This review describes the two main applications of fusion proteins in protein crystallization: the ‘heterologous fusion-protein approach’ and the ‘fusion of interacting proteins approach’.
Keywords: linker, heterologous fusion-protein approach, fusion of interacting proteins approach, membrane-protein crystallization, protein interactions, recombinant fusion protein
Abstract
Fusion proteins can be used directly in protein crystallization to assist crystallization in at least two different ways. In one approach, the ‘heterologous fusion-protein approach’, the fusion partner can provide additional surface area to promote crystal contact formation. In another approach, the ‘fusion of interacting proteins approach’, protein assemblies can be stabilized by covalently linking the interacting partners. The linker connecting the proteins plays different roles in the two applications: in the first approach a rigid linker is required to reduce conformational heterogeneity; in the second, conversely, a flexible linker is required that allows the native interaction between the fused proteins. The two approaches can also be combined. The recent applications of fusion-protein technology in protein crystallization from the work of our own and other laboratories are briefly reviewed.
1. Introduction and overview
Recombinant fusion proteins (also termed chimeric or hybrid proteins) are used widely in a variety of protein-engineering applications ranging from tags to facilitate protein purification and detection to therapeutics and nanotechnology (Yu et al., 2015 ▸; Bell et al., 2013 ▸). Most proteins used for protein crystallization are obtained recombinantly as fusion proteins with tags for affinity chromatography (Derewenda, 2004 ▸). The use of tagged proteins has been further popularized by structural genomics initiatives (Gräslund et al., 2008 ▸).
1.1. The use of fusion proteins for crystallization
Typically, fusion tags are removed from the target protein before crystallization (Derewenda, 2004 ▸; Waugh, 2005 ▸). In the case of short affinity tags (such as the Strep-tag or the polyhistidine tag in the absence of metal ion), tag removal may be favoured owing to the fact that the tags often do not have a defined three-dimensional structure and could represent an entropic impediment to crystallization and shield the protein surface from forming crystal contacts. Limited data exist to establish the general validity of these arguments, however, aside from anecdotal examples showing the requirement of tag removal for crystallization or improved diffraction quality (see, for example, Kim et al., 2001 ▸; Huh et al., 2014 ▸; Sugawara et al., 2005 ▸). Bucher et al. (2002 ▸) examined the effect of different tags on the crystallization of Pyrococcus furiosus maltodextrin-binding protein, demonstrating that the tags can have significant effects on crystallization and diffraction quality. In the proteins crystallized with a short fusion tag present, the fusion tag is rarely observed in the electron-density maps (Carson et al., 2007 ▸). Larger affinity tags [such as glutathione-S-transferase (GST) or maltose-binding protein (MBP)] have a defined three-dimensional structure. However, the protein and its fusion partner will usually not associate in any defined way, causing conformational flexibility and hindering crystallization. These effects are exacerbated by the linker sequences typically containing protease-cleavage sites for tag removal; they are therefore optimized for easy access of the protease through increased length and flexibility. Even though the fusion partner (in particular in the case of large fusion tags) may improve the solubility properties of the protein, which may be advantageous for crystallization, the conformational heterogeneity introduced by the fusion partner will generally have an overwhelmingly negative effect.
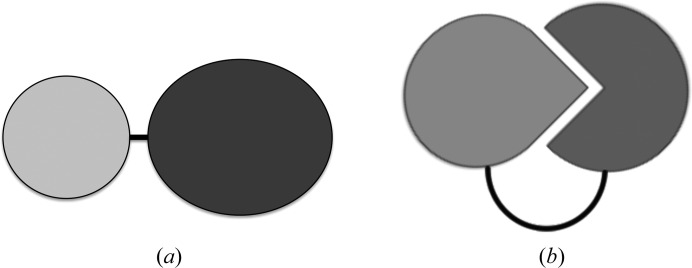
Nevertheless, fusion proteins can be useful in protein crystallization in specific cases. Fundamentally, there are at least two ways in which one can take advantage of intact fusion proteins in the protein crystallization process (Fig. 1 ▸). In one approach, a fusion partner can provide an additional surface area that can contribute to crystal contact formation (Fig. 1 ▸ a); here, we term this approach the ‘heterologous fusion-protein approach’, as the fusion partner will usually be a heterologous fusion tag such as MPB or T4 lysozyme (T4L). This approach can be especially powerful in the case of integral membrane proteins, where polar surface areas that are favourable for crystal contact formation may be scarce. In a fundamentally different application, one can covalently link interacting proteins to promote their interaction by increasing their local concentration and controlling the stoichiometry (Fig. 1 ▸ b). Here, we term this approach the ‘fusion of interacting proteins approach’. As we illustrate below, these applications can also be combined. Whereas these strategies are in general applicable to any protein target, targeted variations have also been developed for specific proteins or protein families. We review the fusion-protein strategies in protein crystallization and illustrate them using specific representative examples. We refer to other recent reviews for more comprehensive coverage of particular aspects of the topic of this article.
Figure 1.
Schematic diagram illustrating (a) the heterologous fusion-protein approach [the protein of interest (dark) is fused to a heterologous fusion partner (light) using a short linker (black line)] and (b) the fusion of interacting proteins approach [the interacting partners are fused covalently using a linker sequence (black line)].
1.2. The role of fusion-protein linkers
The linker plays fundamentally different roles in the heterologous fusion-protein and fusion of interacting proteins approaches. In the heterologous fusion-protein application a rigid connection between the fusion partners is desired to eliminate the conformational heterogeneity caused by the fusion, whereas when fusing interacting proteins the linker has to be of sufficient length and flexible enough not to interfere with the native interaction between the partners (Fig. 1 ▸). To help with the choice of linkers, researchers have analyzed natural linkers that occur between domains in multi-domain proteins. An early analysis suggested that natural interdomain linkers are rich in Thr, Ser, Gly and Ala residues (Argos, 1990 ▸). However, a more recent analysis of a larger set of structures found a very different composition, with Pro, Arg, Phe, Thr, Glu and Gln the preferred amino-acid residues (George & Heringa, 2002 ▸). Just like artificial linkers in recombinant fusion proteins, the natural linkers may obviously play two fundamentally different roles; in some cases they may serve as rigid spacers to prevent unfavourable interactions between domains, whereas in others they may have to be flexible in order to not interfere with interdomain interactions and domain movement. It is likely that the data set in the former study was enriched in flexible linkers, whereas that in the latter study contained more proteins containing rigid linkers, biasing the outcomes in each case. Ideally, such analyses should divide the data depending on the function of the linker, although this may create new challenges. Nevertheless, we can learn from natural linkers when optimizing the composition and length of linkers in recombinant fusion constructs.
