Abstract
Aims
In this paper, we tested the hypothesis that early life lead (Pb) exposure associated DNA methylation (5mC) changes are dependent on the sex of the child and can serve as biomarkers for Pb exposure.
Methods
In this pilot study, we measured the 5mC profiles of DNA extracted from dried blood spots (DBS) in a cohort of 43 children (25 males and 18 females; ages from 3 months to 5 years) from Detroit.
Result & Discussion
We found that the effect of Pb-exposure on the 5-mC profiles can be separated into three subtypes: affected methylation loci which are conserved irrespective of the sex of the child (conserved); affected methylation loci unique to males (male-specific); and affected methylation loci unique to females (female-specific).
Keywords: blood lead levels, differential methylation, DNA methylation, dried blood spots, epigenetics, gene specific, lead, whole blood
Lead (Pb) is an environmental pollutant which has been shown to have acute and chronic effects on human health. The most common routes of exposure to Pb include inhalation of air contaminated by Pb dust, ingestion of contaminated food and direct contact with Pb-tainted soil [1,2]. Pb can directly bind to sulfhydryl-containing compounds like glutathione, and effectively inactivate them thereby reducing its availability as an antioxidant [3]. Pb can also inhibit the enzyme ALAD which results in an increase in the concentration of delta-aminolevulinic acid (ALA) in blood. ALA is a highly unstable compound and its consequent degradation leads to formation of reactive oxygen species (ROS) [4]. Generation of ROS can cause extensive DNA damage and persistent epigenetic changes, which might cause developmental reprogramming and increased susceptibility to adult diseases [5].
Early life exposure to Pb alters underlying mechanisms responsible for biological processes regulating neuronal plasticity, sexual maturity and immune functions [6–8]. This may contribute to developmental defects such as low intelligence quotient (IQ) test scores, cognitive impairment, increased likelihood of childhood delinquency and compromised ability to combat infections. Physiological studies conducted on Pb toxicity in rats have shown conclusively that chronic exposure to low levels of Pb causes a lower density of dendritic spines and synapses and an inhibition of long-term potentiation (a model for learning) in the visual cortex and hippocampus [6]. Studies conducted in our laboratory on Drosophila have shown that exposure to Pb at levels comparable to human exposure levels alters the number of synapses in the neuromuscular junction which leads to impaired locomotor activity [9].
Previous studies in rats have reported that developmental exposure to Pb causes significant changes in the hippocampal transcriptome; however, these changes were not independent of confounding factors like sex, exposure conditions and genetic background [10]. Studies conducted with children exposed to ≤10 μg/dl of Pb from the environment, showed that blood Pb levels in boys were greater than that of girls and was exacerbated by a polymorphism in the gene encoding ALAD or PEPT2 [11].
It is not known how early life exposures to heavy metals can cause neurological disorders that manifest decades later. One of the potential mechanisms is the alteration of the DNA methylation profile of the genome. DNA methylation, or the addition of a methyl (−CH3) group to the 5′ position of a cytosine, is assisted by a group of enzymes called DNA methyltransferases (DNMTs). DNA methylation is a stable epigenetic modification that is involved in the regulation of transcription [12–14]. An increase in the cytosine methylation in the promoter regions of many genes prevents the binding of some transcription factors and promotes recruitment of methyl binding proteins such as MECP2 which, together, contribute to the epigenetic silencing of the gene [15–17]. Conversely, increases in DNA methylation in the gene body are correlated with increased transcription of many genes [18]. We have shown that gene body DNA methylation is correlated with alternative mRNA splicing in the bee brain [19]. DNA methylation mediated transcriptional silencing mechanisms have also been associated with suppression of transposition activity of diverse transposons [20].
Several studies have suggested that exposure to heavy metal toxicants can influence the DNA methylation profile. Kile and colleagues showed a positive correlation between arsenic (As) exposure and DNA methylation of long interspersed element-1 (LINE1) repeat elements in embryonic cord blood leukocytes [21]. Similarly, Wright and colleagues have shown a significant inverse correlation between patella bone Pb levels and DNA methylation of LINE1 repeat elements in UCB, suggesting that methylation might serve as a marker for developmental Pb exposure in the fetus [22].
Studies of global expression patterns and their correlation with DNA methylation in mouse models of prenatal exposure to Pb have revealed a significant association between an increase in DNA methylation and transcriptional repression of genes associated with the immune response, metal binding, metabolism and transcription [23]. While studies in the human cohort or mouse models have shown significant associations between global DNA methylation and heavy metal exposure, there are limited numbers of studies specifically focusing on gene specific DNA methylation changes. Koestler et al. demonstrated that in utero exposure to arsenic exhibited a negative association with DNA methylation levels of the genes encoding ESR1 and PPARGC1A [24]. Hanna and colleagues, reported increase in DNA methylation in the promoter region of GSTM1 on exposure to mercury (Hg) and decrease in DNA methylation in the promoter of the gene COL1A2 on exposure to Pb in whole blood of women undergoing in vitro fertilization (IVF) [25].
