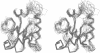Abstract
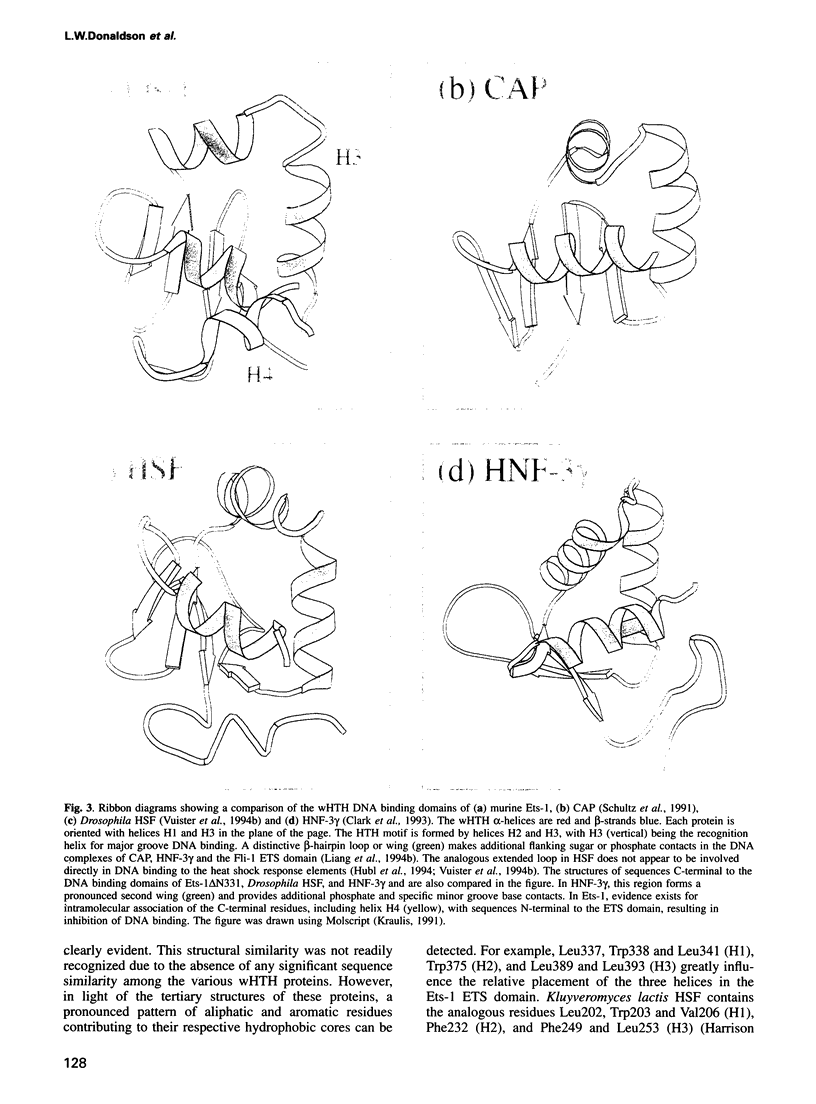
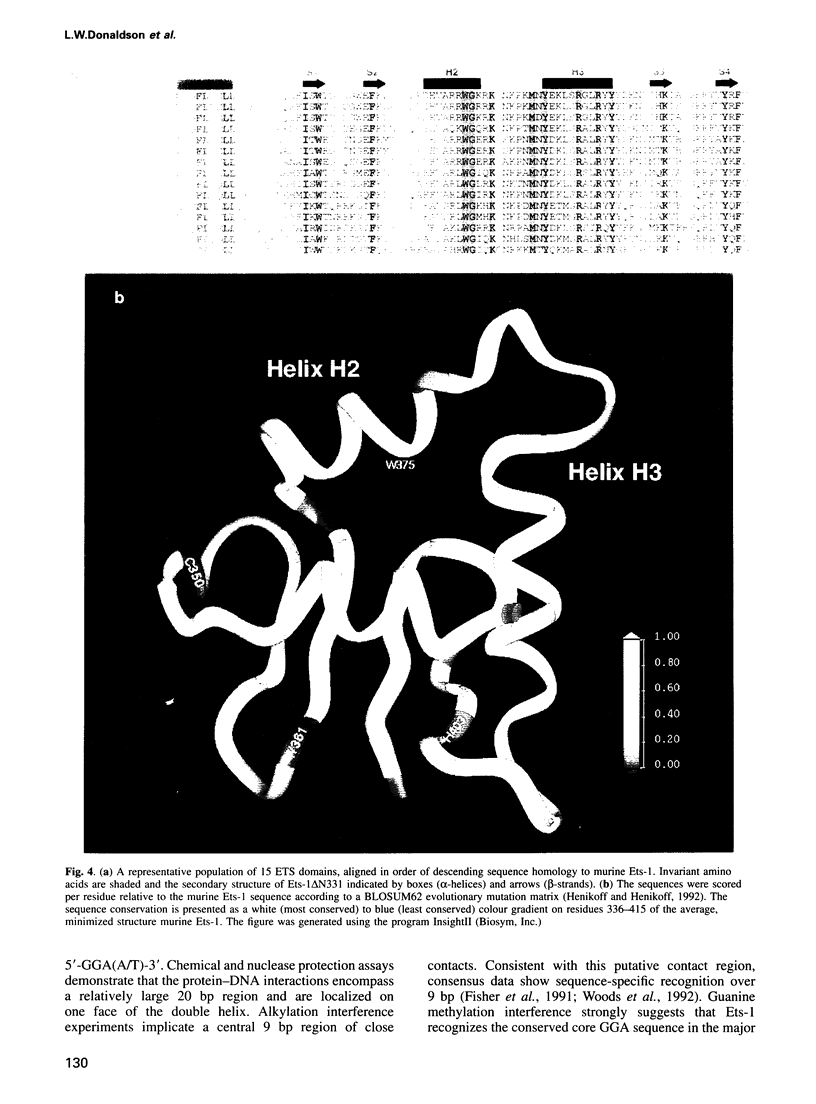
Ets-1 is the prototypic member of the ets family of transcription factors. This family is characterized by the conserved ETS domain that mediates specific DNA binding. Using NMR methods, we have determined the structure of a fragment of murine Ets-1 composed of the 85 residue ETS domain and a 25 amino acid extension that ends at its native C-terminus. The ETS domain folds into a helix-turn-helix motif on a four-stranded anti-parallel beta-sheet scaffold. This structure places Ets-1 in the winged helix-turn-helix (wHTH) family of DNA binding proteins and provides a model for interpreting the sequence conservation of the ETS domain and the specific interaction of Ets-1 with DNA. The C-terminal sequence of Ets-1, which is mutated in the v-Ets oncoprotein, forms an alpha-helix that packs anti-parallel to the N-terminal helix of the ETS domain. In this position, the C-terminal helix is poised to interact directly with an N-terminal inhibitory region in Ets-1 as well as the wHTH motif. This explains structurally the concerted role of residues flanking the ETS domain in the intramolecular inhibition of Ets-1 DNA binding.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bosselut R., Levin J., Adjadj E., Ghysdael J. A single amino-acid substitution in the Ets domain alters core DNA binding specificity of Ets1 to that of the related transcription factors Elf1 and E74. Nucleic Acids Res. 1993 Nov 11;21(22):5184–5191. doi: 10.1093/nar/21.22.5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan R. G. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell. 1993 Sep 10;74(5):773–776. doi: 10.1016/0092-8674(93)90456-z. [DOI] [PubMed] [Google Scholar]
- Bussiere D. E., Bastia D., White S. W. Crystal structure of the replication terminator protein from B. subtilis at 2.6 A. Cell. 1995 Feb 24;80(4):651–660. doi: 10.1016/0092-8674(95)90519-7. [DOI] [PubMed] [Google Scholar]
- Clark K. L., Halay E. D., Lai E., Burley S. K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993 Jul 29;364(6436):412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Clubb R. T., Omichinski J. G., Savilahti H., Mizuuchi K., Gronenborn A. M., Clore G. M. A novel class of winged helix-turn-helix protein: the DNA-binding domain of Mu transposase. Structure. 1994 Nov 15;2(11):1041–1048. doi: 10.1016/s0969-2126(94)00107-3. [DOI] [PubMed] [Google Scholar]
