Motivation
The past decade has seen a surge in the development of multidimensional infrared (IR) spectroscopy. Because IR spectroscopy probes the structural degrees of freedom of a molecule, it directly characterizes the potential energy surface that describes a molecule’s nuclear and electronic structure. Vibrational frequencies are also highly sensitive to molecular environments, and so IR spectroscopy is useful for characterization of local sites and their fluctuations within complex systems. Multidimensional methods provide richer information than is available from linear experiments. They are able, for example, to identify interactions between vibrational modes and to measure the temporal evolution of vibrational frequencies. The experiments of multidimensional IR spectroscopy are akin to those long-established in the field of NMR spectroscopy. Unlike NMR spectroscopy, which is typically limited to detection of species interconverting on millisecond and slower timescales, IR spectroscopy has the advantage of an inherently fast, sub-picosecond timescale that ensures detection of states that rapidly interconvert and enables direct measurement of fast structural and/or environmental fluctuations. IR spectroscopy is thus well suited for the study of condensed phase samples where structural and environmental fluctuations occur on very fast timescales. The combination of high temporal resolution and bond-specific structural detail distinctively provided by IR spectroscopy makes it a powerful experimental approach for the study of molecular structure and dynamics.
The recent development of multidimensional IR spectroscopy has positioned the methods for application in a wide variety of research areas in chemistry and biology. A number of recent reviews and texts offer excellent, comprehensive treatments of the theory and experimental aspects of the methods.1–8 This minireview provides a brief outline of the general principles, experimental implementation, and information available from two-dimensional (2D) IR spectroscopy, one of the most prevalent multidimensional IR experiments. We first highlight recent representative studies of 2D IR spectroscopy applied to small molecule systems to illustrate the principal spectral features of 2D spectra and what they tell us about molecular structure and dynamics. We then present example applications of 2D IR spectroscopy toward characterization of more complex systems, such as of interest in biological and materials chemistry. Our intent is to present only a sample of the possible applications of 2D IR spectroscopy to communicate its advantages for addressing questions in chemistry and biology.
Fundamentals of 2D IR spectroscopy
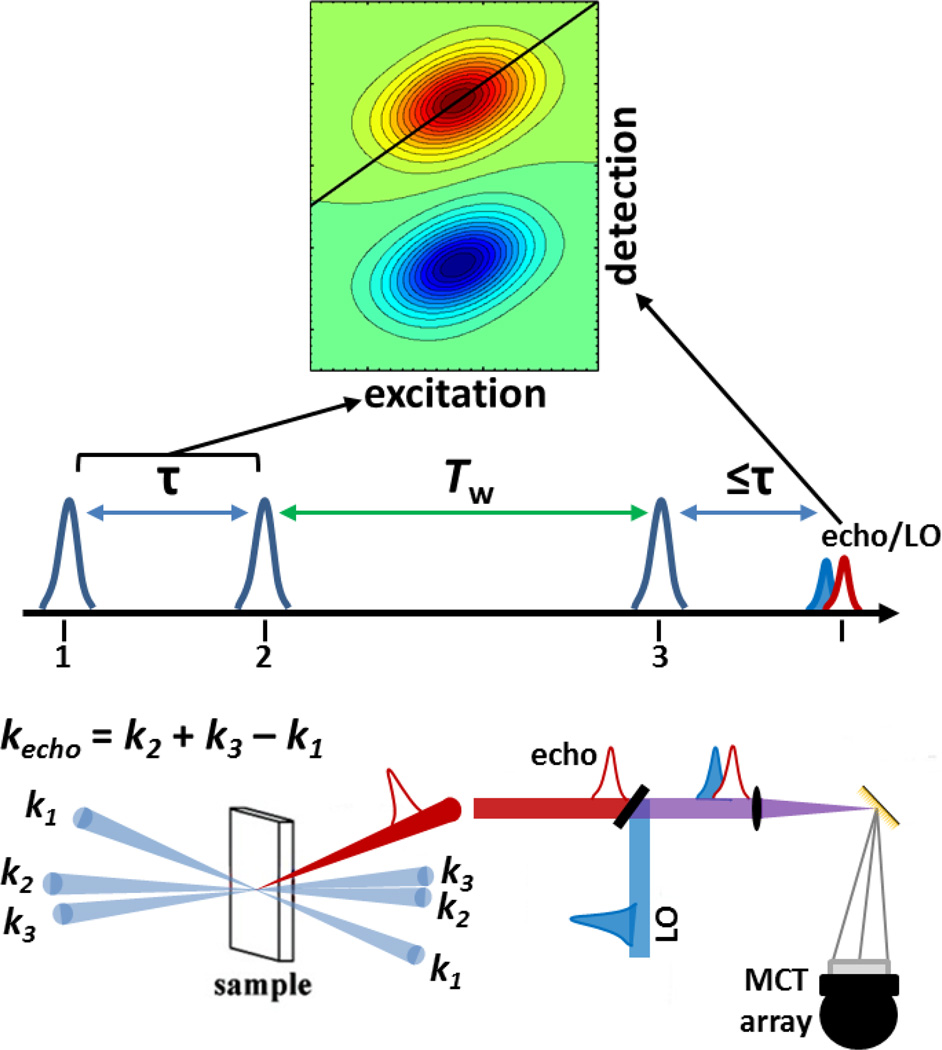
2D IR spectroscopy measures the correlation among the frequencies of the vibrations of a system and their time evolution. The experiment involves application of an IR pulse sequence that labels the system with its initial frequencies, allows the system to evolve, and then reads out the final frequencies. One axis of a 2D spectrum describes initial excited frequencies, while the orthogonal axis associates these frequencies with the final detected frequencies. The simplest case of a 2D spectrum of a single vibrational mode illustrates the additional information available from the second dimension. In the typical situation that the laser bandwidth encompasses both the 0–1 and 1–2 transition frequencies, a single vibrational mode leads to a pair of bands of opposite sign in a 2D spectrum (for example, see Fig. 1). Both of the bands appear along the initial excitation axis at the frequency of the 0–1 transition because they arise from the excitation of an initial ground state population. Along the detection axis, one band arises at the 0–1 frequency due to a combination of stimulated emission and ground state population reduction, while the other arises at the 1–2 frequency due to further excitation of the population in the first excited state. Thus a 2D IR spectrum of a single vibrational mode provides information about the correlation between its initial and final frequencies – in this specific example, between the initial 0–1 transition frequency and both the 0–1 and the 1–2 transition frequencies of the same mode.
Figure 1.
Simulated example 2D IR spectra and schematic illustrating 2D IR experimental pulse timing, geometry, and detection. Adapted with permission from Zheng et al. Accounts of Chemical Research, 2007, 40, 75–83. Copyright © 2007 American Chemical Society.
In a similar manner, the correlations between frequencies reported in 2D spectra elucidate the interaction between vibrational modes, interconversion between chemical species or environments, and intra- or intermolecular vibrational energy relaxation.5–7 For example, cross-bands appear between the frequencies of two coupled modes, indicating that excitation at the frequency of one mode results in detection at the frequency of the other. The cross-band intensities reflect the strength of the coupling interaction, which in turn depends on the proximity and relative orientation of the coupled modes. The cross-bands are therefore highly sensitive to molecular structure and intermolecular interactions, and so are valuable for the assignment and interpretation of IR spectra, which can be very complex for even moderately sized molecules. Moreover, the high sensitivity of vibrational coupling to the distance and relative orientation of the modes enables generation of structural models of molecules.
Multiple bands can appear in a linear IR spectrum not only due to multiple vibrational modes, but also due to a single mode in distinct environments (e.g. resulting from conformational changes or chemical reactions). In these cases, cross-bands arise in a 2D spectrum at the frequencies associated with the different environments or chemical species if they interchange within the time of initial excitation and the time of detection in the acquisition of a 2D spectrum. The cross-bands due to such exchange processes grow in intensity over time on the timescale of the environmental fluctuation or chemical interconversion. In many situations, the different, interconverting species do not lead to distinct bands, but rather manifest as an inhomogeneously broadened band. 2D IR spectroscopy can be used to measure the evolution of these frequencies, referred to as spectral diffusion. Rather than distinct cross-bands, the interconversion of the associated states causes the 2D bands to become progressively less diagonally elongated with increasing time. As we discuss specific applications of 2D IR spectroscopy, we will point out how 2D spectra and their time evolution inform on these processes.
Experimental implementation of 2D IR spectroscopy
The theory and experimental implementation of 2D IR spectroscopy, and multidimensional spectroscopy in general, have been comprehensively treated in recent reviews1–3 and texts,4–7 so a brief outline of a basic, conventional implementation of a 2D IR experiment is presented here. A 2D IR experiment involves temporally-controlled application of three ultrashort mid-IR pulses, which lead to a third-order polarization in the sample (Fig. 1). The times between the first and second excitation pulses, and between the second and third excitation pulses, are commonly referred to as the coherence time (τ) and the waiting time (Tw), respectively. To acquire a single 2D IR spectrum, τ is scanned for a fixed value of Tw. At a time ≤ τ after the third pulse, a third-order signal is generated by the sample in a unique (phase-matched) direction. This IR pulse is spatially filtered and heterodyne-detected by overlapping with another IR pulse, the local oscillator, which provides both signal amplification and phase information. Commonly, the combined pulses are frequency-dispersed onto an array detector, providing a direct measurement of the spectrum along the final detected frequency axis (alternately referred to as the ωm, ω3, or probe axis). At each detected frequency, scanning τ produces an interferogram. Numerical Fourier transformation of these interferograms generates the spectrum along the initial excitation frequency axis (alternately referred to as the ωτ, ω1, or pump axis). (Note that conventions for the placement of excitation and detection frequencies along the horizontal or vertical axes vary among research groups.) The evolution of the system during τ labels the system with the initial frequencies. Detection of the heterodyned signal after the set waiting time Tw interrogates the final frequencies of the system. Thus, the resulting 2D IR spectrum reports on the correlation between the initial and final frequencies of the system after a time separation of Tw.
A number of variations of this conventional implementation of 2D IR spectroscopy have been developed.9–18 For example, control of the polarization of the excitation pulses can provide orientational information about coupled modes or selectively highlight particular cross-bands.11, 12 Upconversion of the mid-IR to visible frequencies enables the use of less expensive, more sensitive CCD array detectors available for visible frequencies.13, 14 Frequency-domain versions of 2D IR spectroscopy are also well developed, including their application to a variety of systems.19–22 An increasingly popular implementation of 2D IR spectroscopy uses pulse shaping to generate and control the timing of the first two excitation pulses.9, 23 In fact, a 2D IR system that utilizes this design is now commercially available.24 Pulse shaping methods enable significantly faster data acquisition, as well as phase-cycling procedures that can decrease data sampling demands or remove scattered light; however, these improvements come with the loss of the background-free signal generation in the conventional experiment. The advantages and disadvantages of different implementations of 2D IR spectroscopy have been discussed by several authors.9, 25–27 These and other advancements in the experimental methods of multidimensional IR spectroscopy over the past decade have established versatile tools for addressing a range of scientific questions.