2. Heterologous fusion-protein approach
The possible benefits of using a fusion protein with a heterologous fusion partner in crystallization include (i) additional surface areas that can help in crystal contact formation (especially if the fusion partners crystallize readily themselves) and (ii) knowledge of the three-dimensional structure of the fusion partner for structure determination by molecular replacement. The approach is therefore analogous to the ‘crystallization chaperone’ approach, in which an interacting partner such as an antibody fragment or a designer non-antibody binding protein such as a DARPin (designed ankyrin-repeat protein) is used to aid in crystal lattice formation (Koide, 2009 ▸; Derewenda, 2010 ▸). Crystallization chaperones have been particularly valuable in cases of integral membrane proteins that lack substantial polar surface areas amenable to crystal contact formation (Hunte & Michel, 2002 ▸). However, unlike the crystallization chaperone approach, the heterologous fusion-protein approach suffers from conformational heterogeneity introduced by the unrestrained relative orientations of the fusion partners. Using fusion to a rigid RNA scaffold to present and help to fold other RNA sequences is a tool that has also been successfully employed in RNA crystallization (Zhang & Ferré-D’Amaré, 2014 ▸).
2.1. Early examples of structures of proteins fused to large fusion partners
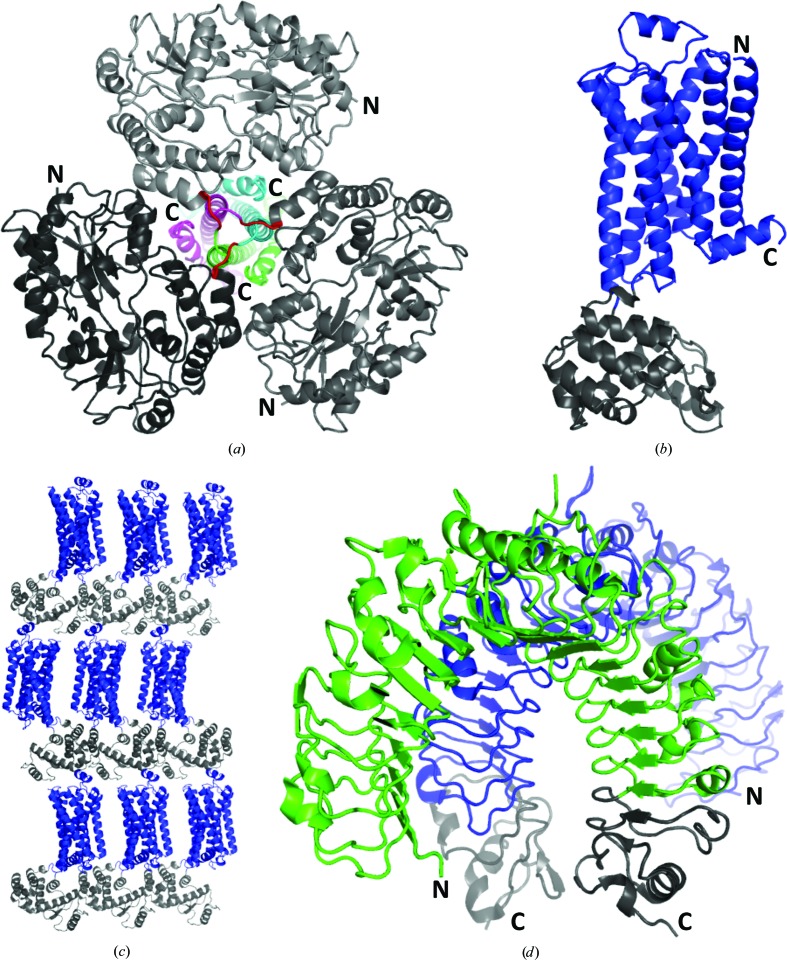
Early applications of the heterologous fusion-protein approach for crystallization explored large fusion partners to facilitate structural studies of small peptides, taking advantage of the crystal lattice created by the fusion partner (Zhan et al., 2001 ▸; Donahue et al., 1994 ▸; Carter et al., 1994 ▸). The first structures of larger proteins containing large fusion partners used MBP as a fusion partner (Table 1 ▸), and most used short linkers between fusion partners to reduce conformational flexibility (Center et al., 1998 ▸; Smyth et al., 2003 ▸); they corresponded to a fragment of the human T-cell leukaemia virus type 1 (HTLV-1) envelope protein gp21 (Kobe et al., 1999 ▸; Fig. 2 ▸ a), the Staphylococcus aureus DNA-binding protein SarR (Liu et al., 2001 ▸) and the Saccharomyces cerevisiae proteins MATa1 (Ke & Wolberger, 2003 ▸) and the ribosomal protein L30 (Chao et al., 2003 ▸) (reviewed by Smyth et al., 2003 ▸).
Table 1. Selected heterologous fusion partners.
| Fusion partner | Size (amino acids) | Biological origin | Selected references |
|---|---|---|---|
| Maltose-binding protein (MBP) | 363 | Escherichia coli | Kobe et al. (1999 ▸), Ullah et al. (2008 ▸) |
| Barnase | 106 | Bacillus amyloliquefaciens | Niemann et al. (2005 ▸) |
| Green fluorescent protein (GFP) | 221 | Aequorea victoria | Suzuki et al. (2010 ▸) |
| Sterile- motif (SAM) from the translocation Ets leukaemia protein | 78 | Homo sapiens | Nauli et al. (2007 ▸) |
| T4 lysozyme (T4L) | 160 | Enterobacteria phage T4 | Rosenbaum et al. (2007 ▸), Thorsen et al. (2014 ▸) |
| Apocytochrome b 562RIL (BRIL) | 104 | Escherichia coli | Liu et al. (2012 ▸) |
Figure 2.
Examples of successful application of the heterologous fusion-protein approach. The structures are not shown on the same scale. (a) Cartoon diagram of the structure of HTLV-1 gp21 (subunits are shown in different colours) fused at the N-terminus to MBP (in different shades of grey) with a three-Ala linker (red; PDB entry 1mg1; Kobe et al., 1999 ▸). (b) Cartoon diagram of the structure of β2-adrenergic receptor (β2AR; blue) with T4 lysozyme (T4L; grey) inserted into a loop in β2AR (Rosenbaum et al., 2007 ▸; PDB entry 2rh1). (c) A view of crystal-packing interactions for the β2AR-T4L fusion protein [shown and coloured as in (b)]. Note the crystal contacts between the fusion partner T4L and the soluble portion of β2AR. (d) Cartoon diagram of the structure of the complex of the extracellular domains of TLR1 (green) and TLR2 (blue) (Jin et al., 2007 ▸; PDB entry 2z7x). Both proteins are fused at the C-terminus to VLR as the fusion partner (grey). All structure figures were produced with PyMOL (Schrödinger).
