Significance
The mitochondrial genomes of flowering plants are characterized by an extreme and often perplexing diversity in size, organization, and mutation rate, but their primary genetic function, in respiration, is extremely well conserved. Here we present the mitochondrial genome of an aerobic parasitic plant, the mistletoe Viscum scurruloideum. This genome is miniaturized, shows clear signs of rapid and degenerative evolution, and lacks all genes for complex I of the respiratory electron-transfer chain. To our knowledge, this is the first report of the loss of this key respiratory complex in any multicellular eukaryote. The Viscum mitochondrial genome has taken a unique overall tack in evolution that, to some extent, likely reflects the progression of a specialized parasitic lifestyle.
Keywords: mitogenome, mutation rate, complex I, parasitic plants, genome reduction
Abstract
Despite the enormous diversity among parasitic angiosperms in form and structure, life-history strategies, and plastid genomes, little is known about the diversity of their mitogenomes. We report the sequence of the wonderfully bizarre mitogenome of the hemiparasitic aerial mistletoe Viscum scurruloideum. This genome is only 66 kb in size, making it the smallest known angiosperm mitogenome by a factor of more than three and the smallest land plant mitogenome. Accompanying this size reduction is exceptional reduction of gene content. Much of this reduction arises from the unexpected loss of respiratory complex I (NADH dehydrogenase), universally present in all 300+ other angiosperms examined, where it is encoded by nine mitochondrial and many nuclear nad genes. Loss of complex I in a multicellular organism is unprecedented. We explore the potential relationship between this loss in Viscum and its parasitic lifestyle. Despite its small size, the Viscum mitogenome is unusually rich in recombinationally active repeats, possessing unparalleled levels of predicted sublimons resulting from recombination across short repeats. Many mitochondrial gene products exhibit extraordinary levels of divergence in Viscum, indicative of highly relaxed if not positive selection. In addition, all Viscum mitochondrial protein genes have experienced a dramatic acceleration in synonymous substitution rates, consistent with the hypothesis of genomic streamlining in response to a high mutation rate but completely opposite to the pattern seen for the high-rate but enormous mitogenomes of Silene. In sum, the Viscum mitogenome possesses a unique constellation of extremely unusual features, a subset of which may be related to its parasitic lifestyle.
Parasitism has evolved at least 12 or 13 times in angiosperms, with parasitic plants comprising about 1% of known angiosperm species (1, 2). Parasitic angiosperms vary enormously in size, life history, anatomy, physiological adaptation to a parasitic lifestyle, and nutritional dependence on host plants (3). Plastid genomes have been sequenced from many parasitic species and lineages and vary enormously in size and gene content, from those that are typical of nonparasitic photosynthetic plants to Rafflesia lagascae, which may not even have a plastid genome (4–6). Despite this great diversity, the mitogenomes of parasitic plants are largely unexplored. Most studies of parasitic plant mtDNAs have dealt primarily with their apparent propensity for uptake of foreign DNA via horizontal gene transfer (HGT). HGT is relatively common in angiosperm mitogenomes, and is especially common in parasitic plants, in which HGT is facilitated by the intimate physical connection between parasites and their host plant (1, 2, 7).
Rafflesiaceae, a small family of endophytic holoparasites famous for possessing the largest flowers in the world, is the only parasitic plant clade for which large-scale mitochondrial genomic sequencing has been undertaken (4, 8). Sequences of 38 genes common to three Rafflesiaceae species and two host species revealed that host-to-parasite HGT has been frequent in Rafflesiaceae mitogenomes, which otherwise are relatively unremarkable with respect to gene content and sequence divergence (8). Depending on the Rafflesiaceae species, 24–41% of protein genes are inferred to have been acquired by HGT. The repetitive nature of Rafflesiaceae mtDNAs and the short reads used in these studies rendered assembly of complete genome sequences impractical, but with a minimum size of 320 kb, the Rafflesia lagascae mitogenome (4) falls within the known angiosperm size range (0.2–11.3 Mb) (9, 10).
To help remedy the lack of knowledge of parasitic plant mitogenomes, we selected the Santalales for comparative sequencing for two reasons. First, the Santalales is the largest group of parasitic plants (with more than 2,000 parasitic species) and is also highly diverse (11, 12). This diversity is manifest in terms of autotrophic autonomy, with members of the order ranging from free-living nonparasitic trees to green, photosynthesizing hemiparasites to highly derived species that lack leaves and maintain only minimal photosynthesis at narrow points in the life cycle. The order also includes holoparasites (completely nonphotosynthetic heterotrophs) if further phylogenetic study confirms the tentative placement of the bizarre holoparasitic family Balanophoraceae as sister to, if not a member of, Santalales (2, 13). Another important aspect of this diversity is that Santalales have undergone multiple transitions from root parasitism (the ancestral parasitic condition in the group) to aerial parasitism, with these “mistletoes” representing the majority of aerial parasitic plants (14). Second, we recently showed (15) that the Santalales are one of the principal donors of foreign mtDNA to the amazingly HGT-rich mitogenome of the basal angiosperm Amborella trichopoda. Accordingly, we wished to determine the number and identity of the Santalales donors and, to the extent possible, the timing and mechanism of these transfers.
Here we present the mitogenome sequence of the hemiparasitic mistletoe Viscum scurruloideum. Unlike Rafflesiaceae mitochondrial genomes, this Viscum genome shows little evidence of HGT. However, it is highly unusual in a number of other ways, including genome size, mutation rate, selective pressure, levels of repeat-mediated recombination and sublimons, and the loss of many genes, including the entire suite of nine mitochondrial genes encoding respiratory complex I. We consider the implications of the unprecedented (for multicellular organisms) loss of complex I and Viscum’s host dependence.
Results and Discussion
The Smallest Angiosperm Mitochondrial Genome.
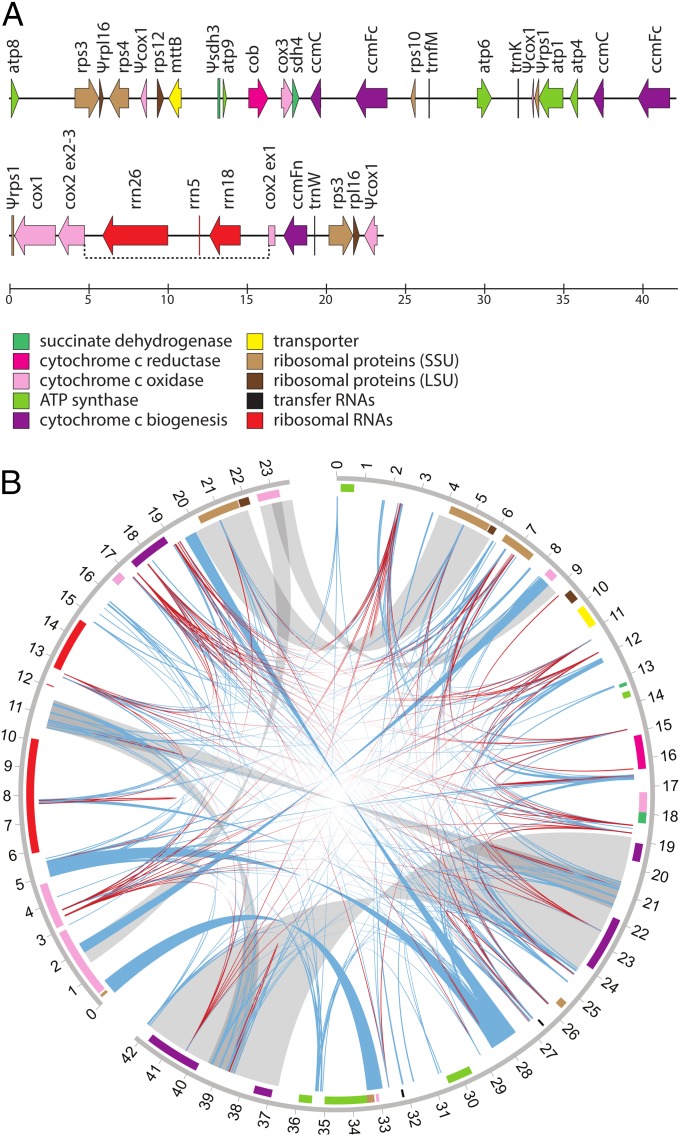
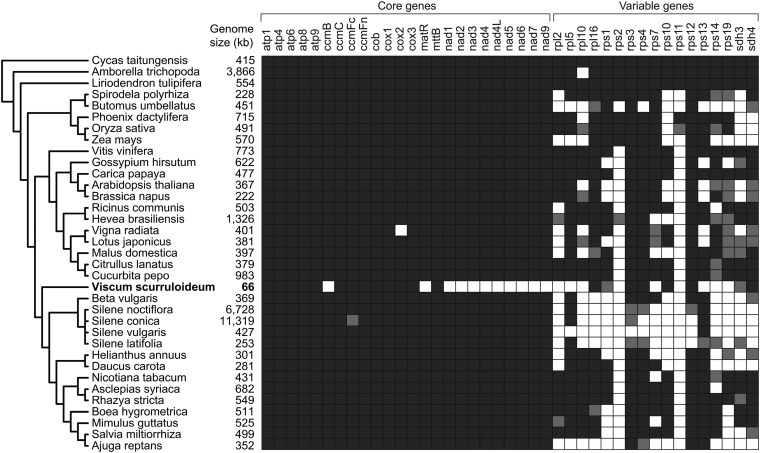
