Abstract
Bacillus cereus is a Gram-positive endospore forming bacterium known for its wide spectrum of phenotypic traits, enabling it to occupy diverse ecological niches. Although the population structure of B. cereus is highly dynamic and rather panmictic, production of the emetic B. cereus toxin cereulide is restricted to strains with specific genotypic traits, associated with distinct environmental habitats. Cereulide is an ionophoric dodecadepsipeptide that is produced non-ribosomally by an enzyme complex with an unusual modular structure, named cereulide synthetase (Ces non-ribosomal peptide synthetase). The ces gene locus is encoded on a mega virulence plasmid related to the B. anthracis toxin plasmid pXO1. Cereulide, a highly thermo- and pH- resistant molecule, is preformed in food, evokes vomiting a few hours after ingestion, and was shown to be the direct cause of gastroenteritis symptoms; occasionally it is implicated in severe clinical manifestations including acute liver failures. Control of toxin gene expression in emetic B. cereus involves central transcriptional regulators, such as CodY and AbrB, thereby inextricably linking toxin gene expression to life cycle phases and specific conditions, such as the nutrient supply encountered in food matrices. While in recent years considerable progress has been made in the molecular and biochemical characterization of cereulide toxin synthesis, far less is known about the embedment of toxin synthesis in the life cycle of B. cereus. Information about signals acting on toxin production in the food environment is lacking. We summarize the data available on the complex regulatory network controlling cereulide toxin synthesis, discuss the role of intrinsic and extrinsic factors acting on toxin biosynthesis in emetic B. cereus and stress how unraveling these processes can lead to the development of novel effective strategies to prevent toxin synthesis in the food production and processing chain.
Keywords: emetic toxin, cereulide, Bacillus cereus, foodborne intoxication, non-ribosomal peptide synthetase, pathometabolism
Introduction
Bacillus cereus is an ubiquitous Gram-positive, facultative anaerobic rod-shaped bacterium that is notorious due to the formation of highly heat, acid, UV, and desiccation resistant endospores. It is increasingly recognized as an opportunistic pathogen causing gastrointestinal as well as local and systemic infections (Bottone, 2010; Ehling-Schulz et al., 2011). Because of its broad spectrum of phenotypic traits, B. cereus can enter food production and processing chains at various points, confronting the food industry with several challenges. Beside its food spoilage potential, it bears a severe health risk for consumers. Several toxins have been described that may cause two types of food borne diseases. The non-hemolytic enterotoxin complex (NHE) and the hemolytic enterotoxin complex (HBL) as well as a variant of the single cytotoxin K have been linked to the diarrheal form of the disease, while the depsipeptide toxin cereulide has been shown to be the causative agent of the emetic form of the disease (Ehling-Schulz et al., 2004; Schoeni and Wong, 2005; Stenfors Arnesen et al., 2008). In the past, diagnostics were hampered by the lack of appropriate detection and identification systems rendering it impossible to estimate the true incidence of food borne diseases related to B. cereus. However, during the last decade several tools, such as multiplex PCR for toxin gene profiling, in situ monitoring of cereulide synthesis with luminescent reporter strains, or mass spectrometry-based methods for toxin quantitation in complex food matrices have been developed (Ehling-Schulz et al., 2006b; Bauer et al., 2010; Dommel et al., 2010; Stark et al., 2013), which facilitate and significantly improve B. cereus diagnostics (Ehling-Schulz and Messelhäusser, 2013). Indeed, the number of reported food poisonings related to B. cereus toxins has shown a steep increase from 2006 onward, and is still increasing (Anonymous, 2009, 2015). Yet, in most countries the reporting of B. cereus-induced gastroenteritis is mandatory only in foodborne outbreak cases, leading to an underestimation of this bacterial incidence on public health. The capacity for enterotoxin production is widely distributed within the B. cereus population. In contrast, emetic toxin production is restricted to strains with specific genotypic and phenotypic traits, although some diversification seems to be ongoing (Prüss et al., 1999; Ehling-Schulz et al., 2005a, 2011; Thorsen et al., 2006; Guinebretiere et al., 2008; Hoton et al., 2009).
Both forms of the disease are normally self-limiting but some more severe forms requiring hospitalization are occasionally reported (Lund et al., 2000; Fricker et al., 2007; Ehling-Schulz and Messelhäusser, 2012). Especially the emetic toxin cereulide is increasingly linked to severe clinical manifestations including acute liver failures and has also been reported as a causative agent of acute encephalopathy (Dierick et al., 2005; Posfay-Barbe et al., 2008; Ichikawa et al., 2010; Naranjo et al., 2011; Tschiedel et al., 2015). Recent results from in vitro tests showed a detrimental effect of cereulide on pancreatic beta cells, even in low concentrations. Since beta cell dysfunction and cell death plays a key role in pathophysiology of diabetes, these results render it possible that a long-termed uptake of cereulide-contaminated food could have some implications in diabetes (Vangoitsenhoven et al., 2014).
The diarrheal type of disease is thought to be the consequence of a foodborne infection with enterotoxic B. cereus and enterotoxin production after outgrowth of bacterial spores in the intestine, while the emetic syndrome is presumably the result of intoxication by cereulide, previously formed in food contaminated with emetic B. cereus. Thus, an understanding of the interplay of food environment and emetic B. cereus is of utmost importance to fully decipher mechanisms of pathogenicity of emetic B. cereus and to develop novel strategies for the prevention of cereulide toxin production in the food production and processing chain. While in recent years considerable progress has been made in the molecular and biochemical characterization of cereulide toxin synthesis, far less is known about the embedment of toxin synthesis in the life cycle of B. cereus and the primary ecological niches of emetic B. cereus are largely unknown.
An Unusual Enzyme Complex for an Unusual Toxin: Architecture of the Genetic Locus for Cereulide Depsipeptide Toxin Production
Intracellular Biochemical Factory for Cereulide Production
In contrast to the enterotoxin genes, which are ‘classical’ protein toxins produced via the ribosomal pathway, the emetic depsipeptide toxin cereulide [D-O-Leu-D-Ala-L-O-Val-L-Val]3 is produced by a non-ribosomal peptide synthetase (NRPS; Ehling-Schulz et al., 2005b). NRPS are large modular multienzyme complexes that act as in vivo biochemical factories for the production of a variety of natural products, such as antibiotics, surfactants, and peptide toxins, following the thiotemplate model instead of the ribosomal route. Each NRPS module incorporates one monomer in the growing peptide chain in a directed manner by the concerted action of conserved domains catalyzing multistep reactions, thereby defining the chemical identity of the final product. An NRPS module comprises at least an adenylation (A) domain that specifically recognizes and activates a certain substrate, its cognate peptidyl carrier protein (PCP; also called T domain) and a condensation (C) domain that catalyzes the chain growth (for review, see Marahiel et al., 1997; Fischbach and Walsh, 2006; Koglin and Walsh, 2009). Beside these ‘core domains’ NRPS often contain ‘auxiliary domains,’ such as epimerization (E) or methylation (MT) domains, contributing to the structural diversity of natural peptide products (Walsh et al., 2001; Sieber and Marahiel, 2005). The genetic locus ces encoding the cereulide synthetase shows the typical architecture of NRPS gene clusters and comprises, beside the structural genes cesA and cesB, up- and downstream genes with known function in NRPS product assembly but also a putative hydrolase (cesH) of unknown function (Figure 1). The A domains of two out of the four modules in the structural genes cesA and cesB are interrupted by unusual insertions showing homologies to ketoreductases (KRs; Ehling-Schulz et al., 2005b, 2006a). Biochemical characterization of CesB1 revealed a novel logic for NRPS product assembly (Magarvey et al., 2006). These A domains recognize and select α-ketocarboxylic acids instead of α-hydroxy acids, which would be the expected precursors according to the toxin sequence. The newly described KR domain embedded in the A domains was shown to carry out chiral reduction on the toxin assembly line (Magarvey et al., 2006). A similar genetic architecture and modular organization with KR domains embedded in A domains was reported for NRPS catalyzing depsipeptides production in Streptomyces and cyanobacteria (Magarvey et al., 2006; Ding et al., 2011), thus suggesting that the chemosynthetic route of cereulide assembly is representative for other NRPS products with alternating ester and peptide bounds.
FIGURE 1.