The cut-off of 5 μg/dl was the maximum safe limit for a blood lead level (BLL) recommended by the Advisory Committee on Childhood Lead Poisoning Prevention (ACCLPP 2012). Therefore, we hypothesize that early life exposure to Pb at concentrations ≥5 μg/dl will cause significant gene-specific changes in DNA methylation in key metabolic and neuronal genes, which will be detectable in DNA extracted from dried whole blood spots (DBS). As male children are known to be more sensitive to Pb exposure [26,27], if the DNA methylation changes associated with Pb exposure are maladaptive, we expect to see more Pb-exposure associated DNA methylation changes in males compared with females. On the other hand, if the DNA methylation changes are adaptive and protective in nature, then females might show a greater number of Pb exposure associated changes in DNA methylation.
It is important to note that epigenetic responses of an individual to environmental toxicants can be confounded by seemingly unrelated conditions and exposures. For this study we identified the specific CpG sites and methylated loci which are different in the DNA for male and female children. Then, using statistical modeling, we segregated the genome into two types of regions: conserved DNA methylated loci and unique loci for male and female children and then tested the effect of exposure on methylation status of the annotated loci. Using this approach we were able to identify gender independent and gender-specific differentially methylated clusters (DMCs) that correlate with Pb exposure. As we hypothesized, mapping of the probes from the DMCs to the genome revealed several interesting target genes associated with significant biological processes. Though we were limited to analyzing only a small sample cohort because of the high cost of the DNA methylation assays, results from this investigation will be useful to determine putative targets for future studies with PCR-based methylation assays.
Methods
Recruitment/consent procedures
Seventy-five children (3 months to 5 years of age) and their biological mothers (75) were enrolled into the study through routine visit at the WIC (Women Infant and Children). Clinics chosen for recruitment from Southwest Detroit, were selected with guidance from DHWP (Detroit Health and Wellness Department). Note: recruitment fliers were placed in each location announcing when the study personnel would be at the location to consent for the study. The biological mothers of the children were asked if they would like to enroll in the study. All of the following are exclusion criteria: mothers born before January 1, 1987; mothers born outside of Michigan; children 6 years of age and older; children who are not the biological children of the mothers; biological children who were born outside of Michigan; non-English speaking individuals and those who are not fluent in English.
Approximately half (38) of the children enrolled had BLL at or above the Center for Disease and Control blood lead level of concern (equal to or greater than 5 μg/dl). The remainder of the children will have lower BLLs.
A finger stick blood draw was taken from the child and mother to determine BLL by placing a sample of the blood in a Lead Care Analyzer. Results were reported to the mother within 15 min of the blood draw. The child’s BLL was reported to the Michigan Department of Community Health within a week of the testing.
The mothers of the children were asked to complete a demographic and environmental questionnaire after enrollment in the study. The questionnaire will be self-administered; staff will be present to assist any participant as needed.
Samples & sample classification
For the study, we selected 43 dried blood spots (DBS) collected from children from Health Fairs ran in three Detroit communities, Rosa Parks, Chene and Kettering-Butzel, because they have a high prevalence (8–11%) of high BLL in children. The study only included mothers born after January 1, 1987 in Michigan with biological children, ages from 3 month to 5 years, also born in Michigan. The final sample used in this study consisted of 25 male children and 18 female children. Among males 15 children had BLL ≥5 μg/dl and among females 11 children had BLL ≥5 μg/dl. The covariate and BLL information is available in Supplementary Table 1 (For Supplementary Data, please see online at: www.futuremedicine.com/doi/full/10.2217/EPI.15.2).
Lead measurements in dried blood spots
The samples were 3-mm punch-outs of blood spots on filter paper. The samples, control filter paper punch-out blanks without blood, blanks without filter paper, and standards were all prepared in parallel at the same time in the same way, except that five-times as much of the standard solutions were made to allow sufficient volume for recalibration every 20 samples. To each container was added ultrahigh purity 0.25 ml of 70% HNO3 (0.25 ml) and 30% H2O2 (0.05 ml). These reagents were purified in an in-house subboiling still from reagent grade materials. After reacting overnight, samples were diluted with 10 ml of 0.5% HNO3 containing 5 ppb each of Ga and Bi as internal standards. Standards (50 ml) were spiked with 0.1 ml of a solution of Fe, Mg, Zn and Pb, prepared from 1000 ppm single element ICP-MS stock solutions (Inorganic Ventures, VA, USA). The final standard concentrations were Fe, 200 ppb, Mg, 20 ppb, Zn 2 ppb, Pb, 1 ppb. All containers were exposed to vacuum (~0.03 bars) for 10 min to reduce the amount of dissolved oxygen from H2O2 dissociation. Even so, some oxygen bubbles were seen during analysis in the sample line to the ICP-MS. Filter paper fibers settled to the container bottoms and did not affect the ICP-MS sample introduction system. Samples were analyzed on a PerkinElmer (MA, USA) Elan 6100 DRC ICP-MS instrument at the Geology Department, Union College. 25Mg, 66Zn and 206,207,208Pb were analyzed in normal mode, and 54Fe was analyzed in DRC mode using NH3 reaction gas at a flow rate of 0.5 ml/min.
Extraction, shearing & denaturation of DNA
DNA was isolated from dried blood spots with Qiagen (Hilden, Germany) EZ1 Advanced® using the DNA Investigator® reagents and protocol card. The ‘Stains on Fabric’ preprocessing and Trace® (tip-dance) instrument protocol was used for isolation. The Quantifiler Human DNA Quantification Kit® (Applied Bio-systems, Inc., MA, USA) was used to determine the amount of amplifiable DNA.
