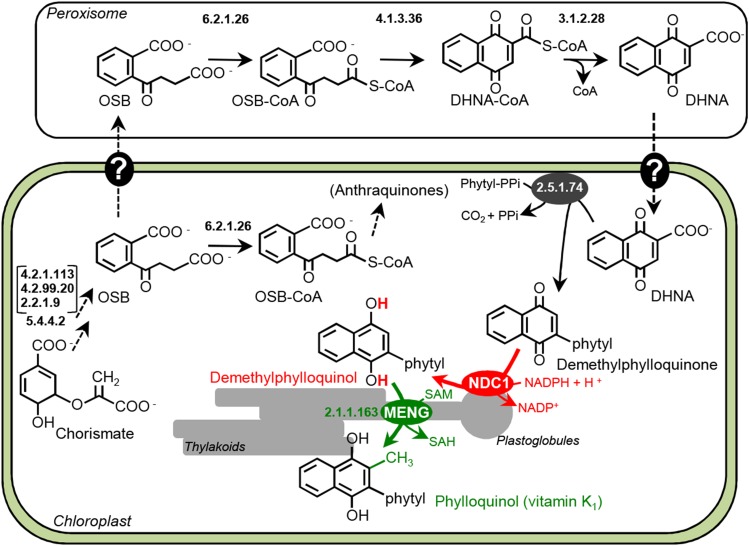
The biosynthesis of vitamin K1 requires an additional bona fide step that consists of the prerequisite reduction of the demethylnaphthoquinone ring prior to its transmethylation.
Abstract
Mutation of Arabidopsis thaliana NAD(P)H DEHYDROGENASE C1 (NDC1; At5g08740) results in the accumulation of demethylphylloquinone, a late biosynthetic intermediate of vitamin K1. Gene coexpression and phylogenomics analyses showed that conserved functional associations occur between vitamin K biosynthesis and NDC1 homologs throughout the prokaryotic and eukaryotic lineages. Deletion of Synechocystis ndbB, which encodes for one such homolog, resulted in the same defects as those observed in the cyanobacterial demethylnaphthoquinone methyltransferase knockout. Chemical modeling and assay of purified demethylnaphthoquinone methyltransferase demonstrated that, by virtue of the strong electrophilic nature of S-adenosyl-l-methionine, the transmethylation of the demethylated precursor of vitamin K is strictly dependent on the reduced form of its naphthoquinone ring. NDC1 was shown to catalyze such a prerequisite reduction by using NADPH and demethylphylloquinone as substrates and flavine adenine dinucleotide as a cofactor. NDC1 displayed Michaelis-Menten kinetics and was markedly inhibited by dicumarol, a competitive inhibitor of naphthoquinone oxidoreductases. These data demonstrate that the reduction of the demethylnaphthoquinone ring represents an authentic step in the biosynthetic pathway of vitamin K, that this reaction is enzymatically driven, and that a selection pressure is operating to retain type II NAD(P)H dehydrogenases in this process.
INTRODUCTION
Vitamin K is a generic term used to designate a class of naphthoquinone derivatives that display some biological activity as cofactors for γ-glutamyl-carboxylases involved in blood coagulation, bone and vascular metabolism, and cell growth regulation (Booth, 2009; Beulens et al., 2013). Natural forms of the vitamin include phylloquinone (2-methyl-3-phytyl-1,4-naphthoquinone), also known as vitamin K1, and menaquinones [2-methyl-3-(all-trans-polyprenyl)-1,4-naphthoquinones], also known as vitamin K2. The occurrence of phylloquinone appears to be restricted to green plants and some species of cyanobacteria, while menaquinones are widespread among archaea, bacteria, red algae, and diatoms (Collins and Jones, 1981; van Oostende et al., 2011). Phylloquinone from plant-based foods is the major source of vitamin K in the Western diet (Booth and Suttie, 1998). Vitamin K-synthesizing organisms use phylloquinone or menaquinone as redox cofactors in their electron transport chains. For instance, in plants, phylloquinone serves as a light-dependent one-electron carrier within photosystem I from chlorophyll a to the iron-sulfur cluster Fx of the heterodimeric PsaA/PsaB reaction center (Brettel, 1997). It also doubles as an electron acceptor for the formation of protein disulfide bonds in the lumen of chloroplasts (Furt et al., 2010; Karamoko et al., 2011).
The biosynthetic pathway of phylloquinone is similar to that of menaquinones in γ-proteobacteria (van Oostende et al., 2011). Chorismate and 2-oxoglutarate serve as precursors for the formation of the naphthalenoid ring, which is prenylated and then methylated. Phylloquinone biosynthetic mutants in the cyanobacterium Synechocystis sp PCC 6803 and the green alga Chlamydomonas reinhardtii are still able to grow photoautotrophically, for these species recruit plastoquinone-9 in place of phylloquinone at the A1 site of photosystem I (Semenov et al., 2000; Lefebvre-Legendre et al., 2007). By contrast, loss of phylloquinone biosynthesis in Arabidopsis thaliana results in seedling lethal phenotype (van Oostende et al., 2011). However, one exception is the Arabidopsis mutant corresponding to demethylnaphthoquinone methyltransferase (MENG; At1g23360; EC 2.1.1.163), which catalyzes the last step of phylloquinone biosynthesis in plastids (Lohmann et al., 2006). The cognate mutant is viable because it accumulates demethylphylloquinone, and the latter can replace phylloquinone in photosystem I, albeit with some loss of photosynthetic efficiency (Lohmann et al., 2006).
Knocking out Arabidopsis NAD(P)H DEHYDROGENASE C1 (NDC1; At5g08740), which encodes a member of the type II NAD(P)H dehydrogenase family, also blocks the methylation of the naphthalenoid ring (Eugeni Piller et al., 2011). NDC1 also affects the redox state of plastoquinone as well as the recycling of α-tocopherol in thylakoid-associated lipoprotein particles called plastoglobules (Eugeni Piller et al., 2011, 2014). However, the reason why the ndc1 knockout accumulates demethylphylloquinone instead of phylloquinone is unknown (Eugeni Piller et al., 2011; Besagni and Kessler, 2013). Because changes in the redox state of plastoquinone impact the expression of photosynthetic genes (Rochaix, 2013), it might seem that the loss of NDC1 simply triggers the downregulation of the transcription of demethylnaphthoquinone methyltransferase. This not the case, however, because the transcript level of this enzyme is unaffected in ndc1 knockout plants (Eugeni Piller et al., 2011). Such a situation is actually reminiscent of that observed in γ-proteobacteria, which can alternate between the accumulation of demethylmenaquinone or menaquinone depending on the growth conditions, but again without apparent changes in the transcription of the cognate methyltransferase (Shestopalov et al., 1997).
Another possibility is that NDC1 is not directly involved in phylloquinone biosynthesis and that loss of function of this gene blocks methylation of the demethylnaphthoquinone intermediate via a general alteration of plastid metabolism. There is a precedent for this: Arabidopsis knockout plants corresponding to gene MUTS-HOMOLOG1, which encodes an endonuclease involved in the maintenance of plastid genome stability, also triggers the accumulation of demethylphylloquinone (Xu et al., 2011). No biochemical work has been done on demethylnaphthoquinone methyltransferases beyond the demonstration that S-adenosyl-l-methionine (AdoMet) is the methyl donor in cell extracts (Kaiping et al., 1984; Koike-Takeshita et al., 1997). Yet, understanding the mechanism of transmethylation of the naphthalenoid ring could be significant, for the accumulation of demethylated naphthoquinone intermediates in ndc1 plants or in γ-proteobacteria suggests a missing step in vitamin K biosynthesis.
Therefore, we examined the connection between type II NAD(P)H dehydrogenases and vitamin K biosynthesis by means of coexpression analyses in Arabidopsis and rice (Oryza sativa), plant-prokaryote comparative genomics, and reverse genetics in a selected cyanobacterial species. We then dissected the molecular mechanism of the transmethylation of the naphthalenoid precursor of vitamin K and established the crucial role of dedicated type II NAD(P)H dehydrogenases in this reaction.
RESULTS
Expression of NDC1 Correlates Strongly with Expression of Phylloquinone Biosynthetic Genes
Searching the GeneCAT coexpression database (Mutwil et al., 2008) with Arabidopsis MENG and NDC1 as queries showed that these two genes share over half of their top 500 coexpressors (Figure 1). Remarkably, such a correlation of expression is significantly higher than that found between NDC1 and tocopherol cyclase (VTE1), which are known functional partners in the recycling of tocopherol; NDC1 and VTE1 share only 30% of their top 500 coexpressors (Figure 1A). Similar results were obtained in rice, where the NDC1 ortholog shares 54% of its top 500 coexpressors with MENG and only 11% with VTE1 (Figure 1A). Expanding these analyses to other phylloquinone and tocopherol biosynthetic genes via hierarchical clustering of coexpression indicated that NDC1 is more closely correlated with genes involved in phylloquinone biosynthesis than with those linked to tocopherol biosynthesis (Figure 1B). All together, these in silico reconstructions indicate that there is a conserved and intimate functional connection between NDC1 and phylloquinone biosynthesis in plants and that the loss of phylloquinone and the accumulation of demethylphylloquinone in the Arabidopsis ndc1 knockout is unlikely to be the result of a non-specific perturbation of plastid metabolism.