- Damberger F. F., Pelton J. G., Harrison C. J., Nelson H. C., Wemmer D. E. Solution structure of the DNA-binding domain of the heat shock transcription factor determined by multidimensional heteronuclear magnetic resonance spectroscopy. Protein Sci. 1994 Oct;3(10):1806–1821. doi: 10.1002/pro.5560031020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan B. M., Degnan S. M., Naganuma T., Morse D. E. The ets multigene family is conserved throughout the Metazoa. Nucleic Acids Res. 1993 Jul 25;21(15):3479–3484. doi: 10.1093/nar/21.15.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
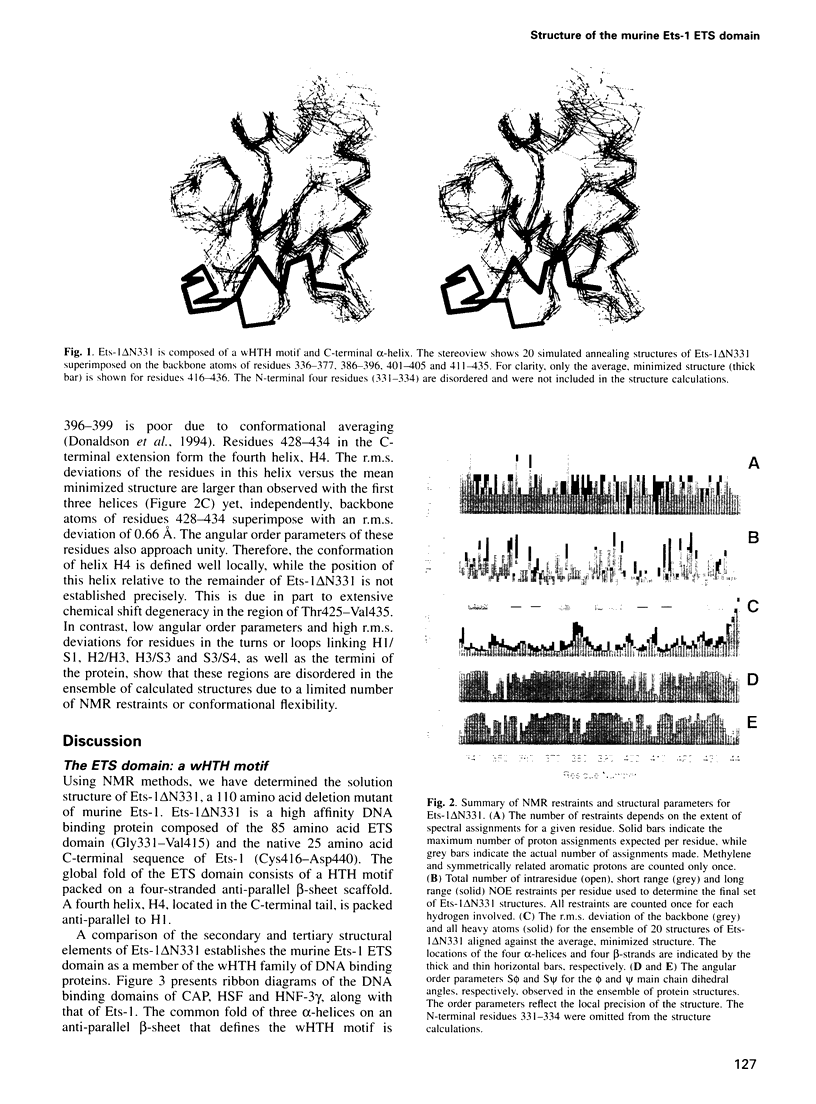
- Donaldson L. W., Petersen J. M., Graves B. J., McIntosh L. P. Secondary structure of the ETS domain places murine Ets-1 in the superfamily of winged helix-turn-helix DNA-binding proteins. Biochemistry. 1994 Nov 22;33(46):13509–13516. doi: 10.1021/bi00250a001. [DOI] [PubMed] [Google Scholar]
- Fisher R. J., Fivash M., Casas-Finet J., Erickson J. W., Kondoh A., Bladen S. V., Fisher C., Watson D. K., Papas T. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 1994 Feb;3(2):257–266. doi: 10.1002/pro.5560030210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. J., Mavrothalassitis G., Kondoh A., Papas T. S. High-affinity DNA-protein interactions of the cellular ETS1 protein: the determination of the ETS binding motif. Oncogene. 1991 Dec;6(12):2249–2254. [PubMed] [Google Scholar]
- Fogh R. H., Ottleben G., Rüterjans H., Schnarr M., Boelens R., Kaptein R. Solution structure of the LexA repressor DNA binding domain determined by 1H NMR spectroscopy. EMBO J. 1994 Sep 1;13(17):3936–3944. doi: 10.1002/j.1460-2075.1994.tb06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golay J., Introna M., Graf T. A single point mutation in the v-ets oncogene affects both erythroid and myelomonocytic cell differentiation. Cell. 1988 Dec 23;55(6):1147–1158. doi: 10.1016/0092-8674(88)90259-0. [DOI] [PubMed] [Google Scholar]
- Hagman J., Grosschedl R. An inhibitory carboxyl-terminal domain in Ets-1 and Ets-2 mediates differential binding of ETS family factors to promoter sequences of the mb-1 gene. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8889–8893. doi: 10.1073/pnas.89.19.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S. L., Wasylyk B. The oncoprotein v-Ets is less selective in DNA binding than c-Ets-1 due to the C-terminal sequence change. Oncogene. 1994 Sep;9(9):2499–2512. [PubMed] [Google Scholar]
- Harrison C. J., Bohm A. A., Nelson H. C. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994 Jan 14;263(5144):224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubl S. T., Owens J. C., Nelson H. C. Mutational analysis of the DNA-binding domain of yeast heat shock transcription factor. Nat Struct Biol. 1994 Sep;1(9):615–620. doi: 10.1038/nsb0994-615. [DOI] [PubMed] [Google Scholar]