The Viscum mitogenome assembly consists of two contigs, a circular contig 42,186 bp in length and a linear contig 23,687 bp in length (Fig. 1A). High-depth contigs (>80×) were constructed from a short (100-bp) paired-end sequencing library (SI Appendix, Fig. S1). Searches for potentially missing mitochondrial sequences did not produce any similarly high-depth contigs (Materials and Methods). The Viscum genome was compared with 33 genomes chosen to sample the phylogenetic diversity of sequenced angiosperm mitogenomes (Fig. 2). With the exception of Silene, for which four species were included to capture the remarkable heterogeneity in genome size and mutation rate within the genus (9), only one species is represented per genus, and at most two per family are included. Angiosperm mitogenomes are exceptional for their large and variable sizes (9, 10). At 66 kb, Viscum is the smallest (by a factor of 3.3) angiosperm mitogenome sequenced to date (Fig. 2), is smaller than any other land plant genome (the next smallest is the moss Bauxbaumia at 101 kb), and among charophytes (the green algal ancestors of land plants) is closest in size to some of the smallest known genomes, those of Entransia (62 kb) and Chara (68 kb) in particular. Despite its small size, Viscum’s guanine-cytosine content of 47.4% is unremarkable compared with that of other angiosperms, which generally fall in the range of 43–45% (10).
Fig. 1.
The mitochondrial genome of V. scurruloideum. (A) Gene map, with gene orientation shown for all genes ≥200 nt in length. Note that the larger of the two contigs is circular but is shown as arbitrarily linearized. The dotted line indicates probable trans-splicing of the first intron in the cox2 gene. (B) Repeat map, for both contigs, with curved lines connecting pairs of repeats and with line width proportional to repeat size. Repeats larger than the library fragment size (800 bp) are in gray, repeats with one or more read pairs consistent with a rearrangement event across the repeat are in blue, and the remaining repeats are in red. Boxes on the outer ring mark the positions of genes, colored as in A. The numbers give genome coordinates in kilobases.
Fig. 2.
Genome size and protein-gene content of 34 sequenced angiosperm mitochondrial genomes and outgroup Cycas taitungensis. Intact genes are dark gray, pseudogenes are light gray, and absent genes are white. The phylogeny shown is the current best estimate of relationships among these plants (ref. 78 and references therein).
Between V. scurruloideum and the astoundingly large mitogenome of Silene conica (11.3 Mb), the known range of angiosperm mitogenome sizes is now on the order of 200-fold. In striking contrast to the highly reduced mitogenome of Viscum, the four species of Viscum examined thus far have exceptionally large nuclear genomes ranging in size from 2C = 122–201 Gb (16). These are the largest nuclear genome sizes reported for eudicots, a group that contains more than 70% of the ca. 350,000 species of angiosperms. No information is available on Viscum plastid genomes.
Dramatically Reduced Core Coding Capacity and Intron Content.
Angiosperm mitogenomes virtually always share the same set of 24 core protein genes, mostly coding for respiratory proteins, but often differ substantially in their inventories of a further 17 protein genes mostly encoding ribosomal proteins (17, 18). Three species of Silene possess the most protein-gene–poor angiosperm mitogenomes described to date (9). Two of these species share the entire core set of 24 protein genes, and the third contains 23 of these genes, with its loss of ccmFc the only known case of such loss in angiosperms. Conversely, the three Silenes have lost all but one or two of the 17 genes comprising the variable set (Fig. 2). The Viscum mitogenome presents a dramatic departure with respect to the core genes. It has lost the entirety of 11 of the 24 core protein genes. These losses include ccmB, matR, and all nine nad genes (nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7, and nad9) (Fig. 2). To check for the possibility that these genes fall within an assembled portion of mtDNA, total genomic reads were searched for evidence of homology with these genes using BLAST. Because 95% of our mitochondrial assembly has a read depth greater than 80 (SI Appendix, Fig. S1), we expect mitochondrial reads matching these genes also to give depths greater than 80. Although eight of these 11 genes yielded a smattering of matches, the low read depths do not support the conclusion that these reads are of mitochondrial origin (SI Appendix, SI Discussion). Because the loss of these three classes of genes is highly unexpected—all 11 genes are found in all 300+ other angiosperm mitogenomes examined (Fig. 2) (10, 17)—they are discussed in separate sections below. The Viscum genome also has lost 11 of the 17 variably present protein genes, with nine of these genes missing entirely and the other two present as obvious pseudogenes (Fig. 2). Altogether, only 19 intact and potentially functional protein genes were identified in the genome assembly. This protein-coding capacity is the smallest identified in an angiosperm mitogenome, five genes fewer than the already highly reduced capacity in Silene.
Three sequences were identified that could be folded into the typical tRNA structure (SI Appendix, Fig. S2). These sequences, which correspond to trnW(cca), trnfM (cau), and trnK (uuu), have well-conserved anticodon and dihydrouridine stem sequences but contain mismatches in other regions of the secondary structure. Although the Viscum mitogenome contains far fewer tRNA genes than most other angiosperms, similarly reduced tRNA gene content (two and three, respectively) has been reported for the S. conica and Silene noctiflora mitogenomes (SI Appendix, Fig. S3) (9).
Large and small subunit rRNA (LSU rRNA and SSU rRNA, respectively) genes were identified also. Although both rRNA gene sequences are exceptionally divergent (Exceptional Divergence of Viscum Protein and rRNA Genes), superimposition onto the Oenothera berteriana mitochondrial SSU rRNA and Zea mays mitochondrial LSU rRNA secondary structures (SI Appendix, Figs. S4 and S5, respectively) revealed significant conservation at the secondary structure level, indicating that these genes are still functional in Viscum. Secondary structure and phylogenetic analyses were not carried out using the 5S rRNA because of its short length (<100 nt), but a pairwise alignment of its sequence to the Liriodendron 5S rRNA yielded 87% identity and 5% gaps.
The ancestral angiosperm mitogenome contained 25 group II introns (19). Most angiosperm genomes examined contain nearly this entire set (SI Appendix, Fig. S6), and none have fewer than 18 of these introns (9). Viscum, however, contains only three group II introns (SI Appendix, Fig. S6). Most of this reduction in intron number can be attributed to the loss of the nad genes, which commonly possess 19 group II introns in seed plants (SI Appendix, Fig. S6). In addition, Viscum has lost rpl2, which normally contains a single group II intron, and a single intron has been lost from each of the otherwise intact genes rps3 and rps10. In addition, the first intron in the Viscum cox2 gene is probably trans-spliced, given the physical separation of the first exon from the rest of the gene (Fig. 1A) and that the exonic boundaries of these two regions do not contain stop and start codons, respectively, as expected under the alternative model of gene fission. Trans-splicing in this intron is known to have arisen in two other lineages of vascular plants (20, 21).