Figure 1.
Correlation Analyses of the Expression of Arabidopsis NDC1 and Its Rice Homolog.
(A) Venn diagrams of the top 500 coexpressing genes of NDC1 versus the top 500 coexpressing genes of MENG (EC2.1.1.163) or of tocopherol cyclase (VTE1; EC5.5.1.24) in Arabidopsis and rice. The annotated and intersecting gene lists, as well as their correlation ranks, are provided in Supplemental Data Set 1.
(B) Hierarchical clustering reconstruction of the coexpression profile of NDC1 (red) with that of phylloquinone (green) and tocopherol (orange) biosynthetic genes in Arabidopsis and rice. Distance matrices of Pearson correlation coefficients were converted to Newick string using the unweighted pair group method with arithmetic mean analysis. Numbers indicate branch lengths. Arabidopsis gene numbers and rice probe sets used as inputs are provided in Methods. HPPD, 4-hydroxyphenylpyruvate dioxygenase (EC1.13.11.27); MENA, 1,4-dihydroxy-2-naphthoate phytyltransferase (EC2.5.1.74); MENB, naphthoate synthase (EC4.1.3.36); MENE, o-succinylbenzoic acid-CoA ligase (EC6.2.1.26); VTE2, homogentisate phytyltransferase (EC2.5.1.115).
Comparative Genomics Identify Conserved Physical Associations between Type II NAD(P)H Dehydrogenase Homologs and Vitamin K Biosynthetic Genes in Bacteria
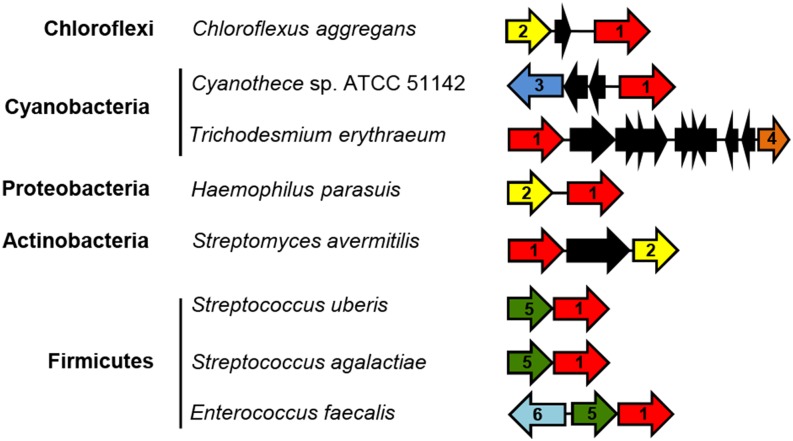
Searches of the STRING (Franceschini et al., 2013) and SEED (http://theseed.uchicago.edu/FIG/index.cgi) databases using Arabidopsis NDC1 as a query identified bacterial type II NAD(P)H dehydrogenases in the neighborhood of naphthoquinone biosynthetic genes (Figure 2). In three notable instances (Chloroflexi, Proteobacteria, and Actinobacteria), the association was detected with demethylnaphthoquinone methyltansferase itself (Figure 2). Furthermore, in Chloroflexi, Proteobacteria, Actinobacteria, and Firmicutes, these gene clusters display operon-like structures (Figure 2), pointing to the existence of a selective pressure to coregulate the expression of the dehydrogenases with that of menaquinone biosynthetic genes. Such arrangements recapitulate the high level of correlation of expression between NDC1 and phylloquinone biosynthetic genes in Arabidopsis and rice and indicate that the functional coupling between type II NAD(P)H dehydrogenases and vitamin K biosynthesis is not restricted to flowering plants.
Figure 2.
Gene Clustering of Type II NAD(P)H Dehydrogenase Homologs with Vitamin K Biosynthetic Genes in Bacterial Genomes.
Homologs display matching color and number. 1, Type II NAD(P)H dehydrogenase (EC 1.6.99.3); 2, demethylnaphthoquinone methyltransferase (EC 2.1.1.163); 3, o-succinylbenzoate synthase (EC 4.2.1.113); 4, 1,4-dihydroxy-2-naphthoyl-CoA synthase (EC 4.1.3.36); 5, 1,4-dihydroxy-2-naphthoate polyprenyl transferase (2.5.1.74); 6, polyprenyl diphosphate synthase (2.5.1.30). Black arrows indicate genes of hypothetical function or of function unrelated to naphthoquinone biosynthesis.
Deletion of Synechocystis slr1743 (ndbB) Blocks the Transmethylation of Demethylphylloquinone
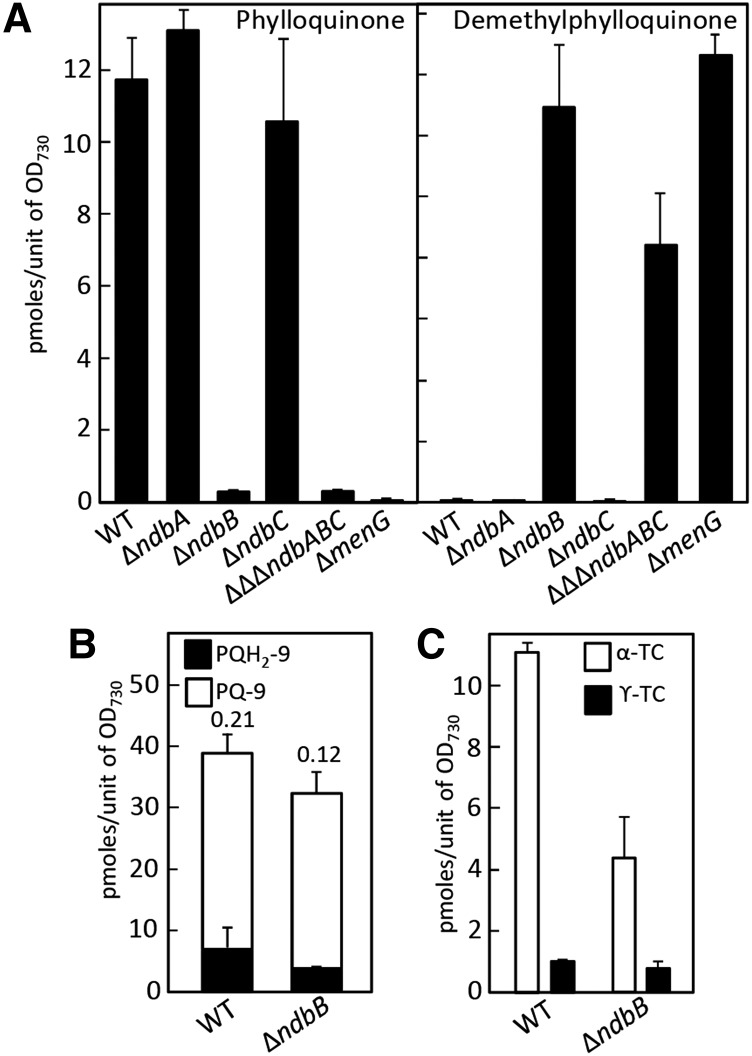
To determine whether prokaryotic type II NAD(P)H dehydrogenases are directly involved in vitamin K biosynthesis, prenylated naphthoquinones were quantified in corresponding knockout strains of the cyanobacterium Synechocystis sp PCC 6803. The genome of this species encodes three type II NAD(P)H dehydrogenases, ndbA, ndbB, and ndbC, which are the gene products of slr0851, slr1743, and sll1484, respectively (Howitt et al., 1999). Because in Synechocystis none of these NDC1 homologs occurs in a cluster with quinone biosynthetic genes, these three genes could be deleted, either singly or in combination, without creating a nonspecific polar effect on naphthoquinone production. Phylloquinone represented most of the pool of prenylated naphthoquinones in the ΔndbA and ΔndbC knockout strains, as is typically observed in wild-type Synechocystis (Figure 3). By contrast, the ΔndbB and ΔΔΔndbABC knockout strains contained only traces of phylloquinone and instead accumulated its demethylnaphthoquinone precursor, just as is observed for the Synechocystis demethylnaphthoquinone methyltransferase mutant (Figure 3A). Synechocystis ΔndbB cells also displayed a lower plastoquinol-9/plastoquinone-9 ratio and a lower tocopherol content than did the wild-type control (Figures 3B and 3C). This suggests that, like its plant homolog NDC1, ndbB contributes to the reduction of plastoquinone-9 and the salvaging of the chromanol ring of tocopherols.
Figure 3.
Synechocystis ndbB Is the Functional Homolog of NDC1.
(A) Levels of phylloquinone (left panel) and of demethylphylloquinone (right panel) in wild-type Synechocystis, type II NAD(P)H dehydrogenase mutant strains ΔndbA, ΔndbB, ΔndbC, and ΔΔΔndbABC, and demethylnaphthoquinone methyltransferase ΔmenG knockout strain. Data are means ± se of three to four replicates.
(B) Plastoquinol-9 (PQH2-9) and plastoquinone-9 (PQ-9) content in wild-type and ΔndbB cells. Numbers above the bars indicate plastoquinol-9/plastoquinone-9 ratios.
