Abstract
Protein phosphatase-1 (PP1) is a key regulator of transcription and is targeted to promoter regions via associated proteins. However, the chromatin binding sites of PP1 have never been studied in a systematic and genome-wide manner. Methylation-based DamID profiling in HeLa cells has enabled us to map hundreds of promoter binding sites of PP1 and three of its major nuclear interactors, i.e. RepoMan, NIPP1 and PNUTS. Our data reveal that the α, β and γ isoforms of PP1 largely bind to distinct subsets of promoters and can also be differentiated by their promoter binding pattern. PP1β emerged as the major promoter-associated isoform and shows an overlapping binding profile with PNUTS at dozens of active promoters. Surprisingly, most promoter binding sites of PP1 are not shared with RepoMan, NIPP1 or PNUTS, hinting at the existence of additional, largely unidentified chromatin-targeting subunits. We also found that PP1 is not required for the global chromatin targeting of RepoMan, NIPP1 and PNUTS, but alters the promoter binding specificity of NIPP1. Our data disclose an unexpected specificity and complexity in the promoter binding of PP1 isoforms and their chromatin-targeting subunits.
INTRODUCTION
Protein phosphatase-1 (PP1) is a member of the PhosphoProtein Phosphatases (PPP) superfamily of Ser/Thr-specific protein phosphatases (1,2). Mammalian genomes harbor three PP1 encoding genes that altogether generate four isozymes, namely PP1α, PP1β and the splice variants PP1γ1 and PP1γ2. These isoforms mainly differ in their extremities and have identical enzymatic properties. Except for PP1γ2, which is only expressed in testis and brain, the other PP1 isoforms appear to be present in all mammalian cells. PP1 dephosphorylates hundreds of proteins. Nevertheless, PP1 acts in a highly specific and timely manner because it forms heterodimeric or heterotrimeric complexes with ≈200 PP1 interacting proteins (PIPs) that determine when and where substrates are dephosphorylated. Recent proteomic data show that the total cellular concentration of PIPs is much higher than that of PP1 (3,4), indicating that PIPs are not constitutively associated with PP1 and compete for binding to the limited cellular pool of PP1. In general, PIPs are structurally unrelated and mainly bind to PP1 via short docking motifs (1,2). The most common PP1-binding sequence is known as the RVxF-motif, which binds to a hydrophobic channel that is remote from the active site and is often essential to anchor PP1 (5–9). Other PP1 binding motifs restrain the activity of PP1, e.g. by occluding a substrate binding groove or the active site (6–7,9), or enhance the activity of PP1 by creating an extended substrate binding site (5). Some PIPs also have a binding region for the N or C-terminus of PP1, accounting for the formation of isoform-specific holoenzymes (5). In addition to their PP1 binding domain, PIPs often also have regions that directly recruit substrates or mediate the targeting of PP1 to a specific subcellular location that contains a subset of substrates (1,8). Finally, some PIPs not only regulate PP1 but are themselves substrates for associated PP1 (1,2).
PP1 has key functions in a variety of cellular processes, including transcription (10–14). However, a detailed map of the genes that are regulated by PP1 is not available. Also, it is often not clear whether transcriptional control is mediated by a pool of PP1 that is associated with specific gene-regulatory elements or is more indirect and involves, for example, the regulation of the concentration, activity or recruitment of specific transcription factors. PP1 itself is not known to bind to DNA or histones, indicating that its targeting to chromatin is mediated by specific PIPs. The quantitatively most important and best characterized nuclear PIPs are NIPP1, PNUTS and RepoMan, which are all three (partially) associated with chromatin (10,12–13,15–22). NIPP1 has been implicated in the silencing of genes via the histone metyltransferase EZH2 (12,13) and the regulation of pre-mRNA splicing (23). PNUTS controls transcription by RNA polymerase II (10,18), but also has a role in DNA repair (17,24) and the regulation of the transcription factors p53 and Rb (25–29). RepoMan has been identified as a mitotic histone targeting subunit of PP1 and as a key regulator of the DNA damage response (15–16,21,30). NIPP1, PNUTS and RepoMan have an RVxF-type PP1 docking motif and mutation of this motif abolishes their binding to PP1, which can be used as a tool to dissect the role of associated PP1.
The two major techniques that are currently used for the mapping of chromatin binding sites of a protein of interest (POI) are chromatin immunoprecipitation (ChIP) and DNA adenine methyltransferase identification (DamID) (31–36). ChIP involves the immunoprecipitation of a POI after its covalent crosslinking to chromatin and shearing of the DNA in ≈500 bp fragments. DamID identifies chromatin interaction sites by mapping adenines in a GATC context that are methylated by the bacterial methyltransferase Dam, which is targeted to specific loci by a fused POI. The co-immunoprecipitated DNA (ChIP) or methylated DNA fragments (DamID) can be identified using DNA microarray technology. ChIP has the advantage that it maps chromatin-binding sites of endogenous proteins. However, it is dependent on the availability of antibodies and suffers from artifacts generated by crosslinking. DamID is antibody-independent and, hence, not limited by epitope-masking in multisubunit complexes like PP1 holoenzymes. Another advantage of DamID is that it samples chromatin binding over a prolonged time, enabling its use for signaling proteins like PP1 that only transiently interact with chromatin. Disadvantages of DamID are that it is somewhat less sensitive than ChIP, requires the generation of stable cell lines and maps chromatin binding of trace amounts of an ectopically expressed fusion protein. However, the latter can also be an advantage as it enables a comparison between a wild-type (WT) and mutant POI.
We have used DamID for the genome-wide mapping of the promoter binding sites of PP1 isoforms and three nuclear PIPs in HeLa cells. In addition, we compared the binding sites of both WT PIPs and their PP1-binding mutants. Our profiling identified hundreds of promoter-binding sites of PP1 and provided insights into the specificity of the chromatin targeting of the PP1 isoforms and their crosstalk with PIPs.
MATERIALS AND METHODS
Plasmids and antibodies
Full-length rabbit PP1α, rabbit PP1β, rat PP1γ1, human PNUTS, bovine NIPP1 and human RepoMan were cloned into the pIND-(V5)-EcoDam vector using the InFusion HD cloning system (Clontech), after removal of the V5-tag. The InFusion system was also adopted for the cloning of NIPP1, PNUTS and RepoMan into the eGFP-N1 vector to generate EGFP-tagged fusion proteins. Antibodies against RNA polymerase II pS2 (pol II-pS2; ab5131), TATA binding protein (TBP; ab51841) and Green Fluorescent Protein (GFP, ChIP grade, ab290) were purchased from Abcam (Cambridge, UK). Anti-PP1α (SC-6107, clone C-19), PP1β (SC-6104, clone N-19), PP1γ (SC-6108, clone C-19), EGFP (SC-8334) and histone H3 (SC-10809) were obtained from Santa-Cruz (Dallas, USA). Alpha tubulin (T6074, clone B-5–1–2) and histone H3 (h0163) antibodies were delivered by Sigma-Aldrich (Saint Louis, USA). Dam antibody was purchased from Acris (AM05338PU-N). The following antibodies were home made: anti-NIPP1, anti-PP1 (13) and anti-PNUTS (37). A synthetic peptide comprising amino acids 581–599 of RepoMan coupled to keyhole limpet haemocyanin was used to generate a rabbit polyclonal antibody. The anti-RepoMan antibodies were affinity-purified on the peptide coupled to bovine serum albumin (BSA) and linked to CNBr-activated Sepharose 4B (GE Healthcare, Buckinghamshire, UK).
Cell culture, fractionation and immunoprecipitations
HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) Low glucose (1 g/l) Glutamax growth medium, supplemented with 10% Fetal Calf Serum (FCS) (Sigma-Aldrich), 100 U/ml penicillin and 100 μg/ml streptomycin. HEK293T cells were cultured in DMEM High glucose (4.5 g/l) Glutamax with the same supplements. Transfection with plasmid DNA was performed using FuGENE 6 HD transfection reagent (Roche, Basel, Switzerland) or Genius DNA Transfection Reagent (Westburg, Leusden, The Netherlands). HEK293T cells were grown in 150 mm plates until confluency and then harvested.
Cytoplasmic and nucleoplasmic fractions were obtained as previously described (38). The chromatin fraction was incubated for 30 min at 37°C in 500 μl nuclease buffer (50 mM Tris/HCl at pH 8, 1.5 mM CaCl2, 25 mM NaF) with 60 units of micrococcal nuclease. The sample was centrifuged for 2 min at 700 x g and the supernatant was used as the ‘soluble’ chromatin fraction. The pellet was dissolved in the same volume of sodium dodecyl sulphate (SDS) sample buffer and used as ‘insoluble’ fraction. Equal volumes of each fraction were loaded on a 10% sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and stained by immunoblotting for PP1α, PP1β, PP1γ, TBP, alpha tubulin and histone H3.
SiRNA duplexes against human PP1α (CCGCAUCUAUGGUUUCUACdTdT), PP1β (UUAUGAGACCUACUGAUGUdTdT), PP1γ (GCAUGAUUUGGAUCUUAUAdTdT), PNUTS (CGAGUAAAUGUGAAUAAGA/GCAGACCCGUUCACCAGAA/GCAAUAGUCAGGAGCGAUA/GCUACAAACUUCUUAACAA), control PP1 (D-001210–02) and control PNUTS (D-001206–13) were obtained from Dharmacon (Chicago, IL, USA). Knockdowns were performed using Genius DNA Transfection Reagent (Westburg) for the PP1 isoforms and DharmaFECT Duo Transfection Reagent (Dharmacon) for PNUTS, and were analyzed after 36 or 16 h, respectively. For cell fractionation after the knockdown of PP1 the micrococcal nuclease treatment was omitted and the chromatin fraction was solubilized by sonication for 15 min at 0°C in SDS lysis buffer (38).