The loss of so many genes and their (mostly accompanying) introns accounts for an appreciable fraction (ca. 25%) of the size reduction in the 66-kb Viscum mitogenome compared with the next smallest sequenced angiosperm mitogenome (222 kb) in our comparative dataset (Fig. 2). Most of the loss in gene space is a consequence of the loss of the intron-rich nad genes, which typically comprise nearly 40 kb of DNA, or more than half the gene space of normal angiosperm mitogenomes (SI Appendix, Fig. S7). The rest of the Viscum size reduction is a consequence of a massive reduction in the amount of intergenic spacer DNA, which totals ca. 36 kb in Viscum compared with ca. 150 kb in the 222-kb Brassica napus genome and more than 11 Mb in S. conica.
Loss of Mitochondrial Maturase Gene matR.
The loss of the maturase gene matR could reflect a very rare case of functional transfer to the nucleus. However, because matR resides within nad1 intron 4, it is more likely that matR was lost along within the entire suite of mitochondrial nad genes in Viscum. MatR is the only mitochondrial-specified maturase homolog present in angiosperms, and although the precise function of MatR in mitochondria is uncertain, it is clearly homologous to proteins known to assist in group II intron splicing in other mitochondrial genomes (22). Furthermore, preliminary data suggest that MatR binds to several group II introns in vivo (23). If MatR is indeed a polyvalent splicing factor, the precise consequences of the loss of this gene for the group II intron-splicing process in Viscum need to be elucidated.
Possible Rare Transfer of ccmB to the Nucleus.
In contrast to the matR situation, functional transfer to the nucleus is the best explanation for the absence of ccmB from Viscum mtDNA. In most plants, mitochondrial biogenesis of cytochrome c is carried out via a maturation pathway encoded by four mitochondrial ccm genes (ccmB, ccmC, ccmFc, and ccmFn) and several nuclear genes (24). Because ccmC, ccmFc, and ccmFn are all present as intact genes in the Viscum mitogenome, albeit in moderately to exceptionally divergent forms (Table 1), the ccm complex is almost certainly functional in Viscum, and therefore ccmB probably has been functionally located to the nucleus. CcmB is a hydrophobic protein with six transmembrane domains that traverse the inner mitochondrial membrane (24). Although functional transfer of ccmB has not been reported previously in plants (Fig. 2) (17), if this gene experienced anything approaching ccmFc- and ccmFn-like levels of divergence while in the mitogenome, this divergence may have altered the hydrophobicity of CcmB to an extent that allowed it to be imported more readily into the mitochondrion following ccmB functional transfer to the nucleus. Such a transfer would be analogous to other cases of rare mitochondrial-to-nuclear gene transfer, which have been accompanied by a reduction in hydrophobicity of the encoded protein (25, 26). Unfortunately, nuclear coverage in our sequence data (ca. 0.1×; Materials and Methods and SI Appendix, SI Discussion) is inadequate to detect a full-length transferred copy of ccmB, and therefore testing of the transfer hypothesis must await nuclear transcriptome or genome sequencing.
Table 1.
Summary of Viscum protein-gene divergence
| Gene | Coding sequence length, nt | % amino acid identity* | dN | dS | dN/dS | ||
| Viscum | Lirio† | Viscum vs. Lirio† | Vitis vs. Lirio† | ||||
| cox1 | 1,611 | 1,572 | 83 | 98 | 0.11 | 0.77 | 0.14 |
| atp9 | 219 | 225 | 81 | 93 | 0.05 | 0.68 | 0.07 |
| atp1 | 1,533 | 1,527 | 72 | 95 | 0.19 | 1.04 | 0.18 |
| atp6 | 912 | 720 | 71 | 95 | 0.21 | 0.91 | 0.24 |
| cox2 | 750 | 762 | 69 | 90 | 0.26 | 1.01 | 0.25 |
| cob | 1,221 | 1,179 | 69 | 97 | 0.19 | 1.05 | 0.18 |
| cox3 | 795 | 795 | 66 | 97 | 0.22 | 0.84 | 0.27 |
| rps12 | 411 | 375 | 61 | 95 | 0.31 | 3.89 | 0.08 |
| ccmC | 654 | 699 | 56 | 91 | 0.39 | 1.06 | 0.37 |
| mttB | 789 | 720 | 46 | 95 | |||
| ccmFn | 1,467 | 1,683 | 43 | 92 | |||
| rps10 | 273 | 357 | 42 | 96 | |||
| rpl16 | 378 | 432 | 44 | 99 | |||
| atp8 | 483 | 474 | 41 | 96 | |||
| rps4 | 1,224 | 1,035 | 39 | 93 | |||
| ccmFc | 1,215 | 1,326 | 38 | 87 | |||
| sdh4 | 390 | 387 | 34 | 88 | |||
| rps3 | 1,518 | 1,554 | 32 | 93 | |||
| atp4 | 474 | 558 | 29 | 93 | |||
Synonymous and nonsynonymous (dS and dN, respectively) divergence and dN/dS ratios are given for the nine best-conserved mitochondrial protein genes of Viscum.
Identity calculation excludes gap positions.
Liriodendron tulipifera.
Unprecedented Loss of Complex I in a Multicellular Organism.
The loss of all mitochondrial nad genes in Viscum is of special interest because it very likely corresponds to the loss of an entire respiratory complex, something that has never been reported before for any plant or alga, including the holoparasite Rafflesia (4). The nad genes contain five or six trans-spliced introns in angiosperms, with the 14 or 15 nad transcription units scattered essentially randomly in the rapidly rearranging mitogenomes of angiosperms (10, 19). Therefore, the eradication of all traces of nad genes from the Viscum mitogenome probably occurred via many separate deletions, presumably occurring over some period of time. This likelihood, combined with the presence of pseudogenes for other “lost” genes (i.e., rps1 and sdh3), suggests that the loss of selection on complex I may have occurred much earlier than the loss of rps1 and sdh3.
ATP formation in mitochondria occurs mainly through oxidative phosphorylation. This is achieved by a process of electron transfer through an assemblage of more than 20 carriers that are organized into complexes I–IV of the electron transport chain (27). With the notable exceptions of four lineages, all respiring eukaryotes contain complex I, a multisubunit, rotenone-sensitive NADH dehydrogenase that is encoded by 5–12 mitochondrial genes and a few dozen nuclear nad genes. The four exceptions, in which all mitochondrial and nuclear nad genes have vanished, include two distinct lineages of yeasts (28), the cryptomycotan fungus Rozella (29), and the lineage comprising dinoflagellates, apicomplexans, Chromera, and Oxyrrhis (30). That these are all unicellular makes the case in Viscum particularly notable.
The loss of the entire suite of mitochondrial nad genes in Viscum almost certainly reflects the loss of complex I itself, as opposed to the functional transfer of all these genes to the nucleus. Computational searches for “nad-like” reads within the total genomic DNA yielded only a small number of matches, most of which corresponded to intronic sequences that are unlikely to be properly expressed in the nucleus (SI Appendix, SI Discussion). Seven of the nine nad genes found in all 300+ other angiosperm mitogenomes examined (Fig. 2) (9, 17) have rarely, if ever, been found functionally transferred to the nucleus in nonangiosperms. Indeed, it is these seven nad genes that are found in all examined animal mitogenomes. Three of the seven (nad1, nad4, and nad5) are universally present in all mitogenomes of eukaryotes that retain complex I, whereas nad2, nad3, nad4L, and nad6 have been reported lost from the mitochondrial genome (and presumably functionally transferred to the nucleus) only one to three times across eukaryotes (31). The difficulty of functionally transferring these seven nad genes to the nucleus is probably the result of severe mechanistic challenges to the successful transport of their hydrophobic products across the outer mitochondrial membrane and insertional assembly within the inner membrane (32, 33). Therefore, even in the absence of any meaningful information on nad gene status in the nucleus of V. scurruloideum, the loss of all nine nad genes from the V. scurruloideum mitogenome most likely reflects the loss of complex I from this plant.