Applications of 2D IR spectroscopy
Applications to small molecule systems
As the techniques of 2D IR spectroscopy have been established over the past decade, a variety of small molecule systems have been investigated, both due to inherent interest, as well as to develop small molecule probes to analyze more complex systems.28–33 In the discussion that follows, we highlight a few examples in order to illustrate the information available from 2D IR spectroscopy.
Cross-band anisotropy of rapidly interconverting species
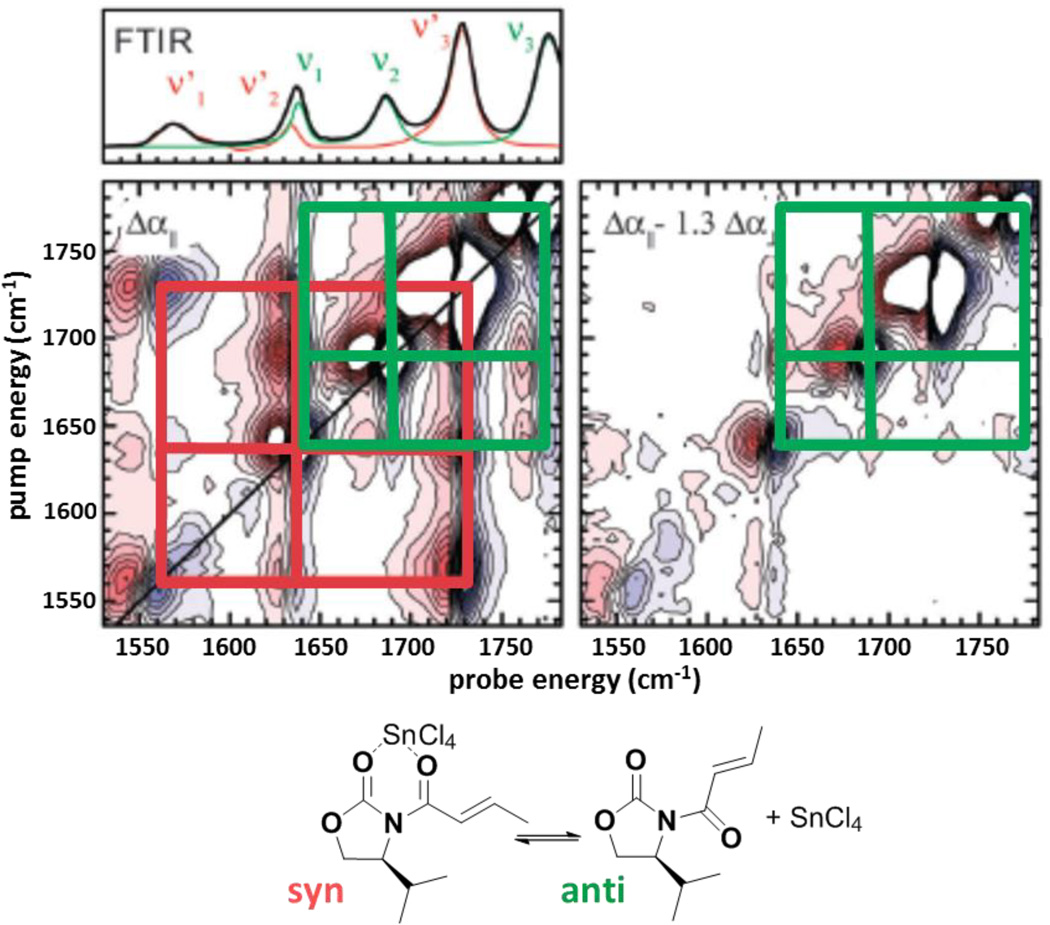
A central advantage of IR spectroscopy is the inherently fast, sub-picosecond timescale associated with the frequencies of IR transitions. 2D IR methods are thus complementary to the more established 2D NMR methods, which are typically limited to resolving structures that interconvert on millisecond or slower timescales. The high temporal resolution, combined with high structural sensitivity, make 2D IR spectroscopy suitable for the analysis of rapidly interconverting molecular structures. An interesting recent application is the structural analysis of molecular conformers in organic chemical reactions.34, 35 Knowledge of the structures adopted by small molecules is valuable for rationalizing and predicting chemical reactions, particularly those involving stereochemical control. For example, 2D IR spectroscopy has been applied to analyze the potential conformations adopted in the stereoselective Diels-Alder reaction of cyclopentadiene and N-crotonyloxazolidinone with Lewis acid catalyst SnCl4.35 2D spectra of equimolar solutions of the dieneophile substrate and SnCl4 are shown in Fig. 2. As in a 2D NMR COSY spectrum, cross-bands indicate coupling interactions, and so identify bands due to a single conformer. The red and green grid lines in the spectra shown in Fig. 2 indicate bands due to the same species. The measurement of the cross-band anisotropy, the difference in the cross-band intensities for spectra obtained with parallel or perpendicularly polarized excitation beams, provides angular information about the transition dipoles of coupled vibrational modes that can be used to assign the structures of the populated conformers. The left panel of Fig. 2 shows the 2D IR spectrum obtained with parallel-polarized excitation bands. The right panel illustrates how select cross-bands can be annihilated by weighted subtraction of parallel- and perpendicular-polarized 2D spectra. In this example, 2D IR spectroscopy yields evidence that the dienophile substrate adopts the anti and syn conformers when free and bound to the SnCl4 catalyst, respectively. The appearance of distinct bands for the two species illustrates the advantage of the fast inherent timescale of IR spectroscopy. In contrast, these conformers are in rapid exchange on the NMR timescale.
Figure 2.
Polarization-dependent 2D IR spectra of an equimolar solution of Diels Alder substrate N-crotonyloxazolidinone and catalyst SnCl4 (see text). Each of the red and green gridlines indicate bands due to a single species. The FTIR spectra are shown above the left panel for the mixture (black), the substrate (green), and the catalyst (red). The structures of the syn conformer of the substrate-catalyst complex and the anti conformer of the substrate are shown below. Adapted from Ref. 35 with permission from the PCCP Owner Societies.
Chemical exchange of ion complexes
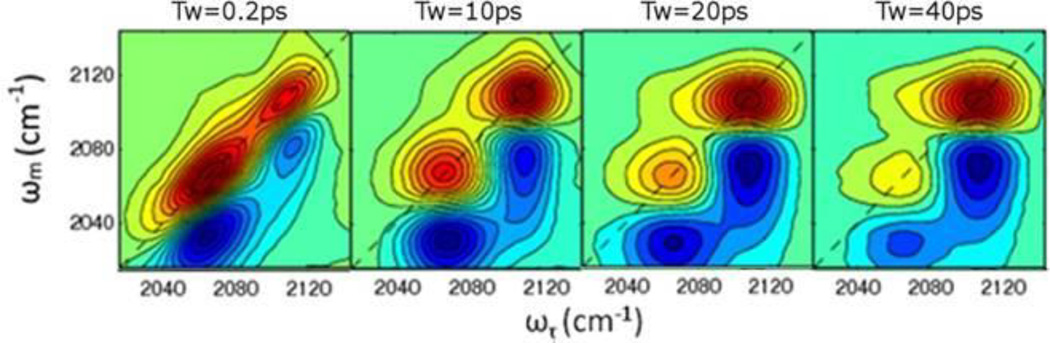
The importance of ions in biology and a wide variety of other systems has prompted extensive investigation into ion pairing and the accompanying changes in solvent coordination.28, 36–40 The fast timescale of 2D IR spectroscopy enables direct, real-time monitoring of chemical exchange reactions when the exchange time is longer than or comparable to the vibrational lifetime of the monitored mode. As an illustration, 2D IR spectroscopy was used to study ion pair formation between magnesium (Mg2+) and calcium (Ca2+) cations with the thiocyanate anion (NCS−) and the anion exchange in the solvation shell.36 The example spectra in Fig. 3 focus on the 2D band for the CN stretching vibration and illustrate the Tw-dependent changes in its shape due to chemical exchange for aqueous solutions of NaNCS and MgI2. At early Tw, two pairs of bands appear at distinct frequencies along the diagonal that reflect the free NCS− and the contact ion pair with Mg2+ (left panel, Fig. 3). As the NCS− exchanges between the two states with increasing Tw, cross-band intensity grows between the diagonal bands because the anion is excited at the frequency of one state but detected at the frequency of the other state (right panels, Fig. 3). Analysis of the time-dependent intensities of the cross-bands gives the timescales of the chemical exchange reaction. In this particular study, measurement of the exchange dynamics revealed different mechanisms for ion complex formation that depend on the identity of the cation, suggesting a dissociative mechanism for Mg2+ but an associative mechanism for Ca2+. 2D IR spectroscopy has been similarly applied to characterize the ligand exchange and solvation dynamics of a wide variety of ion pairs.28, 39, 40
Figure 3.
2D IR spectra of a solution of NaNCS and MgI2 at several waiting times. Reprinted with permission from Sun et al. The Journal of Physical Chemistry B, 2013, 117, 12268 – 12275. Copyright © 2013 American Chemical Society.