EGFP-traps were performed as described previously (12,13). In short, the soluble pool of chromatin-enriched fractions was obtained as described above and the lysates were incubated overnight at 4°C with 25 μl of EGFP-trap beads (1:1 suspension, Chromotek, Planegg-Martinsried, Germany). The beads were spun down for 30 s at 500 x g at 4°C and washed five times with 20 mM Tris/HCl at pH 7.5 plus 0.3 M NaCl. Finally, the pellets were boiled in SDS sample buffer and processed for immunoblotting with EGFP antibodies. Immunoblots were visualized with eCL reagent (PerkinElmer, Waltham, USA) in an ImageQuant LAS4000 imaging system (GE Healthcare) and were quantified using ImageQuant TL software (GE Healthcare).
Immunostaining
For immunofluorescence studies of the localization of the Dam-fusions, HeLa cells were grown on poly-lysine coated coverslips in a 24-well chamber and co-transfected with the pIND-(V5)-EcoDam-PP1 isoforms or the pIND-(V5)-EcoDam-PIP-WT/M vectors and the pVgRXR plasmid encoding the Ecdysone and Retinoic X receptors (Invitrogen, Waltham, USA). Twenty hours post transfection, 2 μM of Ponasterone A (Invitrogen) was added and 24 h later, the cells were treated for 4 min at 4°C with ice cold CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES at pH 6.8) supplemented with 0.2% Triton X-100. Subsequently, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, blocked in 3% BSA-PBS and incubated first overnight in 1% BSA-PBS with the primary Dam antibody and then with Horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody for 2 h. The HRP signal was enhanced by using the TSA-Plus Fluorescien System (PerkinElmer). DNA was stained with DAPI. The cells were visualized with a Leica TCS SPE laser-scanning confocal system mounted on a Leica DMI 4000B microscope, equipped with a Leica ACS APO 40× 1.30NA oil objective.
ChIP and quantitative RT-PCR
ChIP assays were performed as described in (12). Total RNA was isolated using TRIzol® Reagent (Life technologies, Carlsbad, USA) according to the manufacturers’ guidelines. Remnant genomic DNA was removed using the TURBO DNA-free™ Kit (Life technologies). RNA (1–2 μg) was reverse-transcribed with Random Hexamer Primer (Thermo Scientific, Waltham, USA) and oligo dT primer (Sigma-Aldrich) using the RevertAid Premium Reverse Transcriptase and RiboLock RNase inhibitor enzymes (Fermentas, Waltham, USA). About 1.2% of the cDNA was PCR-amplified in duplicate, using SYBR Green qPCR Mix (Invitrogen) and a Rotorgene detection system (Corbett Research, Cambridge, UK), as described by Nuytten et al. (39). Quantitative reverse transcriptase polymerase chain reation (PCR) was performed to check the transcript levels of PP1β (5′-CGAGTTTGATAATGCTGGTGGAATG-3′ and 5′-GCTGTTCGAGTTGGAGTGAC-3′), PNUTS (5′-TCCTCATGAGCCTGATCCT-3′ and 5′-GTCTCAACATACGGAGTCTCATC-3′), SNORD24 (5′-AGAATATTTGCTATCTGAGAGATGGTG-3′ and 5′-TGCATCAGCGATCTTGGT-3′), SNORD28 (5′-TTGATAAGCTGATGTTCTGTGAGG-3′ and 5′-TGCCATCAGAACTCTAACATGC-3′), SNORD33 (5′-TCCCACTCACATTCGAGTTTC-3′ and 5′-CCTCAGATGGTAGTGCATGTG-3′) and HIST1H3D (5′-CGCAGGACTTCAAGACTGAT-3′ and 5′-TAGGTTGGTGTCCTCAAACAG-3′). Data were normalized against the housekeeping gene HPRT (5′-TGACACTGGCAAAACAATGCA-3′ and 5‘-GGTCCTTTCACCAGCAAGCT-3′).
DamID profiling
DamID was performed as described in (31). Briefly, stable polyclonal HeLa cell lines were generated using constructs cloned into the pIND-(V5)-EcoDam vector to express trace amounts of Dam or C-terminal fusions with PP1α, PP1β, PP1γ1, PNUTS, NIPP1 or RepoMan due to leakiness from the uninduced promoter. Cell lines were generated for both the WT PIPs and their PP1-binding mutants (M). Two independent stable cell lines were generated for each construct. Expression of Dam or its fusion proteins leads to methylation of genomic DNA at sites of the fusion proteins’ association with chromatin. Genomic DNA is extracted and processed by methylation-sensitive restriction digests and linker dependent PCR amplification, followed by microarray detection of enriched fragments. To verify the isolated methylated DNA, the PCR-amplified fragments were run on a 2% agarose gel (Supplementary Figure S2).
For DamID it is crucial that only trace amounts of Dam or its fusion proteins are expressed, which cannot be detected by immunoblotting. To verify the expression of the full-length Dam-fusion proteins, HeLa cells were co-transfected with the pIND-(V5)-EcoDam-PP1 isoforms or the pIND-(V5)-EcoDam-PIP-WT/M vectors and the pVg-RXR plasmid encoding the Ecdysone and Retinoid-X receptors as inducible heterodimers that bind to the Ecdysone response element in the pIND vector (Invitrogen). Twenty hours post transfection, 2 μM of the Ecdysone analogue Ponasterone A (Invitrogen) was added and the cells were harvested 24 h later. Cells were lysed in a buffer containing 20 mM Tris/HCl at pH 7.5, 0.3 M NaCl, 0.5% Triton X-100, 25 mM NaF, 1 mM Vanadate, 1 mM PMSF, 1 mM benzamidine and 5 μM leupeptin for 20 min on ice. The lysates were clarified by centrifugation (10 min at 1700 g) and SDS sample buffer was added to the supernatant. Equal amounts (40 μg) were loaded on a 10% SDS-PAGE gel and stained by immunoblotting for endogenous proteins and Dam-fusions with antibodies against the endogenous proteins.
Computational analysis
Tiling-array handling, quality control and preliminary analysis were performed by the VIB MicroArray Facility (www.microarrays.be), as described in (12). In brief, for the genome-wide interaction site profiling, the DamID-DNA was labeled and hybridized to a GeneChip Human Promoter 1.0R Array (Affymetrix, Santa Clara, CA, USA). The tiling array readouts were analyzed with the ‘model-based analysis of tiling arrays’ (MAT) algorithm (version 1.0.0) against the human reference genome (hg19) (40). We normalized the datasets obtained from two independent polyclonal cell lines of each Dam-fusion over two Dam-only datasets. Each dataset consisted of three technical repeats of the same cell line pooled together prior to hybridization to the tiling array in order to reduce possible artifacts. The significant binding peaks at −2/+2 kb relative to all currently annotated transcription start sites (TSS) were selected using a threshold for significance that was set at a P-value of 1 × 10−3–5, depending on the dataset. The resulting set of 78 828 TSSs was derived from UCSC Genes (KnownGenes; Feb. 2009; hg19, GRCh37). Multiple TSSs for one gene were filtered as described in (41). Briefly, if a gene had multiple TSSs assigned to it, we analyzed the RNA Polymerase II signal intensity for a region of 4 kb at either site of the TSS and selected the TSS that was linked to the highest RNA polymerase II signal. This yielded the final list of ∼25 000 TSSs. Further analysis on the significant binding sites was performed using tools linked to the Cistrome-galaxy website (42). All datasets used are available at GEO under the accession number GSE54170.
We created a reference profile of the PP1 isoforms and PIPs by calculating the average signal profile across all previously defined promoter regions, i.e. 2 kb at either side of the TSS, using a window size of 20 bp. The obtained reference list was used to normalize the DamID profiles across their respective genome binding sites within these regions. This strategy was also used to normalize the histone modification ChIP-Seq signal profiles obtained from the UCSC Genome browser (hg19, GRCh37). The average signal profiles were obtained and calculated using the SitePro module of the CEAS package (43). The correlation analyses performed on the DamID datasets and the ENCODE datasets made use of the general correlation tool present on the Cistrome-galaxy portal (42). In addition to the correlation analysis, we also performed co-association analysis using the Genome Structure Correction (GCS) tool (44). Gene Ontology analysis was carried out using DAVID (45). Redundant terms were filtered out and then summarized utilizing the REVIGO tool (46).
Confocal microscopy
The cells were visualized with a Leica TCS SPE laser-scanning confocal system mounted on a Leica DMI 4000B microscope, equipped with a Leica ACS APO 63X 1.30NA oil DIC objective. Z-stacks of 1 μm per slice were made of HeLa cells expressing WT or mutant versions of EGFP-PNUTS, EGFP-NIPP1 or EGFP-RepoMan, and immunostained for Pol II-pS2. Immunostainings of TATA-binding Protein (TBP) and alpha tubulin were used as positive and negative controls, respectively. The co-localization analysis was performed on each Z-stack using the ImageJ plugin Just Another Co-localization Plugin (JACoP) (47), at an image pixel size of 512 × 512. Pearson's Correlation Coefficients (PCCs) were calculated using Costes automatic threshold (48). We have opted for the PCC as this allowed for the co-localization analysis results to be more easily compared to the bioinformatical screening of the DamID versus Encode data, which are also given as correlation co-efficients. The Costes automatic threshold function in the JaCoP plugin also gives this as a standard output. We used Costes automatic threshold as this is more accurate in predicting the background level of fluorescence in both channels. It is also more suitable for high-throughput analysis since the threshold has not to be set manually.