The losses of complex I in two yeast lineages have been accompanied by duplications of nuclear-encoded alternative NADH dehydrogenases that bypass complex I and oxidize NADH without proton translocation (34). Because these alternative means of regenerating NAD+ are unlinked to proton pumping in the mitochondrion, they generate less energy in the form of ATP. Although the evolutionary loss of complex I is unprecedented in plants, mutant lines are informative for understanding the potential consequences of the loss of the nad genes in Viscum. In particular, three mutant lines, Nicotiana sylvestris CMSII lacking the mitochondrial nad7 gene (35, 36), Arabidopsis thaliana ndufs4 lacking a nuclear nad gene (37), and NCS2 maize plants lacking functional nad4, are deficient in complex I (38, 39). Like the complex-I–deficient yeasts, these plant mutants remain viable via use of alternative NADH dehydrogenases (37, 38, 40), albeit with constitutively lower mitochondrial phosphorylation efficiencies. Plants in general have the ability to oxidize NADH and NADPH via several nuclear-encoded alternative dehydrogenases (41). The determination of which of these dehydrogenases Viscum uses to compensate for the lack of complex I awaits experimental investigation and sequencing of its huge nuclear genome.
The plant complex I mutants grow more slowly than their respective wild types and have other deficiencies: N. sylvestris CMSII mutants exhibit male sterility and slow growth (36); A. thaliana ndufs4 shows depressed germination and growth (37); and homoplasmic NC2 maize plants exhibit sterility and severely depressed growth (42). Because Viscum does not exhibit noticeably defective germination or growth, regardless of any deleterious effects caused by loss of complex I in Viscum, the organism has been able to adapt and thrive. Analysis of the CMSII mutant has shown that mitochondrial complex I is essential for optimal photosynthetic performance (43). In particular, steady-state photosynthesis in CMSII is consistently reduced by 20–30% at atmospheric CO2 levels, suggesting that this complex I mutant is not able to use its photosynthetic capacity as well as wild-type N. sylvestris despite the action of alternative dehydrogenases (40, 44). It has long been known that mistletoes typically have lower photosynthetic rates than their hosts (45–48). Thus, the lower rates of photosynthesis and the loss of complex I may be interrelated in Viscum.
The precise molecular interactions underpinning the relationship between reduced photosynthetic efficiency and the mitochondrial electron transport chain have not been described, but it is notable that under conditions in which CO2 fixation (and, hence, sucrose synthesis) is enhanced, photosynthesis in CMSII is less inhibited relative to wild-type N. sylvestris. This observation suggests that the decrease in photosynthesis in CMSII under atmospheric CO2 levels is not caused by insufficient mitochondrial ATP production for sucrose synthesis and is specific to conditions in which the enzymes necessary for photosynthesis are preoccupied with another task (44). We hypothesize that Viscum uses alternative dehydrogenases to compensate for the loss of complex I in maintaining mitochondrial ATP production but suffers an associated reduction in photosynthetic efficiency. Whether cause or cure, parasitism could compensate for some loss of photosynthetic ability.
Recent HGT of the cox1 Intron.
We find little evidence suggestive of HGT in Viscum mtDNA. First, there is the lack of multiple, divergent copies of any genes in the Viscum mitogenome. In contrast, most cases of plant mitochondrial HGT reported thus far have resulted in the maintenance of both horizontally acquired and native gene copies within the mitogenome, with Rafflesiaceae and Amborella especially notable in this regard (8, 15). Second, the extreme divergence of all 21 Viscum protein and rRNA genes (see the next section and SI Appendix, Fig. S8), which confounds phylogenetic detection of HGT, also means that any transfers must be relatively ancient, occurring before the accumulation of a significant fraction of this divergence. Finally, no evidence of gene conversion was found in any Viscum genes using OrgConv with Bonferroni correction for multiple tests (49).
The only clear evidence of HGT in the Viscum mitogenome involves its sole group I intron, located in the cox1 gene, which also is the only group I intron found in any angiosperm mitogenome. This intron has been acquired more or less recently in many angiosperm lineages by hundreds of mitochondrial-to-mitochondrial horizontal transfer events (2, 50, 51). In striking contrast to the extremely divergent gene sequences in Viscum (SI Appendix, Fig. S8), including that of cox1 itself, the Viscum cox1 intron is unexceptional in its level of sequence divergence compared with other angiosperm cox1 introns (SI Appendix, Fig. S9A). This contrast strongly implies that the intron was acquired recently in the Viscum lineage, most likely independently of intron acquisitions in Lepionurus and Dendrophthoe, the two other members of the Santalales known to possess the intron (SI Appendix, SI Discussion). The Viscum cox1 gene contains an unusually lengthy region of putatively converted exonic sequence (known as a coconversion tract), as is also consistent with a recent horizontal acquisition (SI Appendix, Figs. S9B and S10).
Exceptional Divergence of Viscum Protein and rRNA Genes.
Phylogenetic analysis (SI Appendix, Fig. S8) of two of the three rRNA genes and the nine best-conserved protein genes in Viscum shows that they are even more divergent, often considerably so, than genes in the fast-evolving S. conica and S. noctiflora mitogenomes (9). Moreover, the rRNA trees seriously underestimate the extent of rRNA divergence (SI Appendix, legend of Fig. S8), and the 10 most divergent protein genes in Viscum (Table 1) are so divergent that we deemed it imprudent to use them for either phylogenetic analysis or analysis of synonymous (dS) and nonsynonymous (dN) site divergence. Unusually frequent C-to-U mRNA editing in Viscum is highly unlikely to account for its high levels of inferred protein divergence (SI Appendix, SI Discussion); rather, as described below, this divergence appears to be the consequence of major changes in both mutational and selective pressures.
Full alignments for the 10 most divergent Viscum proteins are shown in SI Appendix, Fig. S11. These proteins are characterized by short regions of moderate-to-high sequence similarity interspersed with longer regions exhibiting little to no detectable sequence similarity. These latter regions often include a number of indels, and the reliability of the alignment in these regions is often questionable. Their extraordinary level of divergence raises the question of whether these 10 “genes” code for functional proteins. The most divergent proteins in Viscum are, for the most part, encoded by ORFs of moderate to considerable length (Table 1). Considering Viscum’s highly accelerated evolution, such an accumulation of nucleotide substitutions should have introduced premature stop codons into most if not all of these ORFs by chance. Given the frequency of stop codons in the genome, and assuming a random distribution of point mutations, the probabilities of getting ORFs of lengths 50, 100, 150, and 200 codons by chance alone are 0.14, 0.02, 0.003, and 0.0004, respectively. Thus, it is likely that these proteins are, by and large, functional, especially because the majority of them are of comparable length to those found in other angiosperms (Table 1). It should be noted that the unprecedented level of divergence in Viscum makes it difficult to rule out the possibility that unidentified protein genes exist in the mitogenome but have evolved beyond our ability to recognize them.
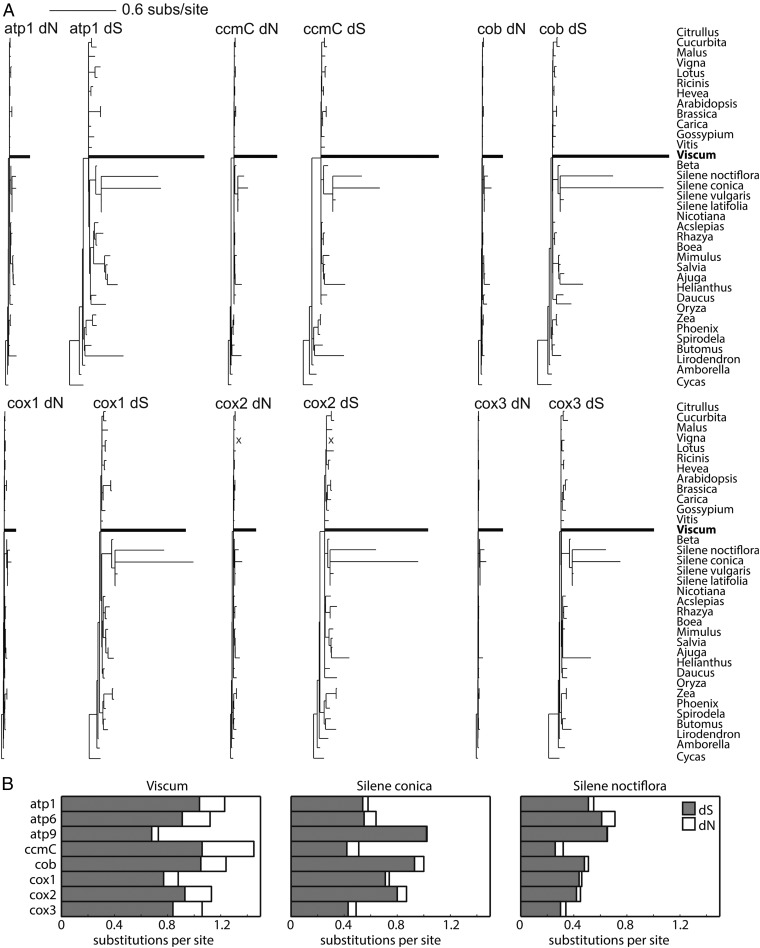
To assess further the level of divergence in Viscum protein genes, dS and dN values were estimated for the nine best-conserved genes (Fig. 3, Table 1, and SI Appendix, Fig. S12). Synonymous substitutions in protein genes often are assumed to be more or less free from natural selection and thus are used frequently to approximate the neutral mutation rate (52). The patterns of divergence in dS trees (Fig. 3A and SI Appendix, Fig. S12) are similar to those in the trees based on all three codon positions (SI Appendix, Fig. S8). For almost all genes, dS branch lengths are longer for Viscum than for any of the other sampled angiosperms, including the high-rate Silene species (Fig. 3). This divergence indicates a highly elevated mutation pressure operating (perhaps still so) across the mitogenome during a portion of the Viscum lineage time. Estimates of the frequency and magnitude of this mutation-rate increase await sampling of mitogenomes from a phylogenetically broad range of Santalales taxa and the construction of molecular chronograms for these taxa. Increased sequence divergence in seven sampled members of the Viscaceae (including one species of Viscum) was observed in a concatenated phylogenetic analysis of one nuclear gene (SSU rDNA) and two plastid genes (rbcL, matK) (11). Although the magnitude of the mitochondrial elevation appears to be significantly higher than for the plastid and nuclear genes, it is possible that multiple factors underlie the mitochondrial situation and that one of these factors is common to all three genomes.
