Abstract
Masson pine is an important timber and resource for oleoresin in South China. Increasing yield of oleoresin in stems can raise economic benefits and enhance the resistance to bark beetles. However, the genetic mechanisms for regulating the yield of oleoresin were still unknown. Here, high-throughput sequencing technology was used to investigate the transcriptome and compare the gene expression profiles of high and low oleoresin-yielding genotypes. A total of 40,690,540 reads were obtained and assembled into 137,499 transcripts from the secondary xylem tissues. We identified 84,842 candidate unigenes based on sequence annotation using various databases and 96 unigenes were candidates for terpenoid backbone biosynthesis in pine. By comparing the expression profiles of high and low oleoresin-yielding genotypes, 649 differentially expressed genes (DEGs) were identified. GO enrichment analysis of DEGs revealed that multiple pathways were related to high yield of oleoresin. Nine candidate genes were validated by QPCR analysis. Among them, the candidate genes encoding geranylgeranyl diphosphate synthase (GGPS) and (-)-alpha/beta-pinene synthase were up-regulated in the high oleoresin-yielding genotype, while tricyclene synthase revealed lower expression level, which was in good agreement with the GC/MS result. In addition, DEG encoding ABC transporters, pathogenesis-related proteins (PR5 and PR9), phosphomethylpyrimidine synthase, non-specific lipid-transfer protein-like protein and ethylene responsive transcription factors (ERFs) were also confirmed to be critical for the biosynthesis of oleoresin. The next-generation sequencing strategy used in this study has proven to be a powerful means for analyzing transcriptome variation related to the yield of oleoresin in masson pine. The candidate genes encoding GGPS, (-)-alpha/beta-pinene, tricyclene synthase, ABC transporters, non-specific lipid-transfer protein-like protein, phosphomethylpyrimidine synthase, ERFs and pathogen responses may play important roles in regulating the yield of oleoresin. These DEGs are worthy of special attention in future studies.
Introduction
Oleoresin, as a mixture of many terpenoids including turpentine (monoterpenes and sesquiterpenes) and rosin (diterpenes), is stored in resin ducts or blisters in stems, roots, needles and cones of conifer trees [1]. Terpenoids as an important natural source for industrial chemicals were used widely in solvents, cleaning agents for varnishes and paints, flavors and fragrances, synthetic rubber, disinfectants, coatings, printing ink resins, and waterproofing materials [2–5]. Terpenoids also can be exploited as advanced liquid biofuels. Biofuels derived from terpenoids have similar properties as diesel and gasoline, and the density and hygroscopicity are amenable to mixing with fossil fuels [6]. In addition, terpenoids also play an important role in defense system of conifer trees. The resin ducts or blisters are severed and oleoresin is released after suffering abiotic or biotic stimuli such as mechanical wounding, insect attack and pathogen invasion. Emitted monoterpenes and sesquiterpenes are toxic volatiles that can directly affect herbivores, while diterpene resin acids provide the protection through forming physical barriers at the point of wound [7–8]. Other studies have found that the trees releasing a large amount of oleoresin upon wounding exhibit the strongest resistance to beetle attack in southern pines [9].
In addition to constitutive oleoresin, new oleoresin can be synthesized and traumatic resin ducts (TRD) can be formed after stem suffering abiotic or biotic stimuli in conifers [10–11]. In conifers, the yield of oleoresin is a quantitative trait under moderate to strong genetic control [12–14]. Important genetic gains can be obtained through the selection of high resin yielders [15]. Based on the traditional breeding, many high oleoresin-yielding families, clones and individuals have been selected [16–18]. Previous studies demonstrate that the yield of oleoresin is strongly correlated with tree diameter, percentage of live crown, radial resin canal number and resin canal volume [15,19]. The relationship between the yield of oleoresin and the concentration of each terpenoid is also investigated and the results show that some terpenoids can be used as the diagnostic marker for high oleoresin-yielding trees [20]. However, molecular studies on genes related to the yield of oleoresin in conifers are still rare. Exploration of the genes for regulating the yield of oleoresin can reduce the breeding cycle and rapidly increase the production of oleoresin in stems of conifers by genetic engineering.
As its well known, the biosynthesis of oleoresin is usually completed through two separate pathways of methyl-erythritol 4-phosphate (MEP) and mevalonate (MVA) pathways in conifers. Monoterpenes and diterpenes are biosynthesized by MEP pathway [21], while the biosynthesis of sesquiterpenes is completed by MVA pathway [22]. During the biosynthesis of oleoresin terpenoids, the enzymes for biosynthesis include 1-deoxyxylulose 5-phosphate reductoisomerase (DXR), prenyltransferases, 1-deoxyxylulose 5-phosphate synthase (DXS), hydroxymethylbutenyl 4-phosphate reductase (HDR), terpenoid synthase (TPS) and cytochrome P450-dependent monooxygenase. In recent decades, many studies on terpenes have focused on the potential role in the resistance against abiotic or biotic stimuli. TPS enzymes as the key enzymes for generating diverse terpenoids have been explored widely. Many TPS genes associated with the resistance have been identified in several coniferous species including loblolly pine (P. taeda), grand fir (Abiesgrandis), and spruce species [23–25]. The genes encoding TPS of genus Pinus belong to TPSd gene families [1].
Masson pine (P. massoniana) is an important timber and resource for oleoresin in South China. Approximately 90% of rosin comes from masson pine. Oleoresin is annually tapped using the bark streak method of wounding, and collected from living pine trees after the diameter at breast height (DBH) reaching a certain size class in China. The State Forestry Administration of China stipulates that pine trees are not used for the tapping of oleoresin until the DBH exceeds 20 cm. The yield of oleoresin varies greatly within masson pine [15]. Many superior provenances, families, and clones with high yield of oleoresin have been selected in the progress of genetic improvement [16–18]. However, the genetic mechanisms for regulating the yield of oleoresin are unknown.
The next-generation sequencing has greatly contributed to biological studies in non-model systems. The de novo transcriptomic analysis has been used to explore the differential gene expression profile among tissues subjected to different treatments. In the present study, the aim is to explore the transcriptome of secondary xylem in masson pine and identify the genes with differential expression between the high and low oleoresin-yielding trees based on high-throughput sequencing technology. These results will be valuable for genomic and genetic studies on high oleoresin-yielding masson pine in the future.
Results
Compositional characterization of oleoresin compounds in masson pine
In order to reveal the compositional properties of oleoresin, we performed gas chromatography-mass spectrometry (GC/MS) analysis for high and low oleoresin-yielding masson pines. Totally 26 oleoresin terpenoids with matching degree more than 90% to NIST and the content higher than 0.01% were identified. As shown in Table 1 and S1 Fig, 26 oleoresin terpenoids were characterized by qualitative and quantitative analysis, including 7 monoterpenes, 11 sesquiterpenes and 8 diterpenes. For both high and low oleoresin-yielding genotypes, the most abundant terpenoid is alpha-pinene, accounting for 85% of all monoterpenes, which is followed by beta-pinene and limonene. Among sesquiterpenes, the most abundant compounds are longifolene and caryophyllene, and the sum of them reaches up to 82% of sesquiterpenes. Among dierpenes, palustric acid reveals the highest level, which is followed by neoabietic acid and abietic acid.