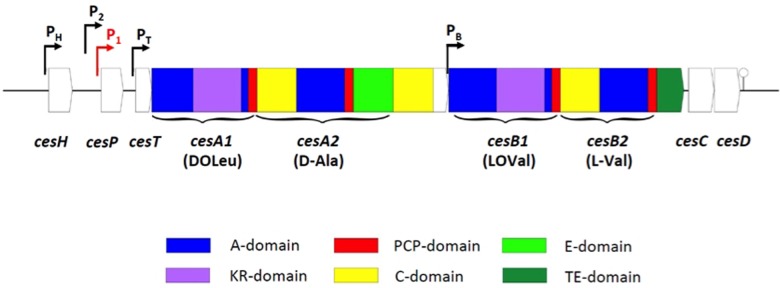
Genetic architecture of the cereulide synthetase gene cluster (according to Ehling-Schulz et al., 2006a; Dommel et al., 2010). Promoters are indicated by bent arrows. The main promoter driving the polycistronic transcription of the ces operon is highlighted in red. A hairpin indicates the terminator.
The order of modules in most NRPS systems is colinear with the product sequences, although it is increasingly becoming evident that the structural diversity of natural products does not always reinforce a strict collinearity (Wenzel and Müller, 2005; Lane and Moore, 2011). Indeed, a mass spectrometric screening and profiling approach of about 80 B. cereus strains demonstrated a high microheterogeneity of cereulide (Stark et al., 2013). In depth analysis of the emetic reference strain F4801/72 by UPLC-TOF MS and ion-trap MSn revealed at least 18 cereulide variants that showed significant differences in their ionophoric properties and toxicity (Marxen et al., 2015a). Commonly, chemical variants of non-ribosomal peptide toxins found in other organisms, such as cyanobacteria, are based on the molecular diversity found within the structural genes of different strains or different species (Dittmann et al., 2013). In contrast, cereulide and its 18 variants are produced by a single NRPS synthetase (Ehling-Schulz et al., 2005b; 2006a) simultaneously in the same strain, interrogating the exact mode of toxin production. Just recently, a novel mechanism for the biosynthesis of cereulide was proposed, based on dipeptide rather than on single monomers as the basic modules in tetradepsipeptide assembly (Marxen et al., 2015b). The emetic strains analyzed so far produce all the same set of isocereulides, although at different concentrations. The contribution of the different cereulide variants to food intoxications caused by emetic B. cereus is hitherto unknown and further studies are needed to decipher the health risk posed by the different isocereulides. Such studies would also provide valuable information for framing and improving knowledge based risk assessments of B. cereus toxins in food production and processing.
pCER270: A pXO1 like Megaplasmid Encoding the Cereulide NRPS
The ces cluster of emetic B. cereus is located on a 270 kb megaplasmid, named pCER270. The pCER270 shares its backbone with the toxin plasmid pXO1 of B. anthracis and the ces genes are inserted in a highly conserved part of these virulence plasmids (Ehling-Schulz et al., 2006a; 2011; Rasko et al., 2007). Although considerable progress has been made in recent years, the regulation of ces gene expression and cereulide production is far from being understood. It is increasingly becoming evident that cereulide formation is tightly linked to the metabolism of the producer strain by complex regulatory interactions between the toxin plasmid and the chromosome. The pCER270 plasmid carries about 10 regulators, including an ortholog of abrB, which might be involved in this interaction network and thus in various physiological processes. A key player in the embedment of cereulide toxin synthesis in the life cycle of emetic B. cereus is CodY, which is sensing the nutrient status of the cell (Frenzel et al., 2012; see next section for details).
Megaplasmids and Pathotype Emergence
Likewise to B. anthracis, emetic B. cereus strains belong to highly clonal lineages in the rather panmictic B. cereus population. In contrast, the chromosomally encoded enterotoxins are distributed over the entire the B. cereus group (Prüss et al., 1999; Ehling-Schulz et al., 2005a; Guinebretiere et al., 2008; Didelot et al., 2009). Thus it is tempting to speculate that the toxin megaplasmids in the B. cereus group play a pivotal role in evolution of specific pathogenic traits. Very recently it has been shown that B. cereus group plasmids are vectors for duplicated non-essential chromosomal genes, which suggests that these plasmids may act as an evolutionary sink tank for emergence of novel eco- and pathotypes (Zheng et al., 2015). For instance, in the pCER270 the pXO1-PAI, bearing the anthrax toxin genes, is replaced by a 77 kb insertion that contains sporulation and germination genes, which may allow emetic strains to rapidly respond to changing environmental conditions such as accounted in foods (Ehling-Schulz et al., 2011). In addition to the Ces NRPS encoding mega plasmid, emetic strains can carry up to nine additional plasmids of variable size with yet unknown functions (Hoton et al., 2005; Ehling-Schulz et al., 2011). There are also some indications for the genetic transferability of the ces genes. In some rare cases the ces gene locus has been found in Bacillus weihenstephanensis (Thorsen et al., 2006; Hoton et al., 2009), either located chromosomally or on a non-pXO1 like plasmid (Mei et al., 2014). In all emetic strains analyzed so far, mobile genetic elements (MGEs) are found in the proximities to the ces locus (Ehling-Schulz et al., 2011; Mei et al., 2014), which requires further attention. The contribution of these MGEs to the dynamics of progression of emetic B. cereus and their putative function in the emergence of novel eco- and pathotypes is hitherto unknown.
Regulatory Circuits in Emetic B. cereus: Who are the Maestros that Orchestrate Metabolism and Virulence Development?
Tight Temporal Control of ces Expression
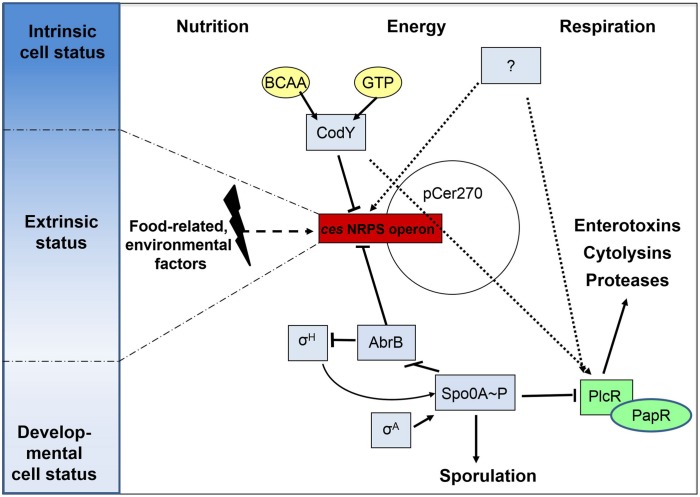
Ces-NRPS gene transcription is initiated during late logarithmic growth, and rapidly switched off after the transition to the stationary phase (Dommel et al., 2011). This strict temporal regulation relies on the tight transcriptional control accomplished by transcriptional regulators that ensure the correct timing and integration of NRPS synthesis in the bacteria’s life cycle (Figure 2). Notable, the transcriptional regulators involved in ces synthetase regulation identified hitherto are all encoded chromosomally, while the ces gene cluster is located on a pXO1-like plasmid, pointing toward an intensive chromosomal – plasmid crosstalk and a tight link between virulence and metabolism. While several promoters were identified within the ces gene cluster, cesP1 was shown to be the σA-dependent main promoter, driving the polycistronic transcription of the cesPTABCD operon (Dommel et al., 2010; Figure 1). This promoter is not subjected to cell density responsive regulation by the PlcR-PapR QS system, but is directly repressed by the chromosomally encoded transition state regulator AbrB (Lücking et al., 2009).
FIGURE 2.
Cereulide toxin synthesis is the result of a complex and multi-layered process. Transcriptional regulation of the ces non-ribosomal peptide synthetase (NRPS) operon is tightly controlled by pleiotropic or metabolite-responsive transcriptional regulators such AbrB, Spo0A, and CodY. These ensure a correct timing of toxin synthesis within the cell cycle by integrating sporulation-inducing trigger factors and sensing the intrinsic nutritional and energy status of the cells by monitoring GTP and branched-chain amino acid (BCAA) levels. Although this ties cereulide formation to ceasing metabolic fitness levels, the unidirectional commitment to the developmental process of sporulation is induced later on by sigma factor H-dependent, self-reinforced levels of Spo0A-P, which are not affecting ces cluster transcription. Since cereulide formation significantly varies dependent on extrinsic stimuli, such as encountered in different food matrices, it is anticipated that additional transcriptional regulators could act on ces gene cluster expression. One prominent factor is oxygen availability, which might alike to CodY form a linker and an oppositely acting strategy for impinging the regulatory networks of virulence determination via PlcR/PapR and for that of cereulide formation. Solid arrows indicate a direct regulation while dashed arrows indicate an indirect regulatory effect.