RESULTS
Mapping of the promoter-binding sites of PP1
To identify the isoforms of PP1 that are associated with chromatin, HeLa (Figure 1A) and HEK293T cell lysates (Supplementary Figure S1) were fractionated by differential centrifugation. Histone H3 (H3) and TATA-binding protein (TBP) served as markers for the chromatin-enriched fractions and tubulin was used as a cytoplasmic marker. All three PP1 isoforms, as visualized by immunoblotting with previously validated isoform-specific antibodies (30), were detected in both the cytoplasmic and chromatin-enriched fractions. PP1 was roughly equally distributed between the ‘soluble’ chromatin fraction, obtained by a nuclease pre-treatment and the remaining ‘insoluble’ fraction, which also comprises nucleoskeletal elements. These data demonstrate that a substantial fraction of PP1α, PP1β and PP1γ is associated with chromatin, consistent with previous immunolocalization data in various cell types (49–51) and the established function of PP1 in transcriptional regulation (10–13,18,52).
Figure 1.
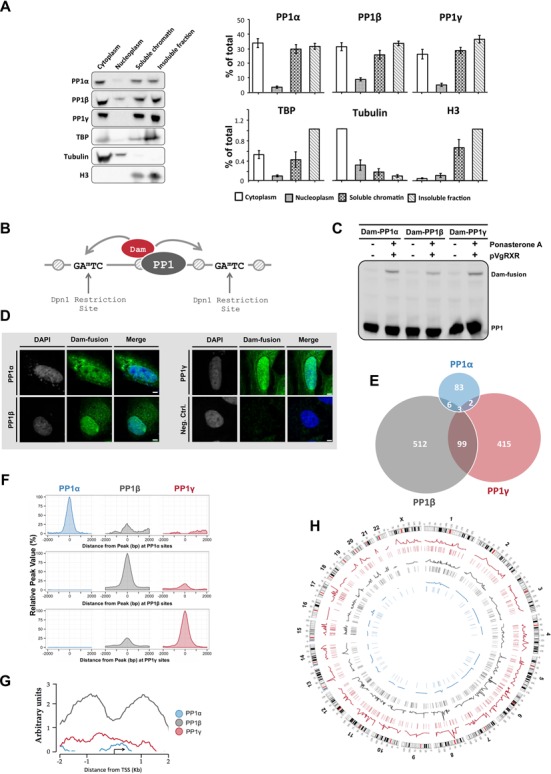
PP1 isoforms bind to distinct promoter subsets. (A) HeLa cells were fractionated by differential centrifugation into cytoplasmic, nucleoplasmic as well as nuclease-solubilized (soluble) and insoluble fractions. All fractions were diluted to the same volume and processed for immunoblotting with isoform-specific PP1 antibodies. TATA-binding protein, α-tubulin and histone H3 served as markers. The left panel shows a representative blot. The right panel shows the average of scans ± S.E.M. of four independent experiments. The protein levels are given as percentages of the total protein pool. (B) Fusions of Dam and PP1 that are targeted to specific chromatin regions methylate flanking adenines in a GATC sequence, which can be mapped using methylation (un)specific restriction enzymes, PCR-amplification and DNA microarray analysis. (C) Expression of the Dam-PP1 fusions after their transient induction with Ponasterone A in HeLa cells. (D) Subcellular localization of the Ponasterone-A induced Dam-PP1 fusions in HeLa cells. (E) Venn diagram showing the significant binding peaks and overlapping peaks of each PP1 isoform on the promoter regions. The area of the circles and overlaps correlates with the number of peaks. In the majority of cases one peak translates to one promoter but multiple peaks can also occupy a single promoter. (F) Global signal profiles of the PP1 isoforms at −2/+2 kb of the peak centers. The percentages of the PP1 signals were calculated according to the highest signal found across the binding sites. The graphs were drawn using the R package ggplot2 (53). (G) The average raw signal profile of the PP1 isoforms on promoter regions, as calculated with the SitePro tool from the CEAS package (43). (H) Circos plot (version 0.61) of the significant PP1 promoter binding peaks across the HeLa genome (54). The line profiles of the PP1 isoforms reflect the intensity of the isoform binding signals. The lines underneath the line profiles denote the position of the significant binding sites.
To gain genome-wide information on the promoter binding sites of PP1, we adopted the DamID technique, in particular because this technique is antibody independent and also detects transient interactions (Figure 1B). First, the expression of the fusion constructs in HeLa cells was verified by their transient co-overexpression with the activator plasmid pVgRXR in the presence of the Ecdysone analogue Ponasterone A (Figure 1C). We also verified that the Dam-PP1 fusions were (partially) targeted to the nucleus (Figure 1D), consistent with the localization of endogenous PP1 isoforms (14). Also, PP1α was excluded from the nucleoli, unlike the other PP1 isoforms. Next, HeLa cell lines were generated that stably express trace amounts of Dam, Dam-PP1α, Dam-PP1β or Dam-PP1γ1 due to leakiness from an uninduced promoter (Supplementary Figure S2). To rule out effects from the random integration of the transgenes, two distinct polyclonal cell lines were generated and analyzed for each construct. From each cell line, three independent genomic samples were isolated and adenine-methylated DNA fragments were prepared, PCR amplified, pooled and hybridized to GenChip Human Promotor 1.0R Arrays (Affymetrix), covering the −7.5/2.5 kb region of ≈25 000 TSS. Using the default threshold of the MAT peak-calling tool we identified 94, 620 and 519 significant promoter-binding peaks for PP1α, PP1β and PP1γ, respectively (Figure 1E). For these analyses the promoter region was defined as −2 kb/+2kb relative to the TSS. The peaks of PP1β and PP1γ showed a moderate overlap (19%), but there was only a weak overlap (<10%) between the peaks of PP1α and the other isoforms. This (lack of) overlap was also confirmed by the signal profiles (53) of the significant binding sites (Figure 1F, Supplementary Figure S3). Thus, although there was some overlap between the binding sites of PP1β and PP1γ, the PP1 isoforms generally bound to distinct loci, hinting at isoform-specific functions.
We also performed promoter profile analyses of the raw signals, which reflect the average binding propensity of the Dam fusions (54). Remarkably, the three PP1 isoforms showed a distinct signal-binding pattern across the TSS region (Figure 1G). The Dam-PP1α signals were low and largely confined to the vicinity of the TSS. However, the Dam-PP1γ signals were more prominent and spread across the entire promoter region. The Dam-PP1β signals were strong and showed a bimodal distribution, with peaks flanking the TSS. Thus, all three PP1 isoforms bind to chromatin but PP1β is quantitatively the most important promoter binding isoform. Finally, we have analyzed the genome-wide distribution of the PP1 binding sites and found that the three isoforms are associated with all chromosomes (Figure 1H).
Mapping of the promoter-binding sites of nuclear PIPs
The major mammalian nuclear PIPs are NIPP1, PNUTS and RepoMan (10,12–13,15–18,24). All three have an RVxF-type PP1 docking site (Figure 2A) and are partially chromatin-associated. To map the promoter-binding sites of these PIPs by DamID, we generated stable HeLa cell lines expressing fusions of Dam and NIPP1, PNUTS or RepoMan (Supplementary Figure S2). The expression, subcellular localization and PP1 binding of the DamID fusion constructs was first validated after their transient induction with Ponasterone A (Supplementary Figure S4). All fusions were nuclear and the NIPP1 fusions were enriched at the nuclear speckles, as previously already shown for endogenous NIPP1 (55). The promoter binding sites were mapped and analyzed as detailed above for the PP1 isoforms. We identified 941, 599 and 330 significant binding peaks for PNUTS-WT, NIPP1-WT and RepoMan-WT, respectively (Figure 2B). The overlaps between the significant binding sites of these PIPs were limited, which was confirmed by the signal profiles of the three PIPs at the PNUTS, NIPP1 and RepoMan promoter binding sites (Figure 2C). The average promoter binding patterns of the PIPs were also strikingly different (Figure 2D). RepoMan showed a relatively weak binding close to the TSS. The binding of NIPP1 was weak and diffuse. However, PNUTS interacted strongly with the promoter and showed a bimodal distribution with the lowest interaction around the TSS. As was the case for the PP1 isoforms, all three targeting subunits had chromosome-binding sites distributed across the entire genome (Figure 2E). Finally, some of the identified DamID targets, namely 5 SNORD target sequences for PNUTS (see below) as well as RPS6KC1 and ATF3 for NIPP1-WT were validated by ChIP (39).
Figure 2.
