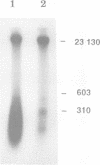
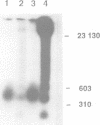
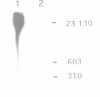Abstract
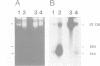
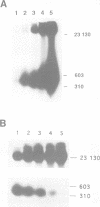
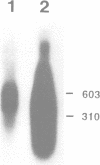
In mammalian cells DNA synthesis is more complicated than in prokaryotes and less well understood. Here we incubated intact mammalian cells (polyamine auxotrophic Chinese hamster ovary cells and primary human fibroblasts) with [32P]orthophosphate and found that, besides high molecular weight DNA, a species of low molecular weight DNA, approximately 450 bp in size, became efficiently labeled. The short DNA was labeled first, and in pulse-chase experiments the labeling was transient. The isolated small DNA fragments (RNase A-treated) were phosphorylated by T4 polynucleotide kinase specific for polynucleotides with 5'-OH ends. A polynucleotide kinase phosphorylating these DNA pieces was also detected in nuclear extracts of the cells. Treatment with alkaline phosphatase removed most of the 32P label incorporated into the small DNA in vivo. Labeling with deoxyribonucleosides did not reveal these fragments. We hypothesize that the low molecular weight DNA represents Okazaki fragments and that the mammalian DNA replication machinery includes a polynucleotide kinase phosphorylating the 5'-termini of Okazaki fragments. This would imply a novel step in DNA synthesis. We also show that depriving cells of polyamines reversibly blocks synthesis of high molecular weight DNA and leads to accumulation of the short DNA pieces, suggesting a role for polyamines in joining the Okazaki fragments.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burhans W. C., Selegue J. E., Heintz N. H. Replication intermediates formed during initiation of DNA synthesis in methotrexate-resistant CHOC 400 cells are enriched for sequences derived from a specific, amplified restriction fragment. Biochemistry. 1986 Jan 28;25(2):441–449. doi: 10.1021/bi00350a025. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Vassilev L. T., Caddle M. S., Heintz N. H., DePamphilis M. L. Identification of an origin of bidirectional DNA replication in mammalian chromosomes. Cell. 1990 Sep 7;62(5):955–965. doi: 10.1016/0092-8674(90)90270-o. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L., Wassarman P. M. Replication of eukaryotic chromosomes: a close-up of the replication fork. Annu Rev Biochem. 1980;49:627–666. doi: 10.1146/annurev.bi.49.070180.003211. [DOI] [PubMed] [Google Scholar]
- Heintz N. H., Stillman B. W. Nuclear DNA synthesis in vitro is mediated via stable replication forks assembled in a temporally specific fashion in vivo. Mol Cell Biol. 1988 May;8(5):1923–1931. doi: 10.1128/mcb.8.5.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölttä E., Pohjanpelto P. Control of ornithine decarboxylase in Chinese hamster ovary cells by polyamines. Translational inhibition of synthesis and acceleration of degradation of the enzyme by putrescine, spermidine, and spermine. J Biol Chem. 1986 Jul 15;261(20):9502–9508. [PubMed] [Google Scholar]
- Levin C. J., Zimmerman S. B. A deoxyribonucleic acid kinase from nuclei of rat liver. Purification and properties. J Biol Chem. 1976 Mar 25;251(6):1767–1774. [PubMed] [Google Scholar]
- Ogawa T., Okazaki T. Discontinuous DNA replication. Annu Rev Biochem. 1980;49:421–457. doi: 10.1146/annurev.bi.49.070180.002225. [DOI] [PubMed] [Google Scholar]
- Oredsson S. M., Nicander B., Heby O. Implications for a reduced DNA-elongation rate in polyamine-depleted cells. Eur J Biochem. 1990 Jul 5;190(3):483–489. doi: 10.1111/j.1432-1033.1990.tb15599.x. [DOI] [PubMed] [Google Scholar]
- Pohjanpelto P., Hölttä E. Deprivation of a single amino acid induces protein synthesis-dependent increases in c-jun, c-myc, and ornithine decarboxylase mRNAs in Chinese hamster ovary cells. Mol Cell Biol. 1990 Nov;10(11):5814–5821. doi: 10.1128/mcb.10.11.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjanpelto P., Hölttä E., Jänne O. A. Mutant strain of Chinese hamster ovary cells with no detectable ornithine decarboxylase activity. Mol Cell Biol. 1985 Jun;5(6):1385–1390. doi: 10.1128/mcb.5.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjanpelto P., Nordling S., Knuutila S. Flow cytometric analysis of the cell cycle in polyamine-depleted cells. Cytometry. 1994 Aug 1;16(4):331–338. doi: 10.1002/cyto.990160407. [DOI] [PubMed] [Google Scholar]
- Pohjanpelto P. Putrescine shortens the S-period in human fibroblasts. Biomedicine. 1975 Nov 10;23(9):350–352. [PubMed] [Google Scholar]
- Probst H., Hofstaetter T., Jenke H. S., Gentner P. R., Müller-Scholz D. Metabolism and non-random occurrence of nonnascent short chains in the DNA of Ehrlich ascites cells. Biochim Biophys Acta. 1983 Jun 24;740(2):200–211. doi: 10.1016/0167-4781(83)90078-7. [DOI] [PubMed] [Google Scholar]
- Tabor C. W., Tabor H. Polyamines. Annu Rev Biochem. 1984;53:749–790. doi: 10.1146/annurev.bi.53.070184.003533. [DOI] [PubMed] [Google Scholar]
- Tsurimoto T., Melendy T., Stillman B. Sequential initiation of lagging and leading strand synthesis by two different polymerase complexes at the SV40 DNA replication origin. Nature. 1990 Aug 9;346(6284):534–539. doi: 10.1038/346534a0. [DOI] [PubMed] [Google Scholar]
- Waga S., Bauer G., Stillman B. Reconstitution of complete SV40 DNA replication with purified replication factors. J Biol Chem. 1994 Apr 8;269(14):10923–10934. [PubMed] [Google Scholar]
- Waga S., Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. Nature. 1994 May 19;369(6477):207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]