Abstract
Inferring the effective population size and the pattern of selection signatures is of interest both from an evolutionary perspective and to improve models for mapping of quantitative trait genes. We used DNA samples of 61 sires and 486 progeny of the Hanwoo, genotyped by the Illumina Bovine SNP50 BeadChip, to analyze the genetic structure. Our study showed a persistent decline in effective population size throughout the period considered, but suggested a marked decline at one distinctive time point (100th generation) and two sharp decline intervals (50th–25th generation and 25th–10th generation). This pattern can be explained by Hanwoo formation and the modern breeding program. Our results revealed 95 regions exhibiting the footprint of recent positive selection at a threshold level of 0.01. We found an overlap of the 11 core regions presenting top P-values and those that had previously been identified as harboring quantitative trait loci from other breeds. The information generated from this study can be used to better understand the mechanism of selection in Hanwoo breeding, and provide important implications for the design and application of association studies in the Hanwoo population.
Keywords: cattle, linkage disequilibrium, effective population size, selection signature
Introduction
Linkage disequilibrium (LD) has recently become the focus of intense study because so many factors affect it and are affected by it. LD describes the nonrandom association of alleles at two or more loci. Information on the structure of LD at the population level is critical for interpreting and applying results of genome-wide association studies (GWAS) and genomic selection for the improvement of economically important traits.1,2 LD can help to unravel the genetic relationships among diverse breeds and the phylogenetic relationships between domestic animals and their wild ancestors. LD can be used to enable effective exploitation of the potentially significant historical events that occurred during domestication, breed formation, and ongoing selection (eg, bottleneck effects, genetic drift, and selective sweeps).
A wide variety of statistics have been proposed to measure the amount of LD, and these have different strengths, depending on the context. In the current literature, the two most popular measures of LD between pairs of biallelic markers are D’ and r2. Typically, r2 is preferred when the focus is on the predictability of one polymorphism given the other, and hence it is often used in power studies for association designs. D’, by contrast, is the measure of choice to assess recombination patterns; thus, haplotype blocks have often been defined on the basis of D’.3
Effective population size (Ne) is widely regarded as one of the most important parameters in both evolutionary biology and conservation biology.4 In particular, the action of selection means that Ne varies across the genome, and advances in genomic techniques give new insight into how selection shapes Ne. There are three different ways to measure Ne: 1) the change in probability of identity by descent (inbreeding effective size); 2) the change in variance in allele frequency (variance effective size); and 3) the rate of loss of heterozygosity (eigenvalue effective size).5 In this study, we estimated Ne by the original single-sample method based on random LD that arises by chance in each generation in a finite population.
Detection of selective sweeps is useful because the effect of selection on the distribution of genetic variation can be difficult to distinguish from the pattern of genetic variation that arises after certain demographic events.6,7 The long-range haplotype (LRH) test examines the relationship between allele frequency and the extent of LD.8 LRH measures the decay of identity, as a function of distance, of haplotypes that carry a specified “core” allele at one end. Alternative methods to detect selective sweeps include Tajima’s D- and Fay and Wu’s H-test.9,10 Both these tests use site frequency spectrum summaries to identify selection signatures. The integrated Haplotype Score (iHS) is an extension of LRH,11 and is more powerful than the Tajima’s D- or Fay and Wu’s H-test for selected mutations that are still segregating in the population. However, iHS is limited by low single nucleotide polymorphism (SNP) density and the inability to completely specify ancestral SNP allele states.12
The Korean native cattle (Hanwoo) is known for its marbled fat, tenderness, juiciness, and characteristic flavor. Hanwoo originated from a hybrid of Bos taurus × Bos zebu that migrated to the Korean peninsula in 4000 BC. Hanwoo has been intensively selected during the last few centuries, especially in the recent decades since the implementation of progeny-test-based breeding programs in the 1980s.13–15 In this study, we use SNP data generated with the Illumina Bovine SNP50K BeadChip to explore some properties of r2 and D’ as the most common measures of LD. The extent of LD is presented along with the estimation of ancestral population size for different generations and the genome-wide footprints of positive selection. This would provide a framework for evaluating the genetic structure and history of this cattle breed, and facilitate detailed studies on the identified candidate genes.
Materials and Methods
DNA samples
SNP markers throughout the Hanwoo genome were genotyped. Samples were collected from 547 bulls born from spring through fall of 2006 in Seosan, South Korea. The bulls were 61 sires and 486 progeny. The size of the 61 sire families ranged from 2 to 18 bulls per sire, with an average of 8 bulls. Pedigree information was obtained from the Hanwoo Improvement Center of National Agricultural Cooperative Federation, Seosan. The mean kinship (coefficient of coancestry) is 0.0119, and the mean inbreeding coefficient is 0.4%, determined using pedigree information from up to four generations. For the purposes of this study, these values were assumed to be zero. Coancestry and inbreeding coefficients were computed using coancestry, inbreeding (F), and contribution (CFC) programs.16
Ethics committee approval for treatment of animals was not required, as all samples used in this study had been collected by veterinarians for routine purposes, and were reused for the research presented here.
Selection and genotyping of markers
A total of 54,001 SNPs were screened using the Illumina Bovine SNP50K BeadChip.17 SNPs were analyzed using an Illumina Bead-Station 5.2 genotyping instrument (Illumina Inc.), and genotyping was performed using BeadStudio 3.0 (Illumina Inc.) software. SNPs were removed on the basis of the following criteria: 1) Markers were filtered to exclude loci assigned to unmapped contigs, or unpositioned according to the latest reference assembly of the bovine genome Btau 4.0, and markers on the X and Y chromosomes. 2) Monomorphic SNPs and those with minor allele frequency smaller than 0.05 were filtered, because it is known that LD between SNPs with a low minor allele frequency is biased upwards, and thus high-frequency polymorphisms are preferable for accurate estimation of LD.18 3) Animals with genotype completeness smaller than 90% were excluded. 4) Markers with significant departure from H–W equilibrium (P < 0.001) were also excluded.
Haplotype phase reconstruction
Haplotype phases were inferred from pedigree by a localized haplotype clustering model (LHCM) method,19 which is a family rule (Mendelian segregation and linkage)-based algorithm. Only maternally inherited haplotypes were used for further detailed analyses, in order to minimize the effect of over-representation of paternally inherited haplotypes within pedigrees of the bulls.
