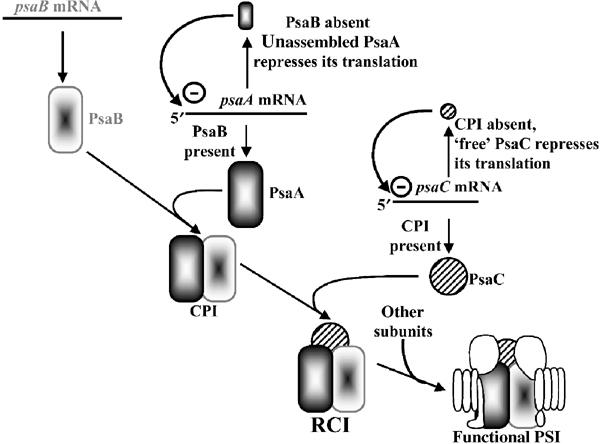
The CES cascade involved in PSI biogenesis.
Depending on light quality and physiological conditions, the amount of PSI (relative to PSII) may vary in chloroplasts (Chow et al, 1990). Translation, the major regulatory step of chloroplast gene expression in C. reinhardtii (Eberhard et al, 2002), is likely involved in this process. One benefit of the CES cascade over independent regulation of translation of each subunit is that only the rate of production of the most upstream subunit, PsaB, has to be controlled in order to determine the stoichiometric expression of all subunits of the protein complex.
Materials and methods
Media, culture conditions and strains
Wild-type and mutant strains of C. reinhardtii were grown on TAP medium, pH 7.2, at 25°C under dim light (5–6 μE m−2 s−1). Recipient strains (all mt+) for chloroplast transformation experiments are listed in Table I. mt− parental strains tab1-F15 (Stampacchia et al, 1997) and maa-F31 (Goldschmidt-Clermont et al, 1990) were used for crosses, performed according to Harris (1989). All tetrad progeny were tested for a 2:2 segregation of mating types. Antibiotic resistance tests were performed according to Choquet et al (1998).
Plasmids and nucleic acid manipulations
Standard nucleic acid manipulations were performed according to Sambrook et al (1989). Details about the construction are presented in Supplementary data. Northern analyses were carried out as described previously (Drapier et al, 1998). DNA probes were as described by Eberhard et al (2002).
Transformation experiments
Cells were transformed by tungsten particle bombardment as previously described (Kuras and Wollman, 1994). Transformants were selected on spectinomycin-supplemented medium (100 μg ml−1) under dim light (5–6 μE m−2 s−1) or for photoautotrophic growth on minimum medium under high light (80 μEm−2s−1). After several rounds of subcloning, correct insertion of transforming DNA and homoplasmy were checked by RFLP analysis of specific PCR amplification products. At least three independent transformants were analysed for each transformation.
Protein isolation, separation and analysis
Pulse-labelling and pulse-chase experiments, protein isolation, separation and immunoblot analysis were carried out on cells grown to a density of 2 × 106 cells ml−1 as described by Kuras and Wollman (1994). Cell extracts were loaded on an equal chlorophyll basis. Quantification of protein synthesis or accumulation was performed according to Choquet et al (2003).
Supplementary Material
Supplementary data
Acknowledgments
We thank Y Takahashi for providing the ΔpsaC strain, D Drapier, R Kuras and T Bollenbach for critical reading of the manuscript, S Bujaldon, D Drapier and L Minai for their help during polysome preparation and analysis. This work was supported by CNRS UPR1261, by Collège de France and by the University of Paris VI. KW was ATER at Collège de France.
References
- Baymann F, Brugna M, Muhlenhoff U, Nitschke W (2001) Daddy, where did (PS)I come from? Biochim Biophys Acta 1507: 291–310 [DOI] [PubMed] [Google Scholar]
- Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD (1997) The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J 16: 6095–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder KM, McEwen JE (1991) Deletion of the COX7 gene in Saccharomyces cerevisiae reveals a role for cytochrome c oxidase subunit VII in assembly of remaining subunits. Mol Microbiol 5: 1769–1777 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Goldschmidt-Clermont M, Girard-Bascou J, Kuck U, Bennoun P, Rochaix JD (1988) Mutant phenotypes support a trans-splicing mechanism for the expression of the tripartite psaA gene in the C. reinhardtii chloroplast. Cell 52: 903–913 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Stern DB, Wostrikoff K, Kuras R, Girard-Bascou J, Wollman FA (1998) Translation of cytochrome f is autoregulated through the 5′ untranslated region of petA mRNA in Chlamydomonas chloroplasts. Proc Natl Acad Sci USA 95: 4380–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet Y, Vallon O (2000) Synthesis, assembly and degradation of thylakoid membrane proteins. Biochimie 82: 615–634 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Wostrikoff K, Rimbault B, Zito F, Girard-Bascou J, Drapier D, Wollman FA (2001) Assembly-controlled regulation of chloroplast gene translation. Biochem Soc Trans 29: 421–426 [DOI] [PubMed] [Google Scholar]
- Choquet Y, Zito F, Wostrikoff K, Wollman FA (2003) Cytochrome f translation in Chlamydomonas chloroplast is autoregulated by its carboxyl-terminal domain. Plant Cell 15: 1443–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow WS, Melis A, Anderson JM (1990) Adjustments of photosystem stoichiometry in chloroplasts improve the quantum efficiency of photosynthesis. Proc Natl Acad Sci USA 87: 7502–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua NH, Matlin K, Bennoun P (1975) A chlorophyll–protein complex lacking in photosystem I mutants of Chlamydomonas reinhardtii. J Cell Biol 67: 361–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauvillée D, Stampacchia O, Girard-Bascou J, Rochaix J-D (2003) Tab2 is a novel conserved RNA-binding protein required for translation of the chloroplast psaB mRNA. EMBO J 22: 6378–6388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepelaire P (1983) Characterization of additional thylakoid membrane polypeptides synthesized inside the chloroplast in Chlamydomonas reinhardtii. Photobiochem Photobiophys 6: 279–291 [Google Scholar]