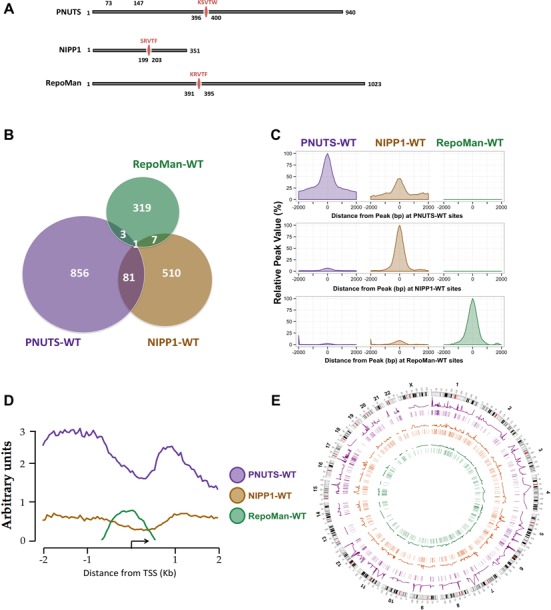
Promoter binding sites of nuclear PIPs. (A) Domain structure of the studied PIPs. (B) Venn diagram showing the significant and overlapping binding peaks of each PIP-WT on the promoter regions. The areas of the circles and overlaps are correlated to the number of peaks. (C) Global signal profiles of the PIPs-WT at −2/+2 kb of the peak centers. The percentages of the PIP-WT signals were calculated according to the highest signal found across the binding sites. The graphs were drawn using the R package ggplot2 (53). (D) The average raw signal profiles of the WT PIPs across all defined promoter regions. (E) Circos plot of the significant promoter binding peaks across the HeLa genome (54). The line profiles of the PIPs indicate the intensity of the PIP binding signal across their significant binding sites. The highlights underneath denote the position of the significant binding sites.
The promoter binding of NIPP1 is regulated by PP1
Mutation of the RVxF-motif (RVxF → RAxA) of PNUTS, NIPP1 or RepoMan abolished their binding to PP1 (Supplementary Figure S4C). To examine the dependency of the promoter binding of these PIPs on associated PP1, we compared the DamID profiles of the Dam-PIP-WT fusions and the corresponding PP1-binding mutants (M). For PNUTS-M the number of significant binding sites was about three times lower than that of PNUTS-WT (313 versus 941; Figure 3A, left panel). However, the percentage of overlapping binding sites between the two fusions was very high (79%). We also computed the DamID signals within a region of 2 kb at either side of the midpoint peaks at both the PNUTS-WT and PNUTS-M binding sites. The signals were normalized to the highest value, which was set at 100%. We found that the average signal profiles and strengths of PNUTS-WT and PNUTS-M were virtually identical at the interaction sites of either the WT or M proteins (Figure 3A, middle panel), suggesting similar promoter-binding of the WT and M proteins. This was also evident from signal profiles at individual promoter regions (Figure 3A, right panel). Taken together, these data indicate that PP1 is not essential for the promoter targeting of PNUTS.
Figure 3.
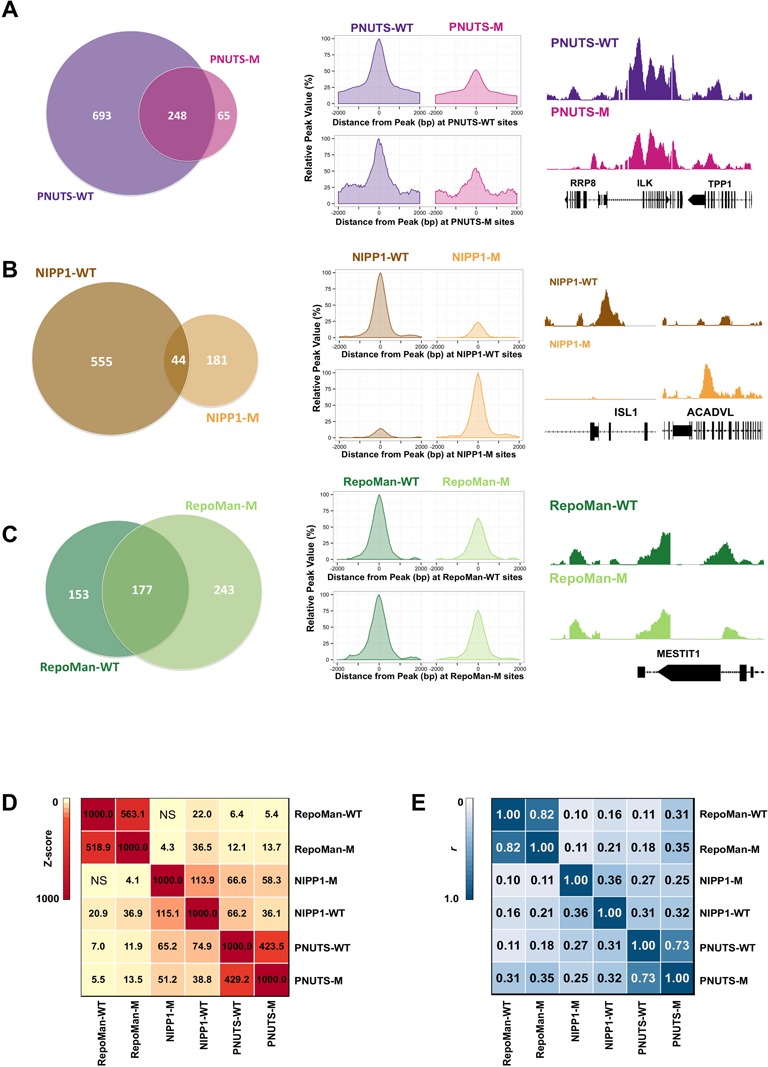
PP1 dependency of the promoter binding of PNUTS, NIPP1 and RepoMan. (A) Venn diagrams representing the overlap of the significant promoter binding peaks between PNUTS-WT and PNUTS-M (left panel). Global signal profiles of PNUTS-WT and PNUTS-M across each others’ binding sites (middle panel). The percentages of the signals were calculated according to the highest signal found across each binding site. A representative peak profile was obtained using the Integrated Genome Browser (IGB 7.0.1), showing the significant peak overlap of PNUTS-WT and PNUTS-M (right panel). The peak profiles for PNUTS-WT and PNUTS-M are on the same scale and the graphs were drawn using the R package ggplot2 (53). (B) Signal profiles for NIPP1-WT and NIPP1-M, as detailed for panel A. (C) Signal profiles for RepoMan-WT and RepoMan-M, as detailed for panel A. (D) Co-association analysis between the significant binding sites of the WT and M variants of the PIPs. Both the color and numbers indicate the Z-score. The binding sites were subjected to genome structure correction and only the promoter regions were used in the analysis (44). NS, not significant (P > 0.001). (E) Correlation analysis between the signal profiles of the R-subunits across the promoter regions. Both the color and the number indicate the correlation co-efficient (r). The correlation co-efficients were calculated using the ‘Multiple wiggle files correlation in given regions (version 1.0.0)’ on the Cistrome-Galaxy portal (42).
The percentage of overlapping binding sites for NIPP1-WT and NIPP1-M only amounted to 20%, indicating that PP1 affects the promoter binding specificity of NIPP1 (Figure 3B, left panel). Consistent with this notion, the signal strength of NIPP1-WT dropped to background levels at the interaction sites of NIPP1-M and vice versa (Figure 3B, middle and right panels). This agrees with previous data showing that NIPP1-WT and NIPP1-M bind to distinct chromatin loci (12,13). The promoter binding sites of RepoMan-WT and RepoMan-M also showed a considerable overlap (42%) (Figure 3C, left panel). The average signal profiles and strengths of RepoMan-WT and RepoMan-M were similar at the interaction sites of either the WT or M proteins (Figure 3C, middle and right panels). Thus, PP1 does not appear to be required for the promoter targeting of RepoMan, in accordance with a recent study showing that the histone binding of RepoMan in mid-mitosis is PP1 independent (15,56).
Next, we calculated the co-association between the significant binding sites of the WT PIPs and their PP1-binding mutants (Figure 3D). These calculations were based on the block_bootstrap.py script created by the ENCODE consortium (44) and compared the proximity of the binding sites of both proteins against the entire genome. The output is a Z-score, which is a measure of co-association. This analysis confirmed a strong link for both RepoMan and PNUTS between the WT proteins and their respective PP1-binding mutants. In contrast, NIPP1-WT correlated only weakly with NIPP1-M. A similar conclusion was drawn from the correlation co-efficients (r) of the raw signal profiles between the WT and M versions of the targeting proteins (Figure 3E). Collectively, these data confirm that NIPP1-WT and NIPP1-M largely bind to distinct loci. This contrasts with PNUTS and RepoMan where the WT and M variants occupy similar loci.
To validate our findings by an independent biochemical approach, we examined the effect of a knockdown of PP1 on the association of PNUTS, RepoMan and NIPP1 with the chromatin-enriched fraction (Supplementary Figure S5). The knockdown of PP1 did not affect the global association of these PIPs with chromatin. These data are consistent with our DamID data, in that they demonstrate that the global targeting of the examined PIPs is PP1-independent. At the same time, these data do not contradict our conclusion that PP1 regulates the promoter binding specificity of NIPP1.
Identification of promoter-associated PP1 holoenzymes
To find out whether PP1 isoforms co-localize with the examined PIPs, we determined the number of overlapping promoter binding sites. Importantly, the majority of PP1 interaction sites were not shared with any of the three examined PIPs (Figures 1F and 4A), pointing to an involvement of other, hitherto unidentified promoter-targeting subunits. However, we noted a striking overlap of 188 binding sites between PNUTS-WT and PP1β (Figure 4A), in accordance with a similar average signal profile of both proteins at the promoter region (Figures 1G and 2D). PNUTS-WT and PP1γ also shared 94 binding sites (Figure 4A), which is consistent with the overlap between the binding peaks of PP1β and PP1γ (Figure 1F). The co-localization of PP1 with PNUTS was observed at individual promoters (Supplementary Figure S6) as well as genome-wide in a Circos plot (Figure 4B). In contrast to PNUTS, RepoMan only had a handful of binding sites in common with PP1 isoforms (Figure 4A and B), in accordance with its PP1-independent chromatin targeting mechanism. The number of overlapping binding sites between NIPP1-WT and PP1 isoforms was intermediate to those of PNUTS and RepoMan (Figure 4A and B).