Table 1. The average content and SD of oleoresin compounds and significant difference between 35 high and 35 low oleoresin-yielding masson pines using non-parametric Mann-Whitney test.
| High oleoresin-yielding trees | Low oleoresin-yielding trees | |||||
|---|---|---|---|---|---|---|
| Compounds | RI | Mean (mg/g) | SD | Mean (mg/g) | SD | Sig. |
| Monoterpenes | 170.53 | 18.97 | 173.79 | 18.96 | 0.5076 | |
| Tricyclene | 920 | 0.27 | 0.08 | 0.34 | 0.15 | 0.0270* |
| alpha-Pinene | 932 | 146.19 | 16.49 | 149.71 | 16.43 | 0.4100 |
| Camphene | 944 | 4.16 | 0.71 | 3.69 | 0.64 | 0.0091** |
| beta-Pinene | 973 | 9.75 | 3.05 | 10.95 | 4.15 | 0.2075 |
| beta-Myrcene | 991 | 3.12 | 0.6 | 2.75 | 0.55 | 0.0146* |
| Limonene | 1025 | 5.51 | 1.07 | 4.74 | 1.44 | 0.0219* |
| Borneol | 1153 | 0.98 | 0.4 | 1.03 | 0.57 | 0.7040 |
| Sesquiterpenes | 130.48 | 31.31 | 106.76 | 36.68 | 0.0092** | |
| Longipinene | 1344 | 5.85 | 2.56 | 4.32 | 1.98 | 0.0122* |
| Longicyclene | 1363 | 5.31 | 2.14 | 4.97 | 2.55 | 0.5790 |
| Ylangene | 1368 | 0.79 | 0.32 | 0.66 | 0.31 | 0.1175 |
| Copaene | 1372 | 0.14 | 0.08 | 0.14 | 0.11 | 0.8415 |
| Sativene | 1383 | 2.8 | 1.14 | 2.58 | 1.33 | 0.4925 |
| Longifolene | 1397 | 88.38 | 23.02 | 66.47 | 26.37 | 0.0011** |
| Caryophyllene | 1413 | 19.79 | 6.87 | 21.57 | 8.73 | 0.3826 |
| Humulene | 1448 | 3.4 | 1.24 | 3.62 | 1.71 | 0.5681 |
| b-Farnesene | 1458 | 1.66 | 1.1 | 1.38 | 1.16 | 0.3394 |
| Germacrene D | 1476 | 2.01 | 2.95 | 0.79 | 0.99 | 0.0951 |
| δ-Cadinene | 1521 | 0.35 | 0.25 | 0.27 | 0.19 | 0.1878 |
| Dierpenes | 523.85 | 24.66 | 540.84 | 25.65 | 0.0114* | |
| Pimaric acid | 2219 | 49.5 | 12.06 | 53.73 | 7.61 | 0.1095 |
| Sandaracopimaric acid | 2227 | 11.15 | 1.43 | 11.35 | 1.54 | 0.6039 |
| Isopimaric acid | 2248 | 21.93 | 10.86 | 15.91 | 12.2 | 0.0481* |
| Palustric acid | 2256 | 268.59 | 31.12 | 268.15 | 27.96 | 0.9541 |
| Dehydroabietic acid | 2273 | 35.58 | 12.97 | 38.61 | 13.53 | 0.3803 |
| 8,12-Abietadienoic acid | 1.89 | 1.1 | 1.66 | 1.05 | 0.4228 | |
| Abietic acid | 2293 | 57.66 | 10.47 | 69.31 | 13.66 | 0.0005** |
| Neoabietic acid | 2341 | 77.55 | 9.15 | 82.11 | 11.23 | 0.0899 |
* and ** show the significant difference at p value less than 0.05 and 0.01, respectively.
Using the non-parametric Mann-Whitney test, the contents of 8 oleoresin terpenoids including tricyclene, camphene, limonene, beta-myrcene, longipinene, longifolene, isopimaric acid and abietic acid, revealed significantly difference between high and low oleoresin-yielding genotypes. For camphene, beta-myrcene, limonene, longipinene, longifolene and isopimaric acid, the contents of these compounds were higher in high oleoresin-yielding trees, while the contents of tricyclene and abietic acid were higher in low oleoresin-yielding trees. Overall, the high oleoresin-yielding trees had a higher content of sesquiterpene and lower content of diterpene when compared with the low oleoresin-yielding trees.
RNA-Seq and de novo transcriptome assembly
In order to obtain information in xylem tissues of masson pine at major oleoresin-secreting stages, total RNA was isolated from twenty high and low oleoresin-yielding clones for RNA-Seq performed on the Illumina Hiseq2000 platform. After quality assessment and data screening, we obtained 40,690,540 high quality reads (8.2 gigabase pairs) with 84.80% (Q≥30 bases). Then these reads were assembled into 3,641,300 contigs using SOAPdenovo (S1 Table). In the following, using Trinity assembly program, short-read sequences were assembled into 137,499 transcripts with a mean length of 904 bp. Finally, 84,842 unigenes were obtained with a mean length of 690 bp. Among these unigenes, 15,713 unigenes (18.52%) were larger than 1 kb. The results suggested that the high quality and throughput sequences were prepared for the further analysis. The length distributions of transcripts and unigenes (S1 Table) have the similar tendency with N50 values of 1,671 bp and 1,291 bp, respectively.
Functional annotation
Functional annotation of retrieved unigenes was carried out by aligning sequence similarity with several public databases with a threshold of E-value less than 10-5, and the annotation results are listed in S2 Table. A total of 35,353 (41.7%) unigenes were annotated in the public databases (S3 Table). Based on the annotation using the NCBI non-redundant protein (Nr) database, 27,147 unigenes were distributed into one or more GO terms, with 28.36% for cellular components, 22.80% for molecular functions and 48.84% for biological processes (S2 Fig). In addition, 9,990 of 84,842 sequences could be assigned to Cluster of Orthologous Groups of proteins (COG) database for functional prediction and classification (S3 Fig).
Analysis of differentially expressed genes (DEGs)
To screen the DEGs related to yield of oleoresin in masson pine, 10 high and 10 low oleoresin-yielding genotypes were used for the establishment of six sequencing libraries. The six libraries generated 9.61–12.40 million high-quality reads. Subsequently, the sequences of these reads were mapped to the transcriptome database described above. More than 80% of these reads matched the sequences in the transcriptome, and 92% of these reads could be identified unequivocally by the unique mapping (S4 Table). The reads were mapped to unigenes and quantified to obtain the abundance of gene expression by Reads per Kilobase per Million mapped Reads (RPKM). In order to explore molecular difference in expression levels between high and low oleoresin-yielding masson pines, the significant DEGs were compared by applying cutoff of p-value less than 0.05 (FDR BH corrected). A total of 649 DEGs were identified (S5 Table), and 343 unigenes were significantly up-regulated and 306 unigenes were significantly down-regulated. These results suggested that the high yield of oleoresin might be due to the different expression levels of these DEGs.