(C) α- and γ-tocopherol (TC) content in wild-type and ΔndbB cells. Data are the means of five to six replicates ± se.
Synechocystis ΔndbB and Arabidopsis ndc1 Mutants Display Increased Photosensitivity to High Light
Whereas under low light intensity the growth of Synechocystis ΔndbB cells was indistinguishable from growth of wild-type cells, this mutant displayed marked growth retardation compared with the wild type when cultured at high light intensity (Figure 4). This result was surprising, for it had been reported that the Arabidopsis ndc1 mutant was not phenotypically different from its wild-type counterpart (Eugeni Piller et al., 2011). To investigate this, Arabidopsis ndc1 knockout and wild-type plants that had been grown under normal light conditions and with day/night cycles were exposed to continuous high light. After 3 d of exposure to high light, ndc1 plants appeared smaller and paler than the wild type (Figure 4B). Chlorophyll quantification confirmed that chlorophyll a and chlorophyll b levels in the leaves of the ndc1 mutant were 19 and 16% lower than those of the wild type, respectively (Table 1).
Figure 4.
Phenotypes of Synechocystis ΔndbB and Arabidopsis ndc1 Mutants.
(A) Growth of wild-type and ΔndbB Synechocystis cells under continuous low (30 μmol photons m−2 s−1) or high (800 μmol photons m−2 s−1) light intensities. Identical numbers of cells were plated on BG-11 medium without antibiotics and were incubated for 1 week at 30°C.
(B) Four-week-old wild-type and ndc1 Arabidopsis plants after 3 d of exposure to continuous high light (800 μmol photons m−2 s−1). Bar = 2 cm.
(C) Efficiency of photosystem II in wild-type and ndc1 Arabidopsis plants after 3 d of high light regime (800 μmol photons m−2 s−1). The open arrowhead indicates the end of actinic illumination.
Table 1. Chlorophyll Levels and Calculated Fluorescence Parameters of Wild-Type and ndc1 Plants Exposed to Continuous High Light Intensity (800 μmol Photons m−2 s−1) for 3 d.
| Level or Parameter | Wild Type | ndc1 |
|---|---|---|
| Chlorophyll a (nmoles g−1 FW) | 1298 ± 38 | 1057 ± 36a |
| Chlorophyll b (nmoles g−1 FW) | 830 ± 15 | 693 ± 53b |
| Fv/Fm | 0.726 ± 0.042 | 0.802 ± 0.038 |
| NPQ | 1.010 ± 0.214 | 1.909 ± 0.429 |
| ϕPSII | 0.321 ± 0.072 | 0.212 ± 0.079 |
Fluorescence parameters were calculated after 5 min of actinic illumination. Data are means of three (chlorophyll) to four\ (fluorescence parameters) biological replicates ± sd. Different superscript letters indicate significant differences from the wild type as determined by Fisher’s test (P < α = 0.05) from an analysis of variance. FW, fresh weight.
Measurements of chlorophyll a fluorescence in response to actinic illumination did not indicate that maximum quantum efficiency of photosystem II (Fv/Fm) or nonphotochemical quenching (NPQ) was decreased in ndc1 plants compared with the wild type (Table 1). However, high light acclimated ndc1 plants displayed lower photosystem II operating efficiency (ϕPSII), which is proportional to the rate of CO2 fixation (Baker, 2008), than the wild type (Figure 4C, Table 1). Such a phenotype of increased photosensitivity, loss of chlorophylls, and decrease of photosystem II efficiency in response to high light treatment is reminiscent of that described for the Arabidopsis demethylnaphthoquinone methyltransferase knockout (Lohmann et al., 2006).
The Reduction of the Demethylnaphthoquinone Ring Is a Prerequisite to Its Transmethylation
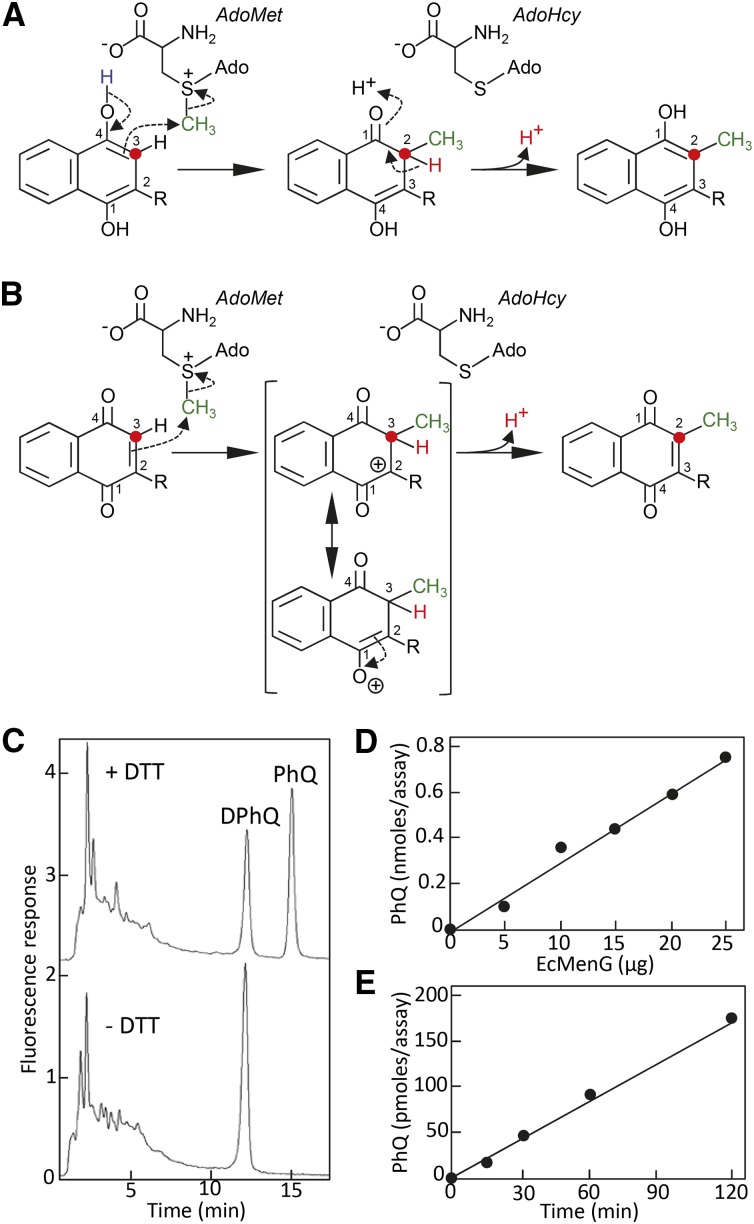
Having shown that the connection between type II NAD(P)H dehydrogenases and naphthoquinone metabolism is not solely specific but widespread throughout the prokaryotic and eukaryotic lineages of vitamin K-synthesizing organisms, we sought to elucidate the molecular mechanism that underlies the apparent epistasis between the loss of function of NDC1/ndbB and that of demethylnaphthoquinone methyltransferase. For that, we first modeled the mechanism of methyl transfer between AdoMet and the demethylated naphthoquinone precursor of vitamin K. The resulting model predicted that the transmethylation reaction strictly requires the reduced form (quinol) of the naphthalenoid ring, where C3 of the demethylnaphthoquinol moiety would act as the attacking nucleophile on the electrophilic methyl group of AdoMet, followed by proton exchanges to reform the C1-C2 double bond and the C1 phenol (Figure 5). By contrast, the alternative scenario starting from the oxidized form (quinone) of the naphthalenoid ring appears improbable because it would require the formation of an unstable cation intermediate (Figure 5B).
Figure 5.
Modeling of the Mechanism of Methyl Transfer between AdoMet and the Naphthalenoid Ring of Vitamin K.
(A) The naphthoquinol ring serves as the methyl group acceptor. The methyl group of AdoMet (green) is bonded to a positively charged sulfur atom and is a strong electrophile; C3 of the naphthoquinol ring (red dot) is the nucleophile. Note that due to the change in numbering priority after the ring methylation, the initial C3 of naphthoquinol becomes C2.
(B) The naphthoquinone ring serves as the acceptor. Ado, adenosine; AdoHcy, S-adenosyl- l-homocysteine; R, prenyl.
(C) Methyl transfer assays using E. coli demethylnaphthoquinone methyltransferase with demethylphylloquinone and AdoMet as substrates, in the presence or absence of DTT. Assays contained 1 μM demethylphylloquinone, 370 μM AdoMet, and 5 μg of MenG and were performed for 1 h at 30°C. The final concentration of DTT in the DTT-containing assay was 3.7 mM. Note that quinol forms are not detected in this assay as they readily reoxidize during solid-phase extraction prior to chromatographic analysis. HPLC traces have been offset for clarity. DPhQ, demethylphylloquinone; PhQ, phylloquinone.
(D) and (E) E. coli MenG (D) and time dependence (E) of phylloquinone formation.