Spectral diffusion due to solvent fluctuations
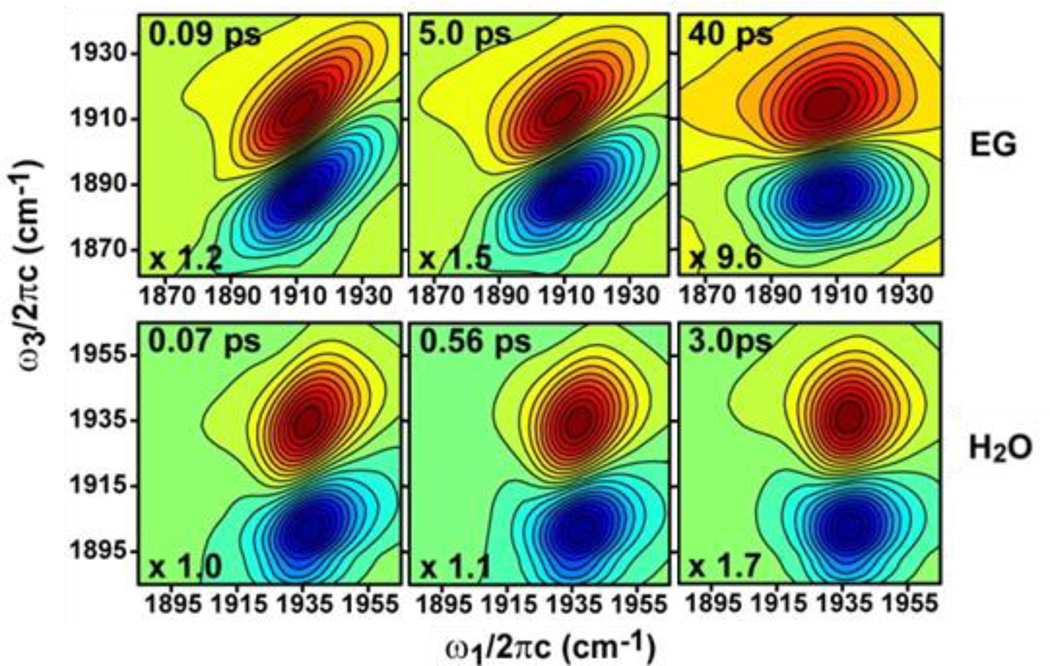
Multidimensional IR spectroscopy is particularly useful for the analysis of condensed phase samples, where the large heterogeneity in the local environments experienced by vibrational modes often leads to a continuous distribution in IR frequencies that result in broad, featureless linear absorption bands. 2D IR spectroscopy facilitates interpretation of such spectra by its ability to extract the underlying inhomogeneous distribution of frequencies that arise from environmental or structural heterogeneity. Furthermore, analysis of the time-dependent changes in the shape of the 2D bands reveals the dynamics of the interconversion within the inhomogeneous distribution of frequencies (spectral diffusion), providing information on the environmental fluctuations or structural dynamics of the molecule. As an example, Fig. 4 shows time-dependent 2D spectra of the NO stretching mode of sodium nitroprusside in ethylene glycol and water solvents.29 At short Tw, the 2D bands appear elongated along the diagonal (left panels, Fig. 4). This occurs because the states associated with the distribution of frequencies are not given sufficient time to interconvert. The frequencies initially excited are the same frequencies detected, and the diagonally elongated 2D bands reflect high correlation between the two frequency axes. As the time (Tw) between labeling and detection of the frequencies is increased, the states have an increasingly longer time to interconvert. When this occurs, the initial and final frequencies of the species are no longer the same, and the loss of their frequency correlation causes the 2D band shape to become less elongated (right panels, Fig. 4). In comparison to the aqueous solution, the 2D bands for the ethylene glycol solution show greater elongation at early Tw times and more slowly changing line shapes, indicating greater heterogeneity and slower dynamics of the NO environment in ethylene glycol. Procedures for quantifying the dynamics of the system by analysis of the 2D band shapes have been described.41 As in this particular example, the measurement of spectral diffusion has been applied to study the dynamics of a variety of systems, including solvent, protein, and membrane environments.42–52
Figure 4.
2D IR spectra of solutions of sodium nitroprusside in ethylene glycol (upper panels) and water (lower panels) at several waiting times. Adapted with permission from Brookes et al. The Journal of Physical Chemistry A, 2013, 117, 6234 – 6243. Copyright © 2013 American Chemical Society.
Vibrational energy transfer
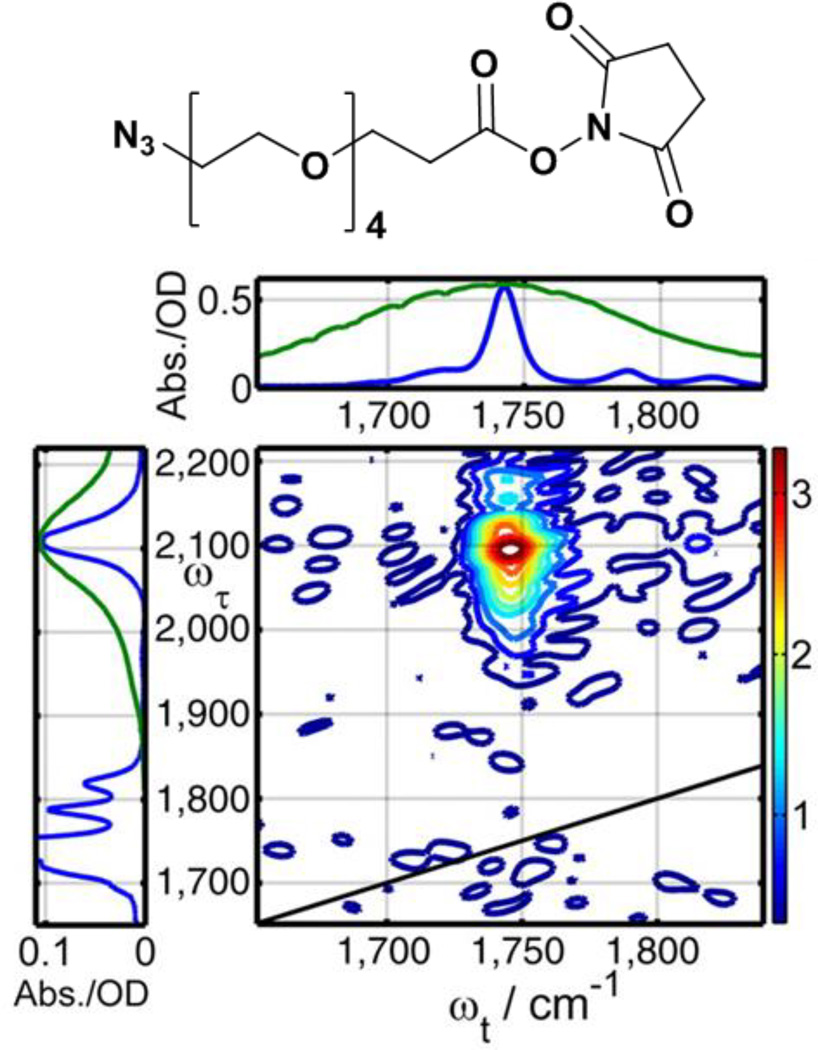
2D IR spectroscopy can be used to analyze molecular structure via measurement of energy transport throughout a molecule.53–56 For example, relaxation-assisted 2D IR spectroscopy, a dual-frequency variation of 2D IR spectroscopy, uses a laser pulse to deposit energy into a vibrational mode at one frequency, then monitors the dissipation of the excess energy through the molecule by detection of its influence on another vibrational mode at a different frequency.54 Unlike cross-bands due to direct, strong coupling between spatially proximal or mechanically connected modes that appear with no waiting time, cross-bands due to relaxation-assisted processes grow in intensity with increasing waiting time as the excess energy deposited at one mode propagates throughout the molecule, typically via anharmonic coupling through the connecting bonds. Arrival of the energy at another mode shifts its frequency or absorption probability and results in cross-band amplitude. The time delay between energy deposition at the first mode and the maximum enhancement of its cross-band with a second mode depends on the bond connectivity between them. Correlation of bond distance and energy transport times can provide a molecular ruler of bond distances in a molecule. As an example, this method was applied to polyethylene glycol (PEG) oligomers of various length bearing azido and succinimide ester moieties as vibrational reporter tags.55 The 2D IR spectrum in Fig. 5 shows a cross-band that arises from pumping at the azido frequency (2107 cm−1) and probing at the frequencies of the three carbonyl modes of the succinimide tag (1724, 1778, 1819 cm−1) 60 ps later. The energy deposited in the azido group dissipates throughout the PEG molecule via anharmonic through-bond coupling to lower frequency modes, which in turn couple to the carbonyl modes of the succinimide group, and contribute to a cross-band. In addition to advancing our fundamental understanding of vibrational energy relaxation mechanisms, relaxation-assisted 2D IR spectroscopy can correlate vibrational frequencies in molecules to assist in spectral assignment and provide bond connectivity and distance information for characterizing molecular structure. Recently, a related experiment has been demonstrated based on through-space resonance energy transfer between vibrational modes.56 Analysis of the appearance of cross-bands due to energy transfer and their polarization dependence can provide angstrom-scale structural models of molecules.
Figure 5.
Structure and 2D IR spectrum of PEG4 with azido and succinimide ester reporter labels (Tw of 60 ps). To the left and above the axes of the 2D spectrum are the FTIR spectra of the labeled PEG4 molecule (blue) and spectra of the laser pulses (green) used to pump and probe the molecule, respectively. Reprinted with permission from Lin, Z. and Rubtsov, I.V. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109, 1413 – 1418.
Applications to complex systems
Nucleic acids
DNA and RNA adopt secondary and tertiary structures that are essential to their function and the regulation of many biological processes. The high sensitivity of IR spectroscopy to structure and interactions, such as hydrogen bonding, and its ability to resolve highly dynamic, flexible structures afford great potential for the characterization of nucleic acids. Unfortunately, like other biomolecules, IR spectra of nucleic acids are highly congested. The spectral complexity hinders interpretation and limits the information attainable from IR spectra. Cross-bands in 2D spectra help alleviate this issue by providing additional information about the coupling between modes to assist spectral assignment. Indeed, 2D IR studies indicate that the vibrational modes of nucleic acids are more highly coupled than previously recognized.57–59 For example, cross-bands in the 2D spectra of the nucleotide monophosphates provide evidence that the commonly analyzed in-plane stretches are highly delocalized over the aromatic rings.57 A better understanding of nucleotide vibrational bands and the factors that contribute to their changes will advance our ability to characterize more complex structures with linear or 2D IR spectroscopy.
Although 2D IR spectroscopy of nucleic acids is less well developed than its application to peptide systems, it has been extended to chemically modified nucleic acid monomers and base pairs, as well as oligomers of poly-adenosine-thymidine (A-T) or guanosine-cytidine (G-C) sequences.57–65 The capability of 2D IR spectroscopy to distinguish and identify vibrational modes, particularly the N-H modes, is critical in these studies. The first study of G-C base pairing found that a variety of factors, including both electrostatics and base stacking, contribute to the observed spectral changes.57, 66, 67 Studies of A-T pairs with 2D IR spectroscopy reveal the presence of not only conventional Watson-Crick base pairing but also reverse and Hoogsteen base pairing interactions, further revealing nucleic acid complexity.58 More complex samples of synthetic oligomers of repeating A-T or G-C at various levels of hydration have been investigated with 2D IR spectroscopy, making possible a more complete understanding of the vibrational modes and couplings.60, 61, 68 These studies also find that water dynamics are influenced by the presence of nucleic acids, showing slower spectral diffusion compared to bulk water.