2.2. Recent examples of heterologous fusion-protein structures of soluble proteins
Despite the potential advantages of the heterologous fusion-protein approach, the number of reported structures obtained using such approaches remains relatively low (for example, less than 100 MBP fusion-protein structures in a total of ∼97 000 crystal structures in the PDB). This suggests that overcoming the conformational heterogeneity problem is difficult and the approach remains limited to niche applications. Following the early examples, most cases of heterologous fusion-protein structures have involved MBP as the fusion partner (for examples, see Moon et al., 2010 ▸). Pedersen and coworkers used surface-entropy reduction to create MBP variants with superior crystallization properties (Cooper et al., 2007 ▸; Moon et al., 2010 ▸) by substituting 2–5 Lys, Asp, Glu and Asn residues by Ala. This strategy allowed the structure determination of several structures by the Pedersen laboratory (see, for example, Ullah et al., 2008 ▸; Bethea et al., 2008 ▸; Mueller et al., 2010 ▸) and others (see, for example, Patrick et al., 2013 ▸; Jin et al., 2013 ▸; Jung et al., 2014 ▸). MBP helps aggregation-prone proteins to become more soluble (Raran-Kurussi et al., 2015 ▸), but not necessarily monodisperse (Nominé et al., 2001 ▸), which could hinder crystallization. Another drawback of using MBP as the fusion partner may also be its relatively large size (over 360 residues), limiting the size of the target protein that can be expressed in Escherichia coli. Fortunately, other smaller fusion partners have also been used successfully. For example, a catalytically inactive variant of the Bacillus amyloliquefaciens ribonuclease barnase was used as a fusion partner to crystallize the small disulfide-rich cysteine-knot protein McoEeT1 (Niemann et al., 2005 ▸), the green fluorescent protein (GFP) allowed the crystallization of ubiquitin and the ubiquitin-binding motif (UBM) of the Y-family polymerase iota (Suzuki et al., 2010 ▸), as well as a fragment of the apoptotic effector Bax (Czabotar et al., 2013 ▸), and an engineered sterile-α motif (SAM) domain module has been shown to drive the crystallization of 11 different proteins (Nauli et al., 2007 ▸). Some attributes of selected fusion partners are summarized in Table 1 ▸. A split GFP system has also been engineered for use as a crystallization partner (Nguyen et al., 2013 ▸). Carrier proteins have even been employed to facilitate the crystallization of antibiotics (Economou et al., 2012 ▸).
2.3. Application of the heterologous fusion-protein approach to integral membrane proteins
As suggested previously, the heterologous fusion-protein approach could be particularly useful for integral membrane proteins, where polar surface areas that can support crystal contact formation are often limited (Privé et al., 1994 ▸; Smyth et al., 2003 ▸). Although it took some time before these suggestions were realised, with the structure of the β2-adrenergic receptor (β2AR; Rosenbaum et al., 2007 ▸), the approach has proven to be particularly useful in one of the most important challenges in structural biology: the crystallization of G protein-coupled receptors (GPCRs). The fusion-protein strategy applied to β2AR involved T4L (Table 1 ▸) inserted into a flexible intracellular loop of the protein (Rosenbaum et al., 2007 ▸; Figs. 2 ▸ b and 2 ▸ c). T4L has been chosen as a well folded soluble protein that crystallizes under many conditions. Most crystal structures of GPCRs published to date have been obtained using the fusion-protein strategy, and the strategy has also yielded the highest resolution structures of the proteins in this family (see, for example, Fenalti et al., 2014 ▸; Thorsen et al., 2014 ▸; Liu et al., 2012 ▸; Cherezov et al., 2007 ▸; Miller-Gallacher et al., 2014 ▸). Clearly, tethering the fusion partner to the GPCR at two ends within a loop could have reduced some of the conformational heterogeneity usually associated with fusion proteins; it has recently been shown, however, that fusing T4L to the N-terminus of a GPCR can also facilitate crystallization (Zou et al., 2012 ▸). Additional GPCR structures have been obtained using another fusion partner, the thermostabilized apocytochrome b 562RIL (Chun et al., 2012 ▸; Table 1 ▸). To further improve the utility of the T4L fusion approach, the T4L moiety has recently been modified to decrease its flexibility and size (Thorsen et al., 2014 ▸). In one variant, the flexibility of the two lobes of T4L was reduced by introducing two disulfide bridges in the interface between the lobes. In another variant, the smaller N-terminal lobe was deleted to create a ‘minimal T4L’. Both variants were shown to improve the diffraction quality of crystals of M3 muscarinic receptor compared with the unmodified T4L fusion (Thorsen et al., 2014 ▸)
2.4. Hybrid LRR approach and grafting
An ingenious approach to address the conformational heterogeneity problem of fusion proteins has been introduced for the class of repeat or solenoid proteins, specifically for leucine-rich repeat (LRR) proteins, and termed the ‘hybrid LRR approach’ (Jin & Lee, 2008 ▸). The approach takes advantage of the repeat structure to form a rigid connection between the fusion partner and the protein of interest. The approach has been applied successfully to obtain structures of the extracellular domains of Toll-like receptors (TLRs) 1, 2 and 4 (Jin et al., 2007 ▸; Kim et al., 2007 ▸; Fig. 2 ▸ d). The approach relies on fusing two structurally compatible LRR proteins. For TLRs, the variable lymphocyte receptor (VLR) from hagfish was chosen as the fusion partner, as this protein is easy to produce and crystallize, and its LRRs are structurally similar to those in TLRs. The two proteins are fused at a conserved motif in the repeat, so that the repeats can form a continuous solenoid, eliminating any conformational heterogeneity at the fusion site. However, choosing the correct fusion site in the repeats of the two partners does not guarantee complementarity at the fusion site; a lack of complementarity may lead to structural collisions or exposure of the hydrophobic core. For this reason, several hybrids should be tested and characterized to find a suitable one (Jin & Lee, 2008 ▸). In related work with a somewhat different objective in mind, namely to create a binding scaffold, a fusion protein was designed from VLR-based consensus LRRs flanked by the N-terminal cap from Listeria monocytogenes internalin B and the C-terminal cap from VLR, and its crystal structure was determined (Lee et al., 2012 ▸).
Another conceptually similar method was used to obtain structural information on the interaction between the LRR proteins in the glycoprotein (GP) Ib–IX–V complex involved in platelet activation and thrombus formation (McEwan et al., 2011 ▸; Kobe, 2011 ▸). In this case, McEwan and coworkers grafted three segments of GPIX onto a portion of the homologous GPIbβ, which had been successfully crystallized previously (McEwan et al., 2011 ▸).