To test the proposal that the transmethylation reaction in vitamin K biosynthesis indeed depends upon the redox state of the naphthoquinone ring, demethylphylloquinone was purified from the leaves of Arabidopsis demethylnaphthoquinone methyltransferase knockout plants and used as a methyl acceptor in assays of purified demethylnaphthoquinone methyltransferase (MenG; 2.1.1.163), with AdoMet as methyl donor and in the presence or absence of DTT as a reductant. Demethylphylloquinone and its methylated product, phylloquinone, were then extracted on solid-phase cartridges and quantified by HPLC with fluorescence detection. Phylloquinone formation was readily detected in the DTT-containing assay (Figure 5C), and the corresponding methyl transferase activity was linear with both the amount of demethylnaphthoquinone methyltransferase and time (Figures 5D and 5E). By contrast, no phylloquinone was detected when DTT was omitted from the assay (Figure 5C).
NDC1 and ndbB Catalyze the Reduction of the Demethylnaphthoquinone Ring
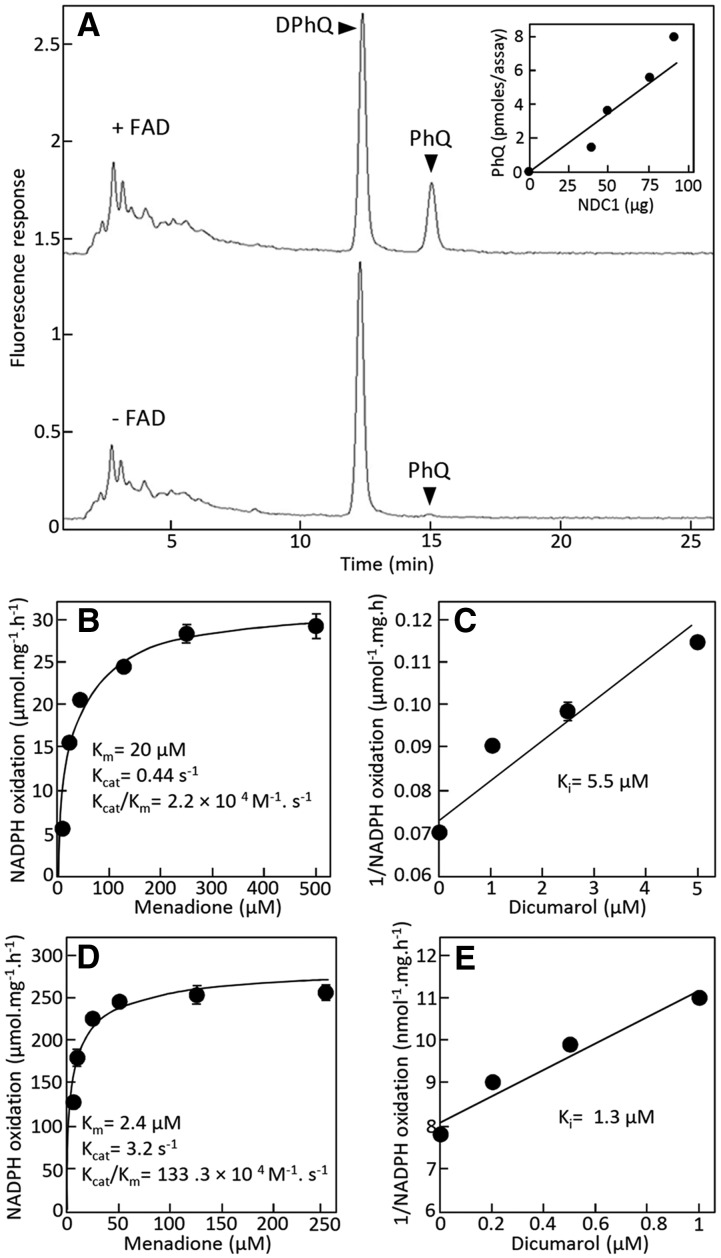
Since the demethylnaphthoquinone methyltransferase-catalyzed reaction strictly requires the quinol form of the methyl-accepting substrate, we examined the possibility that the demethylnaphthoquinone ring could be reduced by NDC1. To do this, purified NDC1 was assayed with demethylphylloquinone and NADPH in a coupled assay with demethylnaphthoquinone methyltransferase and AdoMet. NDC1-dependent demethylnaphthoquinone methyltransferase activity was readily detected, demonstrating that NDC1 and NADPH can effectively replace DTT (Figure 6). Furthermore, consistent with the occurrence of a noncovalently bound flavin cofactor in type II NAD(P)H dehydrogenases (Melo et al., 2004), phylloquinone formation was markedly decreased if purified NDC1 was not preincubated with flavine adenine dinucleotide (FAD) (Figure 6A).
Figure 6.
Assays of NDC1 with Naphthoquinones as Electron Acceptors.
(A) Coupled assays with demethylnaphthoquinone methyltransferase. Assays contained 0.52 μM demethylphylloquinone, 370 μM NADPH, 370 μM AdoMet, 89 μg of NDC1, and 10 μg of MenG and were performed for 3 h at 30°C. Overnight preincubation of NDC1 with FAD prior to the assay stimulated phylloquinone formation ∼25-fold (upper trace) compared with no preincubation (lower trace). Inset shows the formation of phylloquinone as a function of the concentration of NDC1 preincubated with FAD. Note that the point at 0 μg NDC1 also contained FAD. HPLC traces have been offset for clarity. DPhQ, demethylphylloquinone; PhQ, phylloquinone.
(B) NDC1-catalyzed oxidation of NADPH as a function of menadione concentration.
(C) Inhibition of NDC1 by dicumarol. Menadione concentration was 20 μM.
(D) ndbB-catalyzed oxidation of NADPH as a function of menadione concentration.
(E) Inhibition of ndbB by dicumarol.
The menadione concentration was 2.5 μM. The concentration of NADPH was 100 μM, and that of the FAD cofactor was 0.6 μM in (B) and (C) and 1.12 μM in (D) and (E). Data are means ± se of three replicates. Error bars smaller than the points are not visible.
Because demethylphylloquinone has a very low solubility in aqueous solution and could not be dispersed at concentrations greater than a few micromolar, a second assay was developed using menadione, a nonprenylated naphthoquinone derivative, as a substrate and monitoring the oxidation of NADPH spectrophotometrically. NDC1 displayed typical Michaelis-Menten kinetics, and the Km and Kcat of the enzyme for menadione were 20 μM and 0.44 s−1, respectively (Figure 6B). This resulted in a catalytic efficiency of 2.2 × 104 M−1 s−1, which was within the range of values (0.3 × 103 to 145 × 106 M−1 s−1) reported for other eukaryotic flavin oxidoreductases using NADH or NADPH as electron donors and menadione as an electron acceptor (Sánchez et al., 2001; Endo et al., 2008; Dong et al., 2009). Furthermore, the enzyme was markedly inhibited by dicumarol (Figure 6C), a competitive inhibitor of naphthoquinone oxidoreductases (Tie et al., 2011). The inhibition constant (Ki) of NDC1 for dicumarol determined with menadione and NADPH as substrates was 5.5 μM, which was typical of the Ki values reported for naphthoquinone oxidoreductases (4.1 to 10.6 μM) assayed in similar conditions (Sánchez et al., 2001; Wrobel et al., 2002). Similar results were obtained with recombinant ndbB: Its Km, Kcat, and catalytic efficiency values for menadione were 2.4 μM, 3.2 s−1, and 133.3 × 104 M−1 s−1, respectively (Figure 6D). The naphthoquinone oxidoreductase activity of ndbB was also markedly inhibited by dicumarol; the enzyme had a Ki value of 1.3 μM for this inhibitor with menadione and NADPH as substrates (Figure 6E).
DISCUSSION
A Missing Step in Vitamin K Biosynthesis
We establish here that the biosynthesis of vitamin K entails an additional bona fide step that is the enzymatic reduction of the demethylnaphthoquinone ring prior to its AdoMet-dependent transmethylation (Figure 7). Our data explain why the loss of function of NDC1/ndbB is epistatic with that of MENG and why the Arabidopsis ndc1 mutant accumulates the demethylated precursor of phylloquinone rather than phylloquinone itself. It is crucial to the interpretation of our results that AdoMet is strongly electrophilic and, therefore, that the transfer of its methyl group depends upon an electron pair originating from the methyl-accepting naphthalenoid ring. Chemical modeling and in vitro assays demonstrate that such an electron donation occurs exclusively with the quinol, i.e., reduced, form of the naphthalenoid ring. In plants and cyanobacteria, this prerequisite formation of demethylnaphthoquinol is catalyzed by a type II NADPH dehydrogenase, and comparative genomic data indicate that this is most probably also the case for the biosynthesis of menaquinone in Chloroflexi, Proteobacteria, Actinobacteria, and Firmicutes.
Figure 7.
Metabolic Architecture of Phylloquinone Biosynthesis in Plants.
Steps are identified by their recommended Enzyme Commission numbers. Note that in plants, 2.2.1.9, 4.2.99.20, and 4.2.1.113 belong to a single multifunctional enzyme and that OSB-CoA ligase (6.2.1.26) appears to be dual targeted to chloroplasts and peroxisomes (van Oostende et al., 2011). There is also evidence suggesting that OSB-CoA doubles as an intermediate in the biosynthesis of anthraquinones (Heide et al., 1982). Dashed arrows and question marks indicate multiple and hypothetical steps, respectively. DHNA, 1,4-dihydroxynaphthoate; OSB, o-succinylbenzoate.