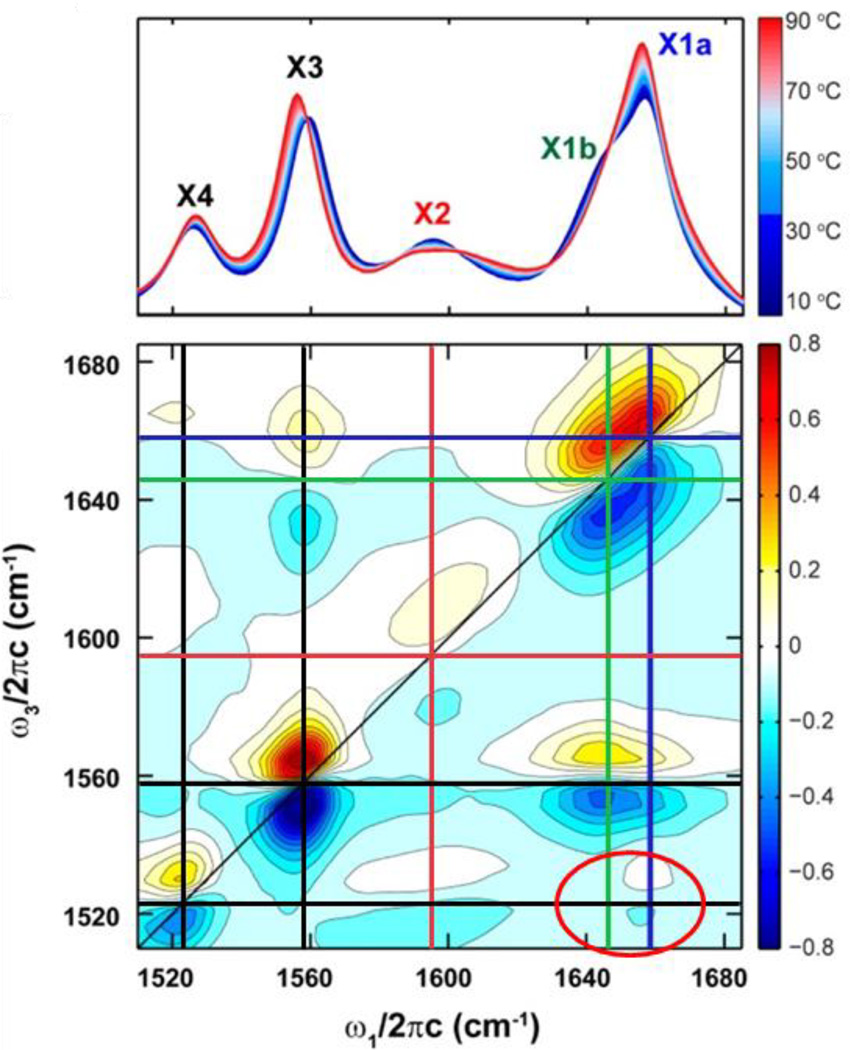
One interesting application of 2D IR spectroscopy is the characterization of different tautomer populations of the nucleic acid bases.69–71 Tautomerization of nucleic acids could be important in mechanisms of mutagenesis and generally in nucleic acid recognition, but its fast timescale has hindered its characterization under physiological conditions. Multiple tautomers have been detected in 2D IR studies of nucleic acid analogues, such as in a recent study of the modified nucleobase oxythiamine.69 Cross-band patterns in 2D spectra due to coupling between vibrational modes permit the discrimination of the three tautomeric states, which is not possible with linear spectroscopy due to spectral congestion, nor with NMR spectroscopy due to their rapid interconversion. To illustrate, the appearance of cross-bands between only one of the two carbonyl stretching modes and a mode associated with a methyl group vibration provides evidence for the presence of two distinct keto forms (Fig. 6). The ability of 2D IR spectroscopy to generate richer spectral data and discern rapidly interconverting states could be very powerful for analysis of nucleic acid biophysics and elucidation of the mechanisms underlying biological function.
Figure 6.
FTIR spectra of oxythiamine taken at various temperatures (upper panel) and 2D IR spectrum taken at 10 °C (lower panel). Blue and green lines in the 2D spectrum are shown along the frequencies of the carbonyl modes of the two keto tautomers (X1a and X1b). The presence of a cross-band between a mode associated with a methyl vibration (X4) at 1525 cm−1 and the X1a mode, and the absence of one with the X1b, is highlighted with a red ellipse. Reprinted with permission from Singh et al. ACS Chemical Biology, 2014, 9, 227 – 236. Copyright © 2014 American Chemical Society.
Peptide and protein structure and dynamics
Backbone structure and dynamics
2D IR spectroscopy has been extensively applied to the characterization of peptide and protein structures via the analysis of backbone vibrations.10, 72–79 Spectral analysis typically focuses on the amide I vibrational mode, which primarily consists of C=O and N-H vibrations, due to its sensitivity to secondary structure and decoupling from protein side chain vibrations. Because the IR spectra of even small peptides are highly complex, site-selective 13C and/or 18O labeling is typically required to spectrally isolate vibrations due to individual residues. Structural information is principally derived from the distance and orientation dependence of transition dipole coupling between the amide I modes. Cross-bands are direct evidence for coupling. Coupling also leads to shifts in the transition frequencies of the coupled modes, which can manifest as band broadening when the bands are unresolved. These spectral changes are frequently analyzed to determine the strength of coupling between particular amide I modes, which in turn reflects their proximity and relative orientation.
An important advantage of 2D IR spectroscopy is the fourth power dependence of the signal intensity on the transition dipole strength, compared to quadratic dependence for linear spectroscopy.80 Because of this, the 2D spectral features of the amide I modes are more intense with respect to the background absorption in comparison with the linear spectrum. 2D IR spectroscopy also enables measurement of the inhomogeneous linewidths, as well as characterization of spectral diffusion, and so provides richer, more rigorous information than is available from linear spectroscopy. For instance, when all the spectral information obtained by 2D IR spectroscopy is combined, it is possible to deduce the three-dimensional structure of a peptide assembly.72 For more routine analysis, secondary structure prediction by 2D IR spectroscopy exhibits better accuracy than analysis with linear spectroscopy.73
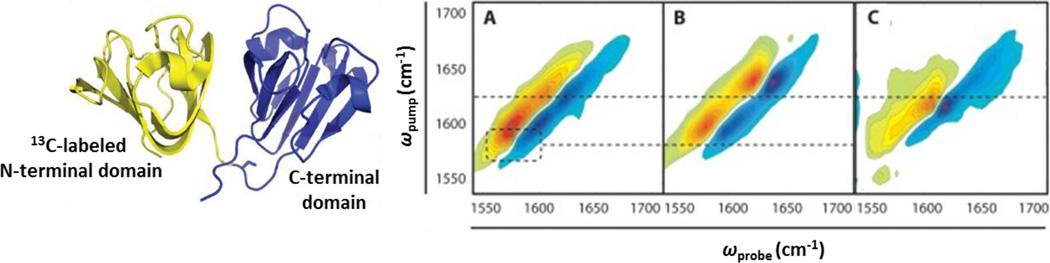
In addition to average structure, 2D IR spectroscopy can be used to characterize structural changes over time to advance our understanding of a variety of processes such as peptide folding and protein oligomerization.10, 74–76, 81–83 For example, 2D IR spectroscopy has been extensively applied toward the study of amyloid fibril formation.77, 78 Figure 7 shows 2D IR spectra from one recent study of human γD-crystallin protein that employed a combination of 2D IR spectroscopy and segmental 13C labeling to elucidate the involvement of the N- and C-terminal domains in the formation of amyloid fibrils.77 These and similar studies take advantage of the band enhancement in the 2D spectra and the characteristic low-energy shift of the amide I mode to monitor beta sheet formation. By investigating proteins with a combination of 13C-labeled N- and/or C-terminal domains, it was possible to show that the mechanism of amyloid formation depends on the route of denaturation. Comparison of the 2D spectra of the thermally induced aggregates and native states of γD-crystallin labeled with 13C at only the N-terminal domain (panels A and B, Fig. 7, respectively) reveals shifts to lower frequency in the amide I bands of both the 13C-labeled N-terminal and unlabeled C-terminal domains (amide I bands observed at low and high frequency, respectively) that indicate beta sheet formation. In contrast, the 2D spectrum of the acid-denatured state of the N-terminally 13C-labeled protein shows frequency shifts only for the high frequency, unlabeled amide bands of the C-terminal domain, not for the 13C-labeled N-terminal domain (panel C, Fig. 7). Thus, the 2D IR data provide evidence that both domains of γD-crystallin participate in amyloid fibril formation upon thermal denaturation, while only the C-terminal domain is involved in acid-induced fibril formation. The highly resolved structural information available from the combination of site-selective labeling and 2D IR spectroscopy should be able to yield similarly detailed insight into the molecular mechanisms underlying many biological processes.
Figure 7.
Structural model of γD-crystallin illustrating 13C-labeled N-terminal (yellow) and unlabeled C-terminal (blue) domains. 2D IR spectra of γD-crystallin labeled with 13C at the N-terminus are shown for (A) thermally-induced aggregates, (B) native, and (C) acid-induced aggregates. Reprinted with permission from Moran et al. Protein Science, 2014, 23, 321–331. Copyright © 2014 John Wiley and Sons.
Ligand probes
In addition to intrinsic vibrations of protein or peptide backbones, small molecule ligands can be used as reporters of their environments when bound to macromolecules. The selection of ligand reporter groups with spectrally isolated vibrational frequencies enables detection of probe absorption bands within the spectral congestion of macromolecular IR spectra. The vibrational mode of carbon monoxide bound to myoglobin was analyzed in the first demonstration of 2D IR spectroscopy to understand the dynamics of any macromolecule and has since been applied toward studying a variety of other heme proteins, such as globins and cytochromes.47, 84–89 Other small molecule reporter ligands, such as nitric oxide, azide, cyanide, and azide- and nitrile-functionalized ligand analogs, permit characterization of the local binding site environment and dynamics in a wider range of proteins.31, 48, 49, 90–92 2D IR spectroscopy can be applied to measure the inhomogeneous line width that reflects the heterogeneity of a probe's local environment and, moreover, to measure spectral diffusion that reflects the dynamics among the different states, providing insight into the protein energy landscape.
For example, 2D IR spectroscopy was applied toward understanding the mechanism underlying the potency of rilpivirine, a non-nucleoside inhibitor of reverse transcriptase from human immunodeficiency virus (HIV).90 Rilpivirine contains two nitrile group substituents that are required for potency, but that also fortuitously act as convenient spectrally isolated reporter vibrations for use with 2D IR spectroscopy. Upon binding reverse transcriptase and two clinically important mutant proteins, the absorption bands of the two nitrile groups become spectrally resolved, indicating that they experience distinct environments in the protein binding pocket. In addition, the observation of a fast rate of spectral diffusion suggests hydrogen bonding occurs between one of the nitriles and a mobile active site water molecule. This interaction might underlie the inhibitor's ability to retain potency against the mutants by providing an anchoring interaction that reduces the impact of other changes. As in this example, the prudent selection of reporter ligands in combination with 2D IR spectroscopy has enabled study of a wide variety of biological processes, such as protein molecular recognition, catalysis, and folding.46, 47, 86, 87, 93
Selective side chain labeling
In principle, 2D IR spectroscopy can be applied to characterize any site in any protein by selective labeling of side chains via incorporation of unnatural amino acids functionalized with spectrally resolved vibrational probes, such as cyano (CN),50, 94–96 thiocyanate (SCN),97 and azido (Az)93, 98 functional groups. A variety of routes exist for their site-selective incorporation into proteins. The introduction of SCN is possible by chemical modification of unique or uniquely reactive cysteine residues. Selective incorporation of cyanophenylalanine (CNPhe) and azidophenylalanine (AzPhe) is possible via the amber codon suppressor approach based on the introduction of unnatural amino acids in response to an amber (stop) codon with evolved tRNA/tRNA synthetase pairs.99 Expression systems for the incorporation of both CNPhe and AzPhe have been developed.100, 101 CNPhe, AzPhe, as well as a number of CN- and Az-substituted amino acids, are also available with standard protecting groups for solid phase peptide synthesis, facilitating selective incorporation via either total synthesis102 or semi-synthesis with expressed protein103 or native chemical ligation.104
The vibrational frequencies of these probe groups are sufficiently isolated for application to larger proteins. Unlike amide backbone modes, the vibrations of the side chain probes are largely localized to the specific probe bond(s). Their sensitivity to the electrostatic field of their local environment and short-range interactions like hydrogen bonding dominate the spectral changes, and so the probes report on their surrounding environment and dynamics when incorporated into proteins. Selection among the Az, CN, and SCN probes involves a compromise among spectral properties. The Az probe shows the most intense signals, which is advantageous for detection in nonlinear experiments of biological samples that are limited in concentration. Az groups often show Fermi resonances that complicate spectral interpretation, but the issue can be avoided with isotopic substitution.105 The short vibrational lifetimes of Az groups also limit the timescale over which they can be used to measure dynamics with 2D experiments. Similarly, a trade-off between intensity and lifetime is found for different CN- and SCN-functionalized amino acids. Aromatic CN groups provide the most intense signals but show the shortest vibrational lifetimes, while alkyl CN groups, and even more so SCN groups, provide weaker signals but show longer vibrational lifetimes. The perturbation to the protein from incorporation of a vibrational probe should also be considered, which will depend on the specific placement of a probe within a protein, e.g. at a surface vs. interior residue.