Multiple pathways are involved in the high yielding of oleoresin identified by GO enrichment analysis of DEGs
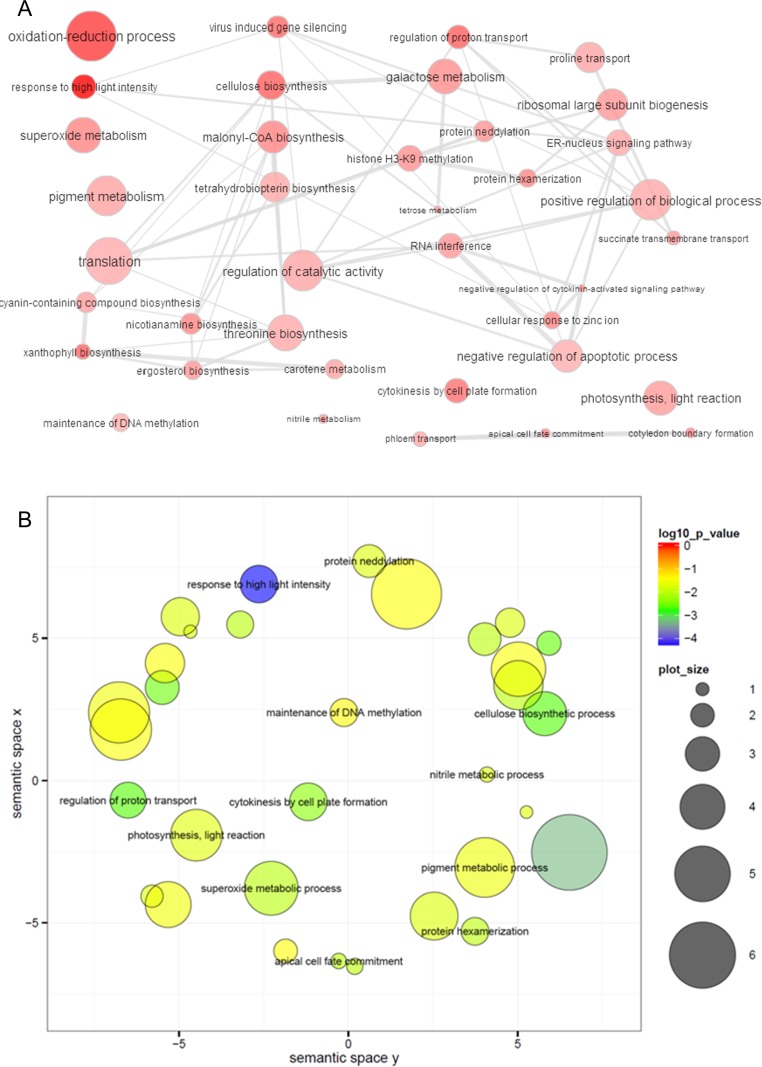
To achieve a functional annotation of the genes related to the yield of oleoresin, we performed GO enrichment analysis of 649 DEGs. In total, we uncovered 109 GO terms that were enriched significantly (p < 0.05, FDR corrected) (S6 Table). In the biological process, the enriched GO terms for DEGs were “oxidation-reduction process”, “regulation of cellular biosynthetic process”, “cellular protein metabolic process”, “regulation of nitrogen compound metabolic process” and “translation” (Fig 1A, S6 Table). However, the most significant GO terms between high and low oleoresin-yielding trees were “response to high light intensity”, “oxidation-reduction process” and “response to far red light”. The enriched GO terms were also visualized by ReviGO [26] (Fig 1B), and pathways such as light response, photosynthesis, and pigment metabolism (Fig 1B), indicating that multiple biological pathways might be involved in regulating the yield of oleoresin.
Fig 1. GO enrichment analysis of DEGs between high and low oleoresin-yielding masson pines using REVIGO.
(A) The interactive graph of the enriched GO terms for DEGs. Bubble color indicates the p-value; bubble size indicates the frequency of the GO term. Highly similar GO terms are linked by edges in the graph, where the line width indicates the degree of similarity. (B) The scatter plot showing the significance of the GO term for DEGs (terms remaining after reducing redundancy) in a two-dimensional space derived by applying multi-dimensional scaling to a matrix of GO term semantic similarities. Bubble color indicates the p-value for the false discovery rates. The circle size represents the frequency of the GO term (more general terms are represented by larger size bubbles).
Validation of candidate gene expression by qRT-PCR
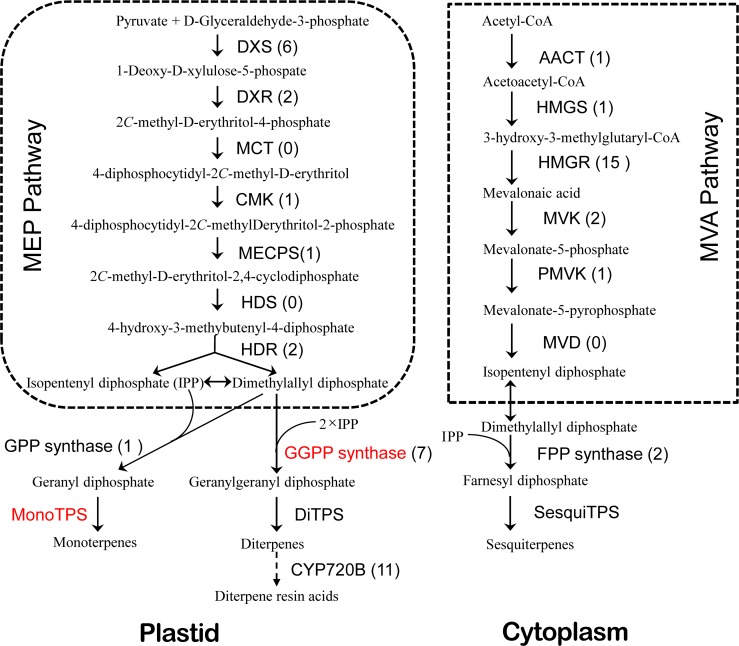
In MEP and MVA pathways, 96 unigenes were associated with the biosynthesis of terpenoid backbone according to transcriptome database. Except 4-diphosphocytidyl-2Cmethyl-D-erythritol synthase (MCT), mevalonate diphosphate decarboxylase (MVD) and 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (HDS), those enzymes participating the biosynthesis of terpenoid backbone in MVA and MEP pathways had one or more corresponding unigenes (Fig 2). Among them, 43 unigenes annotated as TPS by Nr or Swissprot database were obtained, including 14 unigenes for monoTPS, 19 unigenes for sesquiTPS and 10 unigenes for diTPS.