The Redox Properties of the Demethylated Naphthoquinone Ring Impact the Architecture of Phylloquinone Biosynthesis in Plastids
It has been established that the biosynthesis of phylloquinone in plants requires the coordinated synthesis and transport of metabolic intermediates between two subcellular compartments, plastids and peroxisomes (van Oostende et al., 2011; Reumann, 2013). The initial benzenoid intermediate, o-succinylbenzoate, is made from chorismate in plastids (Figure 7). It is then exported to peroxisomes for its cyclization into 1,4-dihydroxynaphthoate, which is transported back into plastids where it is phytylated and methylated (Figure 7). Even within plastids, some phylloquinone biosynthetic intermediates are trafficked, for the demethylnaphthoquinone phytyltransferase (2.5.1.74) and methyltransferase (2.1.1.163) activities are localized in the chloroplast envelope and thylakoid membranes, respectively (Figure 7; Schultz et al., 1981; Kaiping et al., 1984). Our evidence that the plastoglobule-localized NDC1 mediates the mandatory reduction of the naphthoquinone ring between the phytylation and methylation steps not only underscores the intricacy of the reactions of phylloquinone biosynthesis, but also points to a hitherto unsuspected constraint on the architecture of this pathway. Indeed, having low midpoint redox potentials, naphthoquinols are prone to oxidation in vivo (Schoepp-Cothenet et al., 2009). The calculated rate of spontaneous reoxidation for the dihydroxynaphthoate ring gives a half-life of ∼1.5 h at atmospheric O2 concentration and standard ambient temperature and pressure. This is only a minimum estimate; the half-life of demethylnaphthoquinols is most probably much shorter in chloroplasts owing to the oxygenic nature of photosynthesis and to the occurrence of reactive oxygen species. The proximity of demethylnaphthoquinone methyltransferase in thylakoids and of NDC1 in plastoglobules, which are permanently attached to thylakoids (Austin et al., 2006), is therefore not coincidental and is in fact dictated by the need for plants to promptly methylate demethylphylloquinol.
Plants and Cyanobacteria Have Unique Demethylnaphthoquinone Oxidoreductases
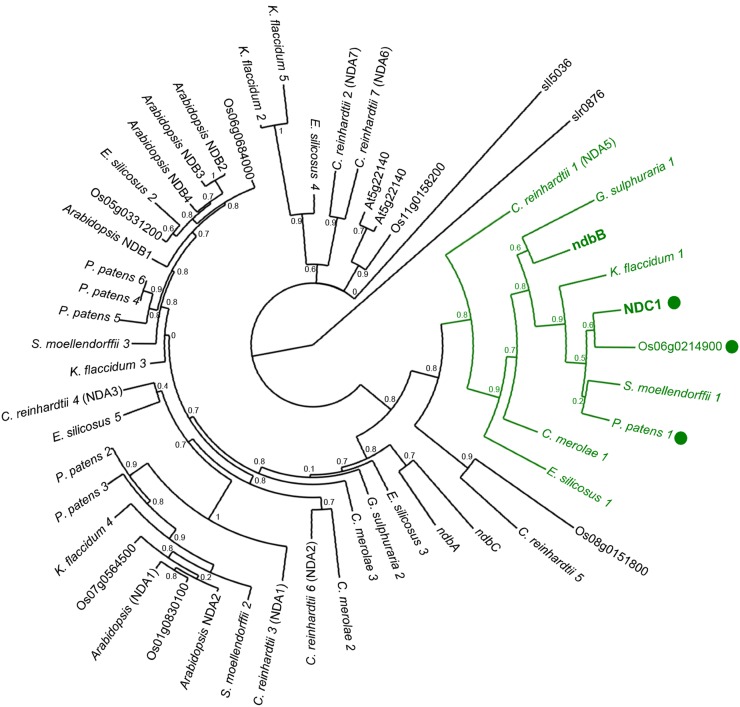
It should be emphasized that our results do not contradict previous reports indicating that NDC1 participates in the redox metabolism of two prenyl benzoquinones, plastoquinone-9 and the tocopherol recycling-intermediate α-tocopherol quinone (Eugeni Piller et al., 2011, 2014). In fact, our data show that ablating ndbB, the Synechocystis ortholog of NDC1, also results in lower plastoquinol-9/plastoquinone-9 ratio and lower α-tocopherol content compared with wild-type cells. It is therefore clear that NDC1 and ndbB are bifunctional oxidoreductases that are able to act both on prenyl naphthoquinones and on prenyl benzoquinones in vitro and in vivo. However, that the ndc1 and ΔndbB mutants contain only traces of phylloquinone and accumulate demethylphylloquinone rules out that other type II NADPH dehydrogenases contribute significantly to phylloquinone biosynthesis. In Arabidopsis, such an absence of functional redundancy is not wholly unexpected because out of seven type II NADPH dehydrogenases, only NDC1 is found in plastids (Carrie et al., 2008; Xu et al., 2013). A similar scenario applies to rice, where the NDC1 ortholog is the sole plastid-targeted type II NADPH dehydrogenase (Xu et al., 2013). By contrast, green fluorescent protein fusion experiments showed that the moss Physcomitrella patens possesses three plastid-targeted type II NADPH dehydrogenases (Xu et al., 2013), whereas two have been identified in the plastid of the green alga Chlamydomonas (Jans et al., 2008; Terashima et al., 2010). However, it would be premature to infer that mosses and green algae have multiple enzymes to reduce the demethylated naphthoquinone intermediate. First, as the case of Synechocystis illustrates, the capacity to reduce demethylphylloquinone is not common to all members of the type II NADPH dehydrogenase family. Second, phylogenetic reconstructions show that species within the Embryophytes, Charophytes, Chlorophytes, Rhodophytes, and Stramenopiles lineages each have a single type II NADPH dehydrogenase that is monophyletic with NDC1 and ndbB (Figure 8). Notably, the P. patens protein is one of the three type II NADPH dehydrogenases that is targeted to plastids (Xu et al., 2013).
Figure 8.
Maximum Likelihood Reconstruction of the Phylogenetic Relationships of Type II NAD(P)H Dehydrogenases within the Plant, Rhodophyte, Stramenopile, and Cyanobacterial Lineages.
Arabidopsis NDC1 closest homologs were identified from fully sequenced prokaryotic and eukaryotic genomes using BLASTp searches. Protein sequences were aligned with MUSCLE (Edgar, 2004), and misalignments and divergent regions were curated with Gblocks (Castresana, 2000). The tree was constructed with PhyML (Guindon et al., 2010) and visualized with TreeDyn (Chevenet et al., 2006). Synechocystis slr0876 was used as an outgroup to root the tree. Numbers next to each branching indicate approximate likelihood ratio test. The NDC1/ndbB clade is displayed in green; green dots indicate proteins for which exists experimental confirmation of subcellular localization in plastids. Arabidopsis NDC1 (AED91343), NDA2 (NP_180560), NDA1 (NP_563783), NDB1 (NP_567801), NDB2 (NP_567283), NDB3 (NP_193880), NDB4 (NP_179673), At5g22140 (BAC42443), and AT3G44190 (NP_190005); rice Os06g0214900 (NP_001057133), Os07g0564500 (NP_001060003), Os01g0830100 (NP_001044694), Os05g0331200 (NP_001055220), Os06g0684000 (NP_001058394), Os11g0158200 (NP_001065804), and Os08g0151800 (NP_001061002); Selaginella moellendorffii 1 (XP_002976807), S. moellendorffii 2 (XP_002978367), and S. moellendorffii 3 (XP_002977754); Physcomitrella patens 1 (XP_001768284), P. patens 2 (XP_001769969), P. patens 3 (XP_001757660), P. patens 4 (XP_001764062), P. patens 5 (XP_001766162), and P. patens 6 (XP_001759207); Chlamydomonas reinhardtii 1 (NDA5) (ABR53723), C. reinhardtii 2 (NDA7) (XP_001703056), C. reinhardtii 3 (NDA1) (XP_001698901), C. reinhardtii 4 (NDA3) (XP_001702271), C. reinhardtii 5 (XP_001696489), C. reinhardtii 6 (NDA2) (XP_001703643), and C. reinhardtii 7 (NDA6) (XP_001703055); Cyanidioschyzon merolae 1 (XP_005537068), C. merolae 2 (XP_005538302), and C. merolae 3 (XP_005535723); Galdieria sulphuraria 1 (XP_005707127), G. sulphuraria 2 (XP_005704866), and G. sulphuraria 3 (XP_005705674); Ectocarpus siliculosus 1 (CBN78924), E. silicosus 2 (CBN78752), E. silicosus 3 (CBN76274), E. silicosus 4 (CBJ25466), and E. silicosus 5 (CBN78336); Synechocystis sp PCC 6803 ndbB (NP_441103), ndbC (WP_014407163), ndbA (NP_441107), sll5036 (WP_011153573), and slr0876 (WP_010872226). Klebsormidium flaccidum protein sequences were retrieved from the K. flaccidum genome project (http://www.plantmorphogenesis.bio.titech.ac.jp/∼algae_genome_project/klebsormidium/): K. flaccidum 1 (kfl00025_0340), K. flaccidum 2 (kfl00064_0190), K. flaccidum 3 (kfl00811_0040), K. flaccidum 4 (kfl00553_0030), and K. flaccidum 5 (kfl00123_0080).