Although cereulide synthesis is independent of later sporulation processes, it requires the early, housekeeping sigma factor A (σA) promoted transcription of spo0A, the master regulator of sporulation in bacilli (Lücking et al., 2009). These initial, low levels of the phosphorylated form of Spo0A release AbrB repression by blocking abrB transcription (Strauch et al., 1990), which is reflected by the cereulide-deficient phenotype of a B. cereus spo0A null mutant (Lücking et al., 2009). The finding that complementation of the spo0A mutant with spo0A under the sole control of the sporulation-associated σH-dependent promoter results in very low cereulide amounts, underpins that ces gene transcription is associated with an early and transient developmental state, when the level of phosphorylated Spo0A is too low to pinch the unidirectional sporulation process. The phosphorylation state of Spo0A is determined by the phosphate flux through a complex phosphorelay network built of several kinases, phosphotransferases, phosphatases, and kinase inhibitors, of which Spo0A is the final receiver protein (Hoch, 1993). In B. subtilis, this signal transduction pathway is fueled by autophosphorylating transmembrane histidine kinases (KinA–KinE), which respond to diverse environmental stresses and metabolic signals (LeDeaux et al., 1995). The identity of specific activating ligands is still elusive and controversially discussed (Tan and Ramamurthi, 2014; Devi et al., 2015). Additionally, the function of KinA–KinE related kinases has not been established in B. cereus yet. However, we could show that an external addition of long-chain polyphosphates (polyPs), which are widely used as additives in the food industry, negatively influences ces gene transcription and cereulide synthesis (see section below). This suggests that imbalances in the intracellular polyP/phosphate household could interfere with the phosphoryl flux through the relay, which results in altered Spo0A activity (Frenzel et al., 2011).
CodY, the Major Conductor in Emetic B. cereus Pathogenesis
As early as with the first description of cereulide, it was apparent that the amino acid composition of the growth medium, in particular the amount of branched-chain amino acids (BCAAs), influences cereulide synthesis levels (Agata et al., 1999). This effect could be explained with the identification of CodY as one of the main regulators directly repressing ces gene transcription, while concomitantly exerting an indirect inducing effect on PlcR-dependent virulence factors (Frenzel et al., 2012). CodY is a pleiotropic transcriptional regulator that controls key metabolic intersections of the carbon and nitrogen metabolism and mediates stationary-phase adaptations in several low G+C Gram-positive bacteria (Sonenshein, 2007). The DNA-binding affinity of CodY is mediated by two types of ligand molecules, GTP and BCAA (Ratnayake-Lecamwasam et al., 2001; Shivers and Sonenshein, 2004). In vivo experiments with B. cereus AH187 showed that enhancing the endogenous CodY effector pool levels by external BCAA surplus feeding significantly reduced cereulide production (Frenzel et al., 2012). In this sense, CodY functions in B. cereus as an adaptor that mediates the regulatory crosstalk between chromosome and megaplasmid, and furthermore links the cell’s nutritional and energy status to pathogenicity. Cereulide synthesis is hence directly coupled to nutrient availability and ces gene expression takes place when levels of the CodY co-repressors pools decline, but the cell’s starvation is not so severe, that the sporulation pathway is already initiated (Figure 2). In this phase of the cell cycle, which is characterized by transient de-repression of CodY regulon members (Belitsky and Sonenshein, 2011; Brinsmade et al., 2014), the cell has to ensure an effective resource allocation and amino acid flow to achieve cellular maintenance, sustain growth, and to allow the energy-intensive formation of secondary molecules such as cereulide. Very logically, relieve of CodY-mediated repression concurrently leads to an enhanced synthesis of immune inhibitor (InhA) metalloproteases to maximize the usage of external nutrients (Frenzel et al., 2012). This indicates that a delicate intrinsic and extrinsic balance of amino acid levels is necessary to allow cereulide formation. The pool of amino acids must be sufficiently low to relieve ces promoter repression, while at the same time levels must be high enough to serve as direct precursors for the cereulide peptide. This is in excellent agreement with the observation at the food–bacteria interface: nutrient rich circumstances, especially proteinaceous food matrices, are less supportive for cereulide biosynthesis as are farinaceous foods (Agata et al., 2002; Dommel et al., 2010).
In opposite to its repressive effect on ces genes transcription, CodY promotes the expression of genes belonging to the PlcR virulence regulon in emetic strains (Frenzel et al., 2012; Figure 2). PlcR, the regulator of the major quorum sensing system in B. cereus, (Slamti and Lereclus, 2002), activates the transcription of a high number of secreted virulence genes encoding for enterotoxins, hemolysins, cytotoxins, proteases, or phospholipases (Gohar et al., 2008). Deletion of codY resulted in a phospholipase- and β-hemolytically inactive phenotype, which was linked to significantly lowered transcription of these genes (Frenzel et al., 2012). Since this phenotype was corroborated in studies with non-emetic strains (Lindbäck et al., 2012), CodY serves as a good example, how well-conserved chromosomal transcriptional regulators are adapted for feeding novel genetic material, such as the pCER270 megaplasmid, into ancient, or previously established, regulatory networks, as part of genetic diversification strategies.
However, gel-shift studies indicated that CodY is not a direct transcriptional activator of PlcR regulon genes, which implies that either an unknown regulator or posttranscriptional regulatory events are likely to mediate its effect (Frenzel et al., 2012). This dual-functioning nature of CodY on the ces genes and on the PlcR regulon could equip emetic strains with a powerful switching tool: when the intracellular levels of BCAAs and GTP is high, reflecting an extracellular milieu rich in proteins and energy sources as is found in animal host, CodY might mediate switches to the transcription of PlcR-dependent genes coding for proteases, phospholipases, enterotoxins, or hemolysins, concomitantly reducing the costly production of cereulide by the NRPS megaenzymes. Vice versa, ceasing amino acid availability that signals a necessity to enter diverging cellular differentiation processes might down regulate the extensive secretome production and thus allows a switch to cereulide production as a defensive strategy against competitors.
In summary, the decision to produce cereulide and PlcR regulon associated virulence factors is substantially mediated by the action of global transcriptional regulators. In emetic B. cereus strains, both virulence circuits are interconnected by the action of CodY and Spo0A. These, on the one hand, sense and respond to the cell’s intrinsic energy and metabolic status, and on the other hand act as timing devices and switching points for cellular differentiation processes, such as sporulation (Figure 2). However, this regulatory framework shows a certain degree of plasticity, which is reflected by the impact of various intrinsic and extrinsic stimuli that modulate the level of toxin production.
Cereulide Synthesis is Shaped by a Complex Panoply of Environmental Cues
External Signals
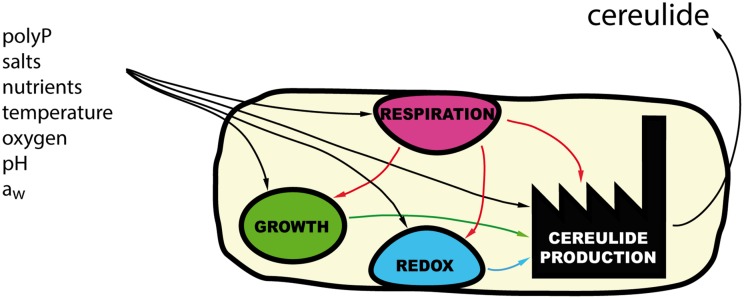
Cereulide production is linked to growth within certain, but not easily to define, limits. For instance, several studies provide evidence that the temperature range for growth of emetic B. cereus strains is broader than the temperature range in which cereulide production is actually detected (Finlay et al., 2000; Carlin et al., 2006; Apetroaie-Constantin et al., 2008). Besides temperature, the availability of oxygen plays a pivotal role in the formation of cereulide. Cereulide biosynthesis requires the presence of oxygen, though it is favored under low oxygen tensions (Finlay et al., 2000; Jääskeläinen et al., 2004). This indicates that channeling respiratory pathways and also the regulation of the redox state of the cell, which has been linked to enterotoxin synthesis (Duport et al., 2006), may also be important for cereulide formation. However, there have been contradicting findings concerning the impact of oxygen on cereulide synthesis at the food–bacteria interface (Agata et al., 2002; Rajkovic et al., 2006; Shaheen et al., 2006), which might be explained by the fact that a plethora of additional stimuli, as such are encountered in the complex nutrient environment, play another determinative role in toxin synthesis. A recent broad-scale, long-term study on cereulide production potentials in food matrices and on cereulide intoxication cases provided deeper insights on the prerequisites for cereulide production in foods (Messelhäusser et al., 2014). This study revealed that a complex interplay of fatty acids, C-sources, N-sources, micro- and macro-nutritional environments in combination with global factors such as pH and water availability determines the risk of food-borne intoxications, either by stimulating or inhibiting cereulide synthesis. For instance, it has been noticed that the ratio of external glycine, Na+ and K+ ions also has an impact on the level of cereulide production (Apetroaie-Constantin et al., 2008). According to our current knowledge on cereulide as being a K+ ionophore, it is tempting to speculate that the K+ homeostasis might be a driving force in cereulide production.