Approximately 3 μg genomic DNA was diluted in 130 μl of buffer TE (10 mM Tris [pH 8.0], and 1 mM EDTA [pH 8.0]) and sheared into ~200–600 bp fragments using microcavitation (Covaris, Inc, MA, USA) setting: Duty Cycle = 5%, intensity = 3, cycles/burst = 200, time = 75 s run at 6–8°C). A 125 μl of the sheared DNA samples were mixed with 330 μl of buffer TE. The sheared DNA was denatured by boiling it in the Thermomixer at 95°C and 700 rpm for 10 min and left on ice for 10 min.
HM450K bead chip array
For this study, we measured the change in DNA methylation on environmental exposure to Pb in dried blood spots using the Illumina (MA, USA) Human Methylation 450K Bead chip array (HM450K). The HM450K assay measures DNA methylation at over ~480,000 CpG and non-CpG sites with single-base resolution [28,29]. The results are represented in the form of beta (β) values ranging from 0 to 1, and provide a quantitative measure of methylation for each queried CG dinucleotide methylation site (CpG site) [28]. DNA methylation changes at CpG sites located close to each other often exhibit common behavior in response to environmental stimuli. These regions show highly correlated changes in methylation signatures and can be defined as co-regulated regions. The co-regulated regions can be assigned to specified clusters and the effect of the exposure on these clusters can be tested using the generalized estimating equation (GEE) [30]. GEE uses a weighted combination of observation to measure the effect of a covariate (in our case Pb exposure) while conserving the correlation structure of the data [31]. Consequently, this approach is much less conservative and yields a greater number of differentially methylated regions compared with the traditional case–control study with a single CpG site β value comparison. Biochemically it makes more sense to study methylation changes as clusters rather than single CG sites because DNA bound DNMT1 and DNMT3a, the maintenance and de novo DNA methyltransferases, respectively, act on multiple sites in a small region when they bind to DNA [32].
Detection of methylated DNA is facilitated by two different probe types (type 1 and type 2 probes). The type 1 probes or the Infinium 1 (Inf1) probes consist of the methylated bead and an unmethylated bead [33]. If the probe for methylated DNA matches the target site there is a single base extension which results in detection which signals into the red channel. Similarly if an unmethylated probe binds to the DNA it signals into the green channel. The type 2 probes or the Infinium 2 (Inf2) queries both methylated and unmethylated DNA on a single bead, and the ratio of incorporation of two differently colored fluorescent nucleotides (signals A and B) determines the methylation signal. The results are represented in the form of β values, specifically, the average β value (AVG_Beta), representative of the average methylation level of the CpG dinucleotide, and a delta β value which signifies the difference in methylation levels between the control and the experimental group. The Beta or β for the ith interrogated CpG nucleotide is:
| Equation 1 |
where yi,methy and yi,unmethy are the intensities measured by the ith methylated and unmethylated probes, respectively. Illumina recommends adding a constant offset α (by default, α = 100) to the denominator to regularize β value when both methylated and unmethylated probe intensities are low. The Beta-value statistic results in a number between 0 and 1, or 0 and 100% [34]. The raw data were retrieved from Genome Studio methylation module version 1.8™ in the form of two files: a sample methylation profile and control probe profile. Quality control, signal correction and normalization of the data was carried out using the HM450K BeadChip data processing pipeline proposed by Teschendorff et al., in R environment (R > 2.13.0) [35]. Several studies have indicated that the Infinium 1 and 2 probes differed in chemistry, henceforth the HM450K are two separate experiment combined as one. The Infinium 1 probes were shown to have a more stable signal and extended dynamic range compared with the Infinium 2 probes [36]. Therefore, a 3-state beta mixture model is utilized to assign methylation values to specific methylation states. Then the probability of assignment to particular state is divided in quantiles and finally a methylation-dependent dilation transformation is performed to preserve sample monotonicity [35]. Prior to analysis the beta values were corrected for batch effect using combat function in R and potential single nucleotide polymorphism (snp)-containing probes were removed from the analysis (>2.15) [37].
The HM450K array is highly reliable for locus specific methylation detection at CpG island associated methylation sites which are frequently associated with dynamic regulation during development and disease states. Bibikova et al., conclusively showed using human sample (lung tissue) that the HM450K array show 95 to 96% correlation which WGBS results [33]. Since then several studies have explored this correlation and found this array to be highly consistent. Therefore, in lieu of recent evidence, we believe that HM450K array can ‘stand on its own’ as an independent system for differential methylation analysis. That said, to look at the whole genome methylation status in an unbiased way sequencing-based approaches are still the best methodology. When we have a much larger cohort, then we could use the less expensive PCR assay to validate the CpG sites that we identify in this pilot study.
Statistical analysis
For studying effects of exposure to Pb on the DNA methylation profile of UCB in male and female samples we used two independent statistical approaches; CpG association analysis [38], which analyzes DNA methylation data using a fixed effect model at single CpG site level and adjacent site clustering algorithm, A-clustering to detect sets of correlated CpG sites and then tested the clusters for multivariate response to environmental exposure to Pb using the generalized estimation equation approach. The aforementioned approach is efficiently implemented using the R-package aclust [30].