Similarly, one can exclude that Arabidopsis LUMEN THIOL OXIDASE1 (LTO1), a thylakoid-localized oxidoreductase, and its cyanobacterial ortholog dsbB, which both couple the formation of disulfide bonds in proteins to the reduction of prenyl naphthoquinones (Bader et al., 1999; Singh et al., 2008; Karamoko et al., 2011) contribute to phylloquinone biosynthesis. HPLC analyses actually confirmed that the Arabidopsis lto1 mutant does not display any significant differences in methylation state and total content of phylloquinone compared with wild-type plants (Supplemental Figure 1).
A Redox Switch Regulates the Output of Demethylmenaquinones versus Menaquinones
Finally, our results call for revisiting the question of how γ-proteobacteria are able to modify their naphthoquinone/demethylnaphthoquinone ratio in response to changes in growth conditions and independently of the transcription of the cognate methyltransferase (Shestopalov et al., 1997). One of the most striking instances of such a case is in Escherichia coli, the menaquinone pool of which switches from mostly demethylated to mostly methylated in a matter of seconds during the aerobic to anaerobic transition (Bekker et al., 2007). We propose that γ-proteobacteria have evolved the ability to make such rapid adjustments in the biosynthetic fluxes of their prenyl naphthoquinones via changes in the redox state of demethylmenaquinone. This also suggests that were strategies developed to engineer vitamin K2 levels in γ-proteobacteria, these cells might be able to defeat attempts to increase the cognate biosynthetic fluxes simply by modifying the redox state of their pool of naphthoquinone intermediates.
METHODS
Chemicals and Reagents
Demethylphylloquinone was purified by HPLC from leaf extracts of the Arabidopsis thaliana demethylnaphthoquinone methyltransferase knockout line GABI_565F06 (Lohmann et al., 2006). Phylloquinone was from MP Biomedicals and menaquinone-4 from Sigma-Aldrich. Unless mentioned otherwise, other reagents were from Fisher Scientific.
Bioinformatics
Microarray rice (Oryza sativa) probe sets were retrieved using the tBLASTn search mode of PLEXdb (http://www.plexdb.org/modules/tools/plexdb_blast.php) and the cognate rice proteins as queries. Coexpressing gene lists were identified in the GeneCAT database (http://genecat.mpg.de; Mutwil et al., 2008) using Arabidopsis genes At1g23360 (MENG), At1g60550 (MENB), At1g30520 (MENE), At1g60600 (MENA), At5g08740 (NDC1), At4g32770 (VTE1), At2g18950 (VTE2), and At1g06570 (HPPD) and rice probe sets Os.49084.1.S1_at (NDC1), Os.6179.1.S1_at (MENG), Os.18701.1.S1_at (VTE1), Os.19161.1.S1_at (MENB), Os.52256.1.S1_at (MENE), Os.35677.1.S1_at (MENA), OsAffx.15929.1.S1_at (VTE2), and Os.11995.1.S1_at (HPPD) as data entries. For Venn analyses of the coexpressing genes of NDC1, MENG, and VTE1, the first 500 coexpressors (top 2.2% of the 22,810 probe sets for Arabidopsis and top 0.9% of the 57,342 probe sets for rice) of each list were aggregated using GeneVenn (http://genevenn.sourceforge.net; Pirooznia et al., 2007). Hierarchical clustering analyses were performed using the expression tree analysis tool of GeneCAT and then visualized with Phy.Fi (http://cgi-www.daimi.au.dk/cgi-chili/phyfi/go; Fredslund, 2006). Comparative genomics searches were performed with the STRING (http://string.embl.de) and SEED (http://theseed.uchicago.edu/FIG/index.cgi) databases using their associated tools and Arabidopsis NDC1 as an initial query.
Biological Material and Growth Conditions
Seeds of Arabidopsis T-DNA insertion line GABI_565F06 (meng) and SALK_151963C (lto1) were germinated on Murashige and Skoog solid medium. Two-week-old seedlings were then transferred to soil and grown at 22°C in 16-h days (110 μmol photons m−2 s−1) for 4 weeks. For high-light treatment, plants were grown at normal light intensity (110 μmol photons m−2 s−1; 16-h days) for 4 weeks and then exposed for 3 d to continuous high light (800 μmol photons m−2 s−1). Synechocystis sp PCC 6803 deletion strains ΔndbA (slr0851), ΔndbB (slr1743), ΔndbC (sll1484), and ΔΔΔndbABC were those described by Howitt et al. (1999), and deletion strain ΔmenG (sll1653) was described by Lohmann et al. (2006). Because the lack of tocopherols impacts macronutrient homeostasis in photomixotrophic conditions (Sakuragi et al., 2006), all Synechocystis mutants were isolated and propagated photoautrophically at a light intensity of 50 to 65 μmol photons m−2 s−1 on solid BG-11 medium (Rippka et al., 1979). Selection and propagation plates contained 25 μg mL−1 erythromycin (ΔndbA), 25 μg mL−1 spectinomycin (ΔndbB and ΔmenG), 15 μg mL−1 zeocin (ΔndbC), or all three antibiotics (ΔΔΔndbABC). Homozygosity for the mutant loci was verified by PCR and the strains were stored at −80°C. For metabolite quantification, wild-type and mutant Synechocystis strains were streaked from their respective original −80°C stocks and grown photoautotrophically at a light intensity of 65 μmol photons m−2 s−1 at 30°C on solid BG-11 medium containing the appropriate antibiotics. For growth assays in normal and high light intensities, cells were scraped from propagation plates, resuspended in liquid BG-11 medium (Rippka et al., 1979), and quantified spectrophotometrically at 730 nm. Identical numbers of wild-type and mutant cells were then spotted on solid BG-11 medium without antibiotics, and the plates were incubated at 30°C under a continuous illumination of 30 or 800 μmol photons m−2 s−1.
Expression, Purification, and Refolding of Recombinant Enzymes
The Escherichia coli menG gene was amplified from strain K12 genomic DNA using the primers 5′-CACCATGGTGGATAAGTCACAAGAAA-3′ (forward) and 5′-TCAGAACTTATAACCACGATGC-3′ (reverse). The Arabidopsis NDC1 cDNA (At5g08740) lacking its predicted N-terminal presequence was amplified from total leaf RNA by RT-PCR using the primers 5′-CACCATGACAGAGATTTCTGATAATGAAACAG-3′ (forward) and 5′-TCAAGAACCAGACAAAACCTT-3′ (reverse); the italicized sequence indicates the initiation codon introduced to truncate the targeting presequence (T60→M). The PCR products were transferred into pDEST 17 (Invitrogen) using Gateway technology. The Synechocystis ndbB gene was amplified from strain PCC 6803 genomic DNA using the primers 5′-GCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACCATGACGGACGCTCGACC-3′ (forward) and 5′-GGAATTCTCAATGATG-ATGATGATGATGGGAAGGTTCATTTTTGAGCCAATCC-3′ (reverse), which contained the XbaI and EcoRI restriction sites (italicized), respectively. The digested PCR fragment was then cloned into the corresponding sites of pET-28b (Novagen). The menG and ndbB constructs were introduced into E. coli BL21 Star (DE3) pLysS (Invitrogen), while the NDC1 construct was introduced into BL21-CodonPlus (DE3)-RIL (Agilent Technologies). Overnight starter cultures (1 mL) were used to inoculate 100 mL of Luria-Bertani medium (Luria and Burrous, 1957) prewarmed at 37°C. Isopropyl β-d-1-thiogalactopyranoside (500 μM) was added when the OD600 reached ∼0.7, and incubation was continued for 4 h. Subsequent operations were at 4°C. Cells were harvested by centrifugation (18,000g for 5 min), resuspended in 2 mL of 50 mM NaH2PO4 (pH 8.0) and 300 mM NaCl, and disrupted with 0.1-mm zirconia/silica beads in a MiniBeadbeater (BioSpec Products). The extract from cells expressing C-terminally 6xHis-tagged ndbB was cleared by centrifugation (18,000g for 15 min), and the recombinant enzyme was purified under native conditions using Ni-NTA His-Bind resin (Novagen), according to the manufacturer's instructions. Extracts of cells harboring the menG or NDC1 constructs were centrifuged (18,000g for 15 min), and the pellets containing the inclusion bodies were washed twice with 2 mL of 50 mM Tris-HCl, pH 7.5, 500 mM NaCl, and 1% (v/v) Triton X-100. Inclusion bodies were then solubilized for 1 h in 2 mL of 50 mM HEPES (pH 7.5), 6 M guanidine-HCl, and 25 mM DTT. After removal of insoluble material (18,000g for 15 min), solubilized proteins were renatured by briskly diluting supernatants 10-fold in 50 mM HEPES (pH 7.5), 0.2 M NaCl, 1 mM DTT, and 1 M 3-(1-pyridino)-1-propane sulfonate (Sigma-Aldrich). Purified NdbB, MenG, and NDC1 were desalted on a PD-10 column (GE Healthcare) equilibrated in 50 mM KH2PO4 (pH 7.5), 150 mM KCl, and 0.2% (v/v) Tween 20. Proteins were quantified using the Bradford method (Bradford, 1976) using BSA as a standard. Aliquots of desalted proteins were flash frozen in liquid N2 and stored at −80°C.