Integration of External Signals
Nonetheless, how these external signals are perceived by the cell and transduced to the ces promoter is unknown. Additionally, how those external cues modulate the cell’s physiology to induce combinatory or opposite stimulating or inhibitory effects on cereulide synthesis, is a very complex question that still awaits elucidation. The conditions permitting cereulide synthesis are far more stringent than the once allowing growth of B. cereus, indicating that specific trigger factors and/or combinations thereof are impacting cereulide production (Figure 3). For instance, increasing sodium chloride concentrations in liquid medium lowers cereulide production at the transcriptional and toxin level, although the growth rate of strains is not affected, rendering it unlikely that the mechanism can be simply related to salt stress (Dommel et al., 2011). A similar observation was made concerning the influence of widely used food additives, long chain polyPs, which are cation-sequestering straight-chain polymers of condensed orthophosphoric acid residues (Frenzel et al., 2011). While treated with non-lytic, non-bacteriostatic polyP concentrations, B. cereus growth rates in liquid culture, or final cell numbers in a model food were not significantly altered. However, polyPs interfered with early stages of the toxin formation process and significantly delayed and reduced ces promoter activity, ces gene transcription, and hence, quenched cereulide synthesis. How external polyP addition affects the intracellular physiology is unclear to date. B. cereus, like many other bacteria, itself endogenously synthesizes and hydrolyzes natural polyP analogs that fulfill manifold physiological functions, ranging from metabolic regulation to protein phosphorylation (Shi et al., 2004; Rao et al., 2009). Thus, external polyP addition obviously leads to imbalances in the internal polyP/Pi ratio, which might not only affect the sporulation process, but also catalytic processes of the NRPS modules.
FIGURE 3.
Environmental signals and cereulide production. An array of external signals, acting on various physiological processes such as respiration, redox potential regulation, or growth, are integrated by the bacterium and modulate cereulide production.
Though some substances, including bacteriocins, organic acids, and nisin, have been reported to inhibit the growth of B. cereus in the food environment (Valero and Salmeron, 2003; Galvez et al., 2007; Ter Beek and Brul, 2010), these studies did not focus on the targeted inhibition of cereulide or enterotoxin formation. Therefore, it is unclear to date, whether other food additives besides polyPs can directly affect virulence regulation networks in B. cereus. Nevertheless, the example of polyPs shows that understanding of how external toxin trigger factors are perceived and integrated in the physiological and metabolic response of B. cereus could pave the way for tailored hurdle concepts in food production and food processing chains.
A Home for Emetic Strains: Seeking for their Natural Habitat
Megaplasmids, Key Determinants of the Ecological Niche for B. cereus
Non-emetic and emetic B. cereus occupy a broad and, especially for emetic strains, ill-defined ecological niche. B. cereus is present in the soil, in the gut of insects and of mammals, on plant roots (Jensen et al., 2003; Ceuppens et al., 2013) or on mycorrhizae-associated fungi (Smith et al., 1999; Toljander et al., 2006; Lakshmanan et al., 2012). Although many B. cereus isolates have been sequenced, strain- or subgroup-specific genetic signatures displaying possible adaptation events to each of these environments could not be identified yet (Ehling-Schulz and Messelhäusser, 2014). In contrast, the ecological niches of B. thuringiensis and B. anthracis, two species genetically close to B. cereus, are far better studied and it is apparent that their lifestyle is defined and shaped by their plasmid-encoded genetic material. B. thuringiensis, which is characterized by expressing Cry toxins via large plasmids, is an entomopathogen, while B. anthracis, carrying the pXO1 and pXO2 mega plasmids with the three-partite Anthrax exotoxin, is a high-potential mammal pathogen. Alike to B. thuringiensis and B. anthracis, the emetic subgroup of B. cereus strains carries the cereulide NRPS cluster encoding megaplasmid pCER270. However, although cereulide seems to act as an effector of ecological competition against a range of Gram-positive bacteria and some fungi (Ladeuze et al., 2011; Tempelaars et al., 2011), it is still unclear, why cereulide producers are frequently associated with human intoxication cases stemming from cereulide production in food matrices.
Since human infections by emetic strains are rare, mammals are probably not the primary target and not the preferred host to thrive and feed on. Interestingly, the prevalence of emetic strains was found to be low in environmental samples, but high in food materials (Table 1). Whereas a high number of soil samples contain B. cereus or B. thuringiensis strains, very few of these strains were emetic, and no emetic strains were found amongst isolates recovered from invertebrates samples. Furthermore, emetic strains were detected in the intestine of small root-, seed-, and insect-consuming mammals, representing as much as 16% of B. cereus isolates, but not in the feces of herbivorous mammals (Table 1). By contrast, the proportion of emetic strains within B. cereus isolates ranged from 1.4 to 10.2% in food samples obtained from retail food shops (random food), and this proportion increased to 17–32% in samples involved in food poisoning cases (non-random food; see Table 1). The higher prevalence of emetic strains in intoxication cases-associated food, compared to randomly sampled food, likely reflects the increased awareness of cereulide intoxications by food authorities and/or diagnostic laboratories and is supported by an optimized screening toolbox for emetic strains via qPCR and cereulide toxin detection per se (Ehling-Schulz and Messelhäusser, 2013). However, the true incidence of emetic strains might still be underestimated since emetic strains are only weakly or non-hemolytic (Ehling-Schulz et al., 2005a; Pirhonen et al., 2005) and thus might be misdiagnosed as being not B. cereus. For instance, in some studies on B. cereus prevalence in food, non-hemolytic strains were discarded and emetic strains were found to be absent or extremely rare (Rosenquist et al., 2005; Ankolekar et al., 2009; Samapundo et al., 2011).
Table 1.
Prevalence of emetic Bacillus cereus strains in samples of various origin.
| Sample origin | n | Bc | Bce % | Reference |
|---|---|---|---|---|
| Soil | nd | 543 | 0.2 | Hoton et al. (2009) |
| Soil | 80 | 61 | 0.0 | Altayar and Sutherland (2006) |
| Soil | nd | 38a | 0.0 | Forghani et al. (2014) |
| Soil | 15 | 12 | 0.0 | Santos et al. (2011) |
| Invertebrates | nd | 58 | 0.0 | Hoton et al. (2009) |
| Shrew or vole intestine | nd | 109 | 16.5 | Hoton et al. (2009) |
| Cow or horse feces | 66 | nd | 0.0 | Altayar and Sutherland, 2006) |
| Food (random) | nd | 582 | 1.4 | Hoton et al. (2009) |
| Food (random) | 742 | 402 | 10.2 | Messelhäusser et al. (2014) |
| Food (non-random) | nd | 95 | 31.6 | Hoton et al. (2009) |
| Food (non-random) | 3654 | 187 | 17.1 | Messelhäusser et al. (2014) |
n, number of samples; Bc, number of B. cereus strains isolated from the samples; Bce %, proportion of emetic B. cereus strains within all B. cereus isolates. aB. cereus and B. thuringiensis strains.
Entrance of Emetic Strains to Food and Food Production Processes
A literature review on the association of the emetic strains with different food components reveals a rather heterogeneous distribution of these strains (Table 2). For instance, emetic strains seem to be rare in meat products, but are occasionally present in potato, rice, mushrooms, or dairy products with such a high prevalence that even out-competes accompanying bacteria. It might be speculated that certain food niches provide an advantageous environment for growth/and or antibiotic production, allowing a selective substrate occupancy by cereulide producers. Elevated temperatures commonly used during food processing might further select for emetic strains, which show an increased heat resistance with respect to vegetative growth and spore D10-values compared to non-emetic strains (Carlin et al., 2006). Thus, the entrance routes into the food chain as well as the modes of selective enrichment of emetic strains within foodstuff are becoming a hotspot of interest during recent years.
Table 2.