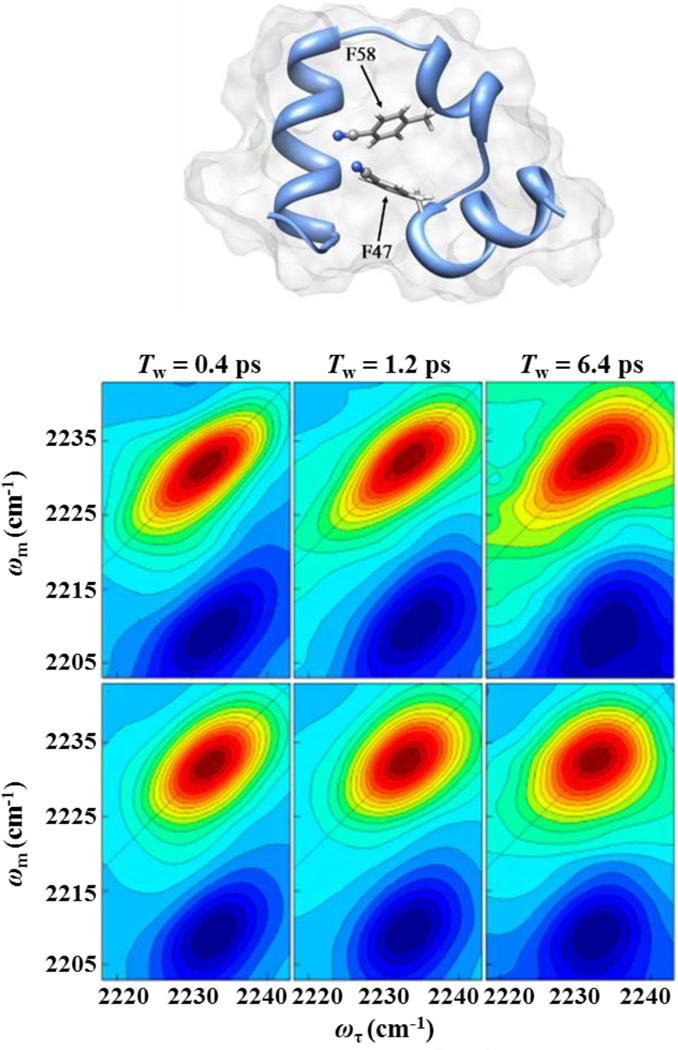
The use of CNPhe as a probe of protein dynamics is demonstrated by several studies of a polypeptide HP35 from chicken villin protein.50, 94–96 As a small system that autonomously folds into a protein-like domain, HP35 is a common subject for the study of protein folding. 2D IR spectroscopy was applied to measure folding-induced changes in dynamics reported at two CNPhe residues incorporated into HP35 via peptide synthesis (Fig. 8).50, 94 Because the CNPhe vibrational frequency is highly sensitive to fluctuations in its environment, the spectral diffusion of the probe reflects the dynamics of its protein or solvent environment. For example, time-dependent 2D spectra of CNPhe in folded and guanidinium-denatured HP35 are shown in Fig. 8.50 The greater elongation of the bands for the folded (upper panel, Fig. 8) compared to the unfolded state (lower panel, Fig. 8) reports on the greater heterogeneity of the environment in the folded protein compared to the homogeneous solvent environment in the unfolded state. Spectral diffusion leads to a reduction in the elongation of the 2D band shapes over time. The 2D band evolution for the folded state is demonstrably slower than for the unfolded state, which reflects the greater restriction of the dynamics in the folded state.
Figure 8.
Structural model of HP35 showing residues labeled with CNPhe and 2D IR spectra of CNPhe in folded (upper panels) and guanidium-denatured (lower panels) HP35 at different waiting times. Structural model: Reprinted with permission from Chung et al. The Journal of Physical Chemistry B, 2012, 116, 11024 – 11031. Copyright © 2012 American Chemical Society. 2D IR spectra: Reprinted with permission from Chung et al. Proceedings of the National Academy of Sciences of the United States of America, 2011, 108, 3578 – 3583.
Water dynamics
Due to its ubiquitous presence in nature, an understanding of water as a dynamic solvent could lend insight into a variety of phenomena from chemistry and biology to geology and astrophysics. 2D IR spectroscopy has been extensively applied toward advancing our fundamental understanding of water and the dynamics of its hydrogen bonding networks. A number of excellent reviews cover the work in this area.38, 106–109 One longstanding question surrounds the nature of water at an interface and the potential differences from bulk water.
Water at interfaces with membranes has attracted interest due to its importance in biology and for artificial energy harvesting devices. Many studies have investigated the nature of membrane-associated water via analysis of O-H or O-D stretching vibrations to directly probe water structure and dynamics.42, 110, 111 Another approach is to characterize the dynamics of interfacial water molecules via their influence on the spectral dynamics of another probe group, such as the phosphate head groups of the constituent lipids or small molecules added to membranes.112, 113 Reverse micelles serve as a useful model system for investigating the differences between bulk and interfacial water, as their preparation with defined size is well established and enables control over the proportion of water in the bulk “nanopool” or at the interface.42, 110, 111 Measurement of the spectral diffusion for water in the two populations with 2D IR spectroscopy shows that the dynamics of interfacial water molecules exhibit markedly slower dynamics than those in bulk solution. Additional 2D IR studies of different membrane structures, such as bicelles and lipid bilayers112, 113 find water dynamics at the interface are generally slower than in bulk solution, supporting the idea that the presence of an interface can impact the nature of water and potentially its behavior in chemical and biological systems.
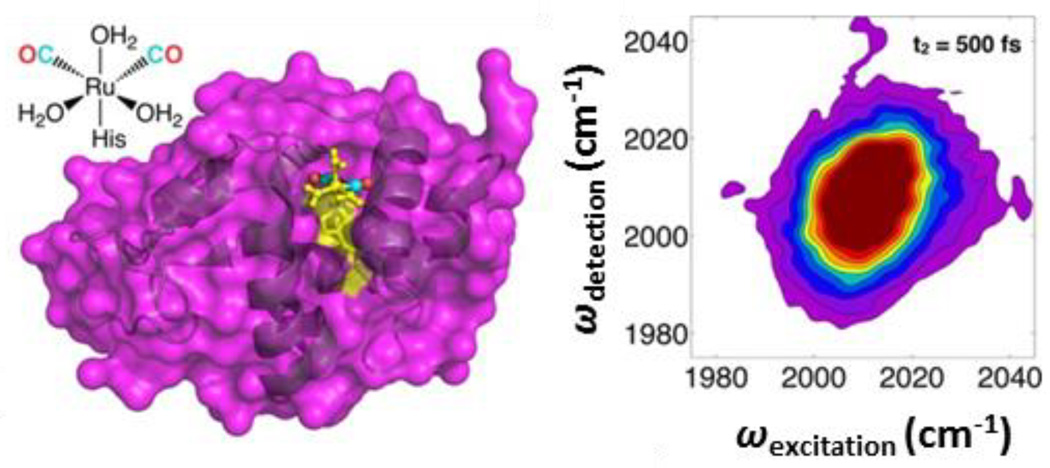
In addition to membranes, 2D IR spectroscopy has been used to probe water dynamics at the interface with DNA68 and proteins/peptides.43, 114–119 Both the O-H stretching vibration of the surrounding water and the N-H vibrations of the nucleotide bases have been used to measure the dynamics of the water shell around DNA oligomers. The hydration layer of proteins can be characterized via the attachment of vibrational probe groups that are sensitive to the dynamics of surrounding water.114, 117 For example, site-selective labeling of lysozyme with a ruthenium carbonyl complex and characterization of the vibrational spectral diffusion with 2D IR spectroscopy permitted measurement of water dynamics within its hydration shell (Fig. 9).114 As observed in studies of membrane-associated water, water at the surface of the protein exhibits markedly slower dynamics than the bulk. Interestingly, the study of lysozyme found that water associated with structured regions of the protein show slower dynamics, while water associated with unstructured, more flexible regions appear to maintain bulk-like behavior.115
Figure 9.
Structural model of a metal-carbonyl vibrational probe attached to hen egg white lysozme and 2D IR spectrum of the metal-carbonyl frequency region. Reprinted with permission from King et al. Journal of the American Chemical Society, 2014, 136, 188 – 194. Copyright © 2014 American Chemical Society.
Another approach to characterize protein-associated water is via the influence of water on spectral dynamics of 13C/18O-shifted vibrations of selectively labeled backbone amide modes.116, 118, 119 For example, the dynamics of water in the M2 proton channel from the influenza A virus have been characterized in this manner and related to pH-dependent conformational changes.118 Perturbation of the ordered water within the channel is observed upon binding of drug molecules, providing molecular insight into the potential entropic gain underlying the favorability of binding.119 These and other studies suggest that the dynamics of protein-associated water are heterogeneous, and that the nature of these dynamics might be important to protein function. The information that can be gained with 2D IR spectroscopy can provide valuable insight into the role water plays in a variety of complex biological settings and processes.
Applications in materials chemistry
In addition to the associated water at the membrane interface, 2D IR spectroscopy can yield insight into interior membrane structure and dynamics.51, 52, 113, 120 The use of nonpolar molecules as vibrational probes permits selective measurement of the spectral dynamics due to fluctuations within the interior hydrophobic bilayer environment.51, 113 For example, recent experiments took advantage of the intense absorption bands of the small, nonpolar molecular probe tungsten hexacarbonyl to measure the dynamics of both planar and vesicular bilayers with 2D IR spectroscopy. Large changes in structural dynamics occur upon bilayer phase transitions induced by dehydration or cholesterol addition, and furthermore, the changes are influenced by the degree of bilayer curvature.51, 52, 120 The combination of similar probes with 2D IR spectroscopy could advance our understanding of membrane biophysics and potentially how variations in membrane properties due to lipid or protein composition might influence biological systems.