For determining the differentially methylated clusters (DMCs) we used the recommended Aclust parameters; Spearman correlation, for calculating the distance between adjacent sites (dist(i,j)= 1 corr(i,j)), average clustering type, which require that mean distance between two sites be at least 0.25, 1000 bp distance restriction for merging of clusters, which ensures that clusters located far away from each other are not merged together based on correlation. The clustering approach is implemented with a 999 bp merge initiation step, which clusters all sites wedged between two high correlated sites within 999 bp of each other together, to reduce the complexity of data and the analysis time for the Aclust step. Finally the data was analyzed using a generalized estimation equation approach and filtered for significant DMCs using False Discovery Rate (FDR) corrected P value cutoff = 0.05 and exposure effect size ≥∣0.02∣. To determine the genomic locations of the probes belonging to individual DMCs, they were annotated using the publicly available Illumina Human Methylation 450k annotation data in R (> 2.15). The target genes mapping to DMCs were individually visualized using UCSC genomic browser. The Delta beta or the beta difference between the median of the beta values for each probe for low BLL samples and high BLL samples were mapped by the chromosomal location of the probes. If A-clustering is an effective technique for DMR identification we hypothesized that the change in methylation status visualized using UCSC genome browser will correspond to the exposure effect (i.e. increase or decrease in methylation) predicted by GEE in the respective regions and might serve as a useful tool for visualization. Gene ontology for mapped DMCs was carried out using hypergeometric testing implemented by the package GO stats in R (> 2.15).
A-clustering takes into consideration that adjacent CpG sites are probably co-regulated by Pb exposure, therefore the differential methylation calls are made based on multiple probes rather than a single CpG site, making it considerably more reliable compared with generalized linear model-based differential methylation calling algorithm such as limma [39]. Moreover, the presence of multiple CpG sites with altered methylation states, in regions such as transcription start sites (TSS), is more likely to cause altered transcription factor binding and affect expression. A significant problem associated with clustering based approach is the possible introduction of gene-specific methylation bias due to varied number of probes mapping to genes. In the A-clustering based approach, the clusters of probes are based on the base-pair distance between adjacent probes, and beta value correlations across samples, but not in the context of the genomic features. Therefore, overall the number of probes in each cluster contributes very little to the differential methylation calls.
Results
Sex-specific differences in DNA methylation regardless of Pb exposure
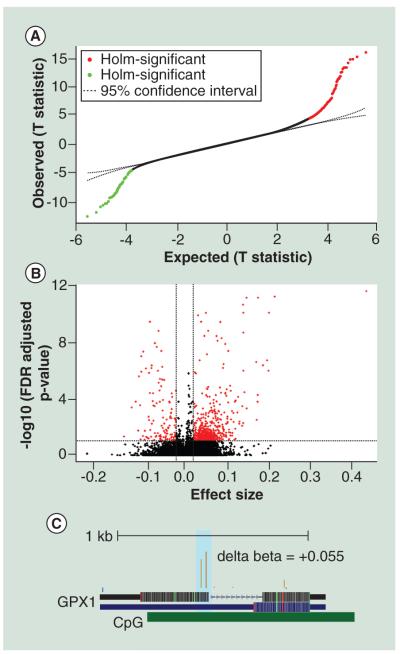
To determine whether sex plays a major role in intersample variations in the DNA methylation profiles, we studied the association between DNA methylation of whole blood with the sex of the children. We restricted these analyses to the autosomes because females have one inactive X chromosome with increased promoter DNA methylation (i.e., the Barr Body [40]). We used a fixed effect model to elucidate the single-nucleotide differences in DNA methylation. There were 456 CpG regions which where differentially methylated between males and females at a FDR corrected P value cutoff of 0.05 and effect size cutoff of ∣0.02∣ or ∣2%∣ (Figure 1A). 365 CpG regions were hyper-methylated and 91 CpG regions were hypo-methylated in females compared with males (Figure 1B). GO mining based on overrepresentation analysis (hypergeometric testing) showed enrichment of hyper-methylated CpG regions in genes associated with neurogenesis (GO:0022008), neuronal differentiation (GO: 0030182) and oxygen and reactive oxygen species metabolic process (GO:0072593) (Supplementary Table 2) such as glutathione peroxidase 1 (GPX1) (Table 1 & Figure 1C), Cytochrome p450 (CYP1A1) (Table 1), Superoxide Dismutase 3 (SOD3) (Table 1) and Solute Carrier Family 6 (Neurotransmitter Transporter) (SLC6A4) (Table 1). Differences in hypermethylation in females was also noticed in nuclear encoded mitochondrial genes such as Glucokinase (GCK) (Table 1), Pyruvate kinase (PKLR) (Table 1), HCLS1 Associated Protein X-1 (HAX1) (Table 1) and DnaJ-Hsp40-Homolog (DNAJA1) (Table 1).
Figure 1. Methylation profiles of DNA extracted from whole blood for male and female children are different from each other.
(A) T-plot of single CpG sites between expected and observed methylation differences which showed significant association with sex of the infant. (B) Volcano plot for significant association between methylation and gender. There were 456 differentially methylated CpG sites at a FDR corrected P value cutoff of 0.05 and beta difference cut-off ≥ ∣0.02∣ or ∣2%∣. (C) The representative University of California, Santa Cruz genome browser plot for the GPX1 gene showing a ~6% increase in DNA methylation around the promoter region. The delta beta value was calculated by subtracting the mean of β values for cases with high BLL samples (≥5 μg/dl) from the mean of β values for controls (BLL ≤ 5μg/dl). All analyses were controlled for potential confounders such as age of the child, gestational age, smoking status of the mothers or immediate family and blood lead levels (BLL).
Table 1.