Prevalence of B. cereus emetic strains in food samples.
| Sample origin | n | Bc | Bce % | Reference |
|---|---|---|---|---|
| Broccoli (raw) | 25 | 8 | 0.0 | Flores-Urban et al. (2014) |
| Carrot (raw) | 25 | 21 | 0.0 | Flores-Urban et al. (2014) |
| Carrot (raw) | 25 | 22 | 0.0 | Altayar and Sutherland, 2006) |
| Potato (raw) | 25 | 21 | 16.0 | Altayar and Sutherland, 2006) |
| Potato (raw) | nd | 8 | 60.0 | Hoornstra et al. (2013) |
| Coriander (raw) | 25 | 11 | 0.0 | Flores-Urban et al. (2014) |
| Lettuce (raw) | 25 | 17 | 0.0 | Flores-Urban et al. (2014) |
| Lettuce (raw) | 48 | 44 | 0.0 | Messelhäusser et al. (2014) |
| Rice (boiled) | 54 | 41 | 17.1 | Delbrassinne et al. (2012) |
| Rice (raw) | nd | 103a | 26.2 | Forghani et al. (2014) |
| Rice (raw) | 78 | 44 | 11.3 | Messelhäusser et al. (2014) |
| Fruits | 27 | 15 | 0.0 | Messelhäusser et al. (2014) |
| Dried mushrooms | 135 | 110 | 10.0 | Messelhäusser et al. (2014) |
| Dairy products | 212 | 119 | 18.5 | Messelhäusser et al. (2014) |
| Dairy products | 809 | 508 | 4.7 | Messelhäusser et al. (2010) |
| Dairy products | nd | 161 | 10.0 | Wijnands et al. (2006) |
| Meat products | 121 | 0 | 0.0 | Messelhäusser et al. (2014) |
| Meat products | nd | 24 | 0.0 | Wijnands et al. (2006) |
n, number of samples; Bc, number of B. cereus strains isolated from the samples; Bce %, proportion of emetic B. cereus strains within all B. cereus group isolates. aB. cereus and B. thuringiensis strains.
Potential Ecological Niches of Emetic Strains
One environmental route for gaining entry into the food chain might be associated with B. cereus thriving in the intestine of small rodents. These feed on plant roots or on invertebrates, which in turn feed on plant roots and tubers. Upon harvest, the root microflora contaminates the crop. In foodborne outbreaks, the incriminated food often contains farinaceous foods, such as rice (Gilbert and Parry, 1977) or pasta (Mahler et al., 1997; Dierick et al., 2005; Naranjo et al., 2011). Emetic strains appear to be preferentially present in specific plants, and their occurrence in dairy products might be a consequence of milk contamination. A survey of B. cereus strains isolated from rice paddy fields showed that 44% of these isolates were capable of producing the emetic toxin (Ueda and Kuwabara, 1993), and emetic strains were found in the extracellular space of potato tubers, behaving as endophytic bacteria (Hoornstra et al., 2013). Emetic strains might be innocuous to potato tubers, because in contrast to most B. cereus strains, they cannot degrade starch and differ in their biofilm production capacities (Ehling-Schulz et al., 2005a; Auger et al., 2009). An endophytic lifestyle of emetic strains in starch-rich plants would explain, why these strains are often found in starch-rich foods (e.g., pasta, rice), despite their inability to hydrolyze starch.
It was postulated that cereulide might provide an adaptive advantage to emetic strains by scavenging potassium within the endophytic and soil-associated environments (Teplova et al., 2006; Ekman et al., 2012) where potassium is rare (Hoornstra et al., 2013). Cereulide might also protect the tuber against attacks by pathogenic fungi by its antifungal activity (Ladeuze et al., 2011), possibly indicating a symbiotic or synergistic effect of emetic strains and plants. Therefore, it seems more reasonable that enrichment of cereulide in certain foods (Dommel et al., 2010; Messelhäusser et al., 2014) might be the consequence of dysregulated cereulide production in non-natural environments, such as encountered in some food matrices, rather than posing an ecological or evolutionary meaning to B. cereus in first instance.
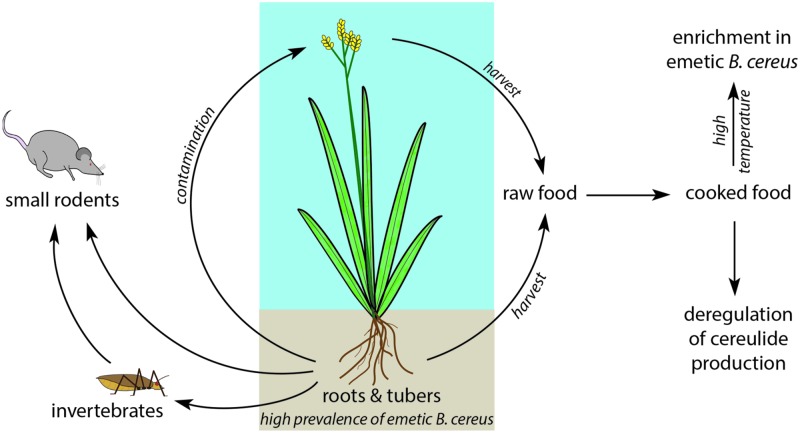
Summarizing the current knowledge on emetic strains, it seems reasonable that they differ concerning their preferred ecological niches compared to their non-emetic siblings. The primary natural habitat of emetic strains might therefore be linked to roots, tubers and mycorrhizae of some plants (Figure 4). Thus, the extremely potent human toxin cereulide might be a key element for their adaptation to the natural (soil) environments and might be a key requisite to thrive in specified niches.
FIGURE 4.
Hypothetical ecological niche of Bacillus cereus emetic strains. Emetic strains of B. cereus are likely to colonize roots and tubers of plants, either as endophytic symbionts or as biofilms. Aerial parts of the plants might then be contaminated by these strains, upon harvest or by small animals feeding on the underground plants parts. The aerial parts of the plants can also, when used as fodder, contaminate cow udder and therefore milk. Since emetic strains spores are more thermoresistant than other B. cereus strains spore, cooked food will then be enriched in emetic strains which, in this unnatural environment, could overproduce the emetic toxin.
Future Perspectives
An in-depth understanding of the molecular mechanisms involved in the specialization of emetic strains for food product colonization is a prerequisite to develop novel prevention and effective counteraction strategies. Toward this end, deciphering the ecological function of the cereulide toxin would be an important step and could also guide efforts to uncover yet unknown environmental sources of emetic strains. Recent reports point toward plants as a potential source for emetic strains, although the nature of this association and its potential contribution to food contamination is hitherto unknown.
In addition, further research is necessary to elucidate the function of the intensive plasmid–chromosomal cross talk for the microevolution and emergence of novel pathotypes in B. cereus sensu lato. The regulatory plasmid – chromosomal interactions are apparently much more pronounced within the B. cereus group than in B. subtilis, underpinning that a simple comparison and/or deduction of the function and regulons of transcriptional regulators is not possible between B. cereus and B. subtilis strains. Results from recent genome studies indicate that the inter-plasmid–chromosomal gene duplications play a pivotal role in intraspecies diversification of B. cereus and it is tempting to speculate that it also plays an important role in the emergence of specific pathogenic traits.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tanja Buchacher for proofreading the manuscript. This work was supported by the German Ministry of Economics and Technology (via AiF) and the FEI (Forschungskreis der Ernährungsindustrie e.V., Bonn); project AiF 16845N.