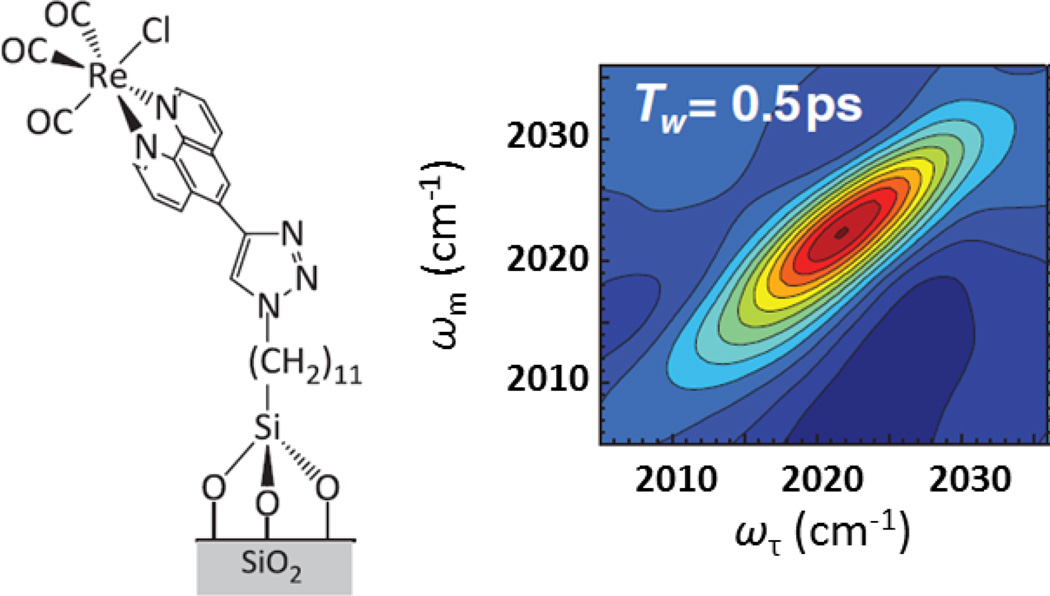
The characterization of materials important for such uses as heterogeneous catalysis, fuel cells, and molecular electronics is another interesting application of 2D IR spectroscopy.44, 121–123 For instance, polymer thin films of varying conductivity have been investigated using embedded metal-carbonyl complex reporter molecules.121 The measurement of spectral diffusion for this system provides evidence that structural dynamics in addition to the static structure contribute to the conductivity of the thin films. The interface with materials can also critically influence chemical behavior, in particular in heterogeneous catalysis. To investigate such systems, 2D IR spectroscopy has been performed on a monolayer of a Re-carbonyl complex covalently attached via a linker to a silica surface, of interest due to the potential of this and similar systems as photoreduction catalysts (Fig. 10).122 The spectral diffusion of the carbonyl vibrations reflects the dynamics of the catalyst and how they are influenced by factors such as solvation, surface density, and linker length. Other studies directed at a similar Re-carbonyl complex-based catalyst, but containing a different linker attachment to a TiO2 surface, found the spectral diffusion to depend on the surface density of the catalyst, indicating coupling between molecules even at low surface densities.44 Because the interfacial heterogeneity and intermolecular interactions can influence the performance of photocatalytic devices, for instance by contributing to energy relaxation pathways that compete with charge transport, understanding their structure and dynamics with 2D IR spectroscopy should be valuable for their future optimization.
Figure 10.
The structure of Re-carbonyl catalyst attached to a monolayer and example 2D IR spectrum. Rosenfeld et al. Science, 2011, 334, 634 – 639. Copyright © 2011 American Association for the Advancement of Science.
Other studies have shown the utility of 2D IR spectroscopy for characterization of nanomaterial surfaces. For example, the three-dimensional molecular conformations of capping molecules on gold nanoparticles have been deduced via structural constraints determined from cross-band intensities in polarization-dependent 2D IR spectra.124 This achievement relied on measurement of cross-band anisotropy, as well as the dependence of the coupling strength on the proximity and relative orientation of the capping molecules, similar in principle to the previously mentioned experiments for determining peptide structure. Finally, an exciting new development is the demonstration of surface-enhanced 2D IR signals, recently observed for vibrational modes of capping ligands on gold nanoparticles.125 Nearly 100-fold signal enhancements promise increased sensitivity in 2D IR spectroscopy for sensor and other applications demanding dilute or weakly absorbing samples. Such surface-enhanced 2D methods could find wide-ranging applications in analytical chemistry similar to linear surface-enhanced Raman and IR spectroscopy.
Summary and outlook
2D IR spectroscopy is a powerful approach to the analysis of molecular structure and dynamics on fast timescales with bond-specific spatial resolution. The additional information provided by the second dimension, as well as the versatility in temporal and polarization control of pulses, facilitate spectral assignment and interpretation, which is particularly critical to studies of complex molecules. We briefly highlighted a few examples that illustrate the potential applications of 2D IR spectroscopy to address chemical and biological questions. 2D IR spectroscopy of nucleic acid systems is yet to be fully developed. Particularly exciting is the further extension of the methods to understand the biophysics of RNA, highly flexible biomolecules with dynamics likely critical to their function, but difficult to study with traditional methods. Although the application of 2D IR spectroscopy to study the biophysics of peptides and proteins is more developed, the scope of the methods has yet to be fully explored. The combination of 2D IR spectroscopy with site-selective labeling with vibrational probes promises to emerge as a particularly powerful approach for the study of side chain dynamics, which are believed to contribute critically to protein function on fast timescales.126–128 Only a handful of proteins thus far have been analyzed via this route. The incorporation of multiple probes in one protein and the potential to obtain distance and orientation information from their coupling, as is currently performed with the 13C/18O-labeled backbone of peptides, is a future possibility. This approach will benefit from further development of spectrally resolved vibrational probes with more intense signals and longer lifetimes, as well as chemical and biological methods for their site-selective incorporation into proteins and nucleic acids.
Only the basic experimental implementation of 2D IR spectroscopy was presented in this minireview. A wide variety of extensions on this experiment have been developed.9–18 For instance, transient 2D IR spectroscopy is possible when combined with laser-induced temperature jump, photo-induced isomerization, or rapid mixing to initiate chemical reactions.10, 71, 76, 81–83, 129 Multidimensional experiments involving a combination of visible and IR excitation beams also have been reported,130, 131 providing information about vibronic coupling that promises to be particularly powerful for advancing our understanding of photosynthetic systems. Frequency-domain versions of 2D IR and visible-IR nonlinear experiments based on Raman processes, which employ narrow band, tunable excitation pulses, have also been well developed and applied in a variety of studies, for example in both nanomaterial and proteomic research.21, 132, 133 In addition, several groups have performed 2D IR experiments using ultra-broadband probe pulses.13, 134–136 Such broadband detection will facilitate analysis of chemical samples with features encompassing wide spectral ranges. Recently, 2D IR spectroscopy was implemented in attenuated total reflection mode,137 which has long been used in linear spectroscopy for its selective sensitivity to molecules at the prism surface. Experiments performed in reflection mode could find wide application in materials science. Finally, the large signal enhancements possible using surface-enhanced 2D IR spectroscopy could significantly improve levels of detection and widely extend the application of 2D IR spectroscopy in the analytical sciences.125 Overall, the rich toolbox of multidimensional IR methods provides a versatile route for characterization of a variety of different samples and for addressing a wide range of scientific questions.
Supplementary Material
Acknowledgements
The authors would like to thank Indiana University and the Department of Energy (DE-FOA-0000751) for funding. R. H. was also supported by the Graduate Training Program in Quantitative and Chemical Biology (T32 GM109825).
Notes and references
- 1.Zheng J, Kwak K, Fayer MD. Acc. Chem. Res. 2007;40:75–83. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]
- 2.Park S, Kwak K, Fayer MD. Laser Phys. Lett. 2007;4:704–718. [Google Scholar]
- 3.Hochstrasser RM. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14190–14196. doi: 10.1073/pnas.0704079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mukamel S. Principles of Nonlinear Optics and Spectroscopy. New York: Oxford University Press; 1995. [Google Scholar]
- 5.Hamm P, Zanni M. Concepts and Methods of 2D Infrared Spectroscopy. Cambridge University Press; 2011. [Google Scholar]
- 6.Cho M. Two-Dimensional Optical Spectroscopy. Boca Raton: CRC Press; 2009. [Google Scholar]
- 7.Fayer MD. Ultrafast Infrared Vibrational Spectroscopy. Boca Raton: CRC Press; 2013. [Google Scholar]
- 8.Cho M. Chem. Rev. 2008;108:1331–1418. doi: 10.1021/cr078377b. [DOI] [PubMed] [Google Scholar]
- 9.Strasfeld DB, Shim SH, Zanni M. In: Advances in Chemical Physics. Rice SA, editor. Vol. 141. Hoboken, New Jersey: John Wiley and Sons, Inc.; 2009. pp. 1–28. [Google Scholar]
- 10.Tucker MJ, Abdo M, Courter JR, Chen J, Brown SP, Smith AB, 3rd, Hochstrasser RM. Proc. Natl. Acad. Sci. U. S. A. 2013;110:17314–17319. doi: 10.1073/pnas.1311876110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalil M, Demirdoven N, Tokmakoff A. J Phys. Chem. A. 2003;107:5258–5279. [Google Scholar]
- 12.Hochstrasser RM. Chem. Phys. 2001;266:273–284. [Google Scholar]
- 13.Baiz CR, Kubarych KJ. Opt. Lett. 2011;36:187–189. doi: 10.1364/OL.36.000187. [DOI] [PubMed] [Google Scholar]
- 14.Nee MJ, McCanne R, Kubarych KJ. Opt. Lett. 2007;32:713–715. doi: 10.1364/ol.32.000713. [DOI] [PubMed] [Google Scholar]
- 15.DeCamp MF, DeFlores LP, Jones KC, Tokmakoff A. Opt. Express. 2007;15:233–241. doi: 10.1364/oe.15.000233. [DOI] [PubMed] [Google Scholar]