Single nucleotide differences in DNA methylation between females and males as estimated by fixed effect model.
| CpG site | Gene | Promoter associated |
CpG island | Effect size | Standard error | P value | Holm.sig | FDR |
|---|---|---|---|---|---|---|---|---|
| cg22584138 | SLC6A4 | No | chr17:28562387- 28563186 |
0.074 | 0.012 | 3.91E-07 | FALSE | 0.001 |
| cg04555966 | GCK | No | chr7:44185961- 44186184 |
0.037 | 0.007 | 5.65E-06 | FALSE | 0.0089 |
| cg09933329 | GCK | No | chr7:44185961- 44186184 |
0.056 | 0.012 | 4.9E-05 | FALSE | 0.041 |
| cg03314840 | PKLR | No | chr1:155264318- 155265536 |
0.053 | 0.011 | 3.93E-05 | FALSE | 0.0357 |
| cg12177922 | HAX1 | Yes | chr1:154244710- 154245289 |
0.052 | 0.007 | 1.06E-08 | TRUE | 5.44E-05 |
| cg21763723 | GPX1 | Yes | chr3:49394855- 49395942 |
0.042 | 0.009 | 2.62E-05 | FALSE | 0.027 |
| cg25187648 | GPX1 | Yes | chr3:49394855- 49395942 |
0.054 | 0.007 | 1.38E-08 | TRUE | 6.88E-05 |
| cg11924019 | CYP1A1 | No | chr15:75018186- 75019336 |
0.053 | 0.011 | 5.6E-05 | FALSE | 0.0448 |
| cg02891686 | SOD3 | No | chr4:24801109- 24801902 |
0.045 | 0.009 | 8.5E-06 | FALSE | 0.0123 |
| cg11304734 | POLR1E | Yes | chr9:37485801- 37486099 |
0.045 | 0.009 | 1.47E-05 | FALSE | 0.0182 |
| cg14268223 | DNAJA1 | Yes | chr9:33025082- 33025797 |
−0.047 | 0.009 | 2.06E-05 | FALSE | 0.0237 |
| cg13479204 | HOXB3 | No | chr17:46641534- 46642110 |
−0.082 | 0.016 | 1.2E-05 | FALSE | 0.0159 |
| cg05323879 | HOXB3 | No | chr17:46641534- 46642110 |
0.476 | −0.071 | 2.39E-05 | FALSE | 0.0257 |
| cg02325951 | FOXN3 | No | chr14:89882421- 89884278 |
−0.062 | 0.007 | 9.55E-10 | TRUE | 7.23E-06 |
| cg26355737 | TFDP1 | No | chr13:114292551- 114292886 |
−0.034 | 0.006 | 6.91E-07 | FALSE | 0.00174 |
| cg19292611 | TFDP1 | N0 | chr13:114292551- 114292886 |
−0.023 | 0.004 | 4.24E-06 | FALSE | 0.00726 |
For the analysis, the females where used as experimental group and males as the control group. All analysis was controlled for covariates such as age of the child, gestational age of the mother, age of the mother, smoking status of the household and blood lead levels.
FDR: False discovery rate; Holm: Holm–Bonferroni statistical method.
Hypo-methylated CpG regions in females were associated with basic regulatory processes such as double-strand break repair (GO:0006302) and aerobic respiration (GO:0009060) (Supplementary Table 2). We hypothesize that these differences in methylation might underlie some of the gender specific differences in the sensitivity to Pb exposure. All of the analyses were controlled for the following potential confounders: BLL, age of the children, gestational age, age of the mother and smoking status of the mother and immediate family.
Pb exposure associated DNA methylation changes in both males & females
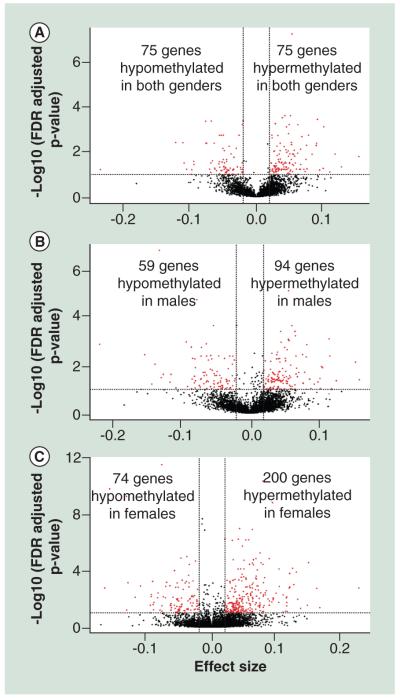
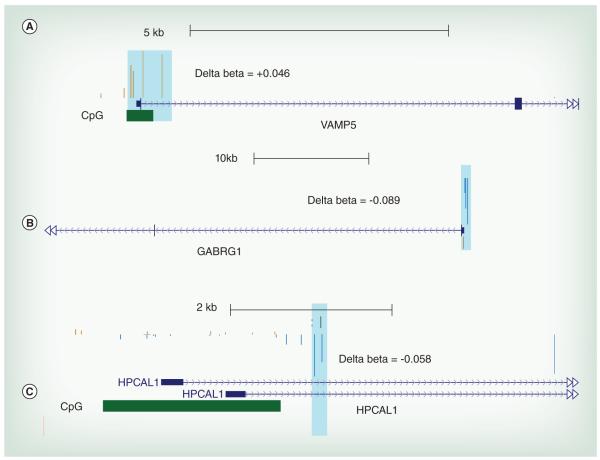
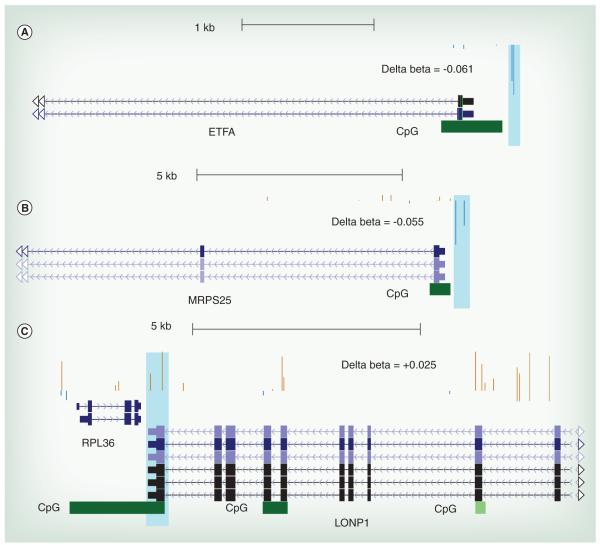
Our analysis revealed large locus-specific and single nucleotide differences in the methylation status of whole blood DNA dependent on the sex of the infant. However, there might be other regions in the human genome which have DNA methylation profiles that correlate with Pb exposure irrespective of sex. We define these clusters of methylated CpG sites as ‘conserved’ regions. Using the GEE model we tested the effect of Pb exposure on DNA methylation status on these regions. At FDR, cut-off of 0.05 and effect size ≥∣0.02∣ or ∣2%∣ we found 75 hyper-methylated Pb-associated DMCs and 38 hypo-methylated Pb-associated DMCs mapping to 75 unique genes as predicted by the GEE model (Figure 2A). Gene ontology analysis of the genes mapping to Pb-associated DMCs (Supplementary Table 3) showed overrepresentation of genes