Fig 2. Biosynthetic pathway of terpenoids in P. massoniana (adapted from Zulak et al [22]).
The number of unigenes homologous to gene families encoding these enzymes was provided in parentheses. The enzymes highlighted with red fonts showed that DEGs were detected in gene families encoding these enzymes. DXS: 1-deoxy-dxylulose-5-phosphate synthase; DXR: 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MCT: 4-diphosphocytidyl-2Cmethyl-D-erythritol synthase; CMK: 4-diphosphocytidyl-2C–methyl-D-erythritol kinase; MECPS: 2C-methyl-D-erythritol 4-phosphate cytidylyltransferase; HDS: 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase; HDR: 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase; AACT: acetoacetyl-CoA thiolase; HMGS: 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR: 3-hydroxy-3-methylglutaryl-CoA reductase; MVK: mevalonate kinase; PMVK: phosphomevalonate kinase; MVD: mevalonate diphosphate decarboxylase; IPPI: isopentenyl-diphosphate isomerase; FPPS: farnesyl diphosphate synthase; GPPS: geranyl diphosphate synthase; SesquiTPS: Sesquiterpene synthase; MonoTPS: Monoterpene synthase; DiTPS: Diterpene synthase; CYP720B: Abietadienol/abietadienal oxidase PtAO.
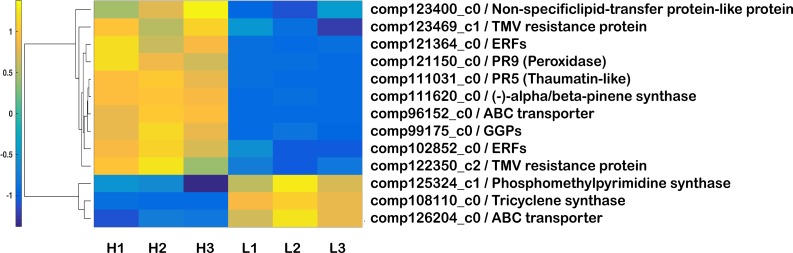
The biosynthesis pathway of terpenoid backbone played central roles in the biosynthesis of oleoresin. Hierarchical clustering of these candidate genes was shown in Fig 3. In high oleoresin-yielding trees, the expression of GGPS and (-)-alpha/beta-pinene synthase was approximately 8 and 14 times higher than that in low oleoresin-yielding trees, respectively. However, the expression of tricyclene synthase was approximately 87 times higher than that in high oleoresin-yielding trees. In addition, many DEGs coding phosphomethylpyrimidine synthase, non-specific lipid-transfer protein-like protein, ethylene responsive transcription factors (ERFs), ATP binding cassette (ABC) transporter, pathogenesis-related proteins (PR5 and PR9) and TMV resistance protein had obvious different expressions between high and low oleoresin-yielding trees. Among these DEGs, two candidate genes of ERFs, two candidate genes of TMV resistance protein, two candidate genes of PRs, one candidate gene of ABC transporter and one candidate gene of non-specific lipid-transfer protein-like protein were up-regulated in high oleoresin-yielding trees (S7 Table). One candidate gene of phosphomethylpyrimidine synthase and one candidate gene of ABC transporter were down-regulated in high oleoresin-yielding trees (S7 Table).
Fig 3. Clustering and heatmap of key DEG related to the yield of oleoresin based on their expression levels.
Three biological replicates were used for high-yielding genotype (H1, H2, H3) and low-yielding genotype (L1, L2, L3). Colored bars on the left of the heatmap mark the major distinct branches in the clustering tree grouping genes with similar expression pattern.
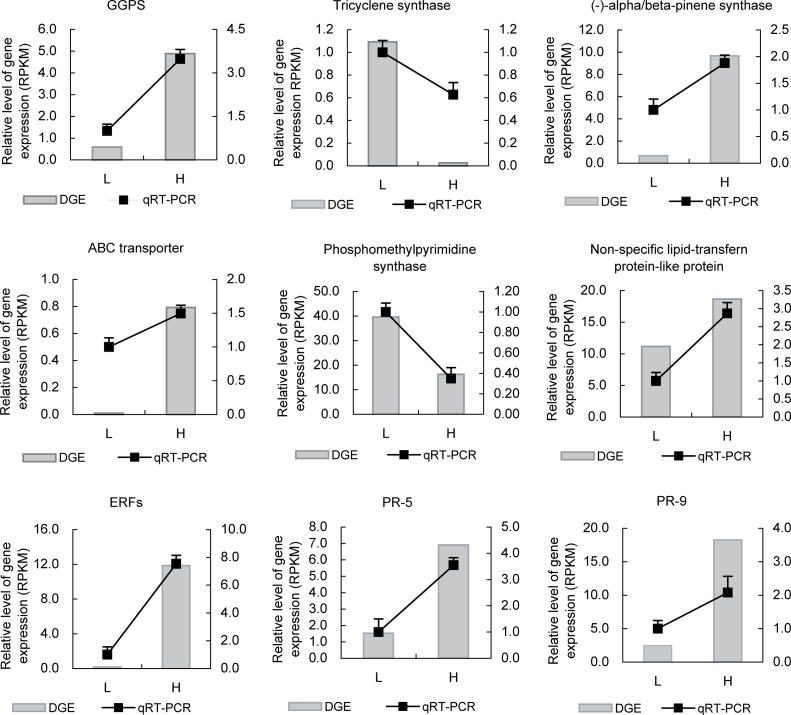
To validate the reliability of the results from DEGs, we selected 9 representative candidate genes for qRT-PCR analysis (Fig 4). Among 9 candidate genes, three candidate genes involve in the biosynthesis of terpeniods including GGPS (comp99175_c0), (-)-alpha/beta-pinene (comp111620_c0) and tricyclene synthase (comp108110_c0), and two candidate genes of PR5 (comp111031_c0) and PR9 (comp121150_c0) involve in defense response. The remaining four candidate genes encoded ABC transporter (comp96152_c0), phosphomethylpyrimidine synthase (comp125324_c1), non-specific lipid-transfer protein-like protein (comp123400_c0) and ERFs (comp121364_c0). Overall, the results of qRT-PCR analysis were consistent with DEG analysis, suggesting that these candidate genes might be the important candidates for controlling the yield of oleoresin (Fig 4).
Fig 4. Quantitative RT-PCR validation of tag-mapped candidate genes associated with the yield of oleoresin.
Relative expression levels of qRT-PCR calculated using Elongation factor 1-alpha as the internal control were shown in the right y-axis. Relative expression levels of DGE were shown in the left y-axis.