- Drapier D, Suzuki H, Levy H, Rimbault B, Kindle KL, Stern DB, Wollman FA (1998) The chloroplast atpA gene cluster in Chlamydomonas reinhardtii. Functional analysis of a polycistronic transcription unit. Plant Physiol 117: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman FA (2002) Searching limiting steps in the expression of chloroplast encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 1–14 [DOI] [PubMed] [Google Scholar]
- Fischer N, Setif P, Rochaix JD (1999) Site-directed mutagenesis of the PsaC subunit of photosystem I. F(b) is the cluster interacting with soluble ferredoxin. J Biol Chem 274: 23333–23340 [DOI] [PubMed] [Google Scholar]
- Fischer N, Stampacchia O, Redding K, Rochaix JD (1996) Selectable marker recycling in the chloroplast. Mol Gen Genet 251: 373–380 [DOI] [PubMed] [Google Scholar]
- Fox TD (1996) Translational control of endogenous and recoded nuclear genes in yeast mitochondria: regulation and membrane targeting. Experientia 52: 1130–1135 [DOI] [PubMed] [Google Scholar]
- Girard-Bascou J, Choquet Y, Schneider M, Delosme M, Dron M (1987) Characterization of a chloroplast mutation in the psaA2 gene of Chlamydomonas reinhardtii. Curr Genet 12: 489–495 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M (1991) Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res 19: 4083–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Girard-Bascou J, Choquet Y, Rochaix JD (1990) Trans-splicing mutants of Chlamydomonas reinhardtii. Mol Gen Genet 223: 417–425 [DOI] [PubMed] [Google Scholar]
- Harris EH (1989) The Chlamydomonas Source Book: A Comprehensive Guide to Biology and Laboratory Use. San Diego: Academic Press [DOI] [PubMed] [Google Scholar]
- Jordan P, Fromme P, Witt HT, Klukas O, Saenger W, Krauss N (2001) Three-dimensional structure of cyanobacterial photosystem I at 2.5 Å resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kuras R, Wollman F-A (1994) The assembly of cytochrome b6f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire C, Wollman FA (1989) The chloroplast ATP synthase in Chlamydomonas reinhardtii. I. Characterization of its nine constitutive subunits. J Biol Chem 264: 10228–10234 [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J 22: 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp JC, Baumgartner BJ, Mullet J (1992) Quantitative analysis of transcription and RNA levels of 15 barley chloroplast genes. Transcription rates and mRNA levels vary over 300-fold; predicted mRNA stabilities vary 30-fold. J Biol Chem 267: 21404–21411 [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. Cold Spring Harbor: Cold Spring Harbor Laboratory Press [Google Scholar]
- Singh M, Boutanaev A, Zucchi P, Bogorad L (2001) Gene elements that affect the longevity of rbcL sequence-containing transcripts in Chlamydomonas reinhardtii chloroplasts. Proc Natl Acad Sci USA 98: 2289–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampacchia O, Girard-Bascou J, Zanasco JL, Zerges W, Bennoun P, Rochaix JD (1997) A nuclear-encoded function essential for translation of the chloroplast psaB mRNA in Chlamydomonas. Plant Cell 9: 773–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Q, Chitnis VP, Jin P, Chitnis PR (1997) Topography of the photosystem I core proteins of the Cyanobacterium synechocystis sp. PCC 6803. J Biol Chem 272: 21793–21802 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Goldschmidt-Clermont M, Soen SY, Franzen LG, Rochaix JD (1991) Directed chloroplast transformation in Chlamydomonas reinhardtii: insertional inactivation of the psaC gene encoding the iron sulfur protein destabilizes photosystem I. EMBO J 10: 2033–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber AN, Bingham SE (1998) Structure and function of photosystem I. In Rochaix J-D, Goldschmidt-Clermont M, Merchant S (eds) The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas, Vol. 7, pp 323–348. Dordrecht/Boston/London: Kluwer Academic Publishers [Google Scholar]
- Wollman FA, Minai L, Nechushtai R (1999) The biogenesis and assembly of photosynthetic proteins in thylakoid membranes. Biochim Biophys Acta 1411: 21–85 [DOI] [PubMed] [Google Scholar]
- Wostrikoff K, Choquet Y, Wollman FA, Girard-Bascou J (2001) TCA1, a single nuclear-encoded translational activator specific for petA mRNA in Chlamydomonas reinhardtii chloroplast. Genetics 159: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Irihimovitch V, Knopf JA, Cohen I, Orr-Dahan I, Nahum E, Keasar C, Shapira M (2004) RNA binding activity of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit from Chlamydomonas reinhardtii. J Biol Chem 279: 10148–10156 [DOI] [PubMed] [Google Scholar]
- Yu J, Smart LB, Jung YS, Golbeck J, McIntosh L (1995) Absence of PsaC subunit allows assembly of photosystem I core but prevents the binding of PsaD and PsaE in synechocystis sp. PCC6803. Plant Mol Biol 29: 331–342 [DOI] [PubMed] [Google Scholar]
- Zerges W (2000) Translation of mRNAs encoded by chloroplast genomes. Biochimie 82: 583–601 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