SNP annotation information
Annotation information was based on genomic positions of Bos taurus genes (Btau 4.0 build) that were obtained from Biomart.20 We divided SNP pairs into the following functional categories: nonsynonymous, synonymous, within an intron or mRNA untranslated region (UTR), or within 2 kb of a gene.11 SNPs in any of these functional classes were the intragenic group, and the remaining SNPs were assigned to the intergenic SNP group.
Measures of linkage disequilibrium
We used the GOLD 1.1.0 program21 to construct LD maps from the maternally inherited haplotypes, thus avoiding the over-representation of paternally inherited haplotypes within the primarily male pedigrees. GOLD computes Lewontin’s disequilibrium coefficient D’ and r2 for all pairwise SNPs.22,23 LD was calculated for intragenic SNPs and intergenic SNPs separately to investigate whether LD within genes is higher or lower than between genes.
Past effective population size using LD
To observe the past effective population size (Ne), Ne was calculated based on the relationship between the effective population size (Ne), the recombination rate, and LD (r2). In the absence of mutation, the function of LD is
where yi = (r2 – 1/n), where n is the number of sampled haplotypes, the recombination rate c is in Morgans, Ne is the population size 1/(2c) generations ago, and ei is the residual.24 The recombination rate (c) was inferred based on the ratio between the physical size of each chromosome and length of the corresponding linkage map (NCBI Map Viewer25). The past effective population size was estimated approximately in each chromosome. Under the assumption of linear population expansion, the effective population size (Ne) was fitted by nonlinear least squares regression in the program R,26 using pairwise r2 and SNP distance bin (0.1 Mb) within ~0.1–15 Mb to estimate Ne of the past generations. However, for the calculation of average r2 for marker distance <100 kb, the SNP distance bin was 0.01 Mb.
Long-range haplotype (LRH) test
Under neutral evolution, positive selection is expected to accelerate the frequency of an advantageous allele faster than recombination can break down LD at the selected haplotype.27
The LRH test calculates the relative extended haplotype homozygosity (REHH), and assesses the significance of REHH by use of simulations, as described in Sabeti et al.8 In brief, core regions (a pair of SNPs to be in strong LD if the upper 95% confidence bound of D’ is between 0.7 and 0.98) were identified throughout the genome. Because a recombination event should have occurred in a two-SNP core region with a core haplotype composed of two derived alleles under an infinite sites model, we selected core regions with at least three SNPs.28 Second, we estimated the haplotypes and haplotype frequencies in these regions. Because of the longer extent of LD in cattle compared with that in humans, 250 kb extended regions adjacent to the core regions were set. Third, extended haplotype homozygosity (EHH) and REHH were calculated in the extended regions based on the estimated haplotypes and haplotype frequencies. Finally, P-values were obtained by log-transforming the EHH and REHH in the bin (20 bins to be all equally frequent haplotypes) to achieve normality. Calculation of LRH was performed using Sweep v.1.1 program. Only maternal haplotypes were loaded.
Results
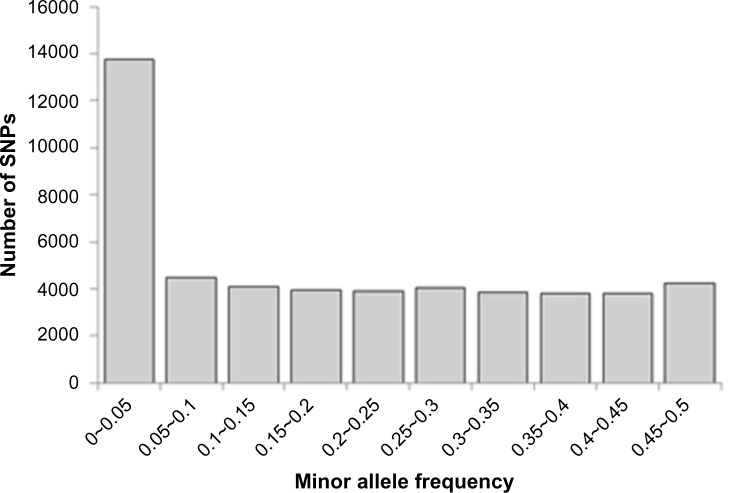
The filtering resulted in 35,968 useful SNPs, which were used for further analysis. More precisely, 3,366 SNPs (6%) did not produce any genotypes, Of the initial SNPs, 7,814 (14.5%) were monomorphic, and 5,953 (11%) were filtered out because of low minor allele frequency (MAF) (<0.05). The fraction of SNPs excluded because of partially (>25%) missing genotypes and deviation of Hardy–Weinberg equilibrium (HWE) was negligible (185 SNPs, <0.1%). This subset of markers covers 2,543.6 Mb of the genome with 70.57 ± 69.0 kb average adjacent marker spacing. The largest gap between SNPs (2081.5 kb) was located on chromosome 10. For the SNPs analyzed in this study, the average observed heterozygosity was estimated at 0.37 ± 0.12. The average MAF of all SNPs before quality control (QC) was 0.20, and using the filtered SNPs, the averaged MAF increased to 0.27, which was higher than the average MAF of SNPs reported on the Illumina Bovine SNP50K BeadChip for Hanwoon (0.198).29 The SNPs genotyped showed an almost uniform distribution across the common frequency classes. This is probably due to the design of the SNP array, which was optimized with respect to a uniform SNP spacing and MAF distribution (Fig. 1).18
Figure 1.
Minor allele frequency of SNPs.
In total, 1,760 core regions were identified, incorporating 5,915 SNPs which corresponded to 7.72% of the combined length of all the autosomes (Table 1). The mean core region length was estimated at 111.53 ± 71.89 kb, with a maximum of 2,081.46 kb. There were 141 core regions spanning 161.02 Mb on chromosome 1, and 20 core regions covering 51.90 Mb on chromosome 29. These were the largest and smallest haplotypic structures in the genome. An additional 1,318 core region consisting of three SNPs were identified (Fig. 2). The maximum number of SNPs in a core region was 14 from chromosome 14. There were 326 core regions with four SNPs, 82 core regions with five SNPs, 18 core regions with six SNPs and 8 core regions with seven SNPs.