- 16.DeFlores LP, Nicodemus RA, Tokmakoff A. Opt. Lett. 2007;32:2966–2968. doi: 10.1364/ol.32.002966. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee SS, Skoff DR, Middleton CT, Zanni MT. J Chem. Phys. 2013;139:144205. doi: 10.1063/1.4824638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helbing J, Hamm P. J Opt. Soc. Am. B. 2011;28:171–178. [Google Scholar]
- 19.Wright JC. Annu. Rev. Phys. Chem. 2011;62:209–230. doi: 10.1146/annurev-physchem-032210-103551. [DOI] [PubMed] [Google Scholar]
- 20.Boyle ES, Neff-Mallon NA, Wright JC. J Phys. Chem. A. 2013;117:12401–12408. doi: 10.1021/jp409377a. [DOI] [PubMed] [Google Scholar]
- 21.Fournier F, Guo R, Gardner EM, Donaldson PM, Loeffeld C, Gould IR, Willison KR, Klug DR. Acc. Chem. Res. 2009;42:1322–1331. doi: 10.1021/ar900074p. [DOI] [PubMed] [Google Scholar]
- 22.Cervetto V, Helbing J, Bredenbeck J, Hamm P. J Chem. Phys. 2004;121:5935–5942. doi: 10.1063/1.1778163. [DOI] [PubMed] [Google Scholar]
- 23.Shim SH, Strasfeld DB, Zanni M. Opt. Express. 2006;14:13120–13130. doi: 10.1364/oe.14.013120. [DOI] [PubMed] [Google Scholar]
- 24.PhaseTech Spectroscopy. [accessed March 19, 2015]; http://phasetechspectroscopy.com. [Google Scholar]
- 25.Zanni M. In: Ultrafast Phenomena. Corkum P, de Silvestri S, Nelson KA, Reidle E, Schoenlein RW, editors. Vol. 92. Berlin: Springer; 2009. pp. 397–399. [Google Scholar]
- 26.Kumar SKK, Tamimi A, Fayer MD. J Chem. Phys. 2012;137:184201. doi: 10.1063/1.4764470. [DOI] [PubMed] [Google Scholar]
- 27.Rock W, Li YL, Pagano P, Cheatum CM. J Phys. Chem. A. 2013;117:6073–6083. doi: 10.1021/jp312817t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park KH, Choi SR, Choi J-H, Park S, Cho M. Chem Phys Chem. 2010;11:3632–3637. doi: 10.1002/cphc.201000595. [DOI] [PubMed] [Google Scholar]
- 29.Brookes JF, Slenkamp KM, Lynch MS, Khalil M. J Phys. Chem. A. 2013;117:6234–6243. doi: 10.1021/jp4005345. [DOI] [PubMed] [Google Scholar]
- 30.Simpson N, Shaw DJ, Frederix PWJM, Gillies AH, Adamczyk K, Greetham GM, Towrie M, Parker AW, Hoskisson PA, Hunt NT. J Phys. Chem. B. 2013;117:16468–16478. doi: 10.1021/jp411091f. [DOI] [PubMed] [Google Scholar]
- 31.Dutta S, Li Y-L, Rock W, Houtman JCD, Kohen A, Cheatum CM. J Phys. Chem. B. 2012;116:542–548. doi: 10.1021/jp208677u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tucker MJ, Gai XS, Fenlon EE, Brewer SH, Hochstrasser RM. Phys. Chem. Chem. Phys. 2011;13:2237–2241. doi: 10.1039/c0cp01625j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Cho M. Chem. Rev. 2013;113:5817–5847. doi: 10.1021/cr3005185. [DOI] [PubMed] [Google Scholar]
- 34.Panman MR, Vos J, Bocokic V, Bellini R, de Bruin B, Reek JHN, Woutersen S. Inorg. Chem. 2013;52:14294–14298. doi: 10.1021/ic402254q. [DOI] [PubMed] [Google Scholar]
- 35.Messmer AT, Lippert KM, Schreiner PR, Bredenbeck J. Phys. Chem. Chem. Phys. 2013;15:1509–1517. doi: 10.1039/c2cp42863f. [DOI] [PubMed] [Google Scholar]
- 36.Sun Z, Zhang W, Ji M, Hartsock R, Gaffney KJ. J Phys. Chem. B. 2013;117:12268–12275. doi: 10.1021/jp407960x. [DOI] [PubMed] [Google Scholar]
- 37.Bian H, Chen H, Zhang Q, Li J, Wen X, Zhuang W, Zheng J. J Phys. Chem. B. 2013;117:7972–7984. doi: 10.1021/jp4016646. [DOI] [PubMed] [Google Scholar]
- 38.Fayer MD. Acc. Chem. Res. 2012;45:3–14. doi: 10.1021/ar2000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuroda DG, Hochstrasser RM. Phys. Chem. Chem. Phys. 2012;14:6219–6224. doi: 10.1039/c2cp23892f. [DOI] [PubMed] [Google Scholar]
- 40.Huerta-Viga A, Domingos SR, Amirjalayer S, Woutersen S. Phys. Chem. Chem. Phys. 2014;16:15784–15786. doi: 10.1039/c4cp00233d. [DOI] [PubMed] [Google Scholar]
- 41.Kwak K, Park S, Finkelstein IJ, Fayer MD. J Chem. Phys. 2007;127:124503. doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 42.Fenn EE, Wong DB, Giammanco CH, Fayer MD. J Phys. Chem. B. 2011;115:11658–11670. doi: 10.1021/jp206903k. [DOI] [PubMed] [Google Scholar]
- 43.Fogarty AC, Laage D. J Phys. Chem. B. 2014;118:7715–7729. doi: 10.1021/jp409805p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laaser JE, Christianson JR, Oudenhoven TA, Joo Y, Gopalan P, Schmidt JR, Zanni MT. J Phys. Chem. C. 2014;118:5854–5861. [Google Scholar]
- 45.King JT, Kubarych KJ. J Am. Chem. Soc. 2012;134:18705–18712. doi: 10.1021/ja307401r. [DOI] [PubMed] [Google Scholar]
- 46.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thielges MC, Fayer MD. Acc. Chem. Res. 2012;45:1866–1874. doi: 10.1021/ar200275k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Proc. Natl. Acad. Sci. U. S. A. 2010;107:17974–17979. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng M, Brookes JF, Montfort WR, Khalil M. J Phys. Chem. B. 2013;117:15804–15811. doi: 10.1021/jp407052a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung JK, Thielges MC, Fayer MD. Proc. Natl. Acad. Sci. U. S. A. 2011;108:3578–3583. doi: 10.1073/pnas.1100587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kel O, Tamimi A, Fayer MD. Proc. Natl. Acad. Sci. U. S. A. 2014;111:918–923. doi: 10.1073/pnas.1323110111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar SKK, Tamimi A, Fayer MD. J Am. Chem. Soc. 2013;135:5118–5126. doi: 10.1021/ja312676e. [DOI] [PubMed] [Google Scholar]
- 53.Hill AD, Zoerb MC, Nguyen SC, Lomont JP, Bowring MA, Harris CB. J Phys. Chem. B. 2013;117:15346–15355. doi: 10.1021/jp403791k. [DOI] [PubMed] [Google Scholar]
- 54.Kurochkin DV, Naraharisetty SRG, Rubtsov IV. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14209–14214. doi: 10.1073/pnas.0700560104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Z, Rubtsov IV. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1413–1418. doi: 10.1073/pnas.1116289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen H, Wen X, Li J, Zheng J. J Phys. Chem. A. 2014;118:2463–2469. doi: 10.1021/jp500586h. [DOI] [PubMed] [Google Scholar]
- 57.Peng CS, Jones KC, Tokmakoff A. J Am. Chem. Soc. 2011;133:15650–15660. doi: 10.1021/ja205636h. [DOI] [PubMed] [Google Scholar]
- 58.Greve C, Preketes NK, Fidder H, Costard R, Koeppe B, Heisler IA, Mukamel S, Temps F, Nibbering ETJ, Elsaesser T. J Phys. Chem. A. 2013;117:594–606. doi: 10.1021/jp310177e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greve C, Preketes NK, Costard R, Koeppe B, Fidder H, Nibbering ETJ, Temps F, Mukamel S, Elsaesser T. J Phys. Chem. A. 2012;116:7636–7644. doi: 10.1021/jp303864m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greve C, Elsaesser T. J Phys. Chem. B. 2013;117:14009–14017. doi: 10.1021/jp408229k. [DOI] [PubMed] [Google Scholar]
- 61.Yang M, Szyc L, Elsaesser T. J Phys. Chem. B. 2011;115:1262–1267. doi: 10.1021/jp1090697. [DOI] [PubMed] [Google Scholar]
- 62.Krummel AT, Mukherjee P, Zanni MT. J Phys. Chem. B. 2003;107:9165–9169. [Google Scholar]
- 63.Krummel AT, Zanni MT. J Phys. Chem. B. 2006;110:13991–14000. doi: 10.1021/jp062597w. [DOI] [PubMed] [Google Scholar]
- 64.Yang M, Szyc L, Rottger K, Fidder H, Nibbering ETJ, Elsaesser T, Temps F. J Phys. Chem. B. 2011;115:5484–5492. doi: 10.1021/jp110561d. [DOI] [PubMed] [Google Scholar]
- 65.Fidder H, Yang M, Nibbering ETJ, Elsaesser T, Rottger K, Temps F. J Phys. Chem. A. 2013;117:845–854. doi: 10.1021/jp309237u. [DOI] [PubMed] [Google Scholar]
- 66.Lee C, Park KH, Cho M. J Chem. Phys. 2006;125:114508. doi: 10.1063/1.2213257. [DOI] [PubMed] [Google Scholar]
- 67.Lee C, Cho M. J Chem. Phys. 2006;125:114509. doi: 10.1063/1.2213258. [DOI] [PubMed] [Google Scholar]
- 68.Yang M, Szyc L, Elsaesser T. J Phys. Chem. B. 2011;115:13093–13100. doi: 10.1021/jp208166w. [DOI] [PubMed] [Google Scholar]
- 69.Singh V, Peng CS, Li D, Mitra K, Silvestre KJ, Tokmakoff A, Essigmann JM. ACS Chem. Biol. 2014;9:227–236. doi: 10.1021/cb400581f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peng CS, Tokmakoff A. J Phys. Chem. Lett. 2012;3:3302–3306. doi: 10.1021/jz301706a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng CS, Baiz CR, Tokmakoff A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9243–9248. doi: 10.1073/pnas.1303235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Remorino A, Korendovych IV, Wu Y, DeGrado WF, Hochstrasser RM. Science. 2011;332:1206–1209. doi: 10.1126/science.1202997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baiz CR, Peng CS, Reppert ME, Jones KC, Tokmakoff A. Analyst. 2012;137:1793–1799. doi: 10.1039/c2an16031e. [DOI] [PubMed] [Google Scholar]
- 74.Ganim Z, Jones KC, Tokmakoff A. Phys. Chem. Chem. Phys. 2012;12:3579–3588. doi: 10.1039/b923515a. [DOI] [PubMed] [Google Scholar]
- 75.Maekawa H, De Poli M, Toniolo and N.-H. Ge C. J Am. Chem. Soc. 2009;131:2042–2043. doi: 10.1021/ja807572f. [DOI] [PubMed] [Google Scholar]