References
- Agata N., Ohta M., Mori M., Shibayama K. (1999). Growth conditions of and emetic toxin production by Bacillus cereus in a defined medium with amino acids. Microbiol. Immunol. 43 15–18. 10.1111/j.1348-0421.1999.tb02367.x [DOI] [PubMed] [Google Scholar]
- Agata N., Ohta M., Yokoyama K. (2002). Production of Bacillus cereus emetic toxin (cereulide) in various foods. Int. J. Food Microbiol. 73 23–27. 10.1016/S0168-1605(01)00692-4 [DOI] [PubMed] [Google Scholar]
- Altayar M., Sutherland A. D. (2006). Bacillus cereus is common in the environment but emetic toxin producing isolates are rare. J. Appl. Microbiol. 100 7–14. 10.1111/j.1365-2672.2005.02764.x [DOI] [PubMed] [Google Scholar]
- Ankolekar C., Rahmati T., Labbe R. G. (2009). Detection of toxigenic Bacillus cereus and Bacillus thuringiensis spores in U.S. rice. Int. J. Food Microbiol. 128 460–466. 10.1016/j.ijfoodmicro.2008.10.006 [DOI] [PubMed] [Google Scholar]
- Anonymous. (2009). The community summary report on food-borne outbreaks in the European union in 2007. EFSA J. 271 54–65. [Google Scholar]
- Anonymous. (2015). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. EFSA J. 13:3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apetroaie-Constantin C., Shaheen R., Andrup L., Smidt L., Rita H., Salkinoja-Salonen M. (2008). Environment driven cereulide production by emetic strains of Bacillus cereus. Int. J. Food Microbiol. 127 60–67. 10.1016/j.ijfoodmicro.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Auger S., Ramarao N., Faille C., Fouet A., Aymerich S., Gohar M. (2009). Biofilm formation and cell surface properties among pathogenic and nonpathogenic strains of the Bacillus cereus group. Appl. Environ. Microbiol. 75 6616–6618. 10.1128/AEM.00155-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer T., Stark T., Hofmann T., Ehling-Schulz M. (2010). Development of a stable isotope dilution analysis for the quantification of the Bacillus cereus toxin cereulide in foods. J. Agric. Food Chem. 58 1420–1428. 10.1021/jf9033046 [DOI] [PubMed] [Google Scholar]
- Belitsky B. R., Sonenshein A. L. (2011). Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J. Bacteriol. 193 473–484. 10.1128/JB.01151-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottone E. J. (2010). Bacillus cereus, a volatile human pathogen. Clin. Microbiol. Rev. 23 382–438. 10.1128/CMR.00073-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinsmade S. R., Alexander E. L., Livny J., Stettner A. I., Segrè D., Rhee K. Y., et al. (2014). Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc. Natl. Acad. Sci. U.S.A. 111 8227–8232. 10.1073/pnas.1321308111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin F., Fricker M., Pielaat A., Heisterkamp S., Shaheen R., Salkinoja-Salonen M., et al. (2006). Emetic toxin-producing strains of Bacillus cereus show distinct characteristics within the Bacillus cereus group Int. J. Food Microbiol. 109 132–138. 10.1016/j.ijfoodmicro.2006.01.022 [DOI] [PubMed] [Google Scholar]
- Ceuppens S., Boon N., Uyttendaele M. (2013). Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol. Ecol. 84 433–450. 10.1111/1574-6941.12110 [DOI] [PubMed] [Google Scholar]
- Delbrassinne L., Andjelkovic M., Dierick K., Denayer S., Mahillon J., Van Loco J. (2012). Prevalence and levels of Bacillus cereus emetic toxin in rice dishes randomly collected from restaurants and comparison with the levels measured in a recent foodborne outbreak. Foodborne Pathog. Dis. 9 809–814. 10.1089/fpd.2012.1168 [DOI] [PubMed] [Google Scholar]
- Devi S. N., Vishnoi M., Kiehler B., Haggett L., Fujita M. (2015). In vivo functional characterization of the transmembrane histidine kinase KinC in Bacillus subtilis. Microbiology 161 (pt 5) 1092–1104. 10.1099/mic.0.000054. [DOI] [PubMed] [Google Scholar]
- Didelot X., Barker M., Falush D., Priest F. G. (2009). Evolution of pathogenicity in the Bacillus cereus group. Syst. Appl. Microbiol. 32 81–90. 10.1016/j.syapm.2009.01.001 [DOI] [PubMed] [Google Scholar]
- Dierick K., Van Coillie E., Swiecicka I., Meyfroidt G., Devlieger H., Meulemans A., et al. (2005). Fatal family outbreak of Bacillus cereus-associated food poisoning. J. Clin. Microbiol. 43 4277–4279. 10.1128/JCM.43.8.4277-4279.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Rath C. M., Bolduc K. L., Håkansson K., Sherman D. H. (2011). Chemoenzymatic synthesis of cryptophycin anticancer agents by an ester bond-forming non-ribosomal peptide synthetase module. J. Am. Chem. Soc. 133 14492–14495. 10.1021/ja204716f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann E., Fewer D. P., Neilan B. A. (2013). Cyanobacterial toxins: biosynthetic routes and evolutionary roots. FEMS Microbiol. Rev. 37 23–43. 10.1111/j.1574-6976.2012.12000.x [DOI] [PubMed] [Google Scholar]
- Dommel M. K., Frenzel E., Strasser B., Blöchinger C., Scherer S., Ehling-Schulz M. (2010). Identification of the main promoter directing cereulide biosynthesis in emetic Bacillus cereus and its application for real-time monitoring of ces gene expression in foods Appl. Environ. Microbiol. 76 1232–1240. 10.1128/AEM.02317-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dommel M., Lücking G., Scherer S., Ehling-Schulz M. (2011). Transcriptional kinetic analyses of cereulide synthetase genes with respect to growth, sporulation and emetic toxin production in Bacillus cereus. Food Microbiol. 28 284–290. 10.1016/j.fm.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Duport C., Zigha A., Rosenfeld E., Schmitt P. (2006). Control of enterotoxin gene expression in Bacillus cereus F4430/73 involves the redox-sensitive ResDE signal transduction system. J. Bacteriol. 188 6640–6651. 10.1128/JB.00702-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling-Schulz M., Fricker M., Grallert H., Rieck P., Wagner M., Scherer S. (2006a). Cereulide synthetase gene cluster from emetic Bacillus cereus: structure and location on a mega virulence plasmid related to Bacillus anthracis toxin plasmid pXO1. BMC Microbiol. 6:20 10.1186/1471-2180-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling-Schulz M., Guinebretiere M. H., Monthan A., Berge O., Fricker M., Svensson B. (2006b). Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol. Lett. 260 232–240. 10.1111/j.1574-6968.2006.00320.x [DOI] [PubMed] [Google Scholar]
- Ehling-Schulz M., Fricker M., Scherer S. (2004). Bacillus cereus, the causative agent of an emetic type of food borne illness. Mol. Nutr. Food Res. 48 479–487. 10.1002/mnfr.200400055 [DOI] [PubMed] [Google Scholar]
- Ehling-Schulz M., Knutsson R., Scherer S. (2011). “Bacillus cereus,” in Genomes of Food- and Water-Borne Pathogens eds Kathariou S., Fratamico P., Liu Y. (Washington DC: ASM Press; ) 147–164. 10.1128/9781555816902.ch11 [DOI] [Google Scholar]
- Ehling-Schulz M., Messelhäusser U. (2012). “One pathogen but two different types of food borne outbreaks; Bacillus cereus in catering facilities in Germany,” in Case Studies in Food Safety and Quality Management: Lessons From Real-life Situations ed. Hoorfar J. (Cambridge: Woodhead Publishing; ) 63–70. [Google Scholar]
- Ehling-Schulz M., Messelhäusser U. (2013). Bacillus ‘next generation’ diagnostics: moving from detection towards sub-typing and risk related strain profiling. Front. Microbiol. 4:32 10.3389/fmicb.2013.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehling-Schulz M., Messelhäusser U. (2014). “The genus Bacillus,” in DNA Methods in Food Safety: Molecular Typing of Foodborne and Waterborne Bacterial Pathogens eds Oyarzabal O., Kathariou S. (West Sussex: Blackwell Publisher; ) 165–184. [Google Scholar]
- Ehling-Schulz M., Svensson B., Guinebretiere M.-H., Lindbäck T., Andersson M., Schulz A., et al. (2005a). Emetic toxin formation of Bacillus cereus is restricted to a single evolutionary lineage of closely related strains. Microbiology 151 183–197. 10.1099/mic.0.27607-0 [DOI] [PubMed] [Google Scholar]
- Ehling-Schulz M., Vukov N., Schulz A., Shaheen R., Andersson M., Märtlbauer E., et al. (2005b). Identification and partial characterization of the nonribosomal peptide synthetase gene responsible for cereulide production in emetic Bacillus cereus. Appl. Environ. Microbiol. 71 105–113. 10.1128/AEM.71.1.105-113.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekman J. V., Kruglov A., Andersson M. A., Mikkola R., Raulio M., Salkinoja-Salonen M. (2012). Cereulide produced by Bacillus cereus increases the fitness of the producer organism in low-potassium environments. Microbiology 158 1106–1116. 10.1099/mic.0.053520-0 [DOI] [PubMed] [Google Scholar]