Table 1.
Genome-wide summary of marker and core region(CR) in Hanwoo cattle.
| CHR | NO. SNP | MEAN MAF | LENGTH (Mb) | LINKAGE MAP1 (cM) | SIZE (kb) | MAX_SIZE (kb) | NO.CR (n) | CR MEAN SIZE ± SE(Kb) | % COVERAGE2 | CR SNPS(n)3 | MAX CR SNPS(N) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2376.00 | 0.27 | 161.02 | 142.10 | 67.75 | 1057.21 | 141 | 113.82 ± 66.66 | 9.97 | 490 | 10 |
| 2 | 1930.00 | 0.27 | 140.67 | 120.40 | 72.91 | 1143.31 | 113 | 116.27 ± 65.15 | 9.34 | 376 | 6 |
| 3 | 1832.00 | 0.27 | 127.85 | 125.20 | 69.40 | 807.55 | 119 | 107.99 ± 68.31 | 10.05 | 386 | 5 |
| 4 | 1736.00 | 0.28 | 124.13 | 101.50 | 71.37 | 853.22 | 92 | 106.23 ± 73.71 | 7.87 | 319 | 10 |
| 5 | 1472.00 | 0.27 | 125.80 | 122.10 | 85.52 | 1115.63 | 70 | 118.57 ± 74.37 | 6.60 | 245 | 13 |
| 6 | 1818.00 | 0.27 | 122.54 | 125.60 | 67.38 | 857.38 | 105 | 108.32 ± 67.59 | 9.28 | 357 | 7 |
| 7 | 1577.00 | 0.27 | 112.06 | 134.10 | 70.87 | 708.37 | 78 | 94.45 ± 60.97 | 6.57 | 255 | 5 |
| 8 | 1673.00 | 0.27 | 116.94 | 116.30 | 69.93 | 768.71 | 109 | 116.98 ± 212.30 | 11.05 | 382 | 9 |
| 9 | 1399.00 | 0.26 | 107.96 | 108.40 | 77.21 | 729.11 | 79 | 117.54 ± 64.4 | 8.60 | 268 | 7 |
| 10 | 1487.00 | 0.27 | 106.20 | 101.40 | 71.03 | 2081.46 | 72 | 92.83 ± 51.20 | 6.29 | 233 | 5 |
| 11 | 1567.00 | 0.28 | 110.17 | 123.50 | 70.27 | 890.68 | 75 | 119.02 ± 85.75 | 8.93 | 263 | 7 |
| 12 | 1145.00 | 0.27 | 85.28 | 105.80 | 74.47 | 948.71 | 55 | 110.82 ± 82.57 | 7.15 | 180 | 5 |
| 13 | 1228.00 | 0.27 | 84.34 | 87.10 | 68.52 | 574.52 | 56 | 104.32 ± 78.77 | 7.65 | 182 | 6 |
| 14 | 1219.00 | 0.27 | 81.30 | 85.70 | 66.71 | 608.29 | 44 | 116.07 ± 94.16 | 6.28 | 156 | 14 |
| 15 | 1160.00 | 0.27 | 84.60 | 93.40 | 72.96 | 761.86 | 49 | 121.21 ± 86.33 | 5.94 | 166 | 6 |
| 16 | 1101.00 | 0.27 | 77.74 | 96.50 | 70.62 | 1015.40 | 45 | 117.07 ± 63.90 | 6.78 | 155 | 7 |
| 17 | 1092.00 | 0.27 | 76.45 | 98.60 | 69.95 | 1388.99 | 43 | 110.10 ± 59.16 | 6.19 | 140 | 5 |
| 18 | 930.00 | 0.28 | 66.12 | 84.70 | 71.08 | 1557.49 | 37 | 116.09 ± 81.81 | 6.96 | 127 | 10 |
| 19 | 993.00 | 0.27 | 65.21 | 99.50 | 65.59 | 585.72 | 42 | 117.80 ± 68.26 | 7.59 | 142 | 6 |
| 20 | 1114.00 | 0.27 | 75.71 | 75.00 | 67.75 | 837.06 | 52 | 106.66 ± 53.92 | 7.33 | 173 | 5 |
| 21 | 997.00 | 0.28 | 69.17 | 87.60 | 69.42 | 849.43 | 45 | 116.32 ± 65.51 | 7.76 | 154 | 6 |
| 22 | 872.00 | 0.27 | 61.83 | 81.10 | 70.75 | 422.76 | 37 | 108.40 ± 63.43 | 6.49 | 120 | 5 |
| 23 | 792.00 | 0.28 | 53.29 | 67.10 | 67.37 | 562.47 | 27 | 127.91 ± 151.04 | 6.48 | 87 | 5 |
| 24 | 918.00 | 0.27 | 64.95 | 62.50 | 70.81 | 531.09 | 38 | 94.26 ± 63.65 | 5.52 | 120 | 4 |
| 25 | 699.00 | 0.29 | 43.86 | 64.91 | 61.98 | 351.37 | 28 | 101.73 ± 75.88 | 6.49 | 88 | 5 |
| 26 | 764.00 | 0.26 | 51.73 | 72.60 | 67.18 | 682.58 | 36 | 111.42 ± 57.57 | 7.75 | 117 | 5 |
| 27 | 694.00 | 0.28 | 48.73 | 64.10 | 70.28 | 1776.78 | 31 | 100.5 ± 56.62 | 6.39 | 100 | 5 |
| 28 | 652.00 | 0.27 | 46.02 | 52.40 | 70.62 | 419.44 | 22 | 103.60 ± 59.54 | 4.95 | 70 | 4 |
| 29 | 731.00 | 0.27 | 51.90 | 65.00 | 70.82 | 1004.44 | 20 | 112.95 ± 77.16 | 4.35 | 64 | 4 |
| Total | 35968.00 | 0.27 | 2543.57 | 2764.21 | 70.57 | 2081.46 | 1760 | 111.53 ± 71.89 | 7.72 | 5915 | 14 |
Notes:
Linkage map, NCBI Map Viewer (http://www.ncbi.nih.gov/mapview).
The proportion of total core region lengths on chromosome length.