2.5. Practical considerations for the heterologous fusion-protein approach and the choice of linkers
Although the developers of the modified MBP variants (Moon et al., 2010 ▸), the SAM-domain modules (Nauli et al., 2007 ▸) and the hybrid LRR approach (Jin & Lee, 2008 ▸) claimed high success rates for their methods, the scarcity of reported heterologous fusion-protein structures suggests that success rates in reality may be low. The key challenges are likely to correspond to finding a suitable linker that would sufficiently reduce the conformational heterogeneity of the fusion protein, and in the case of the hybrid LRR technique the steric incompatibility at the fusion site. Tethering the fusion partner at two sites by insertion into a loop, as demonstrated for GPCRs, may reduce the conformational heterogeneity problem to some extent. Most successful cases used the MBP fusion-protein approach and a linker consisting of three alanine residues (Moon et al., 2010 ▸), following an early successful example (Center et al., 1998 ▸); most of the remaining examples used slightly different sequences of 2–5 residues in length, and no systematic studies have been performed to date to our knowledge. Insufficient data are available to draw any firm conclusions for the other fusion partners. Similarly, other approaches to constructing rigid linkers, such as using proline-rich sequences, appear not to have been trialled extensively. The best strategy for the implementation of the heterologous fusion-protein approach should therefore take into account the nature of the target protein. If the method has previously been applied successfully to a related target protein, for example in the cases of GPCRs and LRR proteins, the most effective approach will be to follow the methods that have proven to be successful for these related proteins. In a more general case, if the protein is smaller than 400–500 residues then the MBP-fusion approach using the modified MBP constructs developed by Pedersen and coworkers (Moon et al., 2010 ▸) and using a short linker of less than five residues in length may currently be the most effective methodology to trial. As it is relatively simple to prepare several constructs in parallel, it should be valuable to systematically trial a number of linkers, and this should simultaneously increase the overall chances of success of finding a crystallizable construct.
3. Fusion of interacting proteins approach
In addition to solubilizing one or both interacting partners when they cannot be expressed on their own, the fusion of interacting partners addresses the problem of achieving adequate local concentrations of the binding partners and maintaining stoichiometry in environments that promote protein crystallization, so that the interacting complex can be captured in the crystals. This strategy has proven to be particularly successful for protein–peptide complexes, although it has also facilitated the crystallization of several protein–protein complexes (reviewed by Reddy Chichili et al., 2013b ▸). One caveat of this fusion-protein strategy is that the linker may prevent the native association mode; for this reason, the linker has to be optimized and the fusion protein has to be characterized, with the observed association mode compared with the native complex in solution, for example by using biophysical techniques and site-directed mutagenesis. An alternative approach to fusing the interacting partners is to cross-link them; however, this alternative approach comes with its own challenges, in particular the heterogeneity introduced by the cross-linking reaction (Reddy Chichili et al., 2013a ▸; Mouradov et al., 2008 ▸; Wine et al., 2007 ▸; Leitner et al., 2010 ▸).
3.1. Fusion of protein–peptide complexes
One of the early applications involved the fusion of an antigen peptide to the N-terminus of the major histocompatibility complex (MHC) class II β1 chain through a 16-residue glycine-rich linker (Fremont et al., 1996 ▸). An analogous approach has subsequently been used for a number of other MHC class II–peptide complexes, as well as T-cell receptor (TCR)–peptide and TCR–peptide–MHC class II ternary complexes (Reddy Chichili et al., 2013b ▸). Another successful example involved the nuclear LIM (Lin-11/Islet-1/Mec-3) domain-containing zinc-binding transcription factors. An 11-residue Gly/Ser-rich linker was used to link the LIM domain of LMO4 to the C-terminus of LDB1 (LIM-domain binding protein 1), facilitating structure determination by both NMR and crystallography (Deane et al., 2003 ▸, 2004 ▸). Again, the strategy has been successfully exploited for a number of LIM-domain complexes (Reddy Chichili et al., 2013b ▸). Further examples of structures of fused protein–peptide complexes include a pregnane X receptor ligand-binding domain complex with a peptide from the steroid receptor activator 1 (SRC-1; Wang et al., 2008 ▸), a paramyxovirus phosphoprotein complex with a peptide from a nucleocapsid protein (Kingston et al., 2004 ▸), a histone chaperone Asf1 (anti-silencing function 1) complex with a peptide from histone H3 (Antczak et al., 2006 ▸), the West Nile virus protease NS3 tethered to a 40-residue portion of NS2B (Erbel et al., 2006 ▸; Robin et al., 2009 ▸), and a calmodulin complex with a peptide from calcineurin (Ye et al., 2006 ▸). In the latter case, the structure was subsequently determined using the unlinked peptide and shown to be identical to the fusion-protein structure (Ye et al., 2008 ▸).
3.2. Fusion of protein–protein complexes
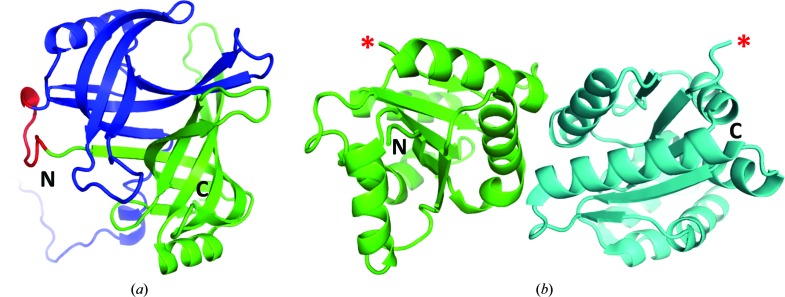
Compared with protein–peptide complexes, fewer cases have been described in which larger interacting protein domains have been linked to each other. Early examples of linking domains included glycyl-tRNA synthetase (Toth & Schimmel, 1986 ▸), immunoglobulin domains (Bird et al., 1988 ▸; Huston et al., 1988 ▸) and an HIV protease homodimer (Cheng et al., 1990 ▸). The main objective for the HIV protease work was to allow modification of one of the subunits so that effects on the enzymatic activity could be assessed. However, the fusion was also found to stabilize the dimer interaction at pH values where the subunits dissociated in the wild-type protein. The structure of the single-chain dimer turned out to be identical to the natural dimer except near the linker region (Bhat et al., 1994 ▸). Two subunits were similarly linked in transthyretin to stabilize its assembly (Foss et al., 2005 ▸). Another example involved the linkage of two domains from the ionotropic glutamate receptor that are separated by transmembrane segments in the wild-type protein; the construct retained its ligand-binding ability (Armstrong & Gouaux, 2000 ▸). The crystal structure of the complex between the Ff bacteriophage minor coat gene 3 protein (g3p) N1 domain and the E. coli TolA C-terminal domain was obtained by fusing these domains with a long flexible linker (Lubkowski et al., 1999 ▸). Park & Hol (2012 ▸) explored various linkages between the OB-fold domain-containing proteins interacting within the editosome complex from Trypanosoma brucei. Testing 25 different expression and co-expression experiments, which included up to four linked domains, resulted in one crystal structure corresponding to two proteins linked through a nine-residue linker (Park & Hol, 2012 ▸; Fig. 3 ▸ a).