- Finlay W. J., Logan N. A., Sutherland A. D. (2000). Bacillus cereus produces most emetic toxin at lower temperatures. Lett. Appl. Microbiol. 31 385–389. 10.1046/j.1472-765x.2000.00835.x [DOI] [PubMed] [Google Scholar]
- Fischbach M. A., Walsh C. T. (2006). Assembly-line enzymology for polyketide and nonribosomal Peptide antibiotics: logic, machinery, and mechanisms. Chem. Rev. 106 3468–3496. 10.1021/cr0503097 [DOI] [PubMed] [Google Scholar]
- Flores-Urban K. A., Natividad-Bonifacio I., Vazquez-Quinones C. R., Vazquez-Salinas C., Quinones-Ramirez E. I. (2014). Detection of toxigenic Bacillus cereus strains isolated from vegetables in Mexico City. J. Food Prot. 77 2144–2147. 10.4315/0362-028X.JFP-13-479 [DOI] [PubMed] [Google Scholar]
- Forghani F., Kim J. B., Oh D. H. (2014). Enterotoxigenic profiling of emetic toxin- and enterotoxin-producing Bacillus cereus, isolated from food, environmental, and clinical samples by multiplex PCR. J. Food Sci. 79 228–2293. 10.1111/1750-3841.12666 [DOI] [PubMed] [Google Scholar]
- Frenzel E., Doll V., Pauthner M., Lücking G., Scherer S., Ehling-Schulz M. (2012). CodY orchestrates the expression of virulence determinants in emetic Bacillus cereus by impacting key regulatory circuits. Mol. Microbiol. 85 67–88. 10.1111/j.1365-2958.2012.08090.x [DOI] [PubMed] [Google Scholar]
- Frenzel E., Letzel T., Scherer S., Ehling-Schulz M. (2011). Inhibition of cereulide toxin synthesis by emetic Bacillus cereus via long-chain polyphosphates. Appl. Environ. Microbiol. 77 1475–1482. 10.1128/AEM.02259-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker M., Messelhäusser U., Busch U., Scherer S., Ehling-Schulz M. (2007). Diagnostic real-time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent foodborne outbreaks. Appl. Environ. Microbiol. 73 1892–1898. 10.1128/AEM.02219-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez A., Abriouel H., Lopez R. L., Ben Omar N. (2007). Bacteriocin based strategies for food biopreservation. Int. J. Food Microbiol. 120 51–70. 10.1016/j.ijfoodmicro.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Gilbert R. J., Parry J. M. (1977). Serotypes of Bacillus cereus from outbreaks of food poisoning and from routine foods. J. Hyg. (Lond.) 78 69–74. 10.1017/S0022172400055947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohar M., Faegri K., Perchat S., Ravnum S., Okstad O. A., Gominet M., et al. (2008). The PlcR virulence regulon of Bacillus cereus. PLoS ONE 3:e2793 10.1371/journal.pone.0002793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinebretiere M. H., Thompson F. L., Sorokin A., Normand P., Dawyndt P., Ehling-Schulz M., et al. (2008). Ecological diversification in the Bacillus cereus group. Environ. Microbiol. 10 851–865. 10.1111/j.1462-2920.2007.01495.x [DOI] [PubMed] [Google Scholar]
- Hoch J. A. (1993). Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu. Rev. Microbiol. 47 441–465. 10.1146/annurev.mi.47.100193.002301 [DOI] [PubMed] [Google Scholar]
- Hoornstra D., Andersson M. A., Teplova V. V., Mikkola R., Uotila L. M., Andersson L. C., et al. (2013). Potato crop as a source of emetic Bacillus cereus and cereulide-induced mammalian cell toxicity. Appl. Environ. Microbiol. 79 3534–3543. 10.1128/AEM.00201-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoton F. M., Andrup L., Swiecicka I., Mahillon J. (2005). The cereulide genetic determinants of emetic Bacillus cereus are plasmid-borne. Microbiology 151 2121–2124. 10.1099/mic.0.28069-0 [DOI] [PubMed] [Google Scholar]
- Hoton F. M., Fornelos N., N’guessan E., Hu X., Swiecicka I., Dierick K., et al. (2009). Family portrait of Bacillus cereus and Bacillus weihenstephanensis cereulide-producing strains. Environ. Microbiol. Rep. 1 177–183. 10.1111/j.1758-2229.2009.00028.x [DOI] [PubMed] [Google Scholar]
- Ichikawa K., Gakumazawa M., Inaba A., Shiga K., Takeshita S., Mori M., et al. (2010). Acute encephalopathy of Bacillus cereus mimicking Reye syndrome. Brain Dev. 32 688–690. 10.1016/j.braindev.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Jääskeläinen E. L., Häggblom M. M., Andersson M. A., Salkinoja-Salonen M. S. (2004). Atmospheric oxygen and other conditions affecting the production of cereulide by Bacillus cereus in food. Int. J. Food Microbiol. 96 75–83. 10.1016/j.ijfoodmicro.2004.03.011 [DOI] [PubMed] [Google Scholar]
- Jensen G. B., Hansen B. M., Eilenberg J., Mahillon J. (2003). The hidden lifestyles of Bacillus cereus and relatives. Environ. Microbiol. 5 631–640. 10.1046/j.1462-2920.2003.00461.x [DOI] [PubMed] [Google Scholar]
- Koglin A., Walsh C. T. (2009). Structural insights into nonribosomal peptide enzymatic assembly lines. Nat. Prod. Rep. 26 987–1000. 10.1039/b904543k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladeuze S., Lentz N., Delbrassinne L., Hu X., Mahillon J. (2011). Antifungal activity displayed by cereulide, the emetic toxin produced by Bacillus cereus. Appl. Environ. Microbiol. 77 2555–2558. 10.1128/AEM.02519-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan V., Kitto S. L., Caplan J. L., Hsueh Y. H., Kearns D. B., Wu Y. S., et al. (2012). Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol. 160 1642–1661. 10.1104/pp.112.200386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane A. L., Moore B. S. (2011). A sea of biosynthesis: marine and natural products meet the molecular age. Nat. Prod. Rep. 28 411–428. 10.1039/C0NP90032J [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDeaux J. R., Yu N., Grossmann A. D. (1995). Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 177 861–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindbäck T., Mols M., Basset C., Granum P. E., Kuipers O. P., Kovács Á. T. (2012). CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol. 14 2233–2246. 10.1111/j.1462-2920.2012.02766.x [DOI] [PubMed] [Google Scholar]
- Lücking G., Dommel M. K., Scherer S., Fouet A., Ehling-Schulz M. (2009). Cereulide synthesis in emetic Bacillus cereus is controlled by the transition state regulator AbrB, but not by the virulence regulator PlcR. Microbiology 155 922–931. 10.1099/mic.0.024125-0 [DOI] [PubMed] [Google Scholar]
- Lund T., De Buyser M. L., Granum P. E. (2000). A new cytotoxin from Bacillus cereus that may cause necrotic enteritis. Mol. Microbiol. 8 254–261. 10.1046/j.1365-2958.2000.02147.x [DOI] [PubMed] [Google Scholar]
- Magarvey N. A., Ehling-Schulz M., Walsh C. T. (2006). Characterization of the cereulide NRPS hydroxy acid specifying modules: activation of keto acids and chiral reduction on the assembly line J. Am. Chem. Soc. 128 10698–10699. 10.1021/ja0640187 [DOI] [PubMed] [Google Scholar]
- Mahler H., Pasi A., Kramer J. M., Schulte P., Scoging A. C., Bar W., et al. (1997). Fulminant liver failure in association with the emetic toxin of Bacillus cereus. N. Engl. J. Med. 336 1142–1148. 10.1056/NEJM199704173361604 [DOI] [PubMed] [Google Scholar]
- Marahiel M. A., Stachelahus T., Mootz H. D. (1997). Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97 2651–2673. 10.1021/cr960029e [DOI] [PubMed] [Google Scholar]
- Marxen S., Stark T. D., Frenzel E., Rütschle A., Lücking G., Pürstinger G., Pohl E. E., et al. (2015a). Chemodiversity of cereulide, the emetic toxin of Bacillus cereus. Anal. Bioanal. Chem. 407 2439–2453. 10.1007/s00216-015-8511-y [DOI] [PubMed] [Google Scholar]
- Marxen S., Stark T. D., Rütschle A., Lücking G., Frenzel E., Scherer S., et al. (2015b). Depsipeptide intermediates interrogate proposed biosynthesis of cereulide, the emetic toxin of Bacillus cereus. Sci. Rep. 5:10637 10.1038/srep10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei X., Xu K., Yang L., Yuan Z., Mahillon J., Hu X. (2014). The genetic diversity of cereulide biosynthesis gene cluster indicates a composite transposon Tnces in emetic Bacillus weihenstephanensis. BMC Microbiol. 14:149 10.1186/1471-2180-14-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messelhäusser U., Frenzel E., Blöchinger C., Zucker R., Kämpf P., Ehling-Schulz M. (2014). Emetic Bacillus cereus are more volatile than thought: recent foodborne outbreaks and prevalence studies in Bavaria (2007-2013). Biomed. Res. Int. 2014:465603 10.1155/2014/465603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messelhäusser U., Kämpf P., Fricker M., Ehling-Schulz M., Zucker R., Wagner B., et al. (2010). Prevalence of emetic Bacillus cereus in different ice creams in Bavaria. J. Food Prot. 73 395–399. [DOI] [PubMed] [Google Scholar]