Figure 4.
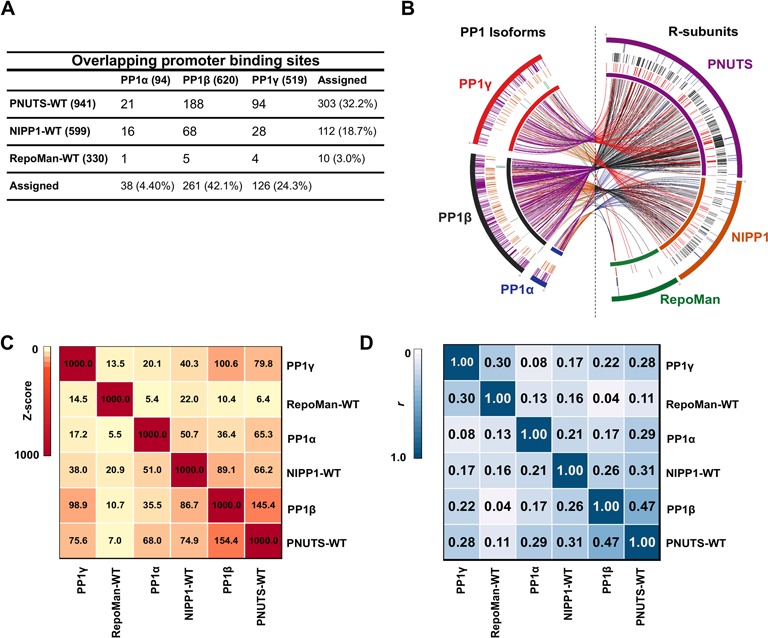
The promoter localization of PNUTS and PP1β are linked. (A) Table showing the overlap of significant promoter binding sites between the PP1 isoforms and PIPs as well as the total number of peaks for each protein overlapping with the PIPs, in the case of the PP1 isoforms, or the PP1 isoforms, in the case of the PIPs. Total number of binding sites are given in brackets next to the protein's name The overlaps were obtained using tools available on the Cistrome webportal (42). (B) Circos plot showing the interactions between the PP1 isoforms and the nuclear PIPs (54). The ‘chromosomes’ consists of all the binding sites of the particular Dam-fusion protein stitched together. The size of the chromosomes is therefore directly correlated with the amount and size of the binding sites of that particular protein on the promoter regions. Every interaction between the PP1 isoforms and PIPs was then mapped onto these chromosomes and the interactions are shown by links as well as the highlights. The colors correspond to the Dam-fusions making the interaction. (C) Co-association analysis between the significant binding sites of PP1 isoforms and nuclear PIPs. Both the color and the numbers indicate the Z-score. The binding sites were subjected to genome structure correction and only the promoter regions were used in the analysis. (D) Correlation analysis between the signal profiles of the PIPs and PP1 isoforms across the promoter regions. Both the color and the numbers indicate the correlation co-efficient (r).
To further explore the isoform binding specificity of the PIPs, we performed co-association and correlation analyses of both sets of proteins (Figure 4C and D). A clear association emerged between the binding sites of PNUTS-WT and PP1β, but not between the binding sites of the PP1 isoforms and either RepoMan-WT or NIPP1-WT. Taken together, our data show that (i) PP1β and PNUTS share many binding sites, (ii) RepoMan and PP1 bind to distinct loci and (iii) PP1 has numerous binding sites that are not shared by the examined PIPs.
PP1β-PNUTS is associated with RNA Polymerase-II transcribed genes
Next, we examined the nature and activity of the genes that are associated with the promoter regions that bound PP1 and/or its chromatin-associated PIPs. We first used publicly available ChIP-Seq datasets of the ENCODE consortium to calculate the signal correlation of our DamID datasets across the promoter region with the corresponding histone modifications and transcription factor (TF) binding sites (Supplementary Figure S7). The binding sites of PP1β and PNUTS were at least two-fold enriched for histone modifications that are associated with actively transcribed genes, including acetylated (ac) H3K9, H3K27ac and trimethylated (me3) H3K4. For PNUTS this applied to both the WT and M fusions, consistent with the view that PP1 does not affect the binding specificity of PNUTS. In contrast, the promoter-binding sites of RepoMan were not significantly enriched for any histone mark. Intriguingly, the histone landscapes of the binding sites of NIPP1-WT and NIPP1-M were very different, in accordance with their association with distinct subsets of promoters. Indeed, the NIPP1-M binding sites were significantly enriched for the active markers H3K9ac and H3K27ac, but this did not apply to NIPP1-WT. This agrees with previous findings that NIPP1-WT, but not NIPP1-M, regulates gene silencing through recruitment of the H3K27 methyltransferase EZH2 (13). Nevertheless, the NIPP1-WT binding sites were not significantly enriched for H3K27me3, which can be explained by published data showing that NIPP1-WT only regulates a subset of EZH2 targets (12,13) and also interacts with actively transcribed genes as a regulator of spliceosome assembly (23). This multifunctional role of NIPP1 impeded the further characterization of its gene targets in a genome-wide analysis.
Our analysis also explored correlations between promoter binding sites of PP1, the examined PIPs and various TFs (Figure 5A). The binding sites of PP1α, PP1γ, RepoMan and NIPP1 did not show a positive correlation with those of the examined TFs. If anything, the RepoMan binding sites were negatively correlated with TF binding sites, consistent with its weak promoter binding activity (Figure 2D) and lack of association with any histone modification (Supplementary Figure S7). In contrast, the binding sites of PP1β and PNUTS showed a positive correlation with most TF binding sites. This applied in particular to General Transcription Factor IIF subunit 1 (GTF2F1; r = 0.42), Brahma-related gene-1 (BRG1; r = 0.44), RNA polymerase II (Pol II; r = 0.40) and RNA polymerase II phosphorylated at Ser2 of its carboxyterminal domain (Pol II-pS2; r = 0.51) (Figure 5B). Similar values were obtained for PNUTS-WT and PNUTS-M. Subcellular localization analysis of pol II-pS2 and ectopically expressed EGFP-tagged fusions of the chromatin targeting subunits by confocal fluorescence microscopy confirmed the co-localization of PNUTS-WT and PNUTS-M with pol II-pS2 (Supplementary Figure S8). For these experiments TBP and α-tubulin were used as positive and negative controls, respectively. The RepoMan fusion did not show a significant co-localization with pol II-pS2, despite its similar diffuse nuclear distribution as PNUTS and therefore serves as an additional negative control. Finally, an interaction between PNUTS and RNA polymerase II was confirmed by co-immunoprecipitation of RNA polymerase II with EGFP-PNUTS-WT/M fusions (Figure 5C).
Figure 5.
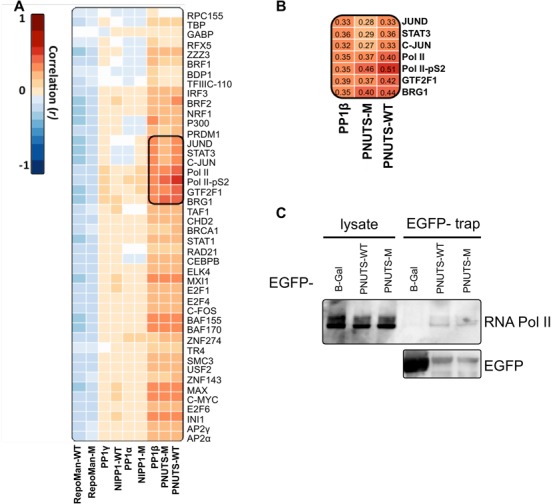
A PP1β/PNUTS holo-enzyme is associated with elongating RNA Polymerase II. (A) Correlation of the DamID signal profiles on the promoter regions and the corresponding transcription factor ChIP-Seq datasets from the ENCODE consortium. The correlation co-efficients were calculated using the ‘Multiple wiggle files correlation in given regions (version 1.0.0)’ found on the Cistrome web portal (42). White boxes indicate non-significant correlations (P > 0.01). (B) Fragment from panel A detailing the correlation coefficients of PP1β, PNUTS-WT and PNUTS-M with Pol II-pS2 and other significant co-associated transcription factors. (C) EGFP-tagged PNUTS-WT or PNUTS-M were transiently expressed in HEK293T cells. EGFP-traps from the cell lysates were immunoblotted for RNA polymerase II.
Out of 941 significant promoter-binding sites of PNUTS-WT (Figure 2B), 782 (83%) overlapped with binding sites of Pol II-pS2 (Figure 6A). Likewise, 175 out of the 188 PP1β-PNUTS holoenzyme-binding sites also bound Pol II-S2 (not shown), indicating that this PP1 holoenzyme is almost exclusively associated with the elongating RNA Polymerase-II complex. On the promoter regions where PNUTS and Pol II-pS2 had overlapping binding sites, Pol II-pS2 binding was, on average, four times above the background level (Figure 6A). These overlapping binding sites were also enriched for GTF2F1 (two-fold) and BRG1 (three-fold), two established interactors of RNA Polymerase II (57,58). Finally, we performed a gene ontology (GO) analysis, using the GREAT tool and filtered for redundant GO terms by REVIGO, to identify the key processes that are regulated by genes that bind both PP1β-PNUTS and Pol II-pS2 (Figure 6B). For this analysis we selected 167 genes that reside within 1 kb from the overlapping interactor sites (Supplementary Table S1). The most enriched gene ontology terms were metabolism and translation/transcription.