Figure 3.
Examples of successful application of the fusion of interacting proteins approach. The structures are not shown on the same scale. (a) Cartoon diagram of the structure of the OB-fold domains from the editosome proteins A3 (blue) and A6 (green) linked with a nine-residue linker (red) (Park & Hol, 2012 ▸; PDB entry 4dni). (b) Cartoon diagram of the structure of the linked TIR domains from RRS1 (green) and RPS4 (cyan) (Williams et al., 2014 ▸; PDB entry 4c6t). There was no interpretable electron density corresponding to the five-residue linker and flanking residues; therefore, it is not clear which domains are linked in the crystal. Nevertheless, the structure revealed a biologically meaningful interdomain interface (Williams et al., 2014 ▸). In the figure, the domain termini that are linked in the fusion protein are labelled with red asterisks.
We have successfully applied the interaction protein-fusion strategy to the family of TIR (Toll/interleukin-1 receptor/resistance protein) domains found in diverse proteins involved in immune signalling from mammals to plants (Ve et al., 2015 ▸). These domains are thought to signal through self-association or homotypic interactions with TIR domains from other proteins. Most reported dissociation constants for TIR–TIR domain interactions are in the micromolar range, and the transient nature of these interactions has made it difficult to define the interfaces between interacting TIR domains using X-ray crystallography. We applied the strategy to a number of TIR domains from both mammalian and plant immune proteins, and found that the addition of linkers between TIR domains facilitated the expression and purification of most of the TIR–TIR domain complexes investigated, improving the yield of soluble protein, and in two cases enabled the production of a soluble TIR domain that could not be produced in a soluble form by itself (Williams et al., 2015 ▸). We obtained crystals of two TIR–TIR domain complexes, one yielding a high-resolution structure of the first heterodimeric TIR-domain complex [of the TIR domains from the Arabidopsis thaliana nucleotide-binding/LRR (NLR) proteins RPS4 and RRS1; Fig. 3 ▸ b; Williams et al., 2014 ▸]. The biological relevance of the observed association was validated through small-angle X-ray scattering and site-directed mutagenesis followed by functional assays (Williams et al., 2014 ▸).
3.3. Practical considerations for the fusion of interacting proteins approach and the choice of linkers
Insufficient successful examples of the fusion of interacting proteins approach exist to extract a generic set of guidelines that could lead to successful application with a high chance of success. As suggested above for the heterologous fusion-protein approach, the most effective strategy in cases where the approach has been successfully applied to related proteins is to follow what worked in these cases; this has been successfully exploited in the cases of MHC class II and LMO-domain proteins. The approach will clearly have to be optimized in each specific case to identify the appropriate length of the linker, so that the native association mode can be retained; any prior knowledge should be used to estimate the approximate length required (considering an ∼3 Å span per residue in an extended structure). Most successful examples have used Gly/Ser/Thr-rich linkers of 2–31 residues in length, with most in the 5–11-residue range (for an extensive list of examples, see Reddy Chichili et al., 2013b ▸). Such a linker composition ensures that the sequence is flexible, hydrophilic and resistant to proteolysis.
Because the linker can potentially interfere with the native association, it is extremely important that the fusion construct is characterized and the observed association compared with the association of unlinked components in solution. This can be achieved through the use of a variety of biophysical and biophysical methods [for example, size-exclusion chromatography (SEC) combined with multi-angle laser light scattering (MALS), small-angle X-ray scattering (SAXS), chemical cross-linking followed by mass spectrometry (MS), hydrogen/deuterium-exchange NMR or mass spectrometry] and the effects of mutations of the residues observed in the interface can be tested by functional assays.
The fusion of interacting proteins approach can clearly lead to an intermolecular, rather than an intramolecular, interaction between the linked partners (equivalent to domain swapping; Gronenborn, 2009 ▸). For example, this occurred in the paramyxovirus phosphoprotein-nucleocapsid protein fusion construct (Kingston et al., 2004 ▸) and is likely to have occurred in the RPS4-RRS1 TIR-domain fusion (Williams et al., 2014 ▸, 2015 ▸). Such an outcome could be promoted through the wrong choice of linker length (in particular if the linker is too short and the native intramolecular association cannot occur). This outcome can easily be detected by measuring the molecular mass of the fusion protein (ideally by methods that determine the mass independent from the shape of the molecule, such as MALS), although domain swapping may also be induced by crystallization itself. It is important to note that such an outcome may not necessarily prevent meaningful biological interpretations about the association mode; however, the observed association needs to be thoroughly validated, as described above.
4. Combination of the two approaches
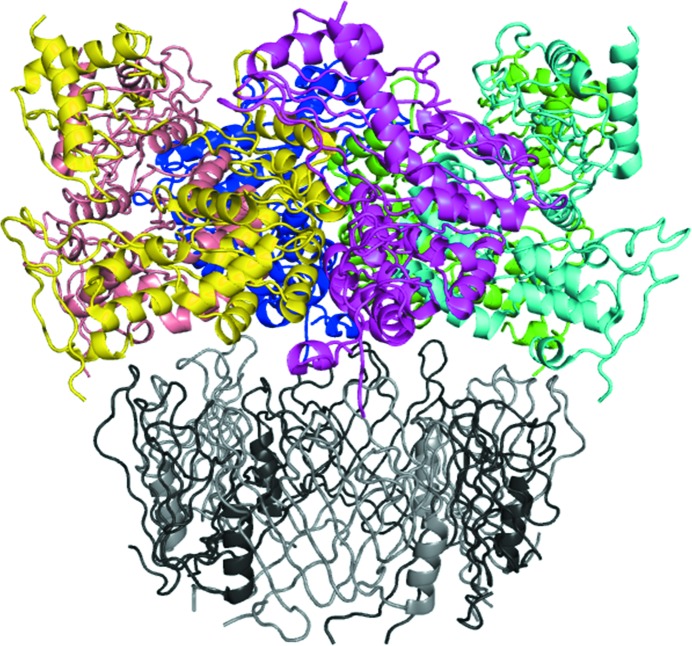
An interesting application of fusion-protein technology, which combines both the heterologous fusion-protein and fusion of interacting proteins approaches discussed above, has been pioneered by Pornillos et al. (2009 ▸). Owing to difficulties in capturing hexamers of the HIV capsid protein in crystals, these researchers fused the hexameric protein CcmK4 to the target protein and successfully determined the crystal structure of the capsid hexamer. This strategy combines the advantages of the two fusion-protein approaches, with the hexameric arrangement of the heterologous fusion partner bringing together the weakly interacting target proteins. Interestingly, in the crystals of the HIV capsid-CcmK4 fusion no interpretable electron density for the CcmK4 moiety was observed, despite an only two-residue linker between the two proteins, suggesting that the fusion partner occupied multiple positions in the crystal. An analogous strategy was used to crystallize the type II secretion system ATPase GspEEpsE from Vibrio cholerae. By fusing GspEEpsE with the Pseudomonas aeruginosa hexamer-forming secretion-system protein Hcp1, the active hexametric structure could be determined (Lu et al., 2013 ▸; Fig. 4 ▸). In this case the fusion protein could be located in the crystals. Fusion of a crystallization chaperone, for example a designer binding protein such as a DARPin, to the protein of interest would be another interesting combination of the two approaches.
