Abstract
Double-stranded RNA (dsRNA)-specific adenosine deaminase (DRADA) has been implicated as an enzyme responsible for the editing of RNA transcripts encoding glutamate-gated ion channel subunits (GLuR) in brain. In one case, the editing alters the gene-encoded glutamine (Q) to an arginine (R) located within the channel-forming domain of the alpha-amino-3-hydroxy-5-methyl-isoxazole-4-propionic acid (AMPA) receptor subunit GLuR-B. The result of editing at this site, called the 'Q/R' site, is a profound alteration of the Ca2+ permeability of the GLuR channel. Using recombinantly expressed DRADA proteins, we now demonstrate in vitro that DRADA is indeed involved in editing of the GLuR-B RNA. In addition to the formation of an RNA duplex structure involving exon and intron sequences, Q/R site-selective editing by DRADA also requires a cofactor protein(s) commonly present even in non-neuronal cells. The accuracy and efficiency of this RNA editing system appear to be determined by the quantitative balance between DRADA, cofactor and substrate GLuR-B RNA.
Full text
PDF







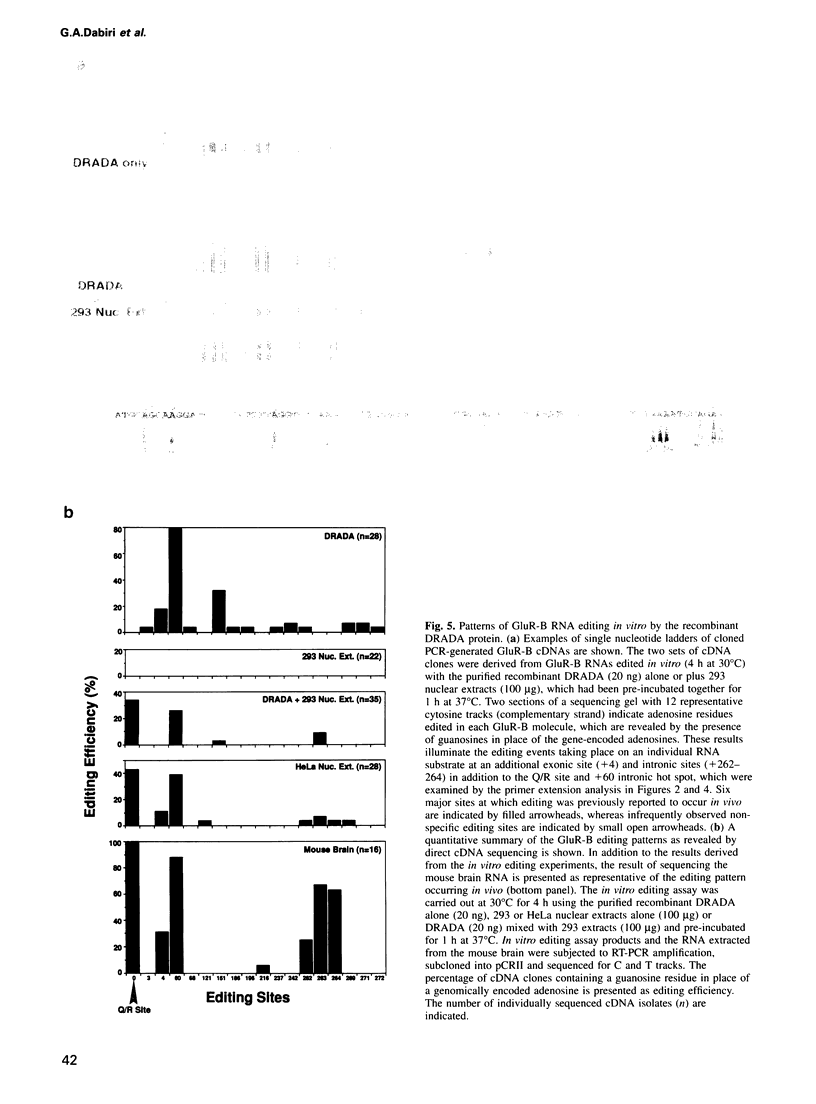



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BASILIO C., WAHBA A. J., LENGYEL P., SPEYER J. F., OCHOA S. Synthetic polynucleotides and the amino acid code. V. Proc Natl Acad Sci U S A. 1962 Apr 15;48:613–616. doi: 10.1073/pnas.48.4.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass B. L. RNA editing. An I for editing. Curr Biol. 1995 Jun 1;5(6):598–600. doi: 10.1016/s0960-9822(95)00119-9. [DOI] [PubMed] [Google Scholar]
- Bass B. L., Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988 Dec 23;55(6):1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Betts L., Xiang S., Short S. A., Wolfenden R., Carter C. W., Jr Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J Mol Biol. 1994 Jan 14;235(2):635–656. doi: 10.1006/jmbi.1994.1018. [DOI] [PubMed] [Google Scholar]
- Burnashev N., Monyer H., Seeburg P. H., Sakmann B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron. 1992 Jan;8(1):189–198. doi: 10.1016/0896-6273(92)90120-3. [DOI] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egebjerg J., Kukekov V., Heinemann S. F. Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10270–10274. doi: 10.1073/pnas.91.22.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995 Sep 8;270(36):20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- Higuchi M., Single F. N., Köhler M., Sommer B., Sprengel R., Seeburg P. H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993 Dec 31;75(7):1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Hollmann M., Hartley M., Heinemann S. Ca2+ permeability of KA-AMPA--gated glutamate receptor channels depends on subunit composition. Science. 1991 May 10;252(5007):851–853. doi: 10.1126/science.1709304. [DOI] [PubMed] [Google Scholar]