Number of SNPs forming core regions.
Figure 2.
(A) Distribution of the length of core regions. (B) The number of SNPs forming the core regions in the Hanwoo genome.
Linkage disequilibrium
Decay of LD (r2) was estimated as a function of distance for each pairwise combination of SNPs on each chromosome, and a total of 24,985,377 SNP pairs were analyzed for all autosomes. To visualize the decay of LD and the proportion of pair markers, we stacked average values of r2 and estimated them as a function of inter-marker distance categories (Table 2). A mean value of r2 = 0.223 ± 0.274 was observed at distances shorter than 40 kb, which decreased to 0.111 ± 0.174 in the third smallest distance bin of ~60–100 kb. In the intervals of 0–40 and 40–60 kb, only 26% and 17% of markers had r2 ≥ 0.3, respectively.
Table 2.
Pairwise linkage disequilibrium (r2) for syntetic SNPs at various distance pooled over all paternal autosomes on Hanwoo.
| DISTANCE | N | MEDIAN | MEAN | SD | %(r2 > 0.2)1 | %(r2 > 0.3)2 |
|---|---|---|---|---|---|---|
| 0–40 kb | 14,603 | 0.102 | 0.2226 | 0.2742 | 35 | 26 |
| 40–60 kb | 11,148 | 0.064 | 0.1583 | 0.2206 | 25 | 17 |
| 60–100 kb | 22,457 | 0.043 | 0.1119 | 0.1735 | 17 | 10 |
| 100–250 kb | 81,413 | 0.024 | 0.0630 | 0.1120 | 7 | 4 |
| 250–500 kb | 132,593 | 0.015 | 0.0355 | 0.0605 | 2 | 1 |
| 0.5–1 Mb | 260,828 | 0.012 | 0.0269 | 0.0418 | 1 | 0 |
| 1–2 Mb | 511,180 | 0.011 | 0.0241 | 0.0359 | 1 | 0 |
| 2–5 Mb | 147,6343 | 0.01 | 0.0215 | 0.0320 | 0 | 0 |
| 5–10 Mb | 2,309,702 | 0.009 | 0.0182 | 0.0270 | 0 | 0 |
| 10–20 Mb | 4,143,933 | 0.007 | 0.0142 | 0.0212 | 0 | 0 |
| 20–50 Mb | 9,006,713 | 0.004 | 0.0097 | 0.0143 | 0 | 0 |
| 50–100 Mb | 6,052,736 | 0.003 | 0.0072 | 0.0103 | 0 | 0 |
| >100 Mb | 961,728 | 0.003 | 0.0068 | 0.0097 | 0 | 0 |
Notes:
Percentage of pairs of SNPs with r2 > 0.2.
Percentage of pairs of SNPs with r2 > 0.3.
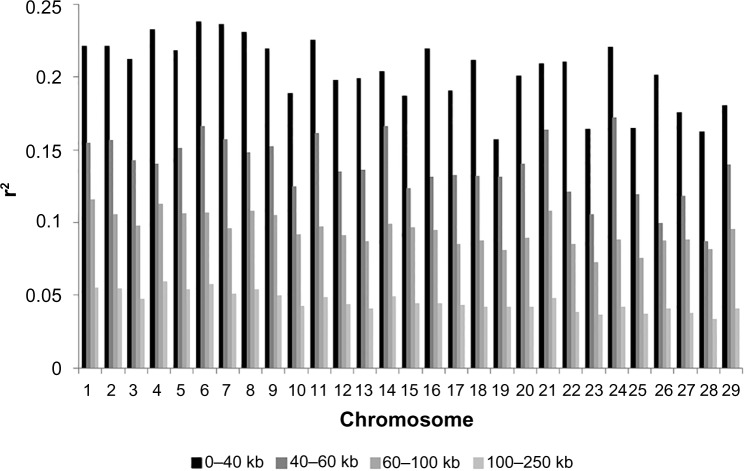
Variation of LD in different distance bins of individual chromosomes combined over the genome was examined (Fig. 3). For marker pairs 0–40 kb apart, the average r2 ranged from <0.18 (BTA19, BTA27, BTA28) to >0.24 (BTA6). Table 3 shows the comparison between the LD of intragenic regions and LD in intergenic regions. There was no difference in the extent of LD or the decline of LD with the distance between intragenic and intergenic regions.
Figure 3.
Chromosome-wise average r2-value and the decay of linkage disequilibrium with increasing physical distance for five smallest bins evaluated.
Table 3.
Linkage disequilibrium (r2) of intrgenic and intragenic pairs of SNP.
| DISTANCE (kb) | INTERGENIC
|
INTRAGENIC
|
P-VALUE1 | ||||
|---|---|---|---|---|---|---|---|
| N | MEAN | SD | N | MEAN | SD | ||
| <50 | 11,670 | 0.05 | 0.16 | 4,892 | 0.05 | 0.16 | >0.05 |
| 50–100 | 15,395 | 0.03 | 0.11 | 5,668 | 0.03 | 0.12 | >0.05 |
| 100–150 | 14,420 | 0.02 | 0.08 | 4,887 | 0.02 | 0.09 | >0.05 |
| 150–200 | 13,986 | 0.02 | 0.06 | 4,407 | 0.02 | 0.08 | >0.05 |
| 200–250 | 13,579 | 0.01 | 0.05 | 3,966 | 0.02 | 0.07 | >0.05 |
Note:
Test of equal variance.