Enzymatic Assays
Prior to assays, NDC1 and NdbB preparations were thawed and incubated overnight at 4°C in the presence of up to a 2-fold molar excess of FAD. For methyl transfer assays, methanolic solutions of demethylphylloquinone (44 to 130 pmoles) were evaporated to dryness in Pyrex screw-cap tubes. Assays (135 μL) contained demethylphylloquinone in 50 mM KH2PO4 (pH7.5), 150 mM KCl, 0.2% (v/v) Tween 20, 0.37 mM AdoMet, 0.37 mM NADPH, 0 to 3.7 mM DTT, 0 to 25 μg of MenG, and 0 to 86 μg of NDC1. Reactions were initiated by the addition of AdoMet and were incubated for 15 to 180 min at 30°C and then stopped with 500 μL 100% methanol and centrifuged (18,000g for 5 min). Supernatants (50 μL) were loaded onto Supelclean LC-18 SPE (Supelco) solid-phase extraction columns, and prenylated naphthoquinones were eluted with 2 mL 100% methanol. Eluates were evaporated to dryness, resuspended in 200 μL methanol/dichloromethane (10:1) and analyzed by HPLC with fluorescence detection as previously described (Widhalm et al., 2012). Spectrophotometric assays (1 mL) contained 50 mM HEPES/KOH (pH 7.5), 0.05% (v/v) Tween 20, 100 μM NADPH, 0 to 500 μM of menadione, and 0 to 31 μg of NDC1 or ndbB. Inhibition assays with dicumarol were performed at a concentration of menadione of 20 μM for NDC1 and to 24 μM for NdbB, so that [dicumarol] = 2Ki for 1/NADPH oxidation = 0. Reactions were performed for 15 min at 30°C, and the oxidation of NADPH was monitored spectrophotometrically at 340 nm.
Metabolite Quantification
Synechocystis cells were directly harvested from BG-11 plates, resuspended in 200 μL of 0.9% (w/v) NaCl, and quantified by measuring the optical density at 730 nm. For naphthoquinone analysis, resuspended cells were transferred to Pyrex screw-cap tubes containing 1 mL of 0.1-mm zirconia/silica beads, 900 μL of 95% (v/v) ethanol, 400 μL of water, and 39 pmoles of menaquinone-4 as an internal standard, and then ruptured by vortexing for 5 min. Prenylated naphthoquinones were phase-partitioned twice with 5 mL hexane and vigorous shaking. Hexane phases were combined and evaporated to dryness under nitrogen. Samples were resuspended in 300 μL of methanol/dichloromethane (10:1) and analyzed by HPLC with fluorescence detection as previously described (Widhalm et al., 2012). Phylloquinone was quantified according to external standards, and data were corrected for recovery of the internal standard of menaquinone-4. For plastoquinone and tocochromanol analysis, 50 μL of resuspended cells were transferred to Pyrex screw-cap tubes containing 400 μL of 0.1-mm zirconia/silica beads, 550 μL of 100% (v/v) ethanol, and then vortexed for 5 min. Extracts were cleared by centrifugation (18,000g for 10 min) and analyzed by HPLC with fluorescence (plastoquinol-9, α- and γ-tocopherol, and plastochromanol-8) or diode array detection (plastoquinone-9) as previously described (Block et al., 2013). Chlorophylls were quantified spectrophotometrically in acetone extracts of Arabidopsis leaves using the molar extinction coefficients of 75.05 mM−1 cm−1 at 663 nm for chlorophyll a, and 47 mM−1 cm−1 at 645 nm for chlorophyll b.
Measurements of Photosynthetic Parameters
Chlorophyll fluorescence was measured on detached and dark-adapted (30 min) leaves using a JTS-10 LED spectrometer (Bio-Logic Scientific Instruments). Actinic illumination (520 μmol photons m−2 s−1) was provided from green LEDs (520 nm; Bio-Logic Scientific Instruments) for 6 min to the adaxial surfaces of the leaves. The maximal fluorescence in the dark-adapted state (Fm) during actinic illumination and dark relaxation (Fm′) were determined using a 250-ms saturating light pulse (3600 μmol photons m−2·s−1). Fo (initial fluorescence of dark-adapted leaves) and Fs (minimal fluorescence during actinic illumination and during dark relaxation) were measured with 10-μs weak detecting flashes. Fluorescence parameters were calculated as follows: Fv/Fm = (Fm − Fo)/Fm, where Fv is the calculated variable fluorescence; NPQ = (Fm − Fm′)/Fm′ ; ΦPSII = (Fm′- Fs )/Fm′.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: Arabidopsis At1g23360 (MENG), At1g60550 (MENB), At1g30520 (MENE), At1g60600 (MENA), At5g08740 (NDC1), At4g32770 (VTE1), At2g18950 (VTE2), and At1g06570 (HPPD); rice CAE04724.1 (MENG), EAY75265 (MENB), EAZ05485 (MENE), NP_00104922 (MENA), NP_001057133 (NDC1), NP_001046545 (VTE1), B7FA90 (VTE2), and BAD26248 (HPPD); Synechocystis NP_441107 (NdbA), NP_441103 (NdbB), and NP_442910 (NdbC); and E. coli YP_026269 (MenG). Germplasm used is as follows: Arabidopsis demethylnaphthoquinone methyltransferase knockout line GABI_565F06; T-DNA insertion lines GABI_565F06 (meng; At1g23360) and SALK_151963C (lto1; At3g50820). Synechocystis sp PCC 6803 deletion strains ΔndbA (slr0851), ΔndbB (slr1743), ΔndbC (sll1484), and ΔΔΔndbABC were those described by Howitt et al. (1999), and deletion strain ΔmenG (sll1653) was that described by Lohmann et al. (2006).
Supplemental Data
Supplemental Figure 1. Quantification of phylloquinone and of demethylphylloquinone in the leaves of wild-type and lto1 knockout plants.
Supplemental Data Set 1. Gene lists, including correlation ranks and functional annotations, of the top 500 coexpressors of MENG, NDC1, and VTE1 in Arabidopsis and rice.
Supplementary Material
Acknowledgments
This work was made possible in part by National Science Foundation Grant MCB-1148968 (to G.J.B.). S.S.M. is supported by the U.S. Department of Energy, Office of Biological and Environmental Research program under Award Number DE-FC02-02ER63421. We thank Peter Dörmann (University of Bonn, Germany), Patrice Hamel (Ohio State University), and Edgar Cahoon (University of Nebraska-Lincoln) for the gift of the Arabidopsis menG, Arabidopsis lto1, and Synechocystis menG knockouts, respectively. We thank Catherine and Steven Clarke (University of California-Los Angeles) for illuminating discussions regarding the mechanism of transmethylation.
AUTHOR CONTRIBUTIONS
A.F., S.L., S.S., A.B., and G.J.B. designed and performed the research, analyzed the data, and wrote the article. P.H.D. designed the research and analyzed the data. W.F.J.V. contributed materials and analyzed the data. S.S.M. analyzed the data.
