Abstract
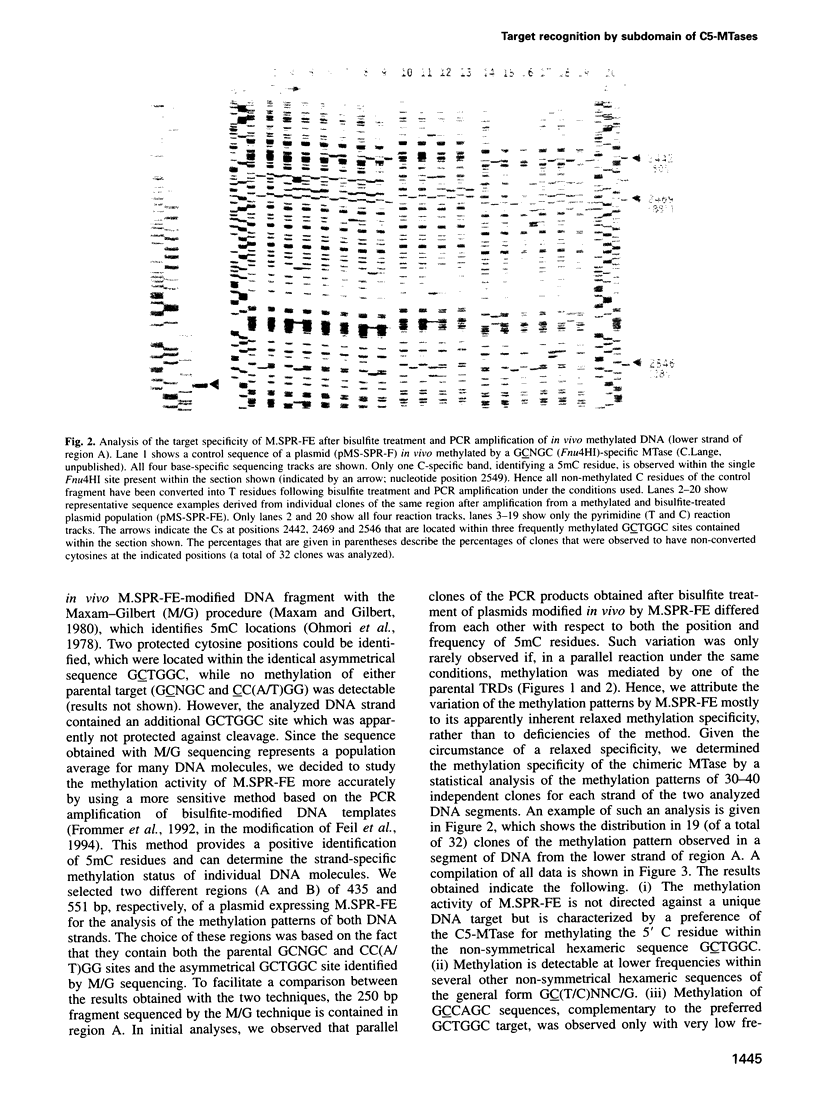
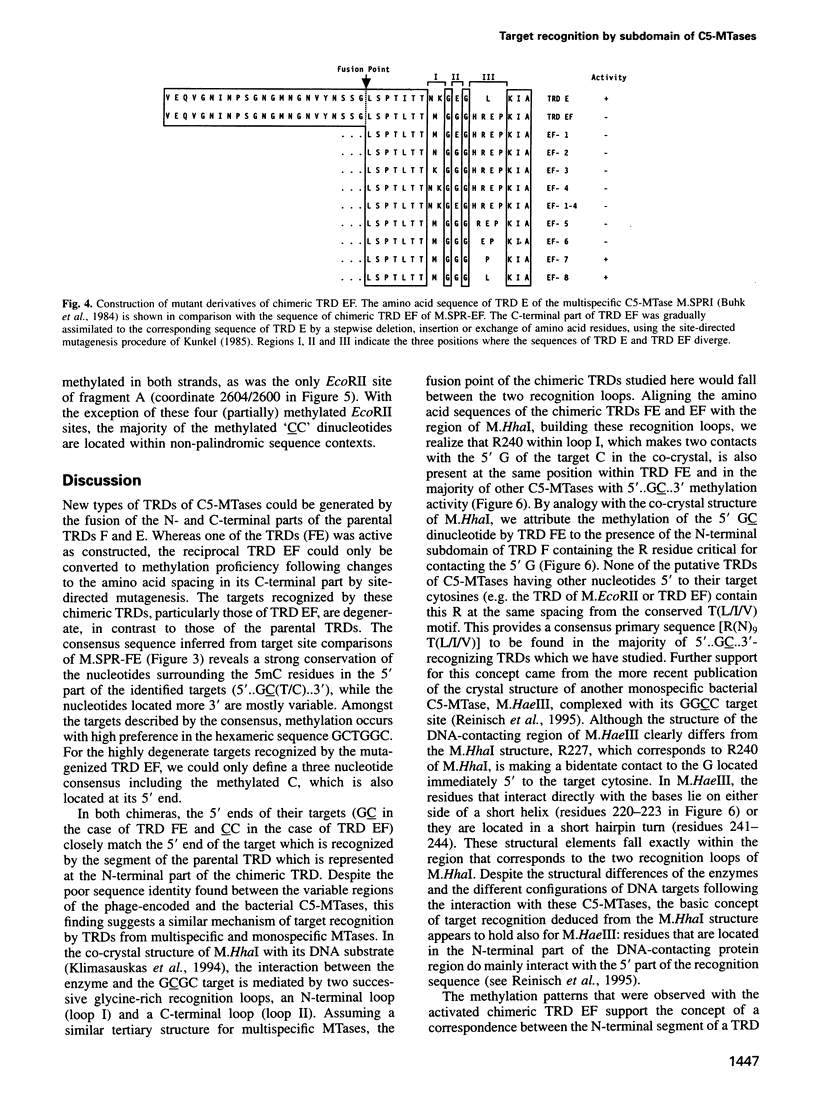
In previous work on DNA-(cytosine-C5)-methyltransferases (C5-MTases), domains had been identified which are responsible for the sequence specificity of the different enzymes (target-recognizing domains, TRDs). Here we have analyzed the DNA methylation patterns of two C5-MTases containing reciprocal chimeric TRDs, consisting of the N- and C-terminal parts derived from two different parental TRDs specifying the recognition of 5'-CC(A/T)GG-3' and 5'-GCNGC-3'. Sequences recognized by these engineered MTases were non-symmetrical and degenerate, but contained at their 5' part a consensus sequence which was very similar to the 5' part of the target recognized by the parental TRD which contributed the N-terminal moiety of the chimeric TRD. The results are discussed in connection with the present understanding of the mechanism of DNA target recognition by C5-MTases. They demonstrate the possibility of designing C5-MTases with novel DNA methylation specificities.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balganesh T. S., Reiners L., Lauster R., Noyer-Weidner M., Wilke K., Trautner T. A. Construction and use of chimeric SPR/phi 3T DNA methyltransferases in the definition of sequence recognizing enzyme regions. EMBO J. 1987 Nov;6(11):3543–3549. doi: 10.1002/j.1460-2075.1987.tb02681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens B., Noyer-Weidner M., Pawlek B., Lauster R., Balganesh T. S., Trautner T. A. Organization of multispecific DNA methyltransferases encoded by temperate Bacillus subtilis phages. EMBO J. 1987 Apr;6(4):1137–1142. doi: 10.1002/j.1460-2075.1987.tb04869.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhk H. J., Behrens B., Tailor R., Wilke K., Prada J. J., Günthert U., Noyer-Weidner M., Jentsch S., Trautner T. A. Restriction and modification in Bacillus subtilis: nucleotide sequence, functional organization and product of the DNA methyltransferase gene of bacteriophage SPR. Gene. 1984 Jul-Aug;29(1-2):51–61. doi: 10.1016/0378-1119(84)90165-3. [DOI] [PubMed] [Google Scholar]