Fig. 3.
Elevated sequence divergence of the best-conserved mitochondrial protein genes in Viscum. (A) Phylograms of dS and dN sequence divergence for six of the nine best-conserved genes (Table 1). All trees are shown to the same scale. The absence of the cox2 gene in Vigna is indicated with an “X.” (B) Levels of dS and dN sequence divergence for eight protein genes in Viscum and two rapidly evolving Silene mitochondrial genomes.
If mutation rates and the effects of selection on synonymous sites were reasonably constant across the Viscum mitogenome, we would expect that synonymous substitution rates should be fairly constant among its protein genes. This expectation is met for eight of the nine best-conserved protein genes (dS = 0.68–1.06), but rps12 is a clear outlier (dS = 3.89) (Table 1; also see SI Appendix, Fig. S12). This observation adds to the growing, and puzzling, recognition that angiosperm mitogenomes can experience substantial heterogeneity in intragenomic rates, although the level found in Viscum pales against that in certain other genomes, especially that of Ajuga (53).
As expected, given the highly elevated mutation rate(s) in Viscum mtDNA, dN branch lengths are highly elevated in Viscum relative to other angiosperms (Fig. 3 and SI Appendix, Fig. S12). Contrary to the pattern seen for high-mutation-rate genomes in Silene (Fig. 3B) and in Plantago and Pelargonium (54), in which dN/dS is depressed (0.04–0.07 for a concatenate of atp1, cob, cox1, cox2, and cox3, five of the nine best-conserved protein genes in Viscum) relative to typically low-mutation angiosperm genomes, the average dN/dS value for these five genes in Viscum is 0.20 (Table 1). These different dN/dS values suggest a different interplay of mutational and selective forces among these independently arisen, high-mutation-rate lineages. In particular, it seems likely that the set of 10 extremely divergent protein genes (SI Appendix, Fig. S11) has and/or is evolving under relaxed, if not to some extent positive, selection compared with other angiosperms.
As well as generally being very large, plant mitogenomes typically are characterized by extremely low rates of point mutation (54). In recent years, the mutational burden hypothesis, which predicts that mitogenome size should be negatively correlated with mutation rate (55), has been adopted by some to explain the origins of variation in mitogenome size. Under this hypothesis, high mutation pressure enhances natural selection against nonessential DNA, and hence against large genomes. The reduced Viscum genome and its highly elevated dS values are consistent with the hypothesis of genomic streamlining in response to a high mutation rate (55) but are in complete contrast to the enormous mitogenomes of S. noctiflora and S. conica, which have experienced both dramatic accelerations in mitochondrial mutation rates and huge expansions of intergenic DNA (9). These entirely opposite findings emphasize the need for further genome sequencing to provide a broader perspective on the remarkable genomic diversity across angiosperms and to enable refined hypotheses regarding the undoubtedly multifaceted relationship among mutation rate, genome size, and other aspects of genome evolution.
Abundant Repeats and Extreme Levels of Recombination and Sublimons.
Repetitive sequences cover 26 kb (39%) of the Viscum mitogenome (Fig. 1B). In light of its small size and gene-dense nature, it is remarkable that large repeats (≥1 kb) cover so much of the Viscum genome (16 kb, 24%), because this is a greater absolute amount than seen in all but two of the 11 much larger (222- to 983-kb) eudicot mitogenomes compared in ref. 56. Also remarkable is that repeat coverage in Viscum (39%), the smallest known angiosperm mitogenome, is proportionally comparable to that of the largest known mitogenome, that of S. conica (40.8%; 4.6 Mb of repeat coverage in a 11.3-Mb genome) (9). In contrast, the also enormous (6.7-Mb) mitogenome of S. noctiflora—which has a high mutation rate—contains a relatively modest proportion of repetitive sequences (10.9%) compared with Viscum and many other angiosperm mtDNAs (9). Thus, there is no strict relationship between repeat content, genome size, and mutation rate in angiosperm mtDNAs.
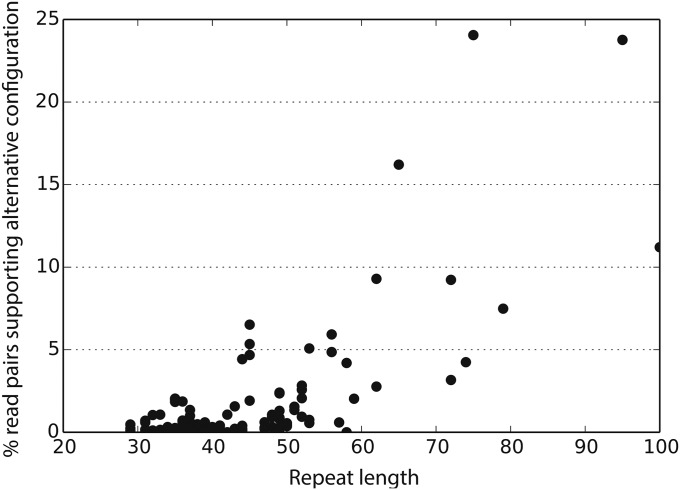
In angiosperm mitochondria, low-abundance substoichiometric DNA molecules (termed “sublimons”) are often present, arising via recombination between short repeats (57–59). Most pairs of repeats in Viscum mtDNA are short; of those ≥30 bp in size, 97% (305 of 315) are ≤100 bp in length, and 86% of these are <50 bp in length. To assess recombination at short Viscum repeats, we counted the number of read pairs that are consistent with the high-depth reference assembly and those that are consistent with the predicted alternative configuration (AC) of the repeat-flanking single-copy sequences that would result from repeat-mediated recombination. To avoid inflating the estimates of ACs, when two or more repeat-pair environments were supported by the same set of spanning reads, only one repeat pair was considered in our analysis. We found evidence for ACs for 127 (78%) of the 162 short-repeat pairs examined (Fig. 4 and SI Appendix, Table S1). There is a strong correlation (r = 0.72, P < 0.001) between repeat length and AC frequency (Fig. 4), but there is no correlation between repeat similarity and AC frequency (SI Appendix, Fig. S13).
Fig. 4.
Alternative configurations for repeat pairs ≤100 bp in length.