associated with differentiation of myeloid lineages. Myeloid cells are precursors to granulocytes, which are involved in the immune response. There was a 3 ±1% increase in DNA methylation in a cluster of 5 CpG sites located in the CpG Island in the promoter region of the gene encoding VAMP5 (Table 2 & Figure 3A), and a 2.4 ± 0.5% increase in DNA methylation in a cluster of three probes located in the promoter associated CpG island in CALN2 (Table 2). There is also a 9 ± 2% increase in DNA methylation in a cluster located in the CpG island near the transcription start site of the gene encoding PF4 (Table 2) and a 3.5 ± 1% increase in methylation in the CpG island near the transcription start site of the gene encoding LEP (Table 2). Additionally, there are changes in DNA methylation of nuclear encoded mitochondrial genes. For example, there is a 4.5 ±1% decrease in DNA methylation in the CpG Island located in the promoter region of the gene encoding ETFA known to be involved in catalyzing the first step of mitochondrial fatty acid beta oxidation (Table 2 & Figure 4A).
Figure 2. Lead causes common and gender-specific changes in DNA methylation.
The Pb-associated differentially methylated clusters (Pb-associated DMCs) selected for further analysis are screened by FDR cut off of 0.05 and effect size ≥∣0.02∣ or ∣2%∣ (A) A-clustering results for conserved region revealed 75 hyper-methylated Pb-associated DMCs and 41 hypo-methylated Pb-associated DMCs mapping to 76 unique genes associated with Pb exposure as predicted by the GEE model. (B) For an effect size cutoff of ∣0.02∣ or ∣2%∣, and FDR corrected P value ≤ 0.05, we found 94 Pb-associated hyper-methylated and Pb-associated 59 hypo-methylated regions mapping to 124 unique genes for males. (C) Analysis results for unique regions in females revealed 200 hyper-methylated and 74 hypo-methylated regions mapping to 201 unique genes.
Table 2.
Representative gene mapping to clusters which show a change in methylation of ≥∣2%∣ or ∣0.02∣, at a FDR cutoff of 0.05.
| Region ID | Gene | CpG island | Promoter associated |
Effect size | Standard error |
P value | FDR | CpG sites/ cluster |
|---|---|---|---|---|---|---|---|---|
| Conserved | VAMP5 | chr2:85811340- 85811855 |
Yes | 0.031 | 0.009 | 0.000839 | 0.0410 | 5 |
| Conserved | CAPN2 | chr1:223936342- 223937044 |
Yes | 0.024 | 0.005 | 5.94E-06 | 0.00174 | 3 |
| Conserved | PF4 | chr4:74847528- 74847830 |
No | 0.0933 | 0.0223 | 2.38E-05 | 0.00395 | 4 |
| Conserved | LEP | chr7:127880750- 127881375 |
No | 0.036 | 0.01 | 0.00045 | 0.0280 | 2 |
| Conserved | ETFA | chr15:76603563- 76604026 |
Yes | −0.046 | 0.013 | 0.000387 | 0.0258 | 2 |
| Male-specific | RUNX1 | chr21:36258952- 36259472 |
No | −0.084 | 0.025 | 0.00082 | 0.0373 | 5 |
| Male-specific | GABRG1 | Non-CpG | No | −0.08 | 0.019 | 4.24E-05 | 0.00534 | 5 |
| Male-specifc | MRPS25 | chr3:15106459- 15106971 |
Yes | −0.064 | 0.020 | 0.00148 | 0.0497 | 2 |
| Female-specific | APP | Non-CpG | No | 0.043 | 0.013 | 0.00131 | 0.0366 | 4 |
| Female-specific | LONP1 | chr19:5690127- 5692213 |
No | 0.038 | 0.012 | 0.00143 | 0.0391 | 2 |
| Female-specific | MTERFD2 | chr2:242041543- 242042026 |
Yes | 0.021 | 0.003 | 8.73E-10 | 5.43E-07 | 7 |
| Female-specific | HPCAL1 | chr2:10442308- 10444509 |
Yes | −0.055 | 0.017 | 0.00111 | 0.0323 | 2 |
| Female-specific | MAP3K6 | chr1:27683277- 27683590 |
Yes | 0.061 | 0.016 | 8.8E-05 | 0.00534 | 5 |
| Female-specific | HIF3A | chr19:46800053- 46800603 |
No | 0.06 | 0.019 | 0.00150 | 0.0405 | 2 |
| Female-specific | RUNX3 | chr1:25255527- 25259005 |
No | 0.045 | 0.012 | 0.00017 | 0.00859 | 2 |
Figure 3. The representative University of California, Santa Cruz genome browser plot for candidate genes.