- Naranjo M., Denayer S., Botteldoorn N., Delbrassinne L., Veys J., Waegenaere J., et al. (2011). Sudden death of a young adult associated with Bacillus cereus food poisoning. J. Clin. Microbiol. 249 4379–4381. 10.1128/JCM.05129-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirhonen T. I., Andersson M. A., Jaaskelainen E., Salkinoja-Salonen M., Honkanen-Buzalski T., Johansson T. M. L. (2005). Biochemical and toxic diversity of Bacillus cereus in a pasta and meat dish associated with a food-poisoning case. Food Microbiol. 22 87–91. 10.1016/j.fm.2004.04.002 [DOI] [Google Scholar]
- Posfay-Barbe K. M., Schrenzel J., Frey J., Studer R., Korff C., Belli D. C., et al. (2008). Food poisoning as a cause of acute liver failure. Pediatr. Infect. Dis. J. 27 846–847. 10.1097/INF.0b013e318170f2ae [DOI] [PubMed] [Google Scholar]
- Prüss B. M., Dietrich R., Nibler B., Märtlbauer E., Scherer S. (1999). The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl. Environ. Microbiol. 65 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkovic A., Uyttendaele M., Ombregt S.-A., Jääskelainen E., Salkinoja-Salonen M., Debevere J. (2006). Influence of type of food on the kinetics and overall production of Bacillus cereus emetic toxin. J. Food Prot. 69 847–852. [DOI] [PubMed] [Google Scholar]
- Rao N. N., Gomez-Garcia M. R., Kornberg A. (2009). Inorganic polyphosphate: essential for growth and survival. Annu. Rev. Biochem. 78 605–647. 10.1146/annurev.biochem.77.083007.093039 [DOI] [PubMed] [Google Scholar]
- Rasko D. A., Rosovitz M. J., Økstad O. A., Fouts D. E., Jiang L., Cer R. Z., et al. (2007). Complete sequence analysis of novel plasmids from emetic and periodontal Bacillus cereus isolates reveals a common evolutionary history among the B. cereus-group plasmids, including Bacillus anthracis pXO1. J. Bacteriol. 189 52–64. 10.1128/JB.01313-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayake-Lecamwasam M., Serror P., Wong K. W., Sonenshein A. L. (2001). Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15 1093–1103. 10.1101/gad.874201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist H., Smidt L., Andersen S. R., Jensen G. B., Wilcks A. (2005). Occurrence and significance of Bacillus cereus and Bacillus thuringiensis in ready-to-eat food. FEMS Microbiol. Lett. 250 129–136. 10.1016/j.femsle.2005.06.054 [DOI] [PubMed] [Google Scholar]
- Samapundo S., Heyndrickx M., Xhaferi R., Devlieghere F. (2011). Incidence, diversity and toxin gene characteristics of Bacillus cereus group strains isolated from food products marketed in Belgium. Int. J. Food Microbiol. 150 34–41. 10.1016/j.ijfoodmicro.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Santos C. A., Almeida F. S., Guimaraes A. G., Abrahao W. M., Arantes O. M., Vilas-Boas G. T. (2011). RE-PCR variability and toxigenic profile of food poisoning, foodborne and soil-associated Bacillus cereus isolates from Brazil. Int. J. Food Microbiol. 151 277–283. 10.1016/j.ijfoodmicro.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Schoeni J., Wong A. C. (2005). Bacillus cereus food poisoning and its toxins. J. Food Prot. 68 636–648. [DOI] [PubMed] [Google Scholar]
- Shaheen R., Andersson M. A., Apetroaie C., Schulz A., Ehling-Schulz M., Ollilainen V. M., et al. (2006). Potential of selected infant food formulas for production of Bacillus cereus emetic toxin, cereulide. Int. J. Food Microbiol. 107 287–294. 10.1016/j.ijfoodmicro.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Shi X., Rao N. N., Kornberg A. (2004). Inorganic polyphosphate in Bacillus cereus: motility, biofilm formation, and sporulation. Proc. Natl. Acad. Sci. U.S.A. 101 17061–17065. 10.1073/pnas.0407787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivers R. P., Sonenshein A. L. (2004). Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53 599–611. 10.1111/j.1365-2958.2004.04135.x [DOI] [PubMed] [Google Scholar]
- Sieber S. A., Marahiel M. A. (2005). Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem. Rev. 105 715–738. 10.1021/cr0301191 [DOI] [PubMed] [Google Scholar]
- Slamti L., Lereclus D. (2002). A cell-cell signaling peptide activates the PlcR virulence regulon in bacteria of the Bacillus cereus group. EMBO J. 21 4550–4559. 10.1093/emboj/cdf450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. P., Handelsman J., Goodman R. M. (1999). Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. U.S.A. 96 4786–4790. 10.1073/pnas.96.9.4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5 917–927. 10.1038/nrmicro1772 [DOI] [PubMed] [Google Scholar]
- Stark T., Marxen S., Rütschle A., Lücking G., Scherer S., Ehling-Schulz M., et al. (2013). Mass spectrometric profiling of Bacillus cereus strains and quantitation of the emetic toxin cereulide by means of stable isotope dilution analysis and HEp-2 bioassay. Anal. Bioanal. Chem. 405 191–201. 10.1007/s00216-012-6485-6 [DOI] [PubMed] [Google Scholar]
- Stenfors Arnesen L. P., Fagerlund A., Granum P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32 579–606. 10.1111/j.1574-6976.2008.00112.x [DOI] [PubMed] [Google Scholar]
- Strauch M., Webb V., Spiegelman G., Hoch J. A. (1990). The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc. Natl. Acad. Sci. U.S.A. 87 1801–1805. 10.1073/pnas.87.5.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan I. S., Ramamurthi K. S. (2014). Spore formation in Bacillus subtilis. Env. Microbiol. Rep. 6 212–225. 10.1111/1758-2229.12130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempelaars M. H., Rodrigues S., Abee T. (2011). Comparative analysis of antimicrobial activities of valinomycin and cereulide, the Bacillus cereus emetic toxin. Appl. Environ. Microbiol. 77 2755–2762. 10.1128/AEM.02671-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova V. V., Mikkola R., Tonshin A. A., Saris N. E., Salkinoja-Salonen M. S. (2006). The higher toxicity of cereulide relative to valinomycin is due to its higher affinity for potassium at physiological plasma concentration. Toxicol. Appl. Pharmacol. 210 39–46. 10.1016/j.taap.2005.06.012 [DOI] [PubMed] [Google Scholar]
- Ter Beek A., Brul S. (2010). To kill or not to kill bacilli: opportunities for food biotechnology. Curr. Opin. Biotechnol. 21 168–174. 10.1016/j.copbio.2010.03.014 [DOI] [PubMed] [Google Scholar]
- Thorsen L., Hansen B. M., Nielsen K. F., Hendriksen N. B., Phipps R. K., Budde B. B. (2006). Characterization of emetic Bacillus weihenstephanensis, a new cereulide-producing bacterium. Appl. Environ. Microbiol. 72 5118–5121. 10.1128/AEM.00170-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toljander J. F., Artursson V., Paul L. R., Jansson J. K., Finlay R. D. (2006). Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol. Lett. 254 34–40. 10.1111/j.1574-6968.2005.00003.x [DOI] [PubMed] [Google Scholar]
- Tschiedel E., Rath P. M., Steinmann J., Becker H., Dietrich R., Paul A., et al. (2015). Lifesaving liver transplantation for multi-organ failure caused by Bacillus cereus food poisoning. Pediatr. Transplant. 19 E11–E14. 10.1111/petr.12378 [DOI] [PubMed] [Google Scholar]
- Ueda S., Kuwabara Y. (1993). An ecological study of Bacillus cereus in rice crop processing. J. Antibact. Antifung. Agents 21 499–502. [Google Scholar]
- Valero M., Salmeron M. C. (2003). Antibacterial activity of 11 essential oils against Bacillus cereus in tyndallized carrot broth. Int. J. Food Microbiol. 85 73–81. 10.1016/S0168-1605(02)00484-1 [DOI] [PubMed] [Google Scholar]
- Vangoitsenhoven R., Rondas D., Cre‘vecoeur I., D’Hertog W., Baatsen P., Masini M., et al. (2014). Foodborne cereulide causes beta-cell dysfunction and apoptosis. PLoS ONE 9:e104866 10.1371/journal.pone.0104866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. T., Chen H., Keating T. A., Hubbard B. K., Losey H. C., Luo L., et al. (2001). Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr. Opin. Chem. Biol. 5 525–534. 10.1016/S1367-5931(00)00235-0 [DOI] [PubMed] [Google Scholar]
- Wenzel S. C., Müller R. (2005). Formation of novel secondary metabolites by bacterial multimodular assembly lines: deviations from textbook biosynthetic logic. Curr. Opin. Chem. Biol. 9 447–458. 10.1016/j.cbpa.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Wijnands L. M., Dufrenne J. B., Rombouts F. M., in’t Veld P. H., Van Leusden F. M. (2006). Prevalence of potentially pathogenic Bacillus cereus in food commodities in The Netherlands. J. Food Prot. 69 2587–2594. [DOI] [PubMed] [Google Scholar]
- Zheng J., Guan Z., Cao S., Peng D., Ruan L., Jiang D., et al. (2015). Plasmids are vectors for redundant chromosomal genes in the Bacillus cereus group. BMC Genomics 16:6 10.1186/s12864-014-1206-5 [DOI] [PMC free article] [PubMed] [Google Scholar]