- Hyberts S. G., Goldberg M. S., Havel T. F., Wagner G. The solution structure of eglin c based on measurements of many NOEs and coupling constants and its comparison with X-ray structures. Protein Sci. 1992 Jun;1(6):736–751. doi: 10.1002/pro.5560010606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R., Nordheim A. Gene regulation by Ets proteins. Biochim Biophys Acta. 1993 Dec 23;1155(3):346–356. doi: 10.1016/0304-419x(93)90014-4. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Zinck R., Ernst W. H., Nordheim A. Functional dissection of the transcription factor Elk-1. Oncogene. 1994 Apr;9(4):1273–1278. [PubMed] [Google Scholar]
- Karim F. D., Urness L. D., Thummel C. S., Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A., Gunther C. V., Nye J. A. The ETS-domain: a new DNA-binding motif that recognizes a purine-rich core DNA sequence. Genes Dev. 1990 Sep;4(9):1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Lautenberger J. A., Burdett L. A., Gunnell M. A., Qi S., Watson D. K., O'Brien S. J., Papas T. S. Genomic dispersal of the ets gene family during metazoan evolution. Oncogene. 1992 Sep;7(9):1713–1719. [PubMed] [Google Scholar]
- Lautenberger J. A., Papas T. S. Inversion of a chicken ets-1 proto-oncogene segment in avian leukemia virus E26. J Virol. 1993 Jan;67(1):610–612. doi: 10.1128/jvi.67.1.610-612.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince D., Crepieux P., Laudet V., Flourens A., Stehelin D. A new mechanism of oncogenic activation: E26 retroviral v-ets oncogene has inverted the C-terminal end of the transcription factor c-ets-1. Virology. 1993 Jun;194(2):855–857. doi: 10.1006/viro.1993.1330. [DOI] [PubMed] [Google Scholar]
- Leprince D., Crepieux P., Stehelin D. c-ets-1 DNA binding to the PEA3 motif is differentially inhibited by all the mutations found in v-ets. Oncogene. 1992 Jan;7(1):9–17. [PubMed] [Google Scholar]
- Leprince D., Gegonne A., Coll J., de Taisne C., Schneeberger A., Lagrou C., Stehelin D. A putative second cell-derived oncogene of the avian leukaemia retrovirus E26. Nature. 1983 Nov 24;306(5941):395–397. doi: 10.1038/306395a0. [DOI] [PubMed] [Google Scholar]
- Liang H., Mao X., Olejniczak E. T., Nettesheim D. G., Yu L., Meadows R. P., Thompson C. B., Fesik S. W. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat Struct Biol. 1994 Dec;1(12):871–875. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- Liang H., Olejniczak E. T., Mao X., Nettesheim D. G., Yu L., Thompson C. B., Fesik S. W. The secondary structure of the ets domain of human Fli-1 resembles that of the helix-turn-helix DNA-binding motif of the Escherichia coli catabolite gene activator protein. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11655–11659. doi: 10.1073/pnas.91.24.