Figure 4.
Example of the combination of the heterologous fusion-protein and fusion of interacting proteins approaches. The type II secretion system ATPase GspEEpsE from V. cholerae was fused at the C-terminus to the P. aeruginosa hexamer-forming secretion-system protein Hcp1 (Lu et al., 2013 ▸; PDB entry 4kss). In the cartoon diagram of the hexameric complex, the individual subunits of GspEEpsE are shown in different colours and individual subunits of Hcp1 are shown in alternating dark and light grey. The linkers between the fusion partners have not been modelled. The N- and C-termini of the proteins are at the top and the bottom of the structure, respectively.
5. Conclusions
Even though most proteins used for protein crystallization are produced as recombinant proteins fused to affinity tags to facilitate efficient purification and to enhance folding and solubility, the tags are generally removed before crystallization and fusion proteins are rarely used in crystallization. However, in this article we illustrate approaches that take advantage of fusion-protein technology in crystallization itself, in particular the heterologous fusion-protein and fusion of interacting proteins approaches. Crystallization is clearly one of the major bottlenecks of macromolecular crystallography, and one must attack the problem on multiple fronts, especially in difficult cases such as membrane proteins and weakly interacting protein complexes. The key to the choice of the most promising approaches for a particular macromolecule or complex is in understanding its chemical and physical properties. We believe appropriate fusion-protein approaches described here should be considered, especially in cases where other approaches have not led to success.
Acknowledgments
The research in the authors’ laboratory has been supported by the National Health and Medical Research Council (NHMRC) Program (1000512, 565526), Project (1003326) and Research Fellowship (1003325), and the Australian Research Council (ARC) Discovery Project (DP120100685) to BK. We acknowledge the use of the University of Queensland Remote Operation Crystallization and X-ray Diffraction Facility (UQ ROCX) and the Australian Synchrotron.
References
- Antczak, A. J., Tsubota, T., Kaufman, P. D. & Berger, J. M. (2006). BMC Struct. Biol. 6, 26. [DOI] [PMC free article] [PubMed]
- Argos, P. (1990). J. Mol. Biol. 211, 943–958. [DOI] [PubMed]
- Armstrong, N. & Gouaux, E. (2000). Neuron, 28, 165–181. [DOI] [PubMed]
- Bell, M. R., Engleka, M. J., Malik, A. & Strickler, J. E. (2013). Protein Sci. 22, 1466–1477. [DOI] [PMC free article] [PubMed]
- Bethea, H. N., Xu, D., Liu, J. & Pedersen, L. C. (2008). Proc. Natl Acad. Sci. USA, 105, 18724–18729. [DOI] [PMC free article] [PubMed]
- Bhat, T. N., Baldwin, E. T., Liu, B., Cheng, Y.-S. E. & Erickson, J. W. (1994). Nature Struct. Mol. Biol. 1, 552–556. [DOI] [PubMed]
- Bird, R. E., Hardman, K. D., Jacobson, J. W., Johnson, S., Kaufman, B. M., Lee, S. M., Lee, T., Pope, S. H., Riordan, G. S. & Whitlow, M. (1988). Science, 242, 423–426. [DOI] [PubMed]
- Bucher, M. H., Evdokimov, A. G. & Waugh, D. S. (2002). Acta Cryst. D58, 392–397. [DOI] [PubMed]
- Carson, M., Johnson, D. H., McDonald, H., Brouillette, C. & DeLucas, L. J. (2007). Acta Cryst. D63, 295–301. [DOI] [PubMed]
- Carter, D. C., Rüker, F., Ho, J. X., Lim, K., Keeling, K., Gilliland, G. & Ji, X. (1994). Protein Pept. Lett. 1, 175–178. [DOI] [PMC free article] [PubMed]
- Center, R. J., Kobe, B., Wilson, K. A., Teh, T., Kemp, B. E., Poumbourios, P. & Howlett, G. J. (1998). Protein Sci. 7, 1612–1619. [DOI] [PMC free article] [PubMed]
- Chao, J. A., Prasad, G. S., White, S. A., Stout, C. D. & Williamson, J. R. (2003). J. Mol. Biol. 326, 999–1004. [DOI] [PubMed]
- Cheng, Y.-S. E., Yin, F. H., Foundling, S., Blomstrom, D. & Kettner, C. A. (1990). Proc. Natl Acad. Sci. USA, 87, 9660–9664. [DOI] [PMC free article] [PubMed]
- Cherezov, V., Rosenbaum, D. M., Hanson, M. A., Rasmussen, S. G. F., Thian, F. S., Kobilka, T. S., Choi, H.-J., Kuhn, P., Weis, W. I., Kobilka, B. K. & Stevens, R. C. (2007). Science, 318, 1258–1265. [DOI] [PMC free article] [PubMed]
- Chun, E., Thompson, A. A., Liu, W., Roth, C. B., Griffith, M. T., Katritch, V., Kunken, J., Xu, F., Cherezov, V., Hanson, M. A. & Stevens, R. C. (2012). Structure, 20, 967–976. [DOI] [PMC free article] [PubMed]
- Cooper, D. R., Boczek, T., Grelewska, K., Pinkowska, M., Sikorska, M., Zawadzki, M. & Derewenda, Z. (2007). Acta Cryst. D63, 636–645. [DOI] [PubMed]
- Czabotar, P. E., Westphal, D., Dewson, G., Ma, S., Hockings, C., Fairlie, W. D., Lee, E. F., Yao, S., Robin, A. Y., Smith, B. J., Huang, D. C. S., Kluck, R. M., Adams, J. M. & Colman, P. M. (2013). Cell, 152, 519–531. [DOI] [PubMed]