Glossary
- AdoMet
S-adenosyl-l-methionine
- NPQ
nonphotochemical quenching
- FAD
flavine adenine dinucleotide
References
- Austin J.R. II, Frost E., Vidi P.A., Kessler F., Staehelin L.A. (2006). Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18: 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M., Muse W., Ballou D.P., Gassner C., Bardwell J.C. (1999). Oxidative protein folding is driven by the electron transport system. Cell 98: 217–227. [DOI] [PubMed] [Google Scholar]
- Baker N.R. (2008). Chlorophyll fluorescence: a probe of photosynthesis in vivo. Annu. Rev. Plant Biol. 59: 89–113. [DOI] [PubMed] [Google Scholar]
- Bekker M., Kramer G., Hartog A.F., Wagner M.J., de Koster C.G., Hellingwerf K.J., de Mattos M.J. (2007). Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology 153: 1974–1980. [DOI] [PubMed] [Google Scholar]
- Besagni C., Kessler F. (2013). A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation. Planta 237: 463–470. [DOI] [PubMed] [Google Scholar]
- Beulens J.W., Booth S.L., van den Heuvel E.G., Stoecklin E., Baka A., Vermeer C. (2013). The role of menaquinones (vitamin K2) in human health. Br. J. Nutr. 110: 1357–1368. [DOI] [PubMed] [Google Scholar]
- Block A., Fristedt R., Rogers S., Kumar J., Barnes B., Barnes J., Elowsky C.G., Wamboldt Y., Mackenzie S.A., Redding K., Merchant S.S., Basset G.J. (2013). Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J. Biol. Chem. 288: 27594–27606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth S.L. (2009). Roles for vitamin K beyond coagulation. Annu. Rev. Nutr. 29: 89–110. [DOI] [PubMed] [Google Scholar]
- Booth S.L., Suttie J.W. (1998). Dietary intake and adequacy of vitamin K. J. Nutr. 128: 785–788. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Brettel K. (1997). Electron transfer and arrangement of the redox cofactors in photosystem I. Biochim. Biophys. Acta 1318: 322–373. [Google Scholar]
- Carrie C., Murcha M.W., Kuehn K., Duncan O., Barthet M., Smith P.M., Eubel H., Meyer E., Day D.A., Millar A.H., Whelan J. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582: 3073–3079. [DOI] [PubMed] [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Chevenet F., Brun C., Bañuls A.L., Jacq B., Christen R. (2006). TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics 7: 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M.D., Jones D. (1981). Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implication. Microbiol. Rev. 45: 316–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C.K., Patel V., Yang J.C., Dvorin J.D., Duraisingh M.T., Clardy J., Wirth D.F. (2009). Type II NADH dehydrogenase of the respiratory chain of Plasmodium falciparum and its inhibitors. Bioorg. Med. Chem. Lett. 19: 972–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S., Matsunaga T., Kitade Y., Ohno S., Tajima K., El-Kabbani O., Hara A. (2008). Human carbonyl reductase 4 is a mitochondrial NADPH-dependent quinone reductase. Biochem. Biophys. Res. Commun. 377: 1326–1330. [DOI] [PubMed] [Google Scholar]
- Eugeni Piller L., Besagni C., Ksas B., Rumeau D., Bréhélin C., Glauser G., Kessler F., Havaux M. (2011). Chloroplast lipid droplet type II NAD(P)H quinone oxidoreductase is essential for prenylquinone metabolism and vitamin K1 accumulation. Proc. Natl. Acad. Sci. USA 108: 14354–14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugeni Piller L., Glauser G., Kessler F., Besagni C. (2014). Role of plastoglobules in metabolite repair in the tocopherol redox cycle. Front. Plant Sci. 5: 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredslund J. (2006). PHY.FI: fast and easy online creation and manipulation of phylogeny color figures. BMC Bioinformatics 7: 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L.J. (2013). STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41: D808–D815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furt F., Oostende Cv., Widhalm J.R., Dale M.A., Wertz J., Basset G.J. (2010). A bimodular oxidoreductase mediates the specific reduction of phylloquinone (vitamin K1) in chloroplasts. Plant J. 64: 38–46. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Heide L., Kolkmann R., Arendt S., Leistner E. (1982). Enzymic synthesis of o-succinylbenzoyl-CoA in cell-free extracts of anthraquinone producing Galium mollugo L. cell suspension cultures. Plant Cell Rep. 1: 180–182. [DOI] [PubMed] [Google Scholar]
- Howitt C.A., Udall P.K., Vermaas W.F.J. (1999). Type 2 NADH dehydrogenases in the cyanobacterium Synechocystis sp. strain PCC 6803 are involved in regulation rather than respiration. J. Bacteriol. 181: 3994–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans F., Mignolet E., Houyoux P.A., Cardol P., Ghysels B., Cuiné S., Cournac L., Peltier G., Remacle C., Franck F. (2008). A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. USA 105: 20546–20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiping S., Soll J., Schultz G. (1984). Site of methylation of 2-phytyl-1,4-naphthoquinol in phylloquinone (vitamin K1) synthesis in spinach chloroplasts. Phytochemistry 23: 89–91. [Google Scholar]
- Karamoko M., Cline S., Redding K., Ruiz N., Hamel P.P. (2011). Lumen Thiol Oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in Arabidopsis. Plant Cell 23: 4462–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Takeshita A., Koyama T., Ogura K. (1997). Identification of a novel gene cluster participating in menaquinone (vitamin K2) biosynthesis. Cloning and sequence determination of the 2-heptaprenyl-1,4-naphthoquinone methyltransferase gene of Bacillus stearothermophilus. J. Biol. Chem. 272: 12380–12383. [DOI] [PubMed] [Google Scholar]
- Lefebvre-Legendre L., Rappaport F., Finazzi G., Ceol M., Grivet C., Hopfgartner G., Rochaix J.D. (2007). Loss of phylloquinone in Chlamydomonas affects plastoquinone pool size and photosystem II synthesis. J. Biol. Chem. 282: 13250–13263. [DOI] [PubMed] [Google Scholar]
- Lohmann A., Schöttler M.A., Bréhélin C., Kessler F., Bock R., Cahoon E.B., Dörmann P. (2006). Deficiency in phylloquinone (vitamin K1) methylation affects prenyl quinone distribution, photosystem I abundance, and anthocyanin accumulation in the Arabidopsis AtmenG mutant. J. Biol. Chem. 281: 40461–40472. [DOI] [PubMed] [Google Scholar]
- Luria S.E., Burrous J.W. (1957). Hybridization between Escherichia coli and Shigella. J. Bacteriol. 74: 461–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo A.M., Bandeiras T.M., Teixeira M. (2004). New insights into type II NAD(P)H:quinone oxidoreductases. Microbiol. Mol. Biol. Rev. 68: 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutwil M., Obro J., Willats W.G., Persson S. (2008). GeneCAT—novel webtools that combine BLAST and co-expression analyses. Nucleic Acids Res. 36: W320–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirooznia M., Nagarajan V., Deng Y. (2007). GeneVenn: A web application for comparing gene lists using Venn diagrams. Bioinformation 1: 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. (2013). Biosynthesis of vitamin K1 (phylloquinone) by plant peroxisomes and its integration into signaling molecule synthesis pathways. Subcell. Biochem. 69: 213–229. [DOI] [PubMed] [Google Scholar]
- Rippka R., Deruelles J., Waterbury J.B., Herdmann M., Stanier R.Y. (1979). Generic assignments, strains histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1–61. [Google Scholar]
- Rochaix J.-D. (2013). Redox regulation of thylakoid protein kinases and photosynthetic gene expression. Antioxid. Redox Signal. 18: 2184–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez L.B., Elmendorf H., Nash T.E., Müller M. (2001). NAD(P)H:menadione oxidoreductase of the amitochondriate eukaryote Giardia lamblia: a simpler homologue of the vertebrate enzyme. Microbiology 147: 561–570. [DOI] [PubMed] [Google Scholar]
- Sakuragi Y., Maeda H., Dellapenna D., Bryant D.A. (2006). α-Tocopherol plays a role in photosynthesis and macronutrient homeostasis of the cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol. 141: 508–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoepp-Cothenet B., Lieutaud C., Baymann F., Verméglio A., Friedrich T., Kramer D.M., Nitschke W. (2009). Menaquinone as pool quinone in a purple bacterium. Proc. Natl. Acad. Sci. USA 106: 8549–8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz G., Ellerbrock B.H., Soll J. (1981). Site of prenylation reaction in synthesis of phylloquinone (vitamin K1) by spinach chloroplasts. Eur. J. Biochem. 117: 329–332. [DOI] [PubMed] [Google Scholar]
- Semenov A.Y., Vassiliev I.R., van Der Est A., Mamedov M.D., Zybailov B., Shen G., Stehlik D., Diner B.A., Chitnis P.R., Golbeck J.H. (2000). Recruitment of a foreign quinone into the A1 site of photosystem I. Altered kinetics of electron transfer in phylloquinone biosynthetic pathway mutants studied by time-resolved optical, EPR, and electrometric techniques. J. Biol. Chem. 275: 23429–23438. [DOI] [PubMed] [Google Scholar]
- Shestopalov A.I., Bogachev A.V., Murtazina R.A., Viryasov M.B., Skulachev V.P. (1997). Aeration-dependent changes in composition of the quinone pool in Escherichia coli. Evidence of post-transcriptional regulation of the quinone biosynthesis. FEBS Lett. 404: 272–274. [DOI] [PubMed] [Google Scholar]
- Singh A.K., Bhattacharyya-Pakrasi M., Pakrasi H.B. (2008). Identification of an atypical membrane protein involved in the formation of protein disulfide bonds in oxygenic photosynthetic organisms. J. Biol. Chem. 283: 15762–15770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M., Specht M., Naumann B., Hippler M. (2010). Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9: 1514–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J.-K., Jin D.-Y., Straight D.L., Stafford D.W. (2011). Functional study of the vitamin K cycle in mammalian cells. Blood 117: 2967–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oostende C., Widhalm J.R., Furt F., Ducluzeau A.-L., Basset G.J. (2011). Vitamin K1 (phylloquinone): Function, enzymes and genes. Adv. Bot. Res. 59: 229–261. [Google Scholar]
- Widhalm J.R., Ducluzeau A.-L., Buller N.E., Elowsky C.G., Olsen L.J., Basset G.J. (2012). Phylloquinone (vitamin K(1) ) biosynthesis in plants: two peroxisomal thioesterases of Lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-CoA. Plant J. 71: 205–215. [DOI] [PubMed] [Google Scholar]
- Wrobel R.L., Matvienko M., Yoder J.I. (2002). Heterologous expression and biochemical characterization of an NAD(P)H quinone oxidoreductase from the hemiparasitic plant Triphysaria versicolor. Plant Physiol. Biochem. 40: 265–272. [Google Scholar]
- Xu L., Law S.R., Murcha M.W., Whelan J., Carrie C. (2013). The dual targeting ability of type II NAD(P)H dehydrogenases arose early in land plant evolution. BMC Plant Biol. 13: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y.Z., et al. (2011). MSH1 is a multi-functional protein in plants that alters mitochondrial and plastid properties and response to high light. Plant Cell 23: 3428–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.