- Cheng X., Kumar S., Posfai J., Pflugrath J. W., Roberts R. J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine. Cell. 1993 Jul 30;74(2):299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- Feil R., Charlton J., Bird A. P., Walter J., Reik W. Methylation analysis on individual chromosomes: improved protocol for bisulphite genomic sequencing. Nucleic Acids Res. 1994 Feb 25;22(4):695–696. doi: 10.1093/nar/22.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace F. V., Bullas L. R., Delius H., Murray N. E. Genetic recombination can generate altered restriction specificity. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6095–6099. doi: 10.1073/pnas.81.19.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- Gann A. A., Campbell A. J., Collins J. F., Coulson A. F., Murray N. E. Reassortment of DNA recognition domains and the evolution of new specificities. Mol Microbiol. 1987 Jul;1(1):13–22. doi: 10.1111/j.1365-2958.1987.tb00521.x. [DOI] [PubMed] [Google Scholar]
- Günthert U., Lauster R., Reiners L. Multispecific DNA methyltransferases from Bacillus subtilis phages. Properties of wild-type and various mutant enzymes with altered DNA affinity. Eur J Biochem. 1986 Sep 15;159(3):485–492. doi: 10.1111/j.1432-1033.1986.tb09912.x. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Klimasauskas S., Nelson J. L., Roberts R. J. The sequence specificity domain of cytosine-C5 methylases. Nucleic Acids Res. 1991 Nov 25;19(22):6183–6190. doi: 10.1093/nar/19.22.6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Cheng X., Klimasauskas S., Mi S., Posfai J., Roberts R. J., Wilson G. G. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res. 1994 Jan 11;22(1):1–10. doi: 10.1093/nar/22.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lange C., Jugel A., Walter J., Noyer-Weidner M., Trautner T. A. 'Pseudo' domains in phage-encoded DNA methyltransferases. Nature. 1991 Aug 15;352(6336):645–648. doi: 10.1038/352645a0. [DOI] [PubMed] [Google Scholar]
- Lauster R., Trautner T. A., Noyer-Weidner M. Cytosine-specific type II DNA methyltransferases. A conserved enzyme core with variable target-recognizing domains. J Mol Biol. 1989 Mar 20;206(2):305–312. doi: 10.1016/0022-2836(89)90480-4. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nagaraja V., Shepherd J. C., Bickle T. A. A hybrid recognition sequence in a recombinant restriction enzyme and the evolution of DNA sequence specificity. Nature. 1985 Jul 25;316(6026):371–372. doi: 10.1038/316371a0. [DOI] [PubMed] [Google Scholar]