- Kim U., Garner T. L., Sanford T., Speicher D., Murray J. M., Nishikura K. Purification and characterization of double-stranded RNA adenosine deaminase from bovine nuclear extracts. J Biol Chem. 1994 May 6;269(18):13480–13489. [PubMed] [Google Scholar]
- Kim U., Nishikura K. Double-stranded RNA adenosine deaminase as a potential mammalian RNA editing factor. Semin Cell Biol. 1993 Aug;4(4):285–293. doi: 10.1006/scel.1993.1034. [DOI] [PubMed] [Google Scholar]
- Kim U., Wang Y., Sanford T., Zeng Y., Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M., Burnashev N., Sakmann B., Seeburg P. H. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993 Mar;10(3):491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Lai F., Drakas R., Nishikura K. Mutagenic analysis of double-stranded RNA adenosine deaminase, a candidate enzyme for RNA editing of glutamate-gated ion channel transcripts. J Biol Chem. 1995 Jul 21;270(29):17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- Lomeli H., Mosbacher J., Melcher T., Höger T., Geiger J. R., Kuner T., Monyer H., Higuchi M., Bach A., Seeburg P. H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994 Dec 9;266(5191):1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Melcher T., Maas S., Higuchi M., Keller W., Seeburg P. H. Editing of alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J Biol Chem. 1995 Apr 14;270(15):8566–8570. doi: 10.1074/jbc.270.15.8566. [DOI] [PubMed] [Google Scholar]
- Navaratnam N., Bhattacharya S., Fujino T., Patel D., Jarmuz A. L., Scott J. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 1995 Apr 21;81(2):187–195. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- Nishikura K., Yoo C., Kim U., Murray J. M., Estes P. A., Cash F. E., Liebhaber S. A. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991 Nov;10(11):3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell M. A., Krause S., Higuchi M., Hsuan J. J., Totty N. F., Jenny A., Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995 Mar;15(3):1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschen W., Hedreen J. C., Ross C. A. RNA editing of the glutamate receptor subunits GluR2 and GluR6 in human brain tissue. J Neurochem. 1994 Nov;63(5):1596–1602. doi: 10.1046/j.1471-4159.1994.63051596.x. [DOI] [PubMed] [Google Scholar]
- Polson A. G., Bass B. L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994 Dec 1;13(23):5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polson A. G., Crain P. F., Pomerantz S. C., McCloskey J. A., Bass B. L. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991 Dec 10;30(49):11507–11514. doi: 10.1021/bi00113a004. [DOI] [PubMed] [Google Scholar]
- Ruano D., Lambolez B., Rossier J., Paternain A. V., Lerma J. Kainate receptor subunits expressed in single cultured hippocampal neurons: molecular and functional variants by RNA editing. Neuron. 1995 May;14(5):1009–1017. doi: 10.1016/0896-6273(95)90339-9. [DOI] [PubMed] [Google Scholar]