Discussion
It is known that the yield of oleoresin is a highly heritable trait, and important genetic gains can be obtained from the selection of high oleoresin yielder by traditional breeding methods [13]. We detected 26 terpenoids from the oleoresin in masson pines using GC/MS in this study. The major compounds of turpentine oil were alpha-pinene, beta-pinene, limonene and longifolene, and the major compounds of rosin were palustric acid, neoabietic acid, abietic acid, pimaric acid, dehydroabietic acid, isopimaric acid and sandaracopimaric acid in masson pines, which were consistent with the results from previous studies [27,28]. Among these terpenoids, the contents of tricyclene, camphene, β-myrcene, limonene, longipinene, longifolene, isopimaric and abietic acid revealed significantly difference between high and low oleoresin-yielding trees. In P. halepensis, myrcene, isopimaric and abietic acids were also reported to be differentiated in high oleoresin-yielding trees [20].
However, our understanding regarding key regulatory genes for the generation of oleoresin in conifers is still fragmental. In order to increase our knowledge of the genes related to the yield of oleoresin, we first analyzed the transcriptome of the secondary xylem that is the major secretion tissue of masson pines. We analyzed the transcriptome of masson pine, and obtained a rich resource of sequences. In the set of highly differentially expressed genes between high and low oleoresin-yielding trees, a number of genes were reported to participate in terpenoid backbone biosynthesis, terpenoid transport and defense response. GGPS, the isoprenyl diphosphate synthases belonging to a class of prenyltransferases, can catalyze the formation of geranylgeranyl diphosphate (GGPP) from isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) via three successive condensation reactions [29,30]. GGPS genes were strongly up-regulated by both insect species and mechanical wounding in Picea sitchensis [31]. The up-regulation of GGPS genes was closely correlated with traumatic resin duct formation, thus producing higher amounts of monoterpenes, sesquiterpenes and diterpene resin acids [32]. In this study, the expression level of GGPS (comp99175_c0) was significantly different between high and low oleoresin-yielding masson pine trees, and had a higher expression in high oleoresin-yielding trees, suggesting the more efficient catalysis during the process of GGPP formation in high oleoresin-yielding trees.
The TPS enzyme generates one or more products using the substrate of GPP, FPP or GGPP by an electrophilic reaction [33–38]. Monoterpenes, sesquiterpenes and diterpenes are the products of GPP, FPP or GGPP, respectively. A large number of known TPS genes are induced for conifers in response to abiotic and biotic stimuli. In our study, the candidate gene of (-)-alpha/beta-pinene synthase (comp111620_c0) had higher expression in high oleoresin-yielding trees, while the candidate gene of tricyclene synthase (comp108110_c0) had higher expression in low oleoresin-yielding trees. Tricyclene, the product of tricyclene synthase, also had significantly different contents between high and low oleoresin-yielding genotypes. Alpha-pinene is a mixture of the isomers (+)-alpha-pinene and (-)-alpha-pinene. The content of (-)-alpha-pinene is approximately 10- to 20-fold higher than (+)-alpha-pinene in masson pines [39]. Although we found the significantly different expression of (-)-alpha/beta-pinene synthases between high and low oleoresin-yielding trees, there was no obvious difference in alpha-pinene using Mann-whitney tests.
GGPP formed by GPPS is the precursor of diterpenes, gibberellins, carotenoids and chlorophylls. Although the candidate gene GPPS had a higher expression level in high oleoresin-yielding trees, the content of diterpenes in oleoresin from high oleoresin-yielding trees was not higher in high oleoresin-yielding trees according to the statistical analysis using Mann-whitney tests. In contrary, oleoresin from high oleoresin-yielding trees had a higher content of monoterpenes and a lower content of diterpenes. After the stem is wounded, diterpene is transported by liquid monoterpenes and sesquiterpenes to the point of the wound, which can polymerize and seal the wound [7,8]. Therefore, we speculate that the increased diterpenes in low oleoresin-yielding trees can not only more rapidly seal the wound, but also can form physical barriers to prevent oleoresin outflowing, which will be the explanation of low oleoresin-yielding trees containing a higher content of diterpenes. The functional studies on the relevant genes are expected to be useful in the improvement of oleoresin yield and the resistance of the population of masson pines.
TPS enzymes producing oleoresin terpenoids have been localized to epithelial cells around resin ducts [35]. To the present, the transport mechanisms are unknown for the terpenoids from living epithelial cells to resin ducts in conifers [22]. The ABC transporters are confirmed to be participated in the active transport of dissimilar compounds using ATP hydrolysis energy [40]. For example, Jasinski et al. [41] have reported that NpABC1 mediates the export of terpenoid from Nicotiana plumbaginfolia cells. In loblolly pine, Westbrook et al. [14] have also found that the expression of ABC transporter is regulated by oleoresin terpenoid. In our study, the variation in oleoresin yield is significantly associated with ABC transporters, which is consistent with the studies from Westbrook et al. [14]. Therefore, ABC transporters are also involved in the transport of conifer oleoresin.
In all differential expression genes associated to oleoresin yield, the candidate genes coding ERFs were involved. ERF containing a highly conserved DNA-binding domain plays an important role in regulating the expression of genes under external stresses [42]. In regulatory pathways of stress signal, ERFs can bind the GCC-box element presenting in many PR genes to control their expression [43]. In our study, the higher expression of ERFs was observed in high oleoresin-yielding trees after stem wounding.
To date, PR genes are commonly considered as the genes involved in systemically acquired resistance (SAR), and are coordinated with a wide range of plant defense genes responding to biotic and abiotic. PR5 family (thaumatin-like proteins) with low molecular weight can cause osmotic rupture [44]. In hybrid poplar, enhanced expression of PR5 is observed in phloem proteomic response to leaf wounding [45]. PR5 is also a hallmark of the resistant cultivar in interactions between Hero A-resistant tomatoes and cyst nematode [46]. In our study, PR5 is induced to a higher level in high oleoresin-yielding genotypes when compared with low oleoresin-yielding genotypes. Therefore, high oleoresin-yielding masson pine may have another characteristics of strong resistance, which has been proven in P. taeda by Storm et al. [9], wherein oleoresin flow is significantly higher in resistant trees than general population trees responding to the southern pine beetle. The deduction for masson pine will be verified in further studies.
Conclusion
In this study, a comprehensive transcriptomic analysis from secondary xylems of masson pine was conducted, which provided a valuable resource for genetic and genomic study in the future. The candidate genes associated with oleoresin biosynthesis and the expression profiles of high and low oleoresin-yielding genotypes were characterized by de novo assembly and DEGs analysis. GO enrichment analysis of DEGs revealed that multiple biological pathways including light signaling response, photosynthesis and lipid metabolism were related to the high yield of oleoresin. Further analysis of candidate genes involved in terpenoid backbone biosynthesis revealed that GGPS and TPS enzymes of (-)-alpha/beta-pinene, and tricyclene synthase were related to the high yield of oleoresin. These results coordinated well with GC/MS data that the products of tricyclene synthases were more enriched in high-yielding genotype than low-yielding genotype. Moreover, DEGs of PRs, ABC transporters, non-specific lipid-transfer protein-like protein, phosphomethylpyrimidine synthase and ERFs might be important for the high yield of oleoresin. Overall, our work provided new insights into the molecular mechanisms for oleoresin production, which may lay a solid foundation for expediting the breeding of high-yield oleoresin pines.