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim F., Kraut N., Framptom J., Graf T. DNA binding by c-Ets-1, but not v-Ets, is repressed by an intramolecular mechanism. EMBO J. 1992 Feb;11(2):643–652. doi: 10.1002/j.1460-2075.1992.tb05096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod K., Leprince D., Stehelin D. The ets gene family. Trends Biochem Sci. 1992 Jul;17(7):251–256. doi: 10.1016/0968-0004(92)90404-w. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Fisher R. J., Smyth F., Watson D. K., Papas T. S. Structural inferences of the ETS1 DNA-binding domain determined by mutational analysis. Oncogene. 1994 Feb;9(2):425–435. [PubMed] [Google Scholar]
- McIntosh L. P., Dahlquist F. W. Biosynthetic incorporation of 15N and 13C for assignment and interpretation of nuclear magnetic resonance spectra of proteins. Q Rev Biophys. 1990 Feb;23(1):1–38. doi: 10.1017/s0033583500005400. [DOI] [PubMed] [Google Scholar]
- McIntosh L. P., Wand A. J., Lowry D. F., Redfield A. G., Dahlquist F. W. Assignment of the backbone 1H and 15N NMR resonances of bacteriophage T4 lysozyme. Biochemistry. 1990 Jul 10;29(27):6341–6362. doi: 10.1021/bi00479a003. [DOI] [PubMed] [Google Scholar]
- Neri D., Szyperski T., Otting G., Senn H., Wüthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989 Sep 19;28(19):7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- Nilges M., Gronenborn A. M., Brünger A. T., Clore G. M. Determination of three-dimensional structures of proteins by simulated annealing with interproton distance restraints. Application to crambin, potato carboxypeptidase inhibitor and barley serine proteinase inhibitor 2. Protein Eng. 1988 Apr;2(1):27–38. doi: 10.1093/protein/2.1.27. [DOI] [PubMed] [Google Scholar]
- Nunn M. F., Hunter T. The ets sequence is required for induction of erythroblastosis in chickens by avian retrovirus E26. J Virol. 1989 Jan;63(1):398–402. doi: 10.1128/jvi.63.1.398-402.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn M. F., Seeburg P. H., Moscovici C., Duesberg P. H. Tripartite structure of the avian erythroblastosis virus E26 transforming gene. Nature. 1983 Nov 24;306(5941):391–395. doi: 10.1038/306391a0. [DOI] [PubMed] [Google Scholar]
- Nye J. A., Petersen J. M., Gunther C. V., Jonsen M. D., Graves B. J. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992 Jun;6(6):975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- Petersen J. M., Skalicky J. J., Donaldson L. W., McIntosh L. P., Alber T., Graves B. J. Modulation of transcription factor Ets-1 DNA binding: DNA-induced unfolding of an alpha helix. Science. 1995 Sep 29;269(5232):1866–1869. doi: 10.1126/science.7569926. [DOI] [PubMed] [Google Scholar]