Figure 6.
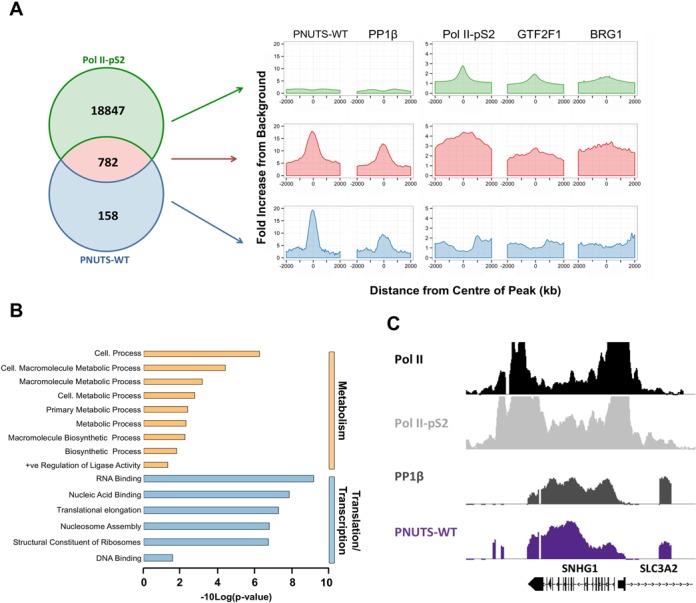
PP1β/PNUTS associates with Pol II-pS2. (A) Venn diagram showing binding site overlaps between PNUTS-WT and Pol II-pS2. The normalized signal profiles of PNUTS-WT, PP1β, Pol II-pS2, GTF2F1 and BRG1 across the binding sites of the (non)overlapping subsets are also included. The signal profiles were normalized using the average background signal of each protein across all the defined promoter regions. (B) Gene Ontology analysis of the genes that lie within 1 kb of regions where PNUTS-WT and Pol II-pS2 overlap. The analysis was performed using DAVID and the redundant terms were filtered out using the REVIGO tool (45,46). The bar chart represents the P-values of the enriched GO terms and are displayed as -10Log(p-value). (C) Representative peak profiles obtained using the Integrated Genome Browser (IGB 7.0.1) showing the overlapping signal profiles of RNA Pol II, Pol II-pS2, PP1β and PNUTS-WT across the SNHG1 gene. All the signal profiles are to scale.
Strikingly, the shared set of targeted genes included numerous genes that encode histones or snoRNAs (Supplementary Table S1), including the Small Nucleolar RNA Host Gene 1 (SNHG1) (Figure 6C), which all generate transcripts that are not adenylated at the 3′ end. ChIP experiments confirmed the association of PNUTS-WT/M with five snoRNA encoding SNORD genes (Supplementary Figure S9). Also, the knockdown of PNUTS reduced the expression of the SNORD and the histone encoding HIST1H3D genes (Figure 7A), whereas the knockdown of PP1β had the opposite effect (Figure 7B). These data functionally validate the DamID-identified targets of PNUTS and PP1β, and suggest that PP1β is an inhibitor of the transcriptional function of PNUTS.
Figure 7.
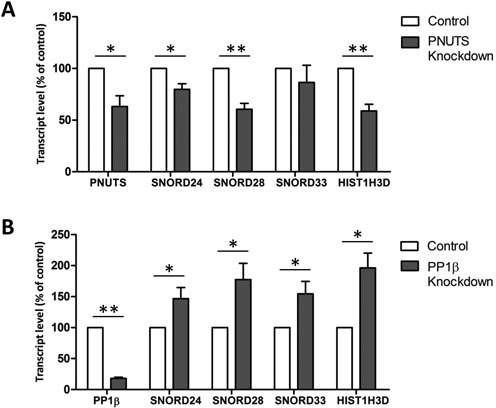
SNORD and HIST1H3D genes are regulated by PNUTS and PP1β (A) The relative transcript levels of SNORD24, SNORD28, SNORD33 and HIST1H3D were measured by qRT-PCR in HeLa cells after knockdown with control or PNUTS siRNA. HPRT was used for normalization and data are presented as percentages of the control ± S.E.M. (n ≥ 3). *, P < 0.05; **, P < 0.01 with the paired Student's t-test. (B) The relative transcript levels of SNORD24, SNORD28, SNORD33 and HIST1H3D were measured by qRT-PCR in HeLa cells after knockdown with control or PP1β siRNA. HPRT was used for normalization and the data are presented as percentages of the control ± S.E.M. (n ≥ 3). *, P < 0.05; **, P < 0.01 with the paired Student's t-test.
DISCUSSION
DamID profiling of PP1
ChIP and DamID represent the major prevailing techniques for the mapping of chromatin–protein interaction sites (31–36). Although they are based on different principles, they generate overlapping data when run in parallel (32). The choice for ChIP is to a large extent determined by the availability of antibodies against an epitope that is accessible in the context of cross-linked chromatin. The commercially available isoform-specific anti-PP1 antibodies can be used for immunoblotting but are not of ChIP grade (our unpublished data). Likewise, various commercial or homemade antibodies against NIPP1, PNUTS and RepoMan are not suited for ChIP experiments (our unpublished data) or give inconsistent results (12). Hence, we adopted the DamID protocol to map the promoter binding sites of PP1 isoforms and three major nuclear PIPs. For these DamID profiling studies we generated 20 stable HeLa cell lines, i.e. two independent cell lines for Dam and each of nine examined Dam-fusions. The DamID tool also enabled us to compare the promoter binding profiles of the WT and PP1 binding mutants of the examined PIPs.
Our DamID profiling studies identified hundreds of promoter binding sites of PP1 and the examined PIPs (Figures 1E and 2B). Various lines of evidence suggest that the results are reliable and biologically relevant. Firstly, we noted a large overlap (79%) between the binding sites of PNUTS-WT and PNUTS-M (Figure 3A), which can serve as an excellent illustration of the reproducibility of the DamID technique. It should be noted that this overlap, if anything, is underestimated since the binding sites of both PNUTS variants do not necessarily have to be identical. We also observed a huge overlap between the binding sites of Pol II-pS2, as mapped by ChIP, either PNUTS (83%) or PP1β-PNUTS (93%) (Figure 6A), as identified by DamID, confirming that both profiling techniques generate equivalent data. Secondly, we obtained very distinct promoter binding peaks and patterns for the PP1 isoforms, but also for the examined PIPs, indicating that DamID signals truly reflect binding affinities and can be detected throughout the promoter region (Figures 1G and 2D). Thirdly, our data are consistent with data from the literature in that they confirm that (i) of all isoforms PP1α is least associated with chromatin (49), (ii) NIPP1-WT and NIPP1-M have a distinct chromatin binding specificity (12,13) and (iii) PP1β-PNUTS is linked to RNA pol II on promoter regions (10).
Isoform specificity of the promoter targeting of PP1
In vitro most PIPs, including NIPP1, PNUTS and RepoMan, bind to PP1 in an isoform-nonspecific manner, which is explained by PP1-anchoring motifs, such as the RVxF motif, that dock to surface grooves that are identical in all PP1 isoforms (19–21). In intact cells, however, RepoMan preferentially interacts with PP1γ (21) and NIPP1 with PP1β (22). Our DamID profiling showed that PNUTS preferentially interacts with PP1β at promoters (Figure 4). Moreover, the observation that the PP1 isoforms have distinct promoter binding peaks and patterns is consistent with the notion that their targeting to promoters is largely mediated by different subsets of PIPs. This isoform selectivity is likely to be accounted for by isoform-specific docking motifs, which have already been identified in some PIPs. For example, the myosin targeting subunit MYPT1 has an ankyrin-repeat domain that specifically binds to the C-terminus of PP1β (5). Our data indicate that isoform-specific docking motifs or domains are much more prevalent than currently appreciated and play a key role in specifying the function of PP1 isoforms. Presumably, these isoform-specific docking motifs/domains do not bind with high affinity to PP1, which explains why they are difficult to detect in vitro in the presence of other, higher-affinity binding motifs, but are sufficient to tilt the balance towards the binding of a specific PP1 isoform in intact cells.
Cross-talk between PP1 and PIPs at promoters
A striking observation was that 73% of the identified PP1 promoter binding sites did not overlap with binding sites of NIPP1, PNUTS and/or RepoMan, demonstrating that the targeting of PP1 is largely mediated by other PIPs (Figure 4A and B). This was somewhat unexpected since NIPP1 and PNUTS together bind to a large fraction of PP1 in nuclear extracts, which did not include, however, the considerable fraction of PP1 that remains associated with chromatin in the presence of 0.3M NaCl (22). A number of additional chromatin-associated PIPs have already been identified (2), and it will be important to delineate their relative contribution to the promoter targeting of PP1. Conversely, numerous promoter-binding sites of NIPP1, PNUTS and RepoMan were not overlapping with PP1 binding sites. This is in accordance with our conclusion that the global chromatin targeting of these PIPs is PP1-independent (this work) and that PIPs exist in a large molar excess to PP1 and hence compete for binding to the limited pool of PP1 (2).