- Deane, J. E., Mackay, J. P., Kwan, A. H. Y., Sum, E. Y. M., Visvader, J. E. & Matthews, J. M. (2003). EMBO J. 22, 2224–2233. [DOI] [PMC free article] [PubMed]
- Deane, J. E., Ryan, D. P., Sunde, M., Maher, M. J., Guss, J. M., Visvader, J. E. & Matthews, J. M. (2004). EMBO J. 23, 3589–3598. [DOI] [PMC free article] [PubMed]
- Derewenda, Z. S. (2004). Methods, 34, 354–363. [DOI] [PubMed]
- Derewenda, Z. S. (2010). Acta Cryst. D66, 604–615. [DOI] [PMC free article] [PubMed]
- Donahue, J. P., Patel, H., Anderson, W. F. & Hawiger, J. (1994). Proc. Natl Acad. Sci. USA, 91, 12178–12182. [DOI] [PMC free article] [PubMed]
- Economou, N. J., Nahoum, V., Weeks, S. D., Grasty, K. C., Zentner, I. J., Townsend, T. M., Bhuiya, M. W., Cocklin, S. & Loll, P. J. (2012). J. Am. Chem. Soc. 134, 4637–4645. [DOI] [PMC free article] [PubMed]
- Erbel, P., Schiering, N., D’Arcy, A., Renatus, M., Kroemer, M., Lim, S. P., Yin, Z., Keller, T. H., Vasudevan, S. G. & Hommel, U. (2006). Nature Struct. Mol. Biol. 13, 372–373. [DOI] [PubMed]
- Fenalti, G., Giguere, P. M., Katritch, V., Huang, X.-P., Thompson, A. A., Cherezov, V., Roth, B. L. & Stevens, R. C. (2014). Nature (London), 506, 191–196. [DOI] [PMC free article] [PubMed]
- Foss, T. R., Kelker, M. S., Wiseman, R. L., Wilson, I. A. & Kelly, J. W. (2005). J. Mol. Biol. 347, 841–854. [DOI] [PubMed]
- Fremont, D. H., Hendrickson, W. A., Marrack, P. & Kappler, J. (1996). Science, 272, 1001–1004. [DOI] [PubMed]
- George, R. A. & Heringa, J. (2002). Protein Eng. Des. Sel. 15, 871–879. [DOI] [PubMed]
- Gräslund, S. et al. (2008). Nature Methods, 5, 135–146. [DOI] [PMC free article] [PubMed]
- Gronenborn, A. M. (2009). Curr. Opin. Struct. Biol. 19, 39–49. [DOI] [PMC free article] [PubMed]
- Huh, I., Gene, R., Kumaran, J., MacKenzie, C. R. & Brooks, C. L. (2014). Acta Cryst. F70, 1532–1535. [DOI] [PMC free article] [PubMed]
- Hunte, C. & Michel, H. (2002). Curr. Opin. Struct. Biol. 12, 503–508. [DOI] [PubMed]
- Huston, J. S., Levinson, D., Mudgett-Hunter, M., Tai, M.-S., Novotný, J., Margolies, M. N., Ridge, R. J., Bruccoleri, R. E., Haber, E., Crea, R. & Oppermann, H. (1988). Proc. Natl Acad. Sci. USA, 85, 5879–5883. [DOI] [PMC free article] [PubMed]
- Jin, T., Huang, M., Smith, P., Jiang, J. & Xiao, T. S. (2013). Acta Cryst. F69, 855–860. [DOI] [PMC free article] [PubMed]
- Jin, M. S., Kim, S. E., Heo, J. Y., Lee, M. E., Kim, H. M., Paik, S.-G., Lee, H. & Lee, J.-O. (2007). Cell, 130, 1071–1082. [DOI] [PubMed]
- Jin, M.-S. & Lee, J.-O. (2008). BMB Rep. 41, 353–357. [DOI] [PubMed]
- Jung, J., Bashiri, G., Johnston, J. M., Brown, A. S., Ackerley, D. F. & Baker, E. N. (2014). J. Struct. Biol. 188, 274–278. [DOI] [PubMed]
- Ke, A. & Wolberger, C. (2003). Protein Sci. 12, 306–312. [DOI] [PMC free article] [PubMed]
- Kim, H. M., Park, B. S., Kim, J.-I., Kim, S. E., Lee, J., Oh, S. C., Enkhbayar, P., Matsushima, N., Lee, H., Yoo, O. J. & Lee, J.-O. (2007). Cell, 130, 906–917. [DOI] [PubMed]
- Kim, K. M., Yi, E. C., Baker, D. & Zhang, K. Y. J. (2001). Acta Cryst. D57, 759–762. [DOI] [PubMed]
- Kingston, R. L., Hamel, D. J., Gay, L. S., Dahlquist, F. W. & Matthews, B. W. (2004). Proc. Natl Acad. Sci. USA, 101, 8301–8306. [DOI] [PMC free article] [PubMed]
- Kobe, B. (2011). Blood, 118, 5065–5066. [DOI] [PubMed]
- Kobe, B., Center, R. J., Kemp, B. E. & Poumbourios, P. (1999). Proc. Natl Acad. Sci. USA, 96, 4319–4324. [DOI] [PMC free article] [PubMed]
- Koide, S. (2009). Curr. Opin. Struct. Biol. 19, 449–457. [DOI] [PMC free article] [PubMed]
- Lee, S. C., Park, K., Han, J., Lee, J. J., Kim, H. J., Hong, S., Heu, W., Kim, Y. J., Ha, J.-S., Lee, S.-G., Cheong, H.-K., Jeon, Y. H., Kim, D. & Kim, H.-S. (2012). Proc. Natl Acad. Sci. USA, 109, 3299–3304. [DOI] [PMC free article] [PubMed]
- Leitner, A., Walzthoeni, T., Kahraman, A., Herzog, F., Rinner, O., Beck, M. & Aebersold, R. (2010). Mol. Cell. Proteomics, 9, 1634–1649. [DOI] [PMC free article] [PubMed]
- Liu, W., Chun, E., Thompson, A. A., Chubukov, P., Xu, F., Katritch, V., Han, G. W., Roth, C. B., Heitman, L. H., IJzerman, A. P., Cherezov, V. & Stevens, R. C. (2012). Science, 337, 232–236. [DOI] [PMC free article] [PubMed]
- Liu, Y. F., Manna, A., Li, R. G., Martin, W. E., Murphy, R. C., Cheung, A. L. & Zhang, G. Y. (2001). Proc. Natl Acad. Sci. USA, 98, 6877–6882. [DOI] [PMC free article] [PubMed]
- Lu, C., Turley, S., Marionni, S. T., Park, Y.-J., Lee, K. K., Patrick, M., Shah, R., Sandkvist, M., Bush, M. F. & Hol, W. G. J. (2013). Structure, 21, 1707–1717. [DOI] [PMC free article] [PubMed]
- Lubkowski, J., Hennecke, F., Plückthun, A. & Wlodawer, A. (1999). Structure, 7, 711–722. [DOI] [PubMed]