Materials and Methods
Plant materials
The experiments were conducted using twenty-seven-year-old masson pine clones cultivated in the first generation seed orchard established in 1986 by Laoshan Forest Farm, Chunan County, Zhejiang, China. The clones were propagated by grafting and no any relationships were existed between their ortets. In 2012, the yield of oleoresin was measured using the bark streak method of wounding for oleoresin tapping [15]. No stimulant to induce oleoresin flow was applied. The yield of oleoresin was calculated as the yield of the individual per day per cm streak length in grams to adjusting stem diameter influence on oleoresin yield. Based on the measurement, 35 high oleoresin-yielding clones (more than 5.0 g d-1 cm-1) and 35 low oleoresin-yielding clones (less than 1.5 g d-1 cm-1) were tapped the xylem oleoresin using the traditional bark streak method of wounding to analyze the compositions of terpenoids. The fresh oleoresin was collected using 1.5 mL polyethylene tube for each sample and the tube was hermitically closed and refrigerated until the analysis.
From these clones, 10 high oleoresin-yielding clones and 10 low oleoresin-yielding clones were selected for the analysis of transcriptome and DEGs. The low oleoresin-yielding clones were served as the control. Three ramets for each clone were selected as three biological replicates. No protected species were involved in this study. After the removal of the bark, phloem, cambium and surface layer of tapping streak last year, approximately 5 mm deep fresh secondary xylem tissues adjoining cambium were harvested from sample trees. These samples were put into liquid nitrogen immediately in field and then stored at -80°C for RNA extraction.
Analysis of oleoresin compositions
For quantitative analysis, the samples for terpenoid identification using gas chromatography-mass spectrometry (GC/MS) were prepared according to Karanikas’ method [20].
Subsequently, terpenoids of masson pine were detected by GC/MS with an Agilent 6890N gas chromatography coupled with a HP-5MS column (ID: 0.25 mm, length: 30 m, film thickness: 0.25 μm), and an Agilent 5975B mass spectrometer. For GC, the program of oven temperature was 60°C for 2 min, increasing 2°C/min to 80°C, 80°C for 5 min, subsequent increase rate of 4°C/min until 280°C and 280°C for 5 min. Helium was used as the carrier gas at a flow rate of 1 mL/min, injector temperature was set up at 260°C, injection volume was 1 μL with a split ratio of 1:50. Electron ionization mass spectrometry analysis was carried out with 70 eV electron energy, 230°C ion source and 280°C connection part temperature. Terpenoids were identified by matching fragmentation of mass spectra with NIST08. In addition, experimental retention indices were also used to match with reference compounds. The concentration of each terpenoid was expressed as the mass of a compound in oleoresin per gram. SPSS 18.0 statistical package was used to perform the statistical analysis. The significant difference between high and low oleoresin-yielding clones was determined by non-parametric Mann-Whitney test.
RNA isolation
Total RNA from each sample of twenty high and low oleoresin-yielding clones was separately extracted using Plant RNA kit of RN38 EASYspin plus (Aidlab Biotech, Beijing, China) containing DNA digestive enzyme. The concentration of total RNA was detected using Ultrasec 2100 pro UV/Visible Spectrophotometer, and the integrity of total RNA was detected using 1% agarose gel electrophoresis. Then, the integrity of RNA was also tested using an Agilent 2100 Bioanalyzer before proceeding. The RNA with concentration more than 400 ng/μL, OD260/280 ranging from 1.8 to 2.2, OD260/230 over 2.0, 28S/18S no less than 1.5 and RIN over 8.0, was defined as high quality RNA. Subsequently, equal quantity of RNA from one ramet of each high oleoresin-yielding clone was pooled for cDNA synthesis. For low oleoresin-yielding clones, the RNAs were pooled using the same method. A total of six RNA samples were obtained, including three biological replicates for high and low oleoresin-yielding genotypes, respectively.
Transcriptome sequencing and assembly of masson pine
To obtain a comprehensive list of transcripts, equal amounts of high-quality RNA from six RNA samples were pooled together. Then, total RNA was delivered to Biomarker Technology Company (Beijing, China) to construct cDNA library and sequencing. The cDNA library was paired-end sequenced on the Illumina HiSeq 2000 sequencing platform, and 2× 101 bp reads were yielded from either end of a cDNA fragment.
After filtering the raw reads, we assembled the high quality reads into contigs using Trinity method [47]. Then, the transcripts were assembled with the paired-end information of contigs. Finally, the longest transcripts from the potential alternative splicing transcripts were selected as the unigenes. The data has been submitted to NCBI, and the accession number is SRP050025.
Functional annotations and classifications
Functional annotations were carried out by matching sequences with public databases. All unigenes were matched with NCBI Nr database, Swiss-Prot database and COG database using BLASTx with the E-value of less than 10-5. GO classification was performed using WEGO software (http://wego.genomics.org.cn/cgibin/wego/index.pl). Pathway assignments were achieved from KEGG web server. RPKM was used to represent the expression abundance of the unigenes.
Differentially expressed genes and GO enrichment
All high quality reads were aligned to the assembled transcripts above using Bowtie, allowing no more than one nucleotide mismatch. In order to compare the expression abundance among six samples, tags were normalized into RPKM values. The differential expression of genes was analyzed by the edgeR package (Version 3.2.4) in BioConductor (release 2.12, R version 2.15.0) [48]. The right-sided hypergeometric enrichment test was performed at a medium network specificity selection and p-value correction was performed using the Benjamini-Hochberg method. The GO terms with p-value less than or equal to 0.05 were considered as the significant enrichment. The biological process of enriched GO terms was visualized by using ReviGO [26]. And the relatives of enriched GO terms were identified and plotted by using Matlab Bioinformatics toolbox (Mathworks Inc.).
Quantitative RT-PCR analysis
The RNA samples used in qRT-PCR and DGE experiments were identical. The primer pairs (S8 Table) for representative candidate genes were designed using Primer 3.0 software with the optimal Tm at 58–62°C, primer length of 19–21 nucleotides, PCR product size of 120–200 bp and GC content of 45–55%.
Quantitative RT-PCR was run in a 7300 Real Time PCR System (Applied Biosystems, CA, USA) with the SYBR green detection method to verify the DGE results. The cDNA was amplified in a total volume of 20 μL including 0.4 μL ROX reference dye, 2 μL cDNA, 0.4 μL primers and 10 μL SYBR Premix Ex Taq. The program of PCR reactions was 95°C for 10 s, next 40 cycles of 95°C for 5 s, 58°C for 30 s and 72°C for 30 s. Three technical replicates for each of three biological replicates were performed. The transcript profiles were normalized using the reference gene of elongation factor 1-alpha, and the relative expression levels of candidate genes were calculated with the 2-ΔΔCt method [49].