- Pongubala J. M., Nagulapalli S., Klemsz M. J., McKercher S. R., Maki R. A., Atchison M. L. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3' enhancer activity. Mol Cell Biol. 1992 Jan;12(1):368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Verlinde C. L., Zhang S., Schmitt M. P., Holmes R. K., Hol W. G. Three-dimensional structure of the diphtheria toxin repressor in complex with divalent cation co-repressors. Structure. 1995 Jan 15;3(1):87–100. doi: 10.1016/s0969-2126(01)00137-x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993 Mar 18;362(6417):219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- Rost B., Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993 Jul 20;232(2):584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Soudant N., Albagli O., Dhordain P., Flourens A., Stéhelin D., Leprince D. A residue of the ETS domain mutated in the v-ets oncogene is essential for the DNA-binding and transactivating properties of the ETS-1 and ETS-2 proteins. Nucleic Acids Res. 1994 Sep 25;22(19):3871–3879. doi: 10.1093/nar/22.19.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T., Neri D., Leiting B., Otting G., Wüthrich K. Support of 1H NMR assignments in proteins by biosynthetically directed fractional 13C-labeling. J Biomol NMR. 1992 Jul;2(4):323–334. doi: 10.1007/BF01874811. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Brown T. A., McKnight S. L. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991 Aug 16;253(5021):762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- Vuister G. W., Kim S. J., Orosz A., Marquardt J., Wu C., Bax A. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol. 1994 Sep;1(9):605–614. [PubMed] [Google Scholar]
- Vuister G. W., Kim S. J., Wu C., Bax A. NMR evidence for similarities between the DNA-binding regions of Drosophila melanogaster heat shock factor and the helix-turn-helix and HNF-3/forkhead families of transcription factors. Biochemistry. 1994 Jan 11;33(1):10–16. doi: 10.1021/bi00167a002. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Hahn S. L., Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993 Jan 15;211(1-2):7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Kerckaert J. P., Wasylyk B. A novel modulator domain of Ets transcription factors. Genes Dev. 1992 Jun;6(6):965–974. doi: 10.1101/gad.6.6.965. [DOI] [PubMed] [Google Scholar]
- Wilson K. P., Shewchuk L. M., Brennan R. G., Otsuka A. J., Matthews B. W. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods D. B., Ghysdael J., Owen M. J. Identification of nucleotide preferences in DNA sequences recognised specifically by c-Ets-1 protein. Nucleic Acids Res. 1992 Feb 25;20(4):699–704. doi: 10.1093/nar/20.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang O., Kay L. E., Olivier J. P., Forman-Kay J. D. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994 Nov;4(6):845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]