Our data have provided unexpected insights in the complex cross-talk between PP1 and the examined PIPs at promoters. RepoMan shared very few promoter-binding sites with PP1 (Figure 4A and B) and the binding specificities of RepoMan-WT and RepoMan-M were similar (Figure 3C–E). This suggests that RepoMan binds to promoters in a PP1-independent manner (Figure 8A), consistent with recent observations that a RepoMan fragment lacking the PP1-binding domain is still correctly targeted to histones (56,59). In contrast, PNUTS had numerous binding sites in common with PP1. Also, the binding specificities of PNUTS-WT and PNUTS-M were very similar but PNUTS-WT had about three times more significant binding sites (Figure 3A, D and E). One interpretation of this result is that PP1 somehow enhances the affinity of PNUTS for an important subset of its promoter binding sites. However, the global binding of PNUTS to chromatin was not affected by the knockdown of PP1 (Supplementary Figure S5). Moreover, PNUTS-WT and PNUTS-M showed the same binding to SNORD genes in ChIP experiments (Supplementary Figure S9). Therefore, the distinct number of chromatin-binding sites for PNUTS-WT and PNUTS-M (Figure 8A) possibly stems from different expression levels of the fusions. Finally, NIPP1 also shared many promoter-binding sites with PP1 (Figure 4C and D), but the binding specificities of NIPP1-WT and NIPP1-M were clearly different (Figure 3B, D and E), consistent with their embedment in a distinct histone landscape. This indicates that PP1 regulates the promoter binding specificity of NIPP1 (Figure 8B), in agreement with recent findings that NIPP1 regulates the chromatin targeting of EZH2 in a PP1-dependent manner (12,13).
Figure 8.
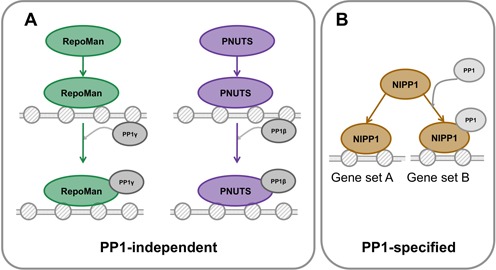
Model on the role of PP1 in the recruitment of PIPs to promoters. (A) The promoter binding sites of the WT and M-versions of RepoMan and PNUTS show a large overlap, indicating that their recruitment is PP1-independent. (B) NIPP1-WT and NIPP1-M bind to distinct subsets of promoters, showing that PP1 regulates the binding specificity of NIPP1.
Collectively, our data indicate that NIPP1, PNUTS and RepoMan fulfill PP1-independent functions at promoters, but that PP1 regulates the binding specificity of NIPP1. We did not find any direct evidence for a role of PP1 in the promoter targeting of PNUTS and RepoMan.
PP1β-PNUTS at active promoters
Our DamID profiling identified a PP1β-PNUTS holoenzyme that is associated with elongating RNA pol II at dozens of active promoters (Figure 5). This agrees with a recent report showing that PP1β-PNUTS regulates RNA pol II mediated transcription in Drosophila through dephosphorylation of the CTD domain of the largest subunit (10). PNUTS was only mapped to about 4% of the RNA-pol II-pS2 regulated promoters. This indicates that the promoter binding of PNUTS is limited to a subset of RNA-pol II regulated genes and/or that its binding to some promoters is too transient to be detected by DamID. In any case, in view of the large overlap between the promoter binding sites of PNUTS-WT and PNUTS-M (79%), it seems unlikely that this low percentage stems from non-saturation of the DamID profiling. Intriguingly, PP1β-PNUTS was enriched at the promoters of gene clusters that encode histones or snoRNAs (Figure 6B and C), which are generated from primary transcripts that do not undergo 3′ end polyadenylation. This leads to the enticing hypothesis that PP1β-PNUTS functions in the uncoupling of the cleavage and polyadenylation steps of 3′ RNA end-processing. Accordingly, PP1 has already been shown to be implicated in 3′ RNA-processing (60,61), and PNUTS is a component of a complex that regulates the processing of 3′ ends of RNA (61). We have confirmed by ChIP analysis that PNUTS is associated with SNORD genes (Supplementary Figure S9). Also, we have found that the expression of the SNORD and HIST1H3D genes is oppositely affected by the knockdown of PNUTS and PP1β (Figure 7A and B), indicating that PP1β downregulates the transcriptional activation of these genes by PNUTS.
In conclusion, we have successfully used the DamID approach to map promoter-binding sites of the PP1 isoforms and three nuclear PIPs in unperturbed, non-synchronized HeLa cells. The data disclosed an unexpected PP1 isoform binding specificity and showed that only a quarter of the promoter binding is mediated by NIPP1, PNUTS or RepoMan. Our data also revealed that PP1 autoregulates its own chromatin targeting by affecting the promoter binding specificity of NIPP1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
Annemie Hoogmartens, Nicole Sente, Gerd Van der Hoeven, Tine Jaspers and Fabienne Withof provided technical assistance.
FUNDING
Fund for Scientific Research-Flanders [G.0473.12, G.0482.12]; Prime Minister's office [IAP7/13]. Funding for open access charge: Fund for Scientific Research-Flanders [G.0473.12, G.0482.12].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bollen M., Peti W., Ragusa M.J., Beullens M. The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 2010;35:450–458. doi: 10.1016/j.tibs.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heroes E., Lesage B., Görnemann J., Beullens M., Van Meervelt L., Bollen M. The PP1 binding code: a molecular-lego strategy that governs specificity. FEBS J. 2013;280:584–595. doi: 10.1111/j.1742-4658.2012.08547.x. [DOI] [PubMed] [Google Scholar]
- 3.Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J., Pääbo S., Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck M., Schmidt A., Malmstroem J., Claassen M., Ori A., Szymborska A., Herzog F., Rinner O., Ellenberg J., Aebersold R. The quantitative proteome of a human cell line. Mol. Syst. Biol. 2011;7:549. doi: 10.1038/msb.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terrak M., Kerff F., Langsetmo K., Tao T., Dominguez R. Structural basis of protein phosphatase 1 regulation. Nature. 2004;429:780–784. doi: 10.1038/nature02582. [DOI] [PubMed] [Google Scholar]
- 6.Hurley T.D., Yang J., Zhang L., Goodwin K.D., Zou Q., Cortese M., Dunker A.K., DePaoli-Roach A.A. Structural basis for regulation of protein phosphatase 1 by inhibitor-2. J. Biol. Chem. 2007;282:28874–28883. doi: 10.1074/jbc.M703472200. [DOI] [PubMed] [Google Scholar]
- 7.Ragusa M.J., Dancheck B., Critton D.A., Nairn A.C., Page R., Peti W. Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat. Struct. Mol. Biol. 2010;17:459–464. doi: 10.1038/nsmb.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Connell N., Nichols S.R., Heroes E., Beullens M., Bollen M., Peti W., Page R. The molecular basis for substrate specificity of the nuclear NIPP1:PP1 holoenzyme. Structure. 2012;20:1746–1756. doi: 10.1016/j.str.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choy M.S., Hieke M., Kumar G.S., Lewis G.R., Gonzalez-Dewhitt K.R., Kessler R.P., Stein B.J., Hessenberger M., Nairn A.C., Peti W., et al. Understanding the antagonism of retinoblastoma protein dephosphorylation by PNUTS provides insights into the PP1 regulatory code. Proc. Natl. Acad. Sci. U.S.A. 2014;111:4097–4102. doi: 10.1073/pnas.1317395111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciurciu A., Duncalf L., Jonchere V., Lansdale N., Vasieva O., Glenday P., Rudenko A., Vissi E., Cobbe N., Alphey L., et al. PNUTS/PP1 regulates RNAPII-mediated gene expression and is necessary for developmental growth. PLoS Genet. 2013;9:e1003885. doi: 10.1371/journal.pgen.1003885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valin A., Ouyang J., Gill G. Transcription factor Sp3 represses expression of p21CIP1 via inhibition of productive elongation by RNA polymerase II. Mol. Cell. Biol. 2013;33:1582–1593. doi: 10.1128/MCB.00323-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minnebo N., Görnemann J., O'Connell N., Van Dessel N., Derua R., Vermunt M.W., Page R., Beullens M., Peti W., Van Eynde A., et al. NIPP1 maintains EZH2 phosphorylation and promoter occupancy at proliferation-related target genes. Nucleic Acids Res. 2013;41:842–854. doi: 10.1093/nar/gks1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Dessel N., Beke L., Görnemann J., Minnebo N., Beullens M., Tanuma N., Shima H., Van Eynde A., Bollen M. The phosphatase interactor NIPP1 regulates the occupancy of the histone methyltransferase EZH2 at Polycomb targets. Nucleic Acids Res. 2010;38:7500–7512. doi: 10.1093/nar/gkq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceulemans H., Bollen M. Functional diversity of protein phosphatase-1, a cellular economizer and reset button. Physiol. Rev. 2004;84:1–39. doi: 10.1152/physrev.00013.2003. [DOI] [PubMed] [Google Scholar]