Supporting Information
The oleoresin from 1102-3-2 clone was as a representative to show the profile for identifying oleoresin terpenoids in masson pine using GC/MS analysis.
(TIF)
The unigenes were annotated in three categories: cellular components, molecular functions and biological processes.
(TIF)
Totally 9,990 of 84,842 sequences with Nr hits were grouped into 25 COG classifications.
(TIF)
(DOC)
(DOC)
(XLS)
(DOC)
(XLS)
(XLS)
(DOC)
Primers were designed from the sequences of masson pine transcriptome library by using Primer Premier 3.0.
(DOC)
Acknowledgments
We greatly thank Hengfu Yin, Xiaojiao Han, Shufeng Wang and Mingying Liu for their assistance in experimental design and implementation and manuscript writing. We also thank the reviewers for their invaluable comments and suggestions for the improvement of the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by funds from: 1) National Science & Technology Pillar Program of China (No. 2012BAD01B02, http://kjzc.jhgl.org/), ZZ. The funders had a role in decision to publish; 2) National Natural Science Foundation of China (31100491), QL. The funders had a role in study design; 3) Major Science & Technology Project of New Agricultural Variety Cultivation of Zhejiang Province, China (2012C12908-12), ZZ. The funders had a role in data collection and analysis, decision to publish.
References
- 1. Trapp S, Croteau R (2001) Defensive resin biosynthesis in conifers. Annu Rev Plant Physiol Plant Mol Biol 52: 689–724. [DOI] [PubMed] [Google Scholar]
- 2. Coppen JJW, Hone GA (1995) Gum Naval Stores: Turpentine and rosin from pine resin Non-wood forest products 2. Natural Resources Institute, FAO, Rome: p62. [Google Scholar]
- 3. Lee HJ, Ravn MM, Coates RM (2001) Synthesis and characterization of abietadiene, levopimaradiene, palustradiene, and neoabietadiene: hydrocarbon precursors of the abietane diterpene resin acids. Tetrahedron 57: 6155–6177. [Google Scholar]
- 4. Kelkar VM, Geils BW, Becker DR, Overby ST, Neary DG (2006) How to recover more value from small pine trees: essential oils and resins. Biomass Bioenerg 30: 316–320. [Google Scholar]
- 5. da Silva Rodrigues-Correa KC, Lima JC, Fett-Neto AG (2013) Oleoresins from pine: production and industrial uses Natural Products. Springer.Germany: Berlin: pp 4037–4060 [Google Scholar]
- 6. Harvey BG, Wright ME, Quintana RL (2010) High-density renewable fuels based on the selective dimerization of pinenes. Energy & Fuels 24: 267–273. [Google Scholar]
- 7. Martin DM, Bohlmann J (2005) Molecular biochemistry and genomics of terpenoid defenses in conifers. Rec Adv Phytochem 39: 29–56. [Google Scholar]
- 8. Miller B, Madilao LL, Ralph S, Bohlmann J (2005) Insect-induced conifer defense: White pine weevil and methyl jasmonate induce traumatic resinosis, de novo formed volatile emissions, and accumulation of terpenoid synthase and putative octadecanoid pathway transcript in Sitka spruce. Plant Physiol 137: 369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Strom BL, Goyer RA, Ingram LL, Boyd GDL, Lott LH (2002) Oleoresin characteristics of progeny of loblolly pines that escaped attack by southern pine beetle. For Ecol Manage 158: 169–178. [Google Scholar]
- 10. Nagy N, Franceschi VR, Krekling T, Solheim H, Christiansen E (2000) Wound-induced traumatic resin duct development in stems of Norway spruce (Pinaceae): anatomy and cytochemical traits. Am J Bot 87: 302–313. [PubMed] [Google Scholar]
- 11. Byun-McKay SAB, Hunter WL, Godard KA, Wang SX, Martin DM, Bohlmann J, et al. (2003) Insect attack and wounding induce traumatic resin duct development and gene expression of (-)-pinene synthase in Sitka spruce. Plant Physiol 133: 368–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tadesse W, Nanos N, Aunon FJ, Alia R, Gil L (2001) Evaluation of high resin yielders of Pinus pinaster AIT. For Genet 8: 271–278. [Google Scholar]
- 13. Roberds JH, Strom BL, Hain FP, Gwaze DP, Mckeand SE, Lott LH (2003) Estimates of genetic parameters for oleoresin and growth traits in juvenile loblolly pine. Can J For Res 33: 2469–2476. [Google Scholar]
- 14. Westbrook JW, Resende MF Jr, Munoz P, Walker AR, Wegrzyn JL, Nelson CD, et al. (2013) Association genetics of oleoresin flow in loblolly pine: discovering genes and predicting phenotype for improved resistance to bark beetles and bioenergy potential. New Phytologist 199: 89–100. 10.1111/nph.12240 [DOI] [PubMed] [Google Scholar]
- 15. Liu QH, Zhou ZC, Fan HH, Liu YR (2013) Genetic Variation and Correlation among Resin Yield, Growth, and Morphologic Traits of Pinus massoniana . Silvae Genetica 62: 38–44. [Google Scholar]
- 16. Weng HL, Chen HW, Duan AA (2010) Selection of plus-tree with high-rosin production for Pinus kesiya var. 1angbianensis, Journal of Northwest Forestry University 25: 71–74. [Google Scholar]
- 17. Li SG, Fu YP, Zhang KF, Jiang YD, Yao ZQ, Li M (2008) Half-sib progeny tests of high-resin-yielding Pinus kesiyavar. langbianensis . Journal of Zhejiang Forestry College 25: 158–162. [Google Scholar]
- 18. Liu YR (2006) Selection of high-resin-yield half-sib Pinus massoniana single tree with superior resin-producing capacity. Forestry Science and Technology 31: 1–4. [Google Scholar]
- 19. Rodriguez-Garcia A, Lopez R, Martín JA, Pinillos F, Gil L (2014) Resin yield in Pinus pinaster is related to tree dendrometry, stand density and tapping-induced systemic changes in xylem anatomy. For Ecol Manage 313: 47–54. [Google Scholar]
- 20. Karanikas C, Walker V, Scaltsoyiannes A, Comte G, Bertrand C (2010) High vs. low yielding oleoresin Pinus halepensis Mill. trees GC terpenoids profiling as diagnostic tool. Ann For Sci 67: 412p1–p8. [Google Scholar]
- 21. Lange BM, Ghassemian M (2003) Genome organization in Arabidopsis thaliana: A survey for genes involved in isoprenoid and chlorophyll metabolism. Plant Mol Biol 58: 925–948. [DOI] [PubMed] [Google Scholar]
- 22. Zulak KG, Bohlmann J (2010) Terpenoid biosynthesis and specialized vascular cells of conifer defense. J Integr Plant Biol 52: 86–97. 10.1111/j.1744-7909.2010.00910.x [DOI] [PubMed] [Google Scholar]