- Rueter S. M., Burns C. M., Coode S. A., Mookherjee P., Emeson R. B. Glutamate receptor RNA editing in vitro by enzymatic conversion of adenosine to inosine. Science. 1995 Mar 10;267(5203):1491–1494. doi: 10.1126/science.7878468. [DOI] [PubMed] [Google Scholar]
- Saccomanno L., Bass B. L. The cytoplasm of Xenopus oocytes contains a factor that protects double-stranded RNA from adenosine-to-inosine modification. Mol Cell Biol. 1994 Aug;14(8):5425–5432. doi: 10.1128/mcb.14.8.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg P. H. The TINS/TiPS Lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993 Sep;16(9):359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- Skeiky Y. A., Iatrou K. Developmental regulation of covalent modification of double-stranded RNA during silkmoth oogenesis. J Mol Biol. 1991 Apr 5;218(3):517–527. doi: 10.1016/0022-2836(91)90698-6. [DOI] [PubMed] [Google Scholar]
- Smith H. C., Kuo S. R., Backus J. W., Harris S. G., Sparks C. E., Sparks J. D. In vitro apolipoprotein B mRNA editing: identification of a 27S editing complex. Proc Natl Acad Sci U S A. 1991 Feb 15;88(4):1489–1493. doi: 10.1073/pnas.88.4.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer B., Köhler M., Sprengel R., Seeburg P. H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991 Oct 4;67(1):11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown N. H., Gall J. G., Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoorn T. A., Burnashev N., Monyer H., Seeburg P. H., Sakmann B. Structural determinants of ion flow through recombinant glutamate receptor channels. Science. 1991 Jun 21;252(5013):1715–1718. doi: 10.1126/science.1710829. [DOI] [PubMed] [Google Scholar]
- Wagner R. W., Smith J. E., Cooperman B. S., Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R. W., Yoo C., Wrabetz L., Kamholz J., Buchhalter J., Hassan N. F., Khalili K., Kim S. U., Perussia B., McMorris F. A. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol Cell Biol. 1990 Oct;10(10):5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zeng Y., Murray J. M., Nishikura K. Genomic organization and chromosomal location of the human dsRNA adenosine deaminase gene: the enzyme for glutamate-activated ion channel RNA editing. J Mol Biol. 1995 Nov 24;254(2):184–195. doi: 10.1006/jmbi.1995.0610. [DOI] [PubMed] [Google Scholar]
- Yamanaka S., Poksay K. S., Balestra M. E., Zeng G. Q., Innerarity T. L. Cloning and mutagenesis of the rabbit ApoB mRNA editing protein. A zinc motif is essential for catalytic activity, and noncatalytic auxiliary factor(s) of the editing complex are widely distributed. J Biol Chem. 1994 Aug 26;269(34):21725–21734. [PubMed] [Google Scholar]
- Yang J. H., Sklar P., Axel R., Maniatis T. Editing of glutamate receptor subunit B pre-mRNA in vitro by site-specific deamination of adenosine. Nature. 1995 Mar 2;374(6517):77–81. doi: 10.1038/374077a0. [DOI] [PubMed] [Google Scholar]