Effective population size
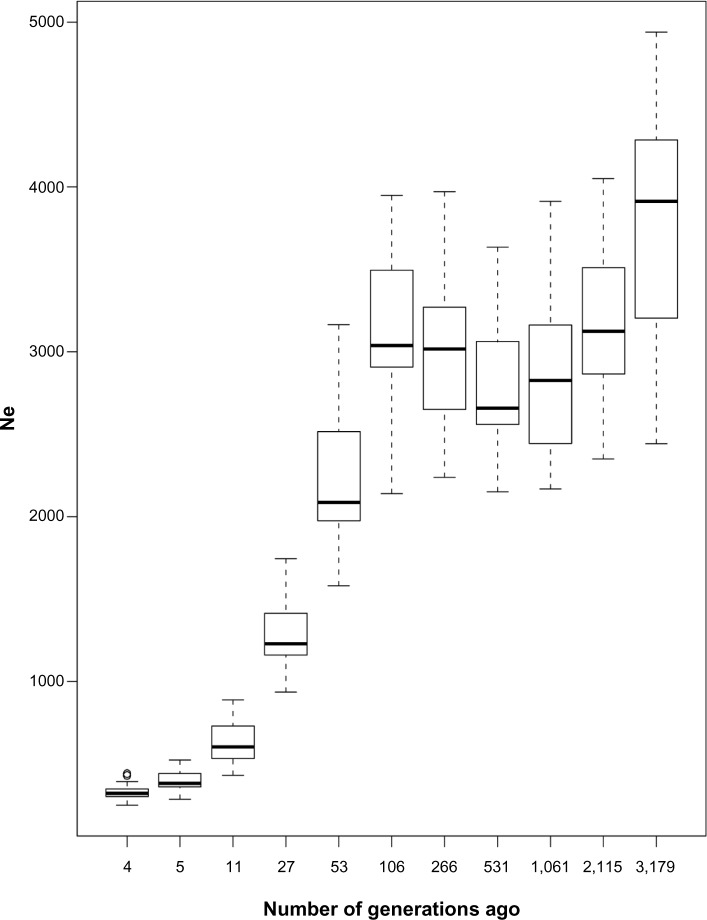
Past effective population size was estimated chromosome-wise using pairwise SNP r2 with inter-marker distance categories (Table 4). The ratio of genetic map distance to physical map distance was between 0.65 and 1.22 across chromosomes. Within a chromosome, the linkage map distance was assumed to be proportional to the physical genomic distance. The coefficient of variation (CV) increased with the length of the SNP interval, indicating that recent population size would be estimated less accurately than population size of many generations ago. Figure 4 shows a decreasing trend over the last 3000 generations, with an increasingly steeper slope since approximately 100 generations ago but a period of increase from 550 to 100 generations ago.
Table 4.
Effective population size(Ne) in the past Hanwoon population.
| FRAGMENT SIZE (kb) | 15 | 25 | 50 | 100 | 200 | 500 | 1,000 | 2,000 | 5,000 | 10,000 | 15,000 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Effective population size1 | |||||||||||
| Mean | 3,802 | 3,210 | 2,886 | 2,771 | 3,008 | 3,130 | 2,206 | 1,284 | 630 | 402 | 327 |
| Median | 700 | 435 | 476 | 363 | 452 | 451 | 391 | 196 | 116 | 59 | 46 |
| SD | 3,913 | 3,125 | 2,827 | 2,659 | 3,017 | 3,038 | 2,087 | 1,229 | 604 | 382 | 322 |
| Min | 2,442 | 2,351 | 2,168 | 2,152 | 2,239 | 2,139 | 1,580 | 936 | 432 | 287 | 250 |
| Max | 4,939 | 4,051 | 3,914 | 3636 | 3,972 | 3,949 | 3,165 | 1,745 | 889 | 524 | 442 |
| CV(%) | 5 | 3 | 4 | 2 | 3 | 8 | 11 | 13 | 16 | 22 | 27 |
| Linkage distance (cM)2 | 0.016 | 0.024 | 0.047 | 0.094 | 0.188 | 0.470 | 0.940 | 1.882 | 4.706 | 9.456 | 14.209 |
| Generation3 | 3,179 | 2,115 | 1,061 | 531 | 266 | 106 | 53 | 27 | 11 | 4 | 4 |
Notes:
Mean, median, standard deviation and 95% CI of past effective population size were calculated from Ne in 29 bovine autosomal chromosomes.
Linkage map distance (c) was inferred approximately by comparison between linkage map and physical genomic information.
The generation in the past population was calculated as 1/2c.
Figure 4.
Estimated effective population size over the past generations from linkage disequilibrium data. The variability at each point time reflects the variation of estimates between the 29 autosomes.
Whole-genome screening for selection signatures
For all 1,760 core regions, a total of 12,186 LRH tests with an average of 6.92 tests per core region were made. Taking this into consideration, we skipped core haplotypes with frequency <25% and plotted the −log10 of the P-values associated with REHH against the chromosomal position to visualize the chromosomal distribution of outlying core haplotypes (Fig. 5). Table 5 presents the genome-wide statistics of the selection signature test, including the number of tests and outlying core haplotypes for each chromosome. Of the 9,028 tests on core haplotypes with frequency >0.1, 95 tests displayed outlying peaks on a threshold level of 0.01.
Figure 5.
Genome-wide map of −log10(REHH P-values) for core haplotypes with frequency >0.1. Dashed lines display the threshold level of 0.01.
Table 5.
Summary statistics of whole genome extent haplotype homozygosity test.
| Chr. | TEST ON CH (n)1 | P-VALUE <0.05 | P-VALUE <0.01 |
|---|---|---|---|
| 1 | 704 | 36 | 8 |
| 2 | 549 | 27 | 5 |
| 3 | 616 | 32 | 9 |
| 4 | 479 | 22 | 5 |
| 5 | 370 | 19 | 4 |
| 6 | 509 | 28 | 4 |
| 7 | 381 | 20 | 3 |
| 8 | 585 | 33 | 10 |
| 9 | 347 | 20 | 5 |
| 10 | 403 | 16 | 6 |
| 11 | 389 | 17 | 4 |
| 12 | 279 | 12 | 1 |
| 13 | 299 | 14 | 7 |
| 14 | 246 | 9 | 2 |
| 15 | 244 | 12 | 2 |
| 16 | 250 | 11 | 2 |
| 17 | 219 | 6 | 1 |
| 18 | 199 | 10 | 1 |
| 19 | 224 | 14 | 1 |
| 20 | 263 | 10 | 3 |
| 21 | 201 | 8 | 1 |
| 22 | 201 | 8 | 2 |
| 23 | 156 | 7 | 2 |
| 24 | 215 | 10 | 3 |
| 25 | 166 | 8 | 3 |
| 26 | 176 | 5 | 1 |
| 27 | 143 | 6 | 0 |
| 28 | 104 | 5 | 0 |
| 29 | 111 | 5 | 0 |
| Total | 9028 | 430 | 95 |
Note:
The number of tests on core haplotypes (both sides) with frequency ≥0.1.