The AC frequency for short repeats in Viscum mtDNA is markedly higher than in the nine other angiosperm mtDNAs for which AC frequency has been measured by high-depth genome sequencing and also is higher than in angiosperms in which non–genome-scale approaches have been used (59). Not even a single AC was detected for the vast number (>300,000 and >70,000) of short-repeat pairs (30–100 bp in length) present in the mitogenomes of S. conica (11.3 Mb) and cucumber (1.7 Mb), respectively (9, 60). Alternative configurations were detected for only one of the ca. 2,400 short repeats in the 6.7-Mb genome of S. noctiflora genome (9, 60) and for only two of 110 short repeats in the 404-kb genome of Vigna angularis (61). AC frequency was not reported for the 526-kb Mimulus mitogenome (62), but the number of AC-consistent reads was given and indicates much lower AC frequency levels than in Viscum. As in Viscum, repeat length correlates with AC frequency in Mimulus. Finally, for four Silene vulgaris mitogenomes (361–429 kb in size) analyzed in aggregate (63), AC frequency averaged only 0.8% (median = 0.7%, range = 0.2–1.5%) for the 13 repeat pairs with a length of 50–100 bp for which at least five ACs were detected. The number of the repeats for which zero to four ACs were recovered was not reported, and therefore AC frequency again is markedly lower than in Viscum.
Only four of the 10 repeat pairs in Viscum with a length ≥100 bp are sufficiently short (387–593 bp) to allow ACs to be detected given the library-fragment size of ∼800 bp. With AC frequencies of 39–58%, all four repeat pairs are essentially at recombinational equilibrium (i.e., a 50:50 ratio of reference configurations and ACs). To our knowledge, these are the smallest repeats at recombinational equilibrium in plant mitogenomes. As with the short repeats, these data are in striking contrast to the extremely low level of recombination detected for the great majority of comparably sized repeats in S. conica, S. noctiflora, and cucumber (9, 60). Repeats of this size either are not present or were not analyzed in Vigna, Mimulus, and the four S. vulgaris mitogenomes (61–63).
Although the Viscum mitogenome assembles as two contigs, the extraordinary abundance and diversity of recombinant read pairs means that this configuration is highly unlikely to exist in vivo. [This consideration is quite apart from the issue of whether any plant mitogenome assembly actually corresponds to the in vivo arrangement of the genome (64–66).] Instead, within an individual and possibly even at the level of a single mitochondrion, the Viscum genome almost certainly exists as an extremely complicated population of recombinationally interrelated, overlapping molecules of a broad range of sizes and stoichiometries. Although the numerous sublimons detected in this study presumably all ultimately arose from repeat-mediated recombination, the extent to which this recombination is actively ongoing is unclear and almost certainly varies among repeats, with ongoing—and also high-frequency/equilibrating—recombination a safe inference for only the 10 large repeat pairs. In contrast, as a consequence of occasional to frequent ongoing repeat-mediated recombination, some of the sublimons associated with the 127 short repeats almost certainly exist in forms independent of the main genome. Indeed, many of the reads indicative of sublimons have polymorphisms relative to the reference assembly, suggesting that many regions are evolving independently of the predominant forms of the genome. The more abundant ACs may even represent multiple independently arisen and replicating sublimons.
All 10 large (387- to 5,439-bp) repeat pairs in Viscum show either 100% identity or, in two cases, 99.9% identity (SI Appendix, Table S1). This level of identity is yet another way in which the highly divergent Viscum genome differs dramatically from the also divergent S. noctiflora and S. conica mitogenomes: In S. noctiflora and S. conica only a tiny fraction of the hundreds to thousands of repeats, respectively, with a length of 400–2,000 bp are 100% identical (most are 75–90% identical) (9). These differences imply much higher rates of gene conversion/concerted evolution in Viscum than in these two Silene species. The greatly reduced levels of both homologous recombination (as measured by AC frequency) and gene conversion in these two Silene genomes were hypothesized to be potentially important factors underlying the highly elevated mutation rates in these genomes (9). Therefore it is all the more intriguing that, compared with these other high-mutation-rate mitogenomes, Viscum has high levels of these two mechanistically related recombinational processes.
Conclusion and Outlook
The mitogenome of the hemiparasitic mistletoe V. scurruloideum has proven to be an unexpected goldmine of surprises, setting new benchmarks for the extremes of genome size, gene content, protein divergence, and recombinational activity and/or sublimon levels in angiosperm mtDNA. The gene repertoire of the Viscum mitogenome may be so specialized to parasitic life that, unlike all other multicellular organisms investigated so far, it can survive without a mitochondrial-encoded respiratory complex I. Recently the intracellular parasitic fungus Rozella allomycis also was found to harbor a very rapidly evolving genome that lacks complex I of the respiratory chain (29). This discovery raises the tantalizing possibility that shared genomic signatures of parasitism may be present in mitogenomes both within and beyond the bounds of the angiosperm clade.
The Viscum findings have begun to illuminate patterns and modes of evolution of angiosperm mitogenomes that previously were unrecognized, raising deep questions about the diversity of gene landscapes, the effect of mutational environments on genomic contraction and expansion, and which genes, if any, are truly essential in angiosperm mitogenomes. We expect that greatly expanded sampling of the speciose and diverse Santalales, as well as the many other lineages of parasitic angiosperms, will provide significant insights into the evolution and function of parasitism in angiosperms, uncover an increasingly fascinating world of mitochondrial diversity, and shed light on the origins of the massive amount of foreign angiosperm mtDNA present in the Amborella mitogenome.
Materials and Methods
Assembly.
The Viscum mitogenome assembly was generated using 100-bp paired-end Illumina HiSeq2000 reads (with 800-bp fragment size) from leaf-derived total genomic DNA (SNP 15591; collected in Sabah, Malaysia, Hamadon, kg. Bundu Tuhan, 1,300 m, by T.J.B., September 22, 2000). The relatively small fraction of reads (<1%) corresponding to the mitochondrial genome was extracted from the pool of 82.5 million total genomic reads by iteratively extending contigs. Reads determined to be similar to known angiosperm mitochondrial genes using BLASTN (67) were used as seeds. These reads then were blasted against all reads, and reads with no or few high-quality base conflicts in the aligned region and which overhung at the end were used to extend a pseudo contig. The ends of nascent contigs were blasted again and extended until no further extension was obtained. To identify identical or highly similar reads, megablast was used with a word size of 32. Pools of reads derived from these iterations were assembled using CAP3 (68) with the following parameter settings: a = 11, b = 16, d = 21, e = 11, o = 50, f = 2, h = 500, i = 60, j = 80, s = 1,800, and t = 300. The majority of the contig ends corresponded to repeats that could be resolved by read depth, read-pair conflicts, and identification of repeat boundaries for which a large portion of reads were chimeric outside the repeat region. Repeat sequences were identified by comparing the mitogenome to itself using ungapped BLASTN with the parameter settings xdrop_gap = 10 and xdrop_gap_final = 10. All alignments with an e-value ≤1e-7 were considered repeats for calculations of repeat number. The repeat map (Fig. 1B) was generated with Circos (69).
The final assembly contained two contigs, a 42,186-bp contig that could be rationalized into a circle based on read-pair information and a 23,687-bp kb contig that was terminated by repeats that matched within the 42-kb contig and whose lengths could not be determined using read-depth and repeat-boundary information. In this sense, therefore, the Viscum mitogenome assembly (Fig. 1A) is incomplete. These repeats may be relatively long and possibly overlap with other repeats, making their boundaries difficult to identify. The read depth adjacent to the corresponding repeat regions in the 42-kb contig is too variable to make a clear judgment as to precisely where the terminal repeats in the 24-kb contig end. The contigs we present are well supported in terms of high-quality read depth and read-pair depth, with the exception of a low-depth region around 16.9 kb in the 42-kb contig (SI Appendix, Fig. S1). This region corresponds to a palindromic sequence. The relative positions of forward- and reverse-oriented reads mapping in this region suggest that the palindrome may have interfered with the paired-end amplification of sequences containing this palindrome.
We looked for potentially missing mitochondrial sequences by using reads that map to the assembly but whose read-pair mates do not (singlets). Although we identified many singlet reads, using their mate pairs as iterative assembly seeds (see above) did not produce any high-depth contigs or contigs that were part of the main mitochondrial genome. On the basis of polymorphisms and homology to sequences in the GenBank nt/nr database, we infer that the majority of these singlets probably correspond to mitochondrial-like sequences in the nucleus. It is conceivable that some nongenic or highly divergent genic regions of the genome are missing from our assembly, but, given its gene density (Fig. 1A) and the high number of singlet partners that did not yield mitochondrial-like contigs, this notion is unlikely.