- McEwan, P. A., Yang, W., Carr, K. H., Mo, X., Zheng, X., Li, R. & Emsley, J. (2011). Blood, 118, 5292–5301. [DOI] [PMC free article] [PubMed]
- Miller-Gallacher, J. L., Nehmé, R., Warne, T., Edwards, P. C., Schertler, G. F. X., Leslie, A. G. W. & Tate, C. G. (2014). PLoS One, 9, e92727. [DOI] [PMC free article] [PubMed]
- Moon, A. F., Mueller, G. A., Zhong, X. & Pedersen, L. C. (2010). Protein Sci. 19, 901–913. [DOI] [PMC free article] [PubMed]
- Mouradov, D., King, G., Ross, I. L., Forwood, J. K., Hume, D. A., Sinz, A., Martin, J. L., Kobe, B. & Huber, T. (2008). Methods Mol. Biol. 426, 459–474. [DOI] [PubMed]
- Mueller, G. A., Edwards, L. L., Aloor, J. J., Fessler, M. B., Glesner, J., Pomés, A., Chapman, M. D., London, R. E. & Pedersen, L. C. (2010). J. Allergy Clin. Immunol. 125, 909–917. [DOI] [PMC free article] [PubMed]
- Nauli, S., Farr, S., Lee, Y.-J., Kim, H.-Y., Faham, S. & Bowie, J. U. (2007). Protein Sci. 16, 2542–2551. [DOI] [PMC free article] [PubMed]
- Nguyen, H. B., Hung, L.-W., Yeates, T. O., Terwilliger, T. C. & Waldo, G. S. (2013). Acta Cryst. D69, 2513–2523. [DOI] [PMC free article] [PubMed]
- Niemann, H. H., Schmoldt, H. U., Wentzel, A., Kolmar, H. & Heinz, D. W. (2005). J. Mol. Biol. 356, 1–8. [DOI] [PubMed]
- Nominé, Y., Ristriani, T., Laurent, C., Lefèvre, J.-F., Weiss, E. & Travé, G. (2001). Protein Eng. 14, 297–305. [DOI] [PubMed]
- Park, Y.-J. & Hol, W. G. J. (2012). J. Struct. Biol. 180, 362–373. [DOI] [PMC free article] [PubMed]
- Patrick, A. N., Cabrera, J. H., Smith, A. L., Chen, X. S., Ford, H. L. & Zhao, R. (2013). Nature Struct. Mol. Biol. 20, 447–453. [DOI] [PMC free article] [PubMed]
- Pornillos, O., Ganser-Pornillos, B. K., Kelly, B. N., Hua, Y., Whitby, F. G., Stout, C. D., Sundquist, W. I., Hill, C. P. & Yeager, M. (2009). Cell, 137, 1282–1292. [DOI] [PMC free article] [PubMed]
- Privé, G. G., Verner, G. E., Weitzman, C., Zen, K. H., Eisenberg, D. & Kaback, H. R. (1994). Acta Cryst. D50, 375–379. [DOI] [PubMed]
- Raran-Kurussi, S., Keefe, K. & Waugh, D. S. (2015). Protein Expr. Purif. 110, 159–164. [DOI] [PMC free article] [PubMed]
- Reddy Chichili, V. P., Kumar, V. & Sivaraman, J. (2013a). Intrinsically Disord. Proteins, 1, e25464. [DOI] [PMC free article] [PubMed]
- Reddy Chichili, V. P., Kumar, V. & Sivaraman, J. (2013b). Protein Sci. 22, 153–167. [DOI] [PMC free article] [PubMed]
- Robin, G., Chappell, K., Stoermer, M. J., Hu, S.-H., Young, P. R., Fairlie, D. P. & Martin, J. L. (2009). J. Mol. Biol. 385, 1568–1577. [DOI] [PubMed]
- Rosenbaum, D. M., Cherezov, V., Hanson, M. A., Rasmussen, S. G. F., Thian, F. S., Kobilka, T. S., Choi, H.-J., Yao, X.-J., Weis, W. I., Stevens, R. C. & Kobilka, B. K. (2007). Science, 318, 1266–1273. [DOI] [PubMed]
- Smyth, D. R., Mrozkiewicz, M. K., McGrath, W. J., Listwan, P. & Kobe, B. (2003). Protein Sci. 12, 1313–1322. [DOI] [PMC free article] [PubMed]
- Sugawara, H., Yamaya, T. & Sakakibara, H. (2005). Acta Cryst. F61, 366–368. [DOI] [PMC free article] [PubMed]
- Suzuki, N., Hiraki, M., Yamada, Y., Matsugaki, N., Igarashi, N., Kato, R., Dikic, I., Drew, D., Iwata, S., Wakatsuki, S. & Kawasaki, M. (2010). Acta Cryst. D66, 1059–1066. [DOI] [PubMed]
- Thorsen, T. S., Matt, R., Weis, W. I. & Kobilka, B. K. (2014). Structure, 22, 1657–1664. [DOI] [PMC free article] [PubMed]
- Toth, M. J. & Schimmel, P. (1986). J. Biol. Chem. 261, 6643–6646. [PubMed]
- Ullah, H., Scappini, E. L., Moon, A. F., Williams, L. V., Armstrong, D. L. & Pedersen, L. C. (2008). Protein Sci. 17, 1771–1780. [DOI] [PMC free article] [PubMed]
- Ve, T., Williams, S. J. & Kobe, B. (2015). Apoptosis, 20, 250–261. [DOI] [PubMed]
- Wang, W., Prosise, W. W., Chen, J., Taremi, S. S., Le, H. V., Madison, V., Cui, X., Thomas, A., Cheng, K.-C. & Lesburg, C. A. (2008). Protein Eng. Des. Sel. 21, 425–433. [DOI] [PubMed]
- Waugh, D. S. (2005). Trends Biotechnol. 23, 316–320. [DOI] [PubMed]
- Williams, S. J. et al. (2014). Science, 344, 299–303.
- Williams, S. J., Ve, T. & Kobe, B. (2015). Protein Eng. Des. Sel. 28, 137–145. [DOI] [PubMed]
- Wine, Y., Cohen-Hadar, N., Freeman, A. & Frolow, F. (2007). Biotechnol. Bioeng. 98, 711–718. [DOI] [PubMed]
- Ye, Q., Li, X., Wong, A., Wei, Q. & Jia, Z. (2006). Biochemistry, 45, 738–745. [DOI] [PubMed]
- Ye, Q., Wang, H., Zheng, J., Wei, Q. & Jia, Z. (2008). Proteins, 73, 19–27. [DOI] [PubMed]
- Yu, K., Liu, C., Kim, B.-G. & Lee, D.-Y. (2015). Biotechnol. Adv. 33, 155–164. [DOI] [PubMed]
- Zhan, Y., Song, X. & Zhou, G. W. (2001). Gene, 281, 1–9. [DOI] [PubMed]
- Zhang, J. & Ferré-D’Amaré, A. R. (2014). Curr. Opin. Struct. Biol. 26, 9–15. [DOI] [PMC free article] [PubMed]
- Zou, Y., Weis, W. I. & Kobilka, B. K. (2012). PLoS One, 7, e46039. [DOI] [PMC free article] [PubMed]