- Noyer-Weidner M., Walter J., Terschüren P. A., Chai S., Trautner T. A. M.phi 3TII: a new monospecific DNA (cytosine-C5) methyltransferase with pronounced amino acid sequence similarity to a family of adenine-N6-DNA-methyltransferases. Nucleic Acids Res. 1994 Oct 11;22(20):4066–4072. doi: 10.1093/nar/22.20.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pósfai J., Bhagwat A. S., Pósfai G., Roberts R. J. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res. 1989 Apr 11;17(7):2421–2435. doi: 10.1093/nar/17.7.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch K. M., Chen L., Verdine G. L., Lipscomb W. N. The crystal structure of HaeIII methyltransferase convalently complexed to DNA: an extrahelical cytosine and rearranged base pairing. Cell. 1995 Jul 14;82(1):143–153. doi: 10.1016/0092-8674(95)90060-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran-Betcke A., Behrens B., Noyer-Weidner M., Trautner T. A. DNA methyltransferase genes of Bacillus subtilis phages: comparison of their nucleotide sequences. Gene. 1986;42(1):89–96. doi: 10.1016/0378-1119(86)90153-8. [DOI] [PubMed] [Google Scholar]
- Trautner T. A., Balganesh T. S., Pawlek B. Chimeric multispecific DNA methyltransferases with novel combinations of target recognition. Nucleic Acids Res. 1988 Jul 25;16(14A):6649–6658. doi: 10.1093/nar/16.14.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautner T. A., Pawlek B., Behrens B., Willert J. Exact size and organization of DNA target-recognizing domains of multispecific DNA-(cytosine-C5)-methyltransferases. EMBO J. 1996 Mar 15;15(6):1434–1442. [PMC free article] [PubMed] [Google Scholar]
- Walter J., Trautner T. A., Noyer-Weidner M. High plasticity of multispecific DNA methyltransferases in the region carrying DNA target recognizing enzyme modules. EMBO J. 1992 Dec;11(12):4445–4450. doi: 10.1002/j.1460-2075.1992.tb05545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke K., Rauhut E., Noyer-Weidner M., Lauster R., Pawlek B., Behrens B., Trautner T. A. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 1988 Aug;7(8):2601–2609. doi: 10.1002/j.1460-2075.1988.tb03110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Murray N. E. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]