- 23. Bohlmann J, Gershenzon J, Aubourg S (2000a) Biochemical, molecular genetic and evolutionary aspects of defense-related terpenoid metabolism in conifers. Rec Adv Phytochem 34: 109–150. [Google Scholar]
- 24. Phillips MA, Wildung MR, Williams DC, Hyatt DC, Croteau RB (2003) cDNA isolation, functional expression, and characterization of (+)-α-pinene synthase from loblolly pine (Pinus taeda): Stereocontrol in pinene biosynthesis. Arch. Biochem. Biophys. 411: 267–276. [DOI] [PubMed] [Google Scholar]
- 25. Byun-McKay A, Godard KA, Toudefallah M, Martin DM, Alfaro R, King J, et al. (2006) Wound-induced terpene synthase gene expression in Sitka spruce that exhibit resistance or susceptibility to attack by the White pine weevil. Plant Physiol 140: 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Supek F, Bosnjak M, Skunca N, Smuc T (2011) REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6: e21800 10.1371/journal.pone.0021800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gu Y, Zhao ZD, Bi LW, Li DM, Wang J (2011) Certification of reference materials of turpentine oil from Pinus massoniana Lamb. Biomass Chemical Engineering 45: 21–24. [Google Scholar]
- 28. Wang ZH, Shang SB, Song ZQ, Cao XM, Ye BH (2007) Preparation of reference materials of the Pinus massoniana Lamb. rosin for gas chromatography analysis. Biomass Chemical Engineering 41: 1–5. [Google Scholar]
- 29. Bouvier F, Rahier A, Camara B (2005) Biogenesis, molecular regulation and function of plant isoprenoids. Prog Lipid Res 44: 357–429. [DOI] [PubMed] [Google Scholar]
- 30. Thabet I, Guirimand G, Guihur A, Lanoue A, Courdavault V, Papon N, et al. (2012) Characterization and subcellular localization of geranylgeranyl diphosphate synthase from Catharanthus roseus . Mol Biol Rep 39: 3235–3243. 10.1007/s11033-011-1091-9 [DOI] [PubMed] [Google Scholar]
- 31. Ralph SG, Yueh H, Friedmann M, Aeschliman D, Zeznik JA, Nelson CC, et al. (2006) Conifer defence against insects: microarray gene expression profiling of Sitka spruce (Picea sitchensis) induced by mechanical wounding or feeding by spruce budworms (Choristoneura occidentalis) or white pine weevils (Pissodes strobi) reveals large-scale changes of the host transcriptome. Plant Cell Environ 29: 1545–1570. [DOI] [PubMed] [Google Scholar]
- 32. Schmidt A, Nagel R, Krekling T, Christiansen E, Gershenzon J, Krokene P (2011) Induction of isoprenyl diphosphate synthases, plant hormones and defense signalling genes correlates with traumatic resin duct formation in Norway spruce (Picea abies) Plant Mol Biol 77: 577–590. 10.1007/s11103-011-9832-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bohlmann J, Meyer-Gauen G, Croteau RB (1998b) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davis EM, Croteau RB (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Top Curr Chem 209: 53–95. [Google Scholar]
- 35. Keeling CI, Bohlmann J (2006) Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol 170: 657–675. [DOI] [PubMed] [Google Scholar]
- 36. Steele CL, Crock J, Bohlmann J, Croteau RB (1998) Sesquiterpene synthases from grand fir (Abies grandis): comparison of constitutive and wound-induced activities, and cDNA isolation, characterization and bacterial expression of δ-selinene synthase and γ-humulene synthase. J Biol Chem 273: 2078–2089. [DOI] [PubMed] [Google Scholar]
- 37. Faldt J, Martin DM, Miller B, Rawat S, Bohlmann J (2003) Traumatic resin defense in Norway spruce (Picea abies): Methyl jasmonate induced terpene synthase gene expression, and cDNA cloning and functional characterization of (+)-3-carene synthase. Plant Mol Biol 51: 119–133. [DOI] [PubMed] [Google Scholar]
- 38. Martin DM, Faldt J, Bohlmann J (2004) Functional characterization of nine Norway spruce TPS genes and evolution of gymnosperm terpene synthases of the TPS-d subfamily. Plant Physiol 135: 1908–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qin XW, Su JW, Ge F, Zhou ZQ (2006) Enantiomeric composition and relative abundance of monoterpene hydrocarbons in volatiles of Pinus massoniana Lamb, Pinus tabulaeformis Carr., and Pinus elliottii Engelm. Journal of Instrumental Analysis 25: 6–9. [Google Scholar]
- 40. Higgin CF (1992) ABC transporters: From microorganisms to man. Annu. Rev Cell Biol 8: 67–113. [DOI] [PubMed] [Google Scholar]
- 41. Jasinski M, Stukkens Y, Degand H, Purnelle B, Marchand-Brynaert J, Boutry M. (2001) A plant plasma membrane ATP binding cassette-type transporter is involved in antifungal terpenoid secretion. Plant Cell 13: 1095–1107. [PMC free article] [PubMed] [Google Scholar]
- 42. Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, et al. (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol 65: 719–732. [DOI] [PubMed] [Google Scholar]
- 43. Guo HW, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin in Plant Biol 7: 40–49. [DOI] [PubMed] [Google Scholar]
- 44. Roberts WK, Selitrennikoff CP (1990) Zeamatin, an antifungal protein from maize with membrane-permeabilizing activity. J Gen Microbiol 136: 1771–1778. [Google Scholar]
- 45. Dafoe NJ, Zamani A, Ekramoddoullah AKM, Lippert D, Bohlmann J, Constabel CP (2009) Analysis of the Poplar phloem proteome and its response to leaf wounding. Proteome Res 8: 2341–2350. [DOI] [PubMed] [Google Scholar]
- 46. Uehara T, Sugiyama S, Matsuura H, Arie T, Masuta C (2010) Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol 51: 1524–1536. 10.1093/pcp/pcq109 [DOI] [PubMed] [Google Scholar]
- 47. Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, et al. (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8: 1494–512. 10.1038/nprot.2013.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The oleoresin from 1102-3-2 clone was as a representative to show the profile for identifying oleoresin terpenoids in masson pine using GC/MS analysis.
(TIF)
The unigenes were annotated in three categories: cellular components, molecular functions and biological processes.
(TIF)
Totally 9,990 of 84,842 sequences with Nr hits were grouped into 25 COG classifications.
(TIF)
(DOC)
(DOC)
(XLS)
(DOC)
(XLS)
(XLS)
(DOC)
Primers were designed from the sequences of masson pine transcriptome library by using Primer Premier 3.0.
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.