We also explored a quantitative trait loci (QTL) database available online (http://www.animalgenome.org/QTLdb/cattle.html)30 to identify any overlapping of the outlying core regions with published QTL in dairy and beef cattle. Additional File 1 lists the traits, approximate position, and reported population of the overlapping QTL for each core region.
Discussion
The pattern of LD in domestic animals means that a marker may be in LD with a QTL some distance away and show an association with the trait affected by the QTL; it also tells us how natural selection, genetic drift, recombination, and mutation all affect the levels of LD.2,31 In this study, we present an analysis of LD of 35,968 SNP genotypes in Hanwoo, in which only maternally inherited haplotypes were used. We elected to use maternal haplotypes in consideration of the complexity of the pedigrees, with dams contributing information to multiple families. Bohmanova et al reported that the use of maternal haplotypes is recommended for analyses of LD in populations consisting of large paternal half-sib families.32
Comparable estimates of extensive LD have been reported in cattle.18,32,33 Consistent with previous analysis in cattle, the decline of LD as a function of distance was rapid, with average r2 declining from 0.1119 to 0.0355 when moving from 60 to 500 kb (Table 2). Recent studies that used SNPs for evaluating the extent of LD revealed that strong LD extends for shorter distances than previously reported. Marques et al used 505 SNPs on BTA14 in Holsteins and found moderate levels of LD (r2 ≥ 0.2) extending up to 100 kb.34 Khatkar et al demonstrated that r2 ≥ 0.2 in Australian Holsteins between SNPs less than 60 kb apart.35 Sargolzaei et al reported useful LD (r2 ≥ 0.2) between markers with intermarker distance ≤100 kb in North American Holsteins.36 In other cattle breeds (Japanese Black, Angus, Brahman, and Holstein), the average LD declined to 0.2 within 100 kb genomic regions.37 Our study shows that moderate levels of r2 (between 0.2 and 0.24) were observed at distances shorter than 40 kb. It is important to note that the level of LD between adjacent markers in Hanwoo was weaker than in other cattle breeds. This indicates that Hanwoo may have a higher actual Ne, probably because of a relatively low selection intensity compared to other breeds.
There were no differences in the extent of LD and the decline of LD with the distance between intragenic and intergenic regions (Table 3). Indeed, knowledge of LD with genes is important. However, noncoding elements such as miRNAs might also play a role in many inherited traits. Therefore, these results suggest that noncoding regions have been an important substrate for adaptive evolution in Hanwoo.
The number of SNPs needed depends on the distance over which LD operates. If SNPs are too far apart, a QTL may not be in sufficient LD with any of the markers, and so will be undetected.2 Meuwissen et al reported the required level of LD (r2) for genomic selection to achieve an accuracy of 0.85 for genomic breeding values to be 0.2 using simulation.38 To achieve this level, our results indicate that the SNP spacing should be ~40 kb in the future population, which corresponds to more than 75,000 evenly distributed SNPs across the genome. LD showed extensive variability between genomic regions and chromosomes. This variation was probably attributed to recombination rates varying between and within chromosomes, heterozygosity, genetic drift, and effects of selection. When the decay of r2 with a distance was plotted separately for each chromosome, the average r2 values are lower than 0.2 on BTA10, 15, 17, 19, 23, 25, 27, 28, and 29 in the 0–40 kb interval when compared to all other autosomes (Fig. 3). This difference could be due to a selection process acting on these chromosomal regions. This suggests that the loci on BTA 10, 15, 17, 19, 23, 25, 27, 28, and 29 need more SNPs to achieve sufficient power.
The pattern of LD observed in a population depends on the history of the population, especially the history of its effective population size.2 This study shows a persistent decline in the effective population size. In particular, there are sharp declines at two time intervals and one distinctive time point (Fig. 4; Table 4). The distinctive point at 100 generations ago shows a sharp decline in population size, which is consistent with previous studies.39,40 This pattern can be explained by breed formation and modern breeding programs.15 The first sharp decline of the effective population size was observed in our study ~25–50 generations ago, and is due to enhancement of selection. A second and more recent sharp decline seems to have occurred ~10–25 generations ago and might thus correspond to several events related to the intensive use of artificial insemination (AI). Overall, the Korean national economy has grown very rapidly since 1970, which has brought about a change in the Korean lifestyle, including significant consumption of meat from cattle. In this regard, improvement of selection methods, together with the adaptation to different agro-ecological constraints, has been necessary and might have had a direct effect on the population structure of cattle.
The estimated effective population size for the most recent time is around 300 individuals. Compared to other reports,18,33,39–41 recent Ne for Hanwoo were much higher than in other livestock populations. This suggests a small genetic drift associated with a small decrease in the population size over generations. Hanwoo values are greater than those recommended by Food and Agriculture Organization (FAO).42 Frankham et al suggest a minimum Ne of 50–100 for sustaining reproductive fitness in the short term (~100 years). However, this is below the value recommended to maintain evolutionary potential for long-term species management (Ne > 500–5000),43 as additive genetic variation may eventually be lost by drift. In addition, a very large Ne indicated genomic selection, which would be difficult to apply to Hanwoo. To overcome these problems, it may be necessary to reduce Ne in a breeding program, for instance, by using only the best families or existing varieties to breed the new strain, where estimation of SNP effects is based on the genotyping of DNA pools and SNP density is reduced by estimating SNP effects within one or a few families.44,45
Positive selection is expected to accelerate the frequency of an advantageous allele faster than recombination can break down LD at the selected haplotype.27 In this report, we employed the LRH test by selecting a “core” haplotype with elevated EHH relative to other core haplotypes, at the locus conditional on haplotype frequency. This was used to identify selection signatures within a single population. Our result revealed 95 regions exhibiting the footprint of recent positive selection at a threshold level of 0.01.