The delta beta value was calculated by subtracting the mean of β values for cases with high BLL samples (≥5 μg/dl) from the mean of β values for controls (BLL ≤ 5 μg/dl). The resulting delta beta was mapped to genome to visualize the estimated changes in methylation in UCSC genome browser for regions mapping to differentially methylated clusters (DMCs). The position of CpG Island is also shown. (A) Representative figure for Vamp5 for DMCs conserved in both males and females. This DMC shows a ~3% increase in DNA methylation in the promoter associated region. (B) Representative figure for GABRG1 for a male-specific Pb-associated DMC. This Pb-associated DMC shows an ~8% decrease in DNA methylation around the transcription start site (TSS) of the gene. (C) Representative figure for HPCAL1 for a female-specific Pb-associated DMC. This Pb-associated DMC shows a ~3.8% decrease in DNA methylation in a promoter associated region.
Figure 4. DNA methylations at nuclear encoded mitochondrial genes are affected by lead.
The delta beta were calculated by subtracting the mean of β values for cases with high BLL samples (≥5 μg/dl) from the mean of β values for controls (BLL ≤ 5 μg/dl). Highlighted regions correspond to the probes mapping to DMCs. The position of the CpG Island is also shown. (A) Representative figure for ETFA for a Pb-associated DMC conserved in both males and females. This DMC shows a ~4.6% increase in DNA methylation in a promoter associated region. (B) Representative figure for MRPS25 for a male-specific Pb-associated DMC. This Pb-associated DMC shows a ~6% decrease in DNA methylation in a promoter associated region. (C) Representative figure for LONP1 for a female-specific Pb-associated DMC. This Pb-associated DMC shows a ~5% decrease around the transcription start site (TSS) of the gene.
Sex-specific effects of Pb exposure on DNA methylation
To determine the gender specific effects of Pb exposure on the DNA methylation profile, we separated the samples based on the sex of the infant before A-clustering. This enabled genome wide identification of methylation loci unique to the sex of the infant. Then we classified the samples into high BLL and low BLL categories based on a cutoff of 5 μg/dl and tested the effect of Pb exposure on the DNA methylation status using the GEE model after controlling for confounding factors such as gestational age, mothers age and smoking status of the household. For an effect size cut-off of ∣0.02∣ or ∣2%∣, and FDR corrected P value ≤ 0.05, we found 94 Pb-associated hyper-methylated and 59 hypomethylated regions (Figure 2B). Gene ontology analysis of genes mapping to male-specific Pb-associated differentially-methylated regions show an enrichment of genes associated with leukocyte proliferation and differentiation (GO: 1902107) and calcium ion transport (GO: 0051924) (Supplementary Table 3). Examples of males specific genes that are differentially methylated by Pb are RUNX1 (Table 2 & Supplementary Figure 1A), GABARG1 (Table 2 & Figure 3B) and MRPS25 (Table 2 & Figure 4B).
We also see female-specific changes in DNA methylation that are associated with Pb exposure. At P value of ≤0.05 and exposure effect size of ≥∣0.02∣ or ∣2%∣ we found 200 hyper-methylated and 74 hypo-methylated regions in females exposed to Pb (Figure 2C). We performed gene ontology analysis and found enrichment of hyper-methylated genes for pathways such as neuron maturation (GO:0042551) and visual learning (GO:0008542) and hypo-methylated genes for pathways such as regulation of type 2 immune response (GO:0002828; Supplementary Table 4). Other interesting genes which also showed a Pb-associated differential methylation included stress response genes such as APP (Table 2) and HIF3A (Table 2), Lonp1 (Table 2 & Figure 4C) and MTERFD2 (Table 2), transcriptional regulator genes like RUNX3 (Table 2 & Supplementary Figure 1B), genes that encode important regulators of signaling pathways such as MAP3K6 (Table 2) and genes that encode neuronal calcium sensors such as HPCAL1 (Table 2 & Figure 3C).