Coverage Estimation.
Nuclear genome coverage was estimated at 0.1× by using the Lander/Waterman equation and conservatively assuming V. scurruloideum to have the same nuclear genome size (2C = 124.6) as Viscum minimum, which has the smallest nuclear genome of the four Viscum species examined thus far (16).
Gene Annotation.
Contig sequences were compared by BLASTX v. 2.2.28 (67) to mitochondrial protein sequences from 55 complete green algal and land plant mitogenomes. Gene boundaries were annotated based on inspection of alignments satisfying e-value <0.001. The locations of rRNA genes were determined similarly by BLASTN searches against the rRNA gene sequences of these genomes. tRNA genes were predicted using tRNAscan version 1.23 (70).
RNA Editing Sites.
C-to-U RNA editing sites were predicted using PREP-Mitochondrial (71) with stringency settings (C-values) of 0.2, 0.6, and 1.0 and PREPACT v. 2.12.3 (72) with default parameter settings. Viscum results are reported (SI Appendix, Table S2) for multiple stringency settings because previous analyses in Silene have suggested that different C-value settings are appropriate for species with different rates of sequence evolution (9). All results reported in the main text were obtained using PREP-Mitochondrial with C-value = 0.2. The predicted editing sites probably underestimate the true extent of editing within Viscum, because the PREP-Mitochondrial approach is blind to synonymous editing sites.
Phylogenetic Analysis.
Protein sequences were extracted from GenBank and aligned using MAFFT v. 7.130b (73) with the L-INS-I option. The resulting amino acid alignments then were computationally converted to nucleotide alignments using PAL2NAL (74), with the resulting arrangement of nucleotide triplets reflecting the protein alignment in the case of each gene set. To avoid bias in phylogenetic analysis and evolutionary rate estimation arising from C-to-U RNA editing, codons known to undergo mitochondrial RNA editing in at least one of eight angiosperms (A. thaliana, Beta vulgaris, B. napus, Citrullus lanatus, Cucurbita pepo, Oryza sativa, Silene latifolia, and Vitis vinifera) (SI Appendix, Table S3) or predicted to undergo RNA editing in V. scurruloideum (SI Appendix, Tables S2 and S3) were excluded from all nucleotide analyses. rRNA gene sequences were aligned similarly using MAFFT L-INS-I, but at the nucleotide level. Ambiguously aligned regions of the rRNA gene alignments were removed using GBLOCKS version 0.91b, with the following parameter settings: t = c, b1 = 11, b2 = 16, b3 = 8, b4 = 5, and b5 = none (75). These stringent settings remove large fractions of most alignments. All phylogenetic trees were estimated with RAxML v. 7.2.8 (76) using the generalized time-reversible (GTR) model with gamma correction for among-site rate variation and 10 starting trees. Support for nodes was assessed using 1,000 bootstrap replicates. The ancestral sequences shown in SI Appendix, Figs. S9, S10, and S11 were reconstructed using baseml and codeml in the PAML package (77).
Evolutionary Rate Estimation.
PAML’s codeml (77) was used to estimate branch dS and dN, using a simplified Goldman–Yang codon model with separate branch dN/dS ratios (ω) that allowed for the following three sets of branches: the Viscum branch; the S. conica and S. noctiflora branches; and all remaining branches. Codon frequencies were computed using the F3 × 4 method. The values for ω and transition/transversion ratio were estimated from the data with start values of 0.4 and 2, respectively.
Supplementary Material
Acknowledgments
We thank Andy Alverson and Virginia Sanchez-Puerta for providing data used in certain analyses and Andy Alverson and Dan Sloan for critical reading of the manuscript. This work was supported in part by National Science Foundation Award IOS 1027529 (to J.D.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database, www.ncbi.nlm.nih.gov/genbank/ (accession nos. KT022222 and KT022223).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504491112/-/DCSupplemental.
References
- 1.Westwood JH, Yoder JI, Timko MP, dePamphilis CW. The evolution of parasitism in plants. Trends Plant Sci. 2010;15(4):227–235. doi: 10.1016/j.tplants.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Barkman TJ, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heide-Jørgensen HS. Parasitic Flowering Plants. Koninklijke Brill NV; Leiden, The Netherlands: 2008. [Google Scholar]
- 4.Molina J, et al. Possible loss of the chloroplast genome in the parasitic flowering plant Rafflesia lagascae (Rafflesiaceae) Mol Biol Evol. 2014;31(4):793–803. doi: 10.1093/molbev/msu051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wicke S, et al. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 2013;25(10):3711–3725. doi: 10.1105/tpc.113.113373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schelkunov MI, et al. Exploring the limits for reduction of plastid genomes: A case study of the mycoheterotrophic orchids Epipogium aphyllum and Epipogium roseum. Genome Biol Evol. 2015;7(4):1179–1191. doi: 10.1093/gbe/evv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mower JP, Jain K, Hepburn NJ. 2012. The role of horizontal transfer in shaping the plant mitochondrial genome. Advances in Botanical Research, ed Maréchal-Drouard L (Academic, New York), Vol 63: Mitochondrial Genome Evolution, pp 41–69.
- 8.Xi Z, et al. Massive mitochondrial gene transfer in a parasitic flowering plant clade. PLoS Genet. 2013;9(2):e1003265. doi: 10.1371/journal.pgen.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10(1):e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mower JP, Sloan DB, Alverson AJ. 2012. Plant mitochondrial diversity—the genomics revolution. Plant Genome Diversity, eds Wendel JF, Greilhuber J, Dolezel, I Leitch IJ (Springer, Berlin), pp 123–144.
- 11.Der JP, Nickrent DL. A molecular phylogeny of Santalaceae (Santalales) Syst Bot. 2008;33(1):107–116. [Google Scholar]
- 12.Nickrent DL, Malécot V, Vidal-Russell R, Der JP. A revised classification of Santalales. Taxon. 2010;59(2):538–558. [Google Scholar]
- 13.Nickrent DL, Der JP, Anderson FE. Discovery of the photosynthetic relatives of the “Maltese mushroom” Cynomorium. BMC Evol Biol. 2005;5:38. doi: 10.1186/1471-2148-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathiasen RL, Nickrent DL, Shaw DC, Watson DM. Mistletoes: Pathology, systematics, ecology, and management. Plant Dis. 2008;92(7):988–1006. doi: 10.1094/PDIS-92-7-0988. [DOI] [PubMed] [Google Scholar]
- 15.Rice DW, et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science. 2013;342(6165):1468–1473. doi: 10.1126/science.1246275. [DOI] [PubMed] [Google Scholar]
- 16.Zonneveld BJM. 2010. New record holders for maximum genome size in eudicots and monocots. J Bot, 10.1155/2010/527357.