We found an overlap of the 11 core regions presenting top P-values and those that had previously been identified as harboring other breeds’ QTL (Supplementary Table 1). In such a case, a possible higher initial frequency of beneficial alleles might be imported in a breed through crosses with other breeds. Selection may have started from a moderate initial frequency, and beneficial alleles may be included in diverse haplotypes. It is important to note the distinct demographic history of Hanwoo: the Ministry of Agriculture, Forestry and Fishery (MAFF) controls the breeding system for improving the productivity of Korean native cattle. From 1955 to 1965, Korean cattle were crossed with beef breeds such as Angus, Hereford, and Brown Swiss to improve the body weight. Since the early 1960s, there has been a program for improvement through pure breeding and crossbreeding. From 1971 to 1975, crosses of Korean cattle and Angus or Charolais were crossed with Holstein, which resulted in an improvement of both body weight and dressing percentage. Since 1978, Korean cattle have been crossed with Charolais bulls to make a new composite breed that is five-eighth Charolais and three-eighth Korean cattle.
Eleven QTL regions, associated not only with production traits but also with meat and carcass traits, have been identified. Within the last century, Hanwoo have been intensively selected for both the quality (marbling, tenderness, and flavor) and the quantity (carcass weight) of meat to meet the growing demand for beef in Korea. Cattle farming methods have changed dramatically, so it is not surprising that QTLs identified are related to meat traits, showing the signatures of recent selection. Especially for BTA14, which harbors known genes and QTL for several economically important traits, we found agreement between the regions (24.3–25.4 Mb) and those that had previously been identified to harbor carcass weight QTL peak on Hanwoo.46 We extended core regions in both directions up to 1 Mb as the length of the core domains. We identified PLAG1, CHCHD7, SDR16C5, SDR16C6, PENK, FAM110B, CYP7A1, SDCBP, and TOX as the positional and functional candidate genes for the carcass weight QTL in cattle. The associated region on bovine chromosome 14 is conserved in human chromosome 8q21, which has been reported to be associated with adult height.47 The positive correlations between REHH values and effects of the SNPs on meat traits suggest co-location of some selected regions of the chromosome and QTL affecting production traits. It is also important to note that the distinct demographic history of Hanwoo, including severe bottlenecks, inbreeding, and nonrandom mating, can produce considerable heterogeneity in genome-wide patterns, confounding inferences of selection. However, the message conveyed by this study has important implications for the design and application of association studies in Hanwoo populations. These results give us the confidence to conduct further in-depth studies.
Conclusion
In this work, the Illumina Bovine SNP50K chips were used for SNP genotyping in an elite Hanwoo population comprising 61 sires and their 486 steers. Haplotype phases were inferred by an LHCM method, and maternal haplotypes were chosen across all autosomal chromosomes. Ne was estimated from the LDs between syntenic SNPs, and the results showed that Ne values started from 4,000 to ~3,200 generations ago, decreasing gradually. However, there were two sharp Ne decline periods, between 25–50 and 10–25 generations ago. In the latter, the Hanwoo breeding program was implemented with the technique of artificial insemination. The Ne of the current Hanwoo population was estimated to be around 300.
To identify selection signatures on any genomic position, an LRH test was applied. Our results revealed 95 regions exhibiting footprints of recent positive selection (P < 0.01). We found an overlap of the 11 core regions presenting top P-values and those that had previously been identified as harboring other breeds’ QTL. Additional studies, especially comparisons of different populations, are needed to confirm and refine our results.
Supplementary Material
Supplementary Table 1. Summary statistics for 11 core haplotypes showing the lowest P-values after the relative extended haplotype homozygosity (REHH) test.
Acknowledgments
Thanks for the academic editor and reviewers for their comments, which were very helpful in improving the manuscript.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
FUNDING: This work was supported by the Shanxi Scholarship Council of China 2013-072 (YL) and by the Natural Science Foundation of Shanxi 2014011030-4 (YL). JJK is supported by the Technology Development Program for Agricultural and Forestry, Ministry of Agriculture, Forestry and Fisheries, Republic of Korea, 2014. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: YL, JJK. Analyzed the data: YL, JJK. Wrote the first draft of the manuscript: YL. Contributed to the writing of the manuscript: YL, JJK. Agree with manuscript results and conclusions: YL, JJK. Jointly developed the structure and arguments for the paper: YL, JJK. Made critical revisions and approved final version: YL, JJK. Both authors reviewed and approved of the final manuscript
REFERENCES
- 1.Habier D. More than a third of the WCGALP presentations on genomic selection. J Anim Breed Genet. 2010;127(5):336–7. doi: 10.1111/j.1439-0388.2010.00897.x. [DOI] [PubMed] [Google Scholar]
- 2.Goddard ME, Hayes BJ. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat Rev Genet. 2009;10(6):381–91. doi: 10.1038/nrg2575. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Lin CH, Sabatti C. Volume measures for linkage disequilibrium. BMC Genet. 2006;7:54. doi: 10.1186/1471-2156-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charlesworth B. Fundamental concepts in genetics: effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10(3):195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 5.Park L. Effective population size of current human population. Genet Res (Camb) 2011;93(2):105–14. doi: 10.1017/S0016672310000558. [DOI] [PubMed] [Google Scholar]
- 6.Hayes BJ, Chamberlain AJ, Maceachern S, et al. A genome map of divergent artificial selection between Bos taurus dairy cattle and Bos taurus beef cattle. Anim Genet. 2009;40(2):176–84. doi: 10.1111/j.1365-2052.2008.01815.x. [DOI] [PubMed] [Google Scholar]
- 7.MacEachern S, Hayes BJ, McEwan J, Goddard ME. An examination of positive selection and changing effective population size in Angus and Holstein cattle populations (Bos taurus) using a high density SNP genotyping platform and the contribution of ancient polymorphism to genomic diversity in domestic cattle. BMC Genomics. 2009;10:181. doi: 10.1186/1471-2164-10-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sabeti PC, Reich DE, Higgins JM, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419(6909):832–7. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 9.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–95. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155(3):1405–13. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4(3):e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bovine HapMap Consortium. Gibbs RA, Taylor JF, et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324(5926):528–32. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee MS, Ryu YC, Kim BC. Comparative studies on metabolic rate and calpain/calpastatin activity between Hanwoo and Holstein beef. Asian Aust J Anim Sci. 2002;15(12):1747–53. [Google Scholar]