Discussion
Exposure-related adverse conditions early in the developmental environment can cause reprogramming of the DNA methylation profile [41]. It has been suggested that these changes can be detected in whole blood [24,42]. This may have potential prognostic value in the determination of the relationship between early life Pb exposure and the susceptibility to adult diseases. Use of whole blood for assessing the DNA methylation levels in the newborn and during early childhood poses several challenges. First, the relationship between DNA methylation and Pb levels can be modulated by several environmental and dietary factors affecting the mothers (e.g., folate deficiency, exposure to other environmental toxicants, and cigarette smoking) [43]. Our study was adjusted for smoking status of the immediate family, gestational age, mother’s age and child’s age. Second, the variance between DNA methylation profiles due to exposure can be the result of a shift in the population of immune cells in the blood [44,45]. For our study we restricted the effect size to at least ∣0.02∣ or ∣2%∣ because we did not want to exclude the possibility that the smaller effect size was due to the variation in cell population in the whole blood [30]. Houseman et al., proposed a statistical model for estimation of blood cell type proportion using methylation data [46]. We performed a similar analysis for our dataset and found that blood cell type only contributed to ≥1% of the total variance in methylation profile (data not shown). The DNA methylation changes in response to Pb can vary depending on sex of the infant. Pilsner and colleagues demonstrated that prenatal exposure to Pb changes global DNA methylation in umbilical cord blood (UCB) in a sex-specific manner [47]. Faulk and colleagues showed significant correlation between birth weight and DNA methylation at the Agoutiviable yellow locus exclusively in yellow agouti (Avy/Avy) male mouse offspring [48]. Recently it was found that there are a large number of gender specific differences in DNA methylation levels in the autosomes. For example, Liu et al. reported over 580 autosomal CpG sites which showed sexspecific differences in DNA methylation in the salivary DNA from healthy subjects [49]. Because Pb increases oxidative stress, we are especially interested in sex-dependent DNA methylation differences for genes involved in oxidative stress response and mitochondrial function. We found significant changes in DNA methylation of several genes associated with detoxification pathways such as GPX1 (4–5% higher promoter methylation in females) and CYP1A1 (5% higher methylation status around the TSS in females). Our data were in line with previous observation; for example, Penaloza et al., showed that the methylation status of the promoter region of CYP1A1 in primary cells cultured from embryos of Swiss Webster CWF mice strain, is sexually dimorphic in nature [50]. The cells from the female embryo had four CYP1A1 promoter-associated methylation sites, whereas the cells cultured from male embryo had three methylation sites, under normal developmental conditions. It is important to note, that the number of gender associated methylation differences for single CpG sites are modest. The sample cohort used for this study was between the ages of a few months to 5 years. We believe that the gender-based differences in methylation are a function of age and are fully established at puberty. That might explain why we observe only 456 CpG sites which differ as a function of gender. Evidence from our study suggest that exposure associated locus specific methylation pattern can be divided into three subtypes: affected methylation loci which are conserved irrespective of the sex of the child (conserved); affected methylation loci unique to males (male-specific); and affected methylation loci unique to females (female-specific). Interestingly, we observed the largest number of unique Pb-associated DMCs for females, followed by the males and the lowest number of Pb-associated DMCs for conserved region. As males have been shown to be more susceptible to Pb exposure compared with females [51], the initial hypothesis for our cohort was that DNA methylation changes are maladaptive in nature. Therefore, we predicted that the number of Pb-associated DMCs seen for males would be greater than for females. Contrary to our expectations, our results show that the number of Pb-associated DMCs are greater for females compared with males. It is possible that these changes are mostly adaptive and protective rather than maladaptive, and that the female-specific changes in DNA methylation after Pb exposure help females to cope with excess ROS production due to Pb exposure. Alternatively, given the small size of our cohort for this preliminary study, our results could be a sample set specfic effect and can be controlled for by analyzing additional cohorts.
Differential methylation analysis also revealed that Pb-exposure associated changes in DNA methylation clusters show a bias toward hypermethylation, that is, increase in methylation associated with high BLL irrespective of gender. This suggest a common mechanism of action by which Pb-exposure cause gene specific changes in DNA methylation clusters. Pb exposure is mainly known to act via oxidative stress. Oxidative stress is known to inhibit α-ketoglutarate dehydrogenase in the mitochondria and this result in the accumulation of Kreb’s cycle intermediate, α-ketoglutarate (α-KG) [52]. As α-KG serve as a co-factor for TET enzymes responsible for DNA hydroxymethylation (5 hmC) [53] (stable intermediate of the demethylation pathway), increase in its level might increase the activity of TET enzyme and cause genome wide increase in 5 hmC levels. As the standard application of the HM450K is unable to differentiate between 5 mC and 5 hmC modification, it is possible that the ‘hyper-methylated’ regions are most likely actively de-methylated regions showing Pb-dependent increase in 5 hmC. Further experiments are required to study ratio of 5 mC/5 hmC modification which might provide useful incites into relationship between environmental exposure and epigenetic regulation.
Our earlier study with the National Health and Nutrition Examination Survey (NHANES) cohort showed a significant negative association between BLL and body mass index (BMI) [54]. We saw an increase in methylation around TSS of LEP in sex-independent regions. Leptin is a protein secreted by white adipose tissues which is implicated in the maintenance of body mass [55]. Studies have shown that increases in DNA methylation in the Leptin promoter is accompanied by recruitment of MBP2 leading to decreased recruitment of RNA polymerase 2 and decreased transcription in adipose tissue of obese mice [56]. This suggests a possible implication of Pb exposure on obesity-associated outcomes. Obesity and other metabolic diseases are closely associated