Abstract
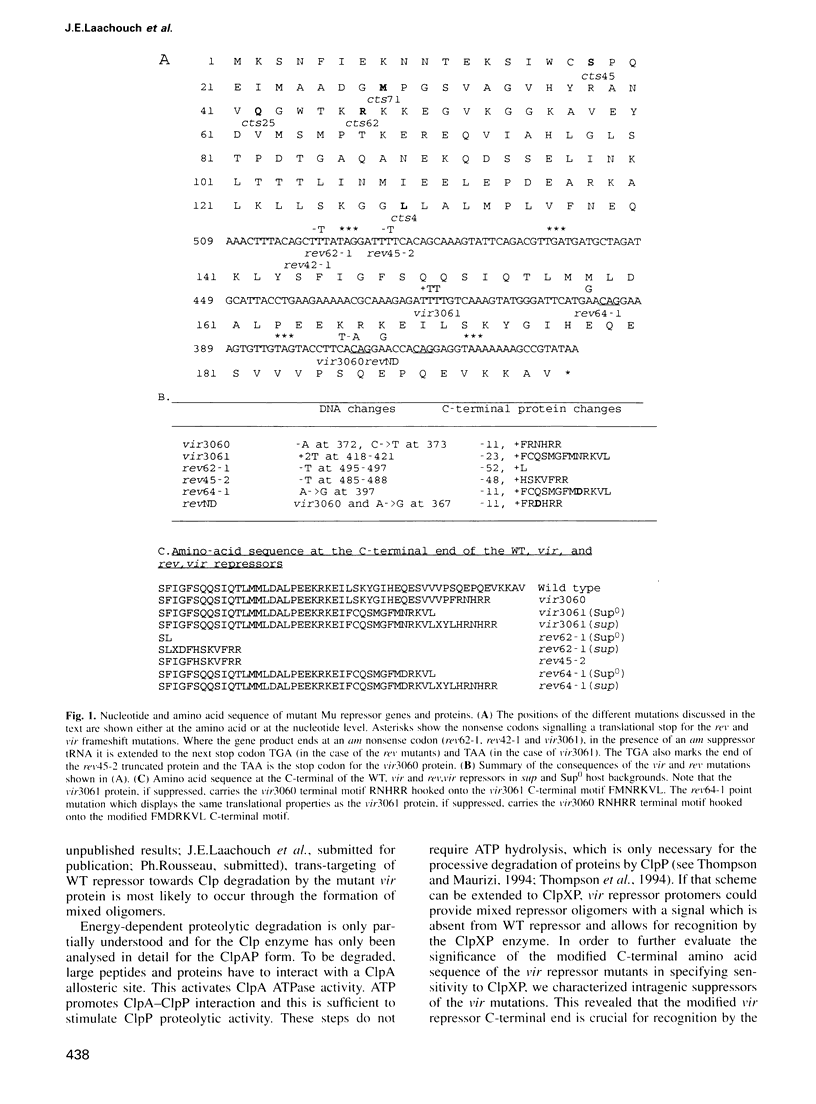
Bacteriophage Mu repressor, which is stable in its wildtype form, can mutate to become sensitive to its Escherichia coli host ATP-dependent ClpXP protease. We further investigated the determinants of the mutant repressor's sensitivity to Clp. We show the crucial importance of a C-terminal, seven amino acid long sequence in which a single change is sufficient to decrease the rate of degradation of the protein. The sequence was fused at the C-terminal end of the CcdB and CcdA proteins encoded by plasmid F. CcdB, which is naturally stable, was unaffected, while CcdA, which is normally degraded by the Lon protease, became a substrate for ClpXP while remaining a substrate for Lon. In agreement with the current hypothesis on the mechanism of recognition of their substrates by energy- dependent proteases, these results support the existence, on the substrate polypeptides, of separate motifs responsible for recognition and cleavage by the protease.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernard P., Couturier M. Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol. 1992 Aug 5;226(3):735–745. doi: 10.1016/0022-2836(92)90629-x. [DOI] [PubMed] [Google Scholar]
- Bernard P., Kézdy K. E., Van Melderen L., Steyaert J., Wyns L., Pato M. L., Higgins P. N., Couturier M. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J Mol Biol. 1993 Dec 5;234(3):534–541. doi: 10.1006/jmbi.1993.1609. [DOI] [PubMed] [Google Scholar]
- CAMPBELL A. Sensitive mutants of bacteriophage lambda. Virology. 1961 May;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chen S., Roseman A. M., Hunter A. S., Wood S. P., Burston S. G., Ranson N. A., Clarke A. R., Saibil H. R. Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature. 1994 Sep 15;371(6494):261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- Cheng H. H., Muhlrad P. J., Hoyt M. A., Echols H. Cleavage of the cII protein of phage lambda by purified HflA protease: control of the switch between lysis and lysogeny. Proc Natl Acad Sci U S A. 1988 Nov;85(21):7882–7886. doi: 10.1073/pnas.85.21.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuskens V., Mhammedi-Alaoui A., Desmet L., Toussaint A. Virulence in bacteriophage Mu: a case of trans-dominant proteolysis by the Escherichia coli Clp serine protease. EMBO J. 1992 Dec;11(13):5121–5127. doi: 10.1002/j.1460-2075.1992.tb05619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuskens V., Vogel J. L., Grimaud R., Desmet L., Higgins N. P., Toussaint A. Frameshift mutations in the bacteriophage Mu repressor gene can confer a trans-dominant virulent phenotype to the phage. J Bacteriol. 1991 Oct;173(20):6578–6585. doi: 10.1128/jb.173.20.6578-6585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S., Clark W. P., de Crecy-Lagard V., Maurizi M. R. ClpX, an alternative subunit for the ATP-dependent Clp protease of Escherichia coli. Sequence and in vivo activities. J Biol Chem. 1993 Oct 25;268(30):22618–22626. [PubMed] [Google Scholar]
- Gottesman S., Gottesman M., Shaw J. E., Pearson M. L. Protein degradation in E. coli: the lon mutation and bacteriophage lambda N and cII protein stability. Cell. 1981 Apr;24(1):225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- Gottesman S., Maurizi M. R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992 Dec;56(4):592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman C., Thévenet D., D'Ari R., Bouloc P. Degradation of sigma 32, the heat shock regulator in Escherichia coli, is governed by HflB. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3516–3520. doi: 10.1073/pnas.92.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Leach D., Symonds N. The isolation and characterisation of a plaque-forming derivative of bacteriophage Mu carrying a fragment of Tn3 conferring ampicillin resistance. Mol Gen Genet. 1979 May 4;172(2):179–184. doi: 10.1007/BF00268280. [DOI] [PubMed] [Google Scholar]