- 14.Lee KT, Chung WH, Lee SY, et al. Whole-genome resequencing of Hanwoo (Korean cattle) and insight into regions of homozygosity. BMC Genomics. 2013;14(1):519. doi: 10.1186/1471-2164-14-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SH, Park BH, Sharma A, et al. Hanwoo cattle: origin, domestication, breeding strategies and genomic selection. J Anim Sci Technol. 2014;56:2. doi: 10.1186/2055-0391-56-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sargolzaei M, Iwaisaki H, Colleau JJ. CFC: a tool for monitoring genetic diversity; Proc. 8th World Congr. Genet. Appl. Livest. Prod., CD-ROM Communication 27–28; Belo Horizonte, Brazil. August 13–18; 2006. [Google Scholar]
- 17.Matukumalli LK, Lawley CT, Schnabel RD, et al. Development and characterization of a high density SNP genotyping assay for cattle. PLoS One. 2009;4(4):e5350. doi: 10.1371/journal.pone.0005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qanbari S, Pimentel EC, Tetens J, et al. The pattern of linkage disequilibrium in German Holstein cattle. Anim Genet. 2010;41(4):346–56. doi: 10.1111/j.1365-2052.2009.02011.x. [DOI] [PubMed] [Google Scholar]
- 19.Druet T, Georges M. A hidden Markov model combining linkage and linkage disequilibrium information for haplotype reconstruction and quantitative trait locus fine mapping. Genetics. 2010;184(3):789–98. doi: 10.1534/genetics.109.108431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smedley D, Haider S, Ballester B, et al. BioMart – biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis GR, Cookson WO. GOLD – graphical overview of linkage disequilibrium. Bioinformatics. 2000;16(2):182–3. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- 22.Hedrick PW. Gametic disequilibrium measures: proceed with caution. Genetics. 1987;117(2):331–41. doi: 10.1093/genetics/117.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29(2):311–22. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- 24.Tenesa A, Navarro P, Hayes BJ, et al. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 2007;17(4):520–6. doi: 10.1101/gr.6023607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2014;42(Database issue):D7–17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 27.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006;22(8):437–46. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C, Bailey DK, Awad T, et al. A whole genome long-range haplotype (WGLRH) test for detecting imprints of positive selection in human populations. Bioinformatics. 2006;22(17):2122–8. doi: 10.1093/bioinformatics/btl365. [DOI] [PubMed] [Google Scholar]
- 29.Decker JE, Pires JC, Conant GC, et al. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci U S A. 2009;106(44):18644–9. doi: 10.1073/pnas.0904691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu ZL, Fritz ER, Reecy JM. AnimalQTLdb: a livestock QTL database tool set for positional QTL information mining and beyond. Nucleic Acids Res. 2007;35:D604–9. doi: 10.1093/nar/gkl946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slatkin M. Linkage disequilibrium – understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9(6):477–85. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bohmanova J, Sargolzaei M, Schenkel FS. Characteristics of linkage disequilibrium in North American Holsteins. BMC Genomics. 2010;11:421. doi: 10.1186/1471-2164-11-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flury C, Tapio M, Sonstegard T, et al. Effective population size of an indigenous Swiss cattle breed estimated from linkage disequilibrium. J Anim Breed Genet. 2010;127(5):339–47. doi: 10.1111/j.1439-0388.2010.00862.x. [DOI] [PubMed] [Google Scholar]
- 34.Marques E, Schnabel RD, Stothard P, et al. High density linkage disequilibrium maps of chromosome 14 in Holstein and Angus cattle. BMC Genet. 2008;9:45. doi: 10.1186/1471-2156-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khatkar MS, Nicholas FW, Collins AR, et al. Extent of genome-wide linkage disequilibrium in Australian Holstein-Friesian cattle based on a high-density SNP panel. BMC Genomics. 2008;9:187. doi: 10.1186/1471-2164-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargolzaei M, Schenkel FS, Jansen GB, Schaeffer LR. Extent of linkage disequlibrium in Holstein cattle in North America. J Dairy Sci. 2008;91:2106–17. doi: 10.3168/jds.2007-0553. [DOI] [PubMed] [Google Scholar]
- 37.McKay SD, Schnabel RD, Murdoch BM, et al. Whole genome linkage disequilibrium maps in cattle. BMC Genet. 2007;8:74. doi: 10.1186/1471-2156-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–29. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Roos AP, Hayes BJ, Spelman RJ, Goddard ME. Linkage disequilibrium and persistence of phase in Holstein-Friesian, Jersey and Angus cattle. Genetics. 2008;179(3):1503–12. doi: 10.1534/genetics.107.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Villa-Angulo R, Matukumalli LK, Gill CA, Choi J, Van Tassell CP, Grefenstette JJ. High-resolution haplotype block structure in the cattle genome. BMC Genet. 2009;10:19. doi: 10.1186/1471-2156-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim ES, Kirkpatrick BW. Linkage disequilibrium in the North American Holstein population. Anim Genet. 2009;40(3):279–88. doi: 10.1111/j.1365-2052.2008.01831.x. [DOI] [PubMed] [Google Scholar]
- 42.Woolliams JA, Gwaze GP, Meuwissen THE, et al., editors. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans Management of Small Populations at Risk. Food and Agriculture Organization of the United Nations; Washington, DC: 1998. [Google Scholar]
- 43.Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Biology. New York: Cambridge University Press; 2002. [Google Scholar]
- 44.Goddard ME, Hayes BJ, Meuwissen TH. Genomic selection in livestock populations. Genet Res (Camb) 2010;92(5–6):413–21. doi: 10.1017/S0016672310000613. [DOI] [PubMed] [Google Scholar]
- 45.Sonesson AK, Meuwissen TH, Goddard ME. The use of communal rearing of families and DNA pooling in aquaculture genomic selection schemes. Genet Sel Evol. 2010;42:41. doi: 10.1186/1297-9686-42-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, Kim JJ. Multiple linkage disequilibrium mapping methods to validate additive QTL in Korean native cattle (Hanwoo) Asian Aust J Anim Sci. 2015;28(7):926–35. doi: 10.5713/ajas.15.0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gudbjartsson DF, Walters GB, Thorleifsson G, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40(5):609–15. doi: 10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Summary statistics for 11 core haplotypes showing the lowest P-values after the relative extended haplotype homozygosity (REHH) test.