- 17.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA. 2002;99(15):9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adams KL, Palmer JD. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29(3):380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 19.Richardson AO, Rice DW, Young GJ, Alverson AJ, Palmer JD. The “fossilized” mitochondrial genome of Liriodendron tulipifera: Ancestral gene content and order, ancestral editing sites, and extraordinarily low mutation rate. BMC Biol. 2013;11:29. doi: 10.1186/1741-7007-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Yoon MK. Comparison of mitochondrial and chloroplast genome segments from three onion (Allium cepa L.) cytoplasm types and identification of a trans-splicing intron of cox2. Curr Genet. 2010;56(2):177–188. doi: 10.1007/s00294-010-0290-6. [DOI] [PubMed] [Google Scholar]
- 21.Hecht J, Grewe F, Knoop V. Extreme RNA editing in coding islands and abundant microsatellites in repeat sequences of Selaginella moellendorffii mitochondria: The root of frequent plant mtDNA recombination in early tracheophytes. Genome Biol Evol. 2011;3:344–358. doi: 10.1093/gbe/evr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wahleithner JA, MacFarlane JL, Wolstenholme DR. A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc Natl Acad Sci USA. 1990;87(2):548–552. doi: 10.1073/pnas.87.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown GG, Colas des Francs-Small C, Ostersetzer-Biran O. Group II intron splicing factors in plant mitochondria. Front Plant Sci. 2014;5:35. doi: 10.3389/fpls.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rurek M. Proteins involved in maturation pathways of plant mitochondrial and plastid c-type cytochromes. Acta Biochim Pol. 2008;55(3):417–433. [PubMed] [Google Scholar]
- 25.Daley DO, Clifton R, Whelan J. Intracellular gene transfer: Reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci USA. 2002;99(16):10510–10515. doi: 10.1073/pnas.122354399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daley DO, Whelan J. Why genes persist in organelle genomes. Genome Biol. 2005;6(5):110. doi: 10.1186/gb-2005-6-5-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;283(5407):1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 28.Gabaldón T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) J Mol Biol. 2005;348(4):857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- 29.James TY, et al. Shared signatures of parasitism and phylogenomics unite Cryptomycota and microsporidia. Curr Biol. 2013;23(16):1548–1553. doi: 10.1016/j.cub.2013.06.057. [DOI] [PubMed] [Google Scholar]
- 30.Flegontov P, et al. Divergent mitochondrial respiratory chains in phototrophic relatives of apicomplexan parasites. Mol Biol Evol. 2015;32(5):1115–1131. doi: 10.1093/molbev/msv021. [DOI] [PubMed] [Google Scholar]
- 31.Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 32.Popot JL, de Vitry C. On the microassembly of integral membrane proteins. Annu Rev Biophys Biophys Chem. 1990;19:369–403. doi: 10.1146/annurev.bb.19.060190.002101. [DOI] [PubMed] [Google Scholar]
- 33.Claros MG, et al. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur J Biochem. 1995;228(3):762–771. [PubMed] [Google Scholar]
- 34.Marcet-Houben M, Marceddu G, Gabaldón T. Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol Biol. 2009;9:295. doi: 10.1186/1471-2148-9-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutierres S, et al. Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA. 1997;94(7):3436–3441. doi: 10.1073/pnas.94.7.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pineau B, Mathieu C, Gérard-Hirne C, De Paepe R, Chétrit P. Targeting the NAD7 subunit to mitochondria restores a functional complex I and a wild type phenotype in the Nicotiana sylvestris CMS II mutant lacking nad7. J Biol Chem. 2005;280(28):25994–26001. doi: 10.1074/jbc.M500508200. [DOI] [PubMed] [Google Scholar]
- 37.Meyer EH, et al. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009;151(2):603–619. doi: 10.1104/pp.109.141770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karpova OV, Newton KJ. A partially assembled complex I in NAD4-deficient mitochondria of maize. Plant J. 1999;17(5):511–521. [Google Scholar]
- 39.Marienfeld JR, Newton KJ. The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 mitochondrial gene and is associated with reduced complex I function. Genetics. 1994;138(3):855–863. doi: 10.1093/genetics/138.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sabar M, De Paepe R, de Kouchkovsky Y. Complex I impairment, respiratory compensations, and photosynthetic decrease in nuclear and mitochondrial male sterile mutants of Nicotiana sylvestris. Plant Physiol. 2000;124(3):1239–1250. doi: 10.1104/pp.124.3.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasmusson AG, Soole KL, Elthon TE. Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol. 2004;55:23–39. doi: 10.1146/annurev.arplant.55.031903.141720. [DOI] [PubMed] [Google Scholar]
- 42.Yamato KT, Newton KJ. Heteroplasmy and homoplasmy for maize mitochondrial mutants: A rare homoplasmic nad4 deletion mutant plant. J Hered. 1999;90(3):369–373. [Google Scholar]
- 43.Noctor G, Dutilleul C, De Paepe R, Foyer CH. Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J Exp Bot. 2004;55(394):49–57. doi: 10.1093/jxb/erh021. [DOI] [PubMed] [Google Scholar]
- 44.Dutilleul C, et al. Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol. 2003;131(1):264–275. doi: 10.1104/pp.011155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hollinger DY. Photosynthesis and water relations of the mistletoe, Phoradendron villosum, and its host, the California calley oak, Quercus lobata. Oecologia. 1983;60(3):396–400. doi: 10.1007/BF00376858. [DOI] [PubMed] [Google Scholar]
- 46.Ehleringer JR, Cook CS, Tieszen LL. Comparative water-use and nitrogen relationships in a mistletoe and its host. Oecologia. 1986;68(2):279–284. doi: 10.1007/BF00384800. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JM, Choinski JS. Photosynthesis in the Tapinanthus-Diplorhynchus mistletoe host relationship. Ann Bot (Lond) 1993;72(2):117–122. [Google Scholar]
- 48.Marshall JD, Ehleringer JR, Schulze ED, Farquhar G. Carbon-isotope composition, gas-exchange and heterotrophy in Australian mistletoes. Funct Ecol. 1994;8(2):237–241. [Google Scholar]
- 49.Hao W. OrgConv: Detection of gene conversion using consensus sequences and its application in plant mitochondrial and chloroplast homologs. BMC Bioinformatics. 2010;11:114. doi: 10.1186/1471-2105-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho Y, Qiu YL, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci USA. 1998;95(24):14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol Biol Evol. 2008;25(8):1762–1777. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge Univ Press; Cambridge, UK: 1984. [Google Scholar]
- 53.Zhu A, Guo W, Jain K, Mower JP. Unprecedented heterogeneity in the synonymous substitution rate within a plant genome. Mol Biol Evol. 2014;31(5):1228–1236. doi: 10.1093/molbev/msu079. [DOI] [PubMed] [Google Scholar]
- 54.Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311(5768):1727–1730. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- 56.Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS ONE. 2011;6(1):e16404. doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Small ID, Isaac PG, Leaver CJ. Stoichiometric differences in DNA molecules containing the atpA gene suggest mechanisms for the generation of mitochondrial genome diversity in maize. EMBO J. 1987;6(4):865–869. doi: 10.1002/j.1460-2075.1987.tb04832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Small I, Suffolk R, Leaver CJ. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell. 1989;58(1):69–76. doi: 10.1016/0092-8674(89)90403-0. [DOI] [PubMed] [Google Scholar]
- 59.Woloszynska M. Heteroplasmy and stoichiometric complexity of plant mitochondrial genomes—though this be madness, yet there’s method in’t. J Exp Bot. 2010;61(3):657–671. doi: 10.1093/jxb/erp361. [DOI] [PubMed] [Google Scholar]
- 60.Alverson AJ, Rice DW, Dickinson S, Barry K, Palmer JD. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant Cell. 2011;23(7):2499–2513. doi: 10.1105/tpc.111.087189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Naito K, Kaga A, Tomooka N, Kawase M. De novo assembly of the complete organelle genome sequences of azuki bean (Vigna angularis) using next-generation sequencers. Breed Sci. 2013;63(2):176–182. doi: 10.1270/jsbbs.63.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mower JP, Case AL, Floro ER, Willis JH. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol Evol. 2012;4(5):670–686. doi: 10.1093/gbe/evs042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sloan DB, Müller K, McCauley DE, Taylor DR, Storchová H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 2012;196(4):1228–1239. doi: 10.1111/j.1469-8137.2012.04340.x. [DOI] [PubMed] [Google Scholar]
- 64.Sloan DB. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013;200(4):978–985. doi: 10.1111/nph.12395. [DOI] [PubMed] [Google Scholar]
- 65.Bendich AJ. Reaching for the ring: The study of mitochondrial genome structure. Curr Genet. 1993;24(4):279–290. doi: 10.1007/BF00336777. [DOI] [PubMed] [Google Scholar]
- 66.Backert S, Börner T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.) Curr Genet. 2000;37(5):304–314. doi: 10.1007/s002940050532. [DOI] [PubMed] [Google Scholar]
- 67.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9(9):868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mower JP. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009;37(Web Server issue):W253-9. doi: 10.1093/nar/gkp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lenz H, Knoop V. PREPACT 2.0: Predicting C-to-U and U-to-C RNA editing in organelle genome sequences with multiple references and curated RNA editing annotation. Bioinform Biol Insights. 2013;7:1–19. doi: 10.4137/BBI.S11059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34(Web Server issue):W609-12. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 76.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 77.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 78. Stevens PF (2014) Angiosperm Phylogeny Website. Available at www.mobot.org/mobot/research/apweb/welcome.html. Accessed March 1, 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.