- Lehnherr H., Yarmolinsky M. B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc Natl Acad Sci U S A. 1995 Apr 11;92(8):3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I., Luo L., Baker T. A. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995 Oct 1;9(19):2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- Maurizi M. R. Proteases and protein degradation in Escherichia coli. Experientia. 1992 Feb 15;48(2):178–201. doi: 10.1007/BF01923511. [DOI] [PubMed] [Google Scholar]
- Mhammedi-Alaoui A., Pato M., Gama M. J., Toussaint A. A new component of bacteriophage Mu replicative transposition machinery: the Escherichia coli ClpX protein. Mol Microbiol. 1994 Mar;11(6):1109–1116. doi: 10.1111/j.1365-2958.1994.tb00387.x. [DOI] [PubMed] [Google Scholar]
- Mizusawa S., Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of sulA protein. Proc Natl Acad Sci U S A. 1983 Jan;80(2):358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J. M. Proteasomes: protein degradation machines of the cell. Trends Biochem Sci. 1994 Sep;19(9):377–382. doi: 10.1016/0968-0004(94)90115-5. [DOI] [PubMed] [Google Scholar]
- Roberts J. W., Roberts C. W. Proteolytic cleavage of bacteriophage lambda repressor in induction. Proc Natl Acad Sci U S A. 1975 Jan;72(1):147–151. doi: 10.1073/pnas.72.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. A role for the Clp protease in activating Mu-mediated DNA rearrangements. J Bacteriol. 1993 May;175(9):2625–2631. doi: 10.1128/jb.175.9.2625-2631.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. K., Maurizi M. R. Mutational analysis demonstrates different functional roles for the two ATP-binding sites in ClpAP protease from Escherichia coli. J Biol Chem. 1994 Nov 25;269(47):29537–29545. [PubMed] [Google Scholar]
- Squires C., Squires C. L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992 Feb;174(4):1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. W., Maurizi M. R. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J Biol Chem. 1994 Jul 8;269(27):18201–18208. [PubMed] [Google Scholar]
- Thompson M. W., Singh S. K., Maurizi M. R. Processive degradation of proteins by the ATP-dependent Clp protease from Escherichia coli. Requirement for the multiple array of active sites in ClpP but not ATP hydrolysis. J Biol Chem. 1994 Jul 8;269(27):18209–18215. [PubMed] [Google Scholar]
- Tobias J. W., Shrader T. E., Rocap G., Varshavsky A. The N-end rule in bacteria. Science. 1991 Nov 29;254(5036):1374–1377. doi: 10.1126/science.1962196. [DOI] [PubMed] [Google Scholar]
- Torres-Cabassa A. S., Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. J Bacteriol. 1987 Mar;169(3):981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L., Bernard P., Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol Microbiol. 1994 Mar;11(6):1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- Vogel J. L., Li Z. J., Howe M. M., Toussaint A., Higgins N. P. Temperature-sensitive mutations in the bacteriophage Mu c repressor locate a 63-amino-acid DNA-binding domain. J Bacteriol. 1991 Oct;173(20):6568–6577. doi: 10.1128/jb.173.20.6568-6577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawrzynow A., Wojtkowiak D., Marszalek J., Banecki B., Jonsen M., Graves B., Georgopoulos C., Zylicz M. The ClpX heat-shock protein of Escherichia coli, the ATP-dependent substrate specificity component of the ClpP-ClpX protease, is a novel molecular chaperone. EMBO J. 1995 May 1;14(9):1867–1877. doi: 10.1002/j.1460-2075.1995.tb07179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M. R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowiak D., Georgopoulos C., Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J Biol Chem. 1993 Oct 25;268(30):22609–22617. [PubMed] [Google Scholar]
- Yoo S. J., Seol J. H., Kang M. S., Ha D. B., Chung C. H. clpX encoding an alternative ATP-binding subunit of protease Ti (Clp) can be expressed independently from clpP in Escherichia coli. Biochem Biophys Res Commun. 1994 Sep 15;203(2):798–804. doi: 10.1006/bbrc.1994.2253. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]