- 76.Jones KC, Peng CS, Tokmakoff A. Proc. Natl. Acad. Sci. U. S. A. 2013;110:2828–2833. doi: 10.1073/pnas.1211968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moran SD, Zhang TO, Zanni MT. Protein Sci. 2014;23:321–331. doi: 10.1002/pro.2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moran SD, Woys AM, Buchanan LE, Bixby E, Decatur SM, Zanni MT. Proc. Natl. Acad. Sci. U. S. A. 2012;109:3329–3334. doi: 10.1073/pnas.1117704109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buchanan LE, Dunkelberger EB, Tran HQ, Cheng P-N, Chiu C-C, Cao P, Raleigh DP, de Pablo JJ, Nowick JS, Zanni MT. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19285–19290. doi: 10.1073/pnas.1314481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grechko M, Zanni MT. J Chem. Phys. 2012;137:184202. doi: 10.1063/1.4764861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chung HS, Ganim Z, Jones KC, Tokmakoff A. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14237–14342. doi: 10.1073/pnas.0700959104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hamm P, Helbing J, Bredenbeck J. Annu. Rev. Phys. Chem. 2008;59:291–317. doi: 10.1146/annurev.physchem.59.032607.093757. [DOI] [PubMed] [Google Scholar]
- 83.Tucker MJ, Abdo M, Courter JR, Chen J, Smith AB, 3rd, Hochstrasser RM. J Photochem. Photobiol. A. 2012;234:156–163. doi: 10.1016/j.jphotochem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Merchant KA, Thompson DE, Xu Q-H, Williams RB, Loring RF, Fayer MD. Biophys. J. 2002;82:3277–3288. doi: 10.1016/S0006-3495(02)75669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Thielges MC, Chung JK, Fayer MD. J Am. Chem. Soc. 2011;133:3995–4004. doi: 10.1021/ja109168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chung JK, Thielges MC, Bowman SEJ, Bren KL, Fayer MD. J Am. Chem. Soc. 2011;133:6681–6691. doi: 10.1021/ja111009s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ishikawa H, Kwak K, Chung JK, Kim S, Fayer MD. Proc. Natl. Acad. Sci. U. S. A. 2008;105:8619–8624. doi: 10.1073/pnas.0803764105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Proc. Natl. Acad. Sci. U. S. A. 2007;104:16116–16121. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Finkelstein IJ, Zheng J, Ishikawa H, Kim S, Kwak K, Fayer MD. Phys. Chem. Chem. Phys. 2007;9:1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 90.Kuroda DG, Bauman JD, Challa JR, Patel D, Troxler T, Das K, Arnold E, Hochstrasser RM. Nat. Chem. 2013;5:174–181. doi: 10.1038/nchem.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adamczyk K, Candelaresi M, Kania R, Robb K, Bellota-Anton C, Greetham GM, Pollard MR, Towrie M, Parker AW, Hoskisson PA, Tucker NP, Hunt NT. Phys. Chem. Chem. Phys. 2012;14:7411–7419. doi: 10.1039/c2cp23568d. [DOI] [PubMed] [Google Scholar]
- 92.Maj M, Oh Y, Park K, Lee J, Kwak KW, Cho M. J Chem. Phys. 2014;140:235104. doi: 10.1063/1.4883505. [DOI] [PubMed] [Google Scholar]
- 93.Bloem R, Koziol K, Waldauer SA, Buchli B, Walser R, Samatanga B, Jelesarov I, Hamm P. J Phys. Chem. B. 2012;116:13705–13712. doi: 10.1021/jp3095209. [DOI] [PubMed] [Google Scholar]
- 94.Chung JK, Thielges MC, Fayer MD. J Am. Chem. Soc. 2012;134:12118–12124. doi: 10.1021/ja303017d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Urbanek DC, Vorobyev DY, Serrano AL, Gai F, Hochstrasser RM. J Phys. Chem. Lett. 2010;1:3311–3315. doi: 10.1021/jz101367d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bagchi S, Boxer SG, Fayer MD. J Phys. Chem. B. 2012;116:4034–4042. doi: 10.1021/jp2122856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Wilderen LJGW, Kern-Michler D, Muller-Werkmeister HM, Bredenbeck J. Phys. Chem. Chem. Phys. 2014;16:19643–19653. doi: 10.1039/c4cp01498g. [DOI] [PubMed] [Google Scholar]
- 98.Thielges MC, Axup JY, Wong D, Lee HS, Chung JK, Schultz PG, Fayer MD. J Phys. Chem. B. 2011;115:11294–11304. doi: 10.1021/jp206986v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu CC, Schultz PG. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 100.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J Am. Chem. Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 101.Ohno S, Yokogawa T, Nishikawa K. J Biochem. 2001;130:417–423. doi: 10.1093/oxfordjournals.jbchem.a003001. [DOI] [PubMed] [Google Scholar]
- 102.Dawson PE, Fitzgerald MC, Muir TW, Kent SBH. J Am. Chem. Soc. 1997;119:7917–7927. [Google Scholar]
- 103.Muir TW. Annu. Rev. Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- 104.Dawson PE, Kent SBH. Annu. Rev. Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 105.Lipkin JS, Song R, Fenlon EE, Brewer SH. J Phys. Chem. Lett. 2011;2:1672–1676. doi: 10.1021/jz2006447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Laage D, Stirnemann G, Sterpone F, Hynes JT. Acc. Chem. Res. 2012;45:53–62. doi: 10.1021/ar200075u. [DOI] [PubMed] [Google Scholar]
- 107.Roberts ST, Ramasesha K, Tokmakoff A. Acc. Chem. Res. 2009;42:1239–1249. doi: 10.1021/ar900088g. [DOI] [PubMed] [Google Scholar]
- 108.Bakker HJ, Skinner JL. Chem. Rev. 2010;110:1498–1517. doi: 10.1021/cr9001879. [DOI] [PubMed] [Google Scholar]
- 109.Fayer MD, Moilanen DE, Wong D, Rosenfeld DE, Fenn EE, Park S. Acc. Chem. Res. 2009;42:1210–1219. doi: 10.1021/ar900043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bakulin AA, Cringus D, Pieniazek PA, Skinner JL, Jansen TLC, Pshenichnikov MS. J Phys. Chem. B. 2013;117:15545–15558. doi: 10.1021/jp405853j. [DOI] [PubMed] [Google Scholar]
- 111.Costard R, Greve C, Heisler IA, Elsaesser T. J Phys. Chem. Lett. 2012;3:3646–3651. doi: 10.1021/jz3018978. [DOI] [PubMed] [Google Scholar]
- 112.Costard R, Heisler IA, Elsaesser T. J Phys. Chem. Lett. 2014;5:506–511. doi: 10.1021/jz402493b. [DOI] [PubMed] [Google Scholar]
- 113.Osborne DG, Dunbar JA, Lapping JG, White AM, Kubarych KJ. J Phys. Chem. B. 2013;117:15407–15414. doi: 10.1021/jp4049428. [DOI] [PubMed] [Google Scholar]
- 114.King JT, Arthur EJ, Brooks CL, 3rd, Kubarych KJ. J Am. Chem. Soc. 2014;136:188–194. doi: 10.1021/ja407858c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.King JT, Arthur EJ, Brooks CL, 3rd, Kubarych KJ. J Phys. Chem. B. 2012;116:5604–5611. doi: 10.1021/jp300835k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ghosh A, Hochstrasser RM. Chem. Phys. 2011;390:1–13. doi: 10.1016/j.chemphys.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Woys AM, Mukherjee SS, Skoff DR, Moran SD, Zanni MT. J Phys. Chem. B. 2013;117:5009–5018. doi: 10.1021/jp402946c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ghosh A, Qiu J, DeGrado WF, Hochstrasser RM. Proc. Natl. Acad. Sci. U. S. A. 2011;108:6115–6120. doi: 10.1073/pnas.1103027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghosh A, Wang J, Moroz YS, Korendovych IV, Zanni M, DeGrado WF, Gai F, Hochstrasser RM. J Chem. Phys. 2014;140:235105. doi: 10.1063/1.4881188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kel O, Tamimi A, Fayer MD. J Phys. Chem. B. 2014 doi: 10.1021/jp503940k. [DOI] [PubMed] [Google Scholar]
- 121.Eigner AA, Jones BH, Koprucki BW, Massari AM. J Phys. Chem. B. 2011;115:4583–4591. doi: 10.1021/jp1113009. [DOI] [PubMed] [Google Scholar]
- 122.Rosenfeld DE, Gengeliczki Z, Smith BJ, Stack TD, Fayer MD. Science. 2011;334:634–639. doi: 10.1126/science.1211350. [DOI] [PubMed] [Google Scholar]
- 123.Rosenfeld DE, Nishida J, Yan C, Gengeliczki Z, Smith BJ, Fayer MD. J Phys. Chem. C. 2012;116:23428–23440. doi: 10.1021/jp307677b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bian H, Li J, Chen H, Yuan K, Wen X, Li Y, Sun Z, Zheng J. J Phys. Chem. C. 2012;116:7913–7924. doi: 10.1021/jp306369w. [DOI] [PubMed] [Google Scholar]
- 125.Donaldson PM, Hamm P. Angew. Chem. Int. Ed. Engl. 2013;52:634–638. doi: 10.1002/anie.201204973. [DOI] [PubMed] [Google Scholar]
- 126.Henzler-Wildman K, Kern D. Nature. 2007;450:964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 127.Jarmowycz VA, Stone MJ. Chem. Rev. 2006;106:1624–1671. doi: 10.1021/cr040421p. [DOI] [PubMed] [Google Scholar]
- 128.Marlow MS, Dogan J, Frederick KK, Valentine KG, Wand AJ. Nat. Chem. Biol. 2010;6:352–358. doi: 10.1038/nchembio.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cervetto V, Hamm P, Helbing J. J Phys. Chem. B. 2008;112:8398–8405. doi: 10.1021/jp801166q. [DOI] [PubMed] [Google Scholar]
- 130.Oliver TAA, Lewis NHC, Fleming GR. Proc. Natl. Acad. Sci. U. S. A. 2014;111:10061–10066. doi: 10.1073/pnas.1409207111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Donaldson PM, Strzalka H, Hamm P. Opt. Express. 2012;20:12761–12770. doi: 10.1364/OE.20.012761. [DOI] [PubMed] [Google Scholar]
- 132.Fournier F, Gardner EM, Kedra DA, Donaldson PM, Guo R, Butcher SA, Gould IR, Willison KR, Klug DR. Proc. Natl. Acad. Sci. U. S. A. 2008;105:15352–15357. doi: 10.1073/pnas.0805127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pakoulev AV, Rickard MA, Kornau KM, Mathew NA, Yurs LA, Block SB, Wright JA. Acc. Chem. Res. 2009;42:1310–1321. doi: 10.1021/ar900032g. [DOI] [PubMed] [Google Scholar]
- 134.Stingel AM, Calabrese C, Petersen PB. J Phys. Chem. B. 2013;117:15714–15719. doi: 10.1021/jp406441r. [DOI] [PubMed] [Google Scholar]
- 135.De Marco L, Ramasesha K, Tokmakoff A. J Phys. Chem. B. 2013;117:15319–15327. doi: 10.1021/jp4034613. [DOI] [PubMed] [Google Scholar]
- 136.Cheng M, Reynolds A, Widgren H, Khalil M. Opt. Lett. 2012;37:1787–1789. doi: 10.1364/OL.37.001787. [DOI] [PubMed] [Google Scholar]
- 137.Kraack JP, Lotti D, Hamm P. J Phys. Chem. Lett. 2014;5:2325–2329. doi: 10.1021/jz500978z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.