- 15.Vagnarelli P., Ribeiro S., Sennels L., Sanchez-Pulido L., de Lima Alves F., Verheyen T., Kelly D.A., Ponting C.P., Rappsilber J., Earnshaw W.C. Repo-Man coordinates chromosomal reorganization with nuclear envelope reassembly during mitotic exit. Dev. Cell. 2011;21:328–342. doi: 10.1016/j.devcel.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng A., Lewellyn A.L., Schiemann W.P., Maller J.L. Repo-man controls a protein phosphatase 1-dependent threshold for DNA damage checkpoint activation. Curr. Biol. 2010;20:387–396. doi: 10.1016/j.cub.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landsverk H.B., Mora-Bermúdez F., Landsverk O.J., Hasvold G., Naderi S., Bakke O., Ellenberg J., Collas P., Syljuåsen R.G., Küntziger T. The protein phosphatase 1 regulator PNUTS is a new component of the DNA damage response. EMBO Rep. 2010;11:868–875. doi: 10.1038/embor.2010.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jerebtsova M., Klotchenko S.A., Artamonova T.O., Ammosova T., Washington K., Egorov V.V., Shaldzhyan A.A., Sergeeva M.V., Zatulovskiy E.A., Temkina O.A., et al. Mass spectrometry and biochemical analysis of RNA polymerase II: targeting by protein phosphatase-1. Mol. Cell. Biochem. 2011;347:79–87. doi: 10.1007/s11010-010-0614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allen P.B., Kwon Y.G., Nairn A.C., Greengard P. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J. Biol. Chem. 1998;273:4089–4095. doi: 10.1074/jbc.273.7.4089. [DOI] [PubMed] [Google Scholar]
- 20.Beullens M., Van Eynde A., Vulsteke V., Connor J., Shenolikar S., Stalmans W., Bollen M. Molecular determinants of nuclear protein phosphatase-1 regulation by NIPP-1. J. Biol. Chem. 1999;274:14053–14061. doi: 10.1074/jbc.274.20.14053. [DOI] [PubMed] [Google Scholar]
- 21.Trinkle-Mulcahy L., Andersen J., Lam Y.W., Moorhead G., Mann M., Lamond A.I. Repo-Man recruits PP1 gamma to chromatin and is essential for cell viability. J. Cell Biol. 2006;172:679–692. doi: 10.1083/jcb.200508154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jagiello I., Beullens M., Stalmans W., Bollen M. Subunit structure and regulation of protein phosphatase-1 in rat liver nuclei. J. Biol. Chem. 1995;270:17257–17263. doi: 10.1074/jbc.270.29.17257. [DOI] [PubMed] [Google Scholar]
- 23.Beullens M., Bollen M. The protein phosphatase-1 regulator NIPP1 is also a splicing factor involved in a late step of spliceosome assembly. J. Biol. Chem. 2002;277:19855–19860. doi: 10.1074/jbc.M200847200. [DOI] [PubMed] [Google Scholar]
- 24.Bounaix Morand du Puch C., Barbier E., Kraut A., Couté Y., Fuchs J., Buhot A., Livache T., Sève M., Favier A., Douki T., et al. TOX4 and its binding partners recognize DNA adducts generated by platinum anticancer drugs. Arch. Biochem. Biophys. 2011;507:296–303. doi: 10.1016/j.abb.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 25.De Leon G., Sherry T.C., Krucher N.A. Reduced expression of PNUTS leads to activation of Rb-phosphatase and caspase-mediated apoptosis. Cancer Biol. Ther. 2008;7:833–841. doi: 10.4161/cbt.7.6.5839. [DOI] [PubMed] [Google Scholar]
- 26.Kavela S., Shinde S.R., Ratheesh R., Viswakalyan K., Bashyam M.D., Gowrishankar S., Vamsy M., Pattnaik S., Rao S., Sastry R.A., et al. PNUTS functions as a proto-oncogene by sequestering PTEN. Cancer Res. 2013;73:205–214. doi: 10.1158/0008-5472.CAN-12-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krucher N.A., Rubin E., Tedesco V.C., Roberts M.H., Sherry T.C., De Leon G. Dephosphorylation of Rb (Thr-821) in response to cell stress. Exp. Cell Res. 2006;312:2757–2763. doi: 10.1016/j.yexcr.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Vietri M., Bianchi M., Ludlow J.W., Mittnacht S., Villa-Moruzzi E. Direct interaction between the catalytic subunit of Protein Phosphatase 1 and pRb. Cancer Cell Int. 2006;6:3. doi: 10.1186/1475-2867-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S.J., Lim C.J., Min J.K., Lee J.K., Kim Y.M., Lee J.Y., Won M.H., Kwon Y.G. Protein phosphatase 1 nuclear targeting subunit is a hypoxia inducible gene: its role in post-translational modification of p53 and MDM2. Cell Death Differ. 2007;14:1106–1116. doi: 10.1038/sj.cdd.4402111. [DOI] [PubMed] [Google Scholar]
- 30.Qian J., Lesage B., Beullens M., Van Eynde A., Bollen M. PP1/Repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Curr. Biol. 2011;21:766–773. doi: 10.1016/j.cub.2011.03.047. [DOI] [PubMed] [Google Scholar]
- 31.Vogel M.J., Peric-Hupkes D., van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- 32.Orian A. Chromatin profiling, DamID and the emerging landscape of gene expression. Curr. Opin. Genet. Dev. 2006;16:157–164. doi: 10.1016/j.gde.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 33.van Steensel B., Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- 34.Southall T.D., Brand A.H. Chromatin profiling in model organisms. Brief. Funct. Genomic Proteomic. 2007;6:133–140. doi: 10.1093/bfgp/elm013. [DOI] [PubMed] [Google Scholar]
- 35.Haring M., Offermann S., Danker T., Horst I., Peterhansel C., Stam M. Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park P.J. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesage B, Beullens M., Nuytten M., Van Eynde A., Keppens S., Himpens B., Bollen M. Interactor-mediated nuclear translocation and retention of protein phosphatase-1. J. Biol. Chem. 2004;279:55978–55984. doi: 10.1074/jbc.M411911200. [DOI] [PubMed] [Google Scholar]
- 38.Méndez J., Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nuytten M., Beke L., Van Eynde A., Ceulemans H., Beullens M., Van Hummelen P., Fuks F., Bollen M. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene. 2008;27:1449–1460. doi: 10.1038/sj.onc.1210774. [DOI] [PubMed] [Google Scholar]
- 40.Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Göke J., Chan Y.S., Yan J., Vingron M., Ng H.H. Genome-wide kinase-chromatin interactions reveal the regulatory network of ERK signaling in human embryonic stem cells. Mol. Cell. 2013;50:844–855. doi: 10.1016/j.molcel.2013.04.030. [DOI] [PubMed] [Google Scholar]
- 42.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y., et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin H., Liu T., Manrai A.K., Liu X.S. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 44.Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M., Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 46.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolte S., Cordelières F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- 48.Costes S.V., Daelemans D., Cho E.H., Dobbin Z., Pavlakis G., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trinkle-Mulcahy L., Sleeman J.E., Lamond A.I. Dynamic targeting of protein phosphatase 1 within the nuclei of living mammalian cells. J. Cell Sci. 2001;114:4219–4228. doi: 10.1242/jcs.114.23.4219. [DOI] [PubMed] [Google Scholar]
- 50.Haneji T., Morimoto H., Morimoto Y., Shirakawa S., Kobayashi S., Kaneda C., Shima H., Nagao M. Subcellular localization of protein phosphatase type 1 isotypes in mouse osteoblastic cells. Biochem. Biophys. Res. Commun. 1998;248:39–43. doi: 10.1006/bbrc.1998.8913. [DOI] [PubMed] [Google Scholar]
- 51.Morimoto H., Okamura H., Haneji T. Interaction of protein phosphatase 1 delta with nucleolin in human osteoblastic cells. J. Histochem. Cytochem. 2002;50:1187–1193. doi: 10.1177/002215540205000905. [DOI] [PubMed] [Google Scholar]
- 52.Gräff J., Koshibu K., Jouvenceau A., Dutar P., Mansuy I.M. Protein phosphatase 1-dependent transcriptional programs for long-term memory and plasticity. Learn. Mem. 2010;17:355–363. doi: 10.1101/lm.1766510. [DOI] [PubMed] [Google Scholar]
- 53.Wickham H. ggplot2: elegant graphics for data analysis. NY: Springer; 2009. [Google Scholar]
- 54.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jagiello I., Van Eynde A., Vulsteke V., Beullens M., Boudrez A., Keppens S., Stalmans W., Bollen M. Nuclear and subnuclear targeting sequences of the protein phosphatase-1 regulator NIPP1. J. Cell Sci. 2000;113:3761–3768. doi: 10.1242/jcs.113.21.3761. [DOI] [PubMed] [Google Scholar]
- 56.Qian J., Beullens M., Lesage B., Bollen M. Aurora B defines its own chromosomal targeting by opposing the recruitment of the phosphatase scaffold Repo-Man. Curr. Biol. 2013;23:1136–1143. doi: 10.1016/j.cub.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 57.Cho H., Orphanides G., Sun X., Yang X.J., Ogryzko V., Lees E., Nakatani Y., Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol. Cell. Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naito M., Zager R.A., Bomsztyk K. BRG1 increases transcription of proinflammatory genes in renal ischemia. J. Am. Soc. Nephrol. 2009;20:1787–1796. doi: 10.1681/ASN.2009010118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vagnarelli P. Chromatin reorganization through mitosis. Adv. Protein Chem. Struct. Biol. 2013;90:179–224. doi: 10.1016/B978-0-12-410523-2.00006-7. [DOI] [PubMed] [Google Scholar]
- 60.He X., Moore C. Regulation of yeast mRNA 3′ end processing by phosphorylation. Mol. Cell. 2005;19:619–629. doi: 10.1016/j.molcel.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 61.Shi Y., Di Giammartino D.C., Taylor D., Sarkeshik A., Rice W.J., Yates J.R., Frank J., Manley J.L. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell. 2009;33:365–376. doi: 10.1016/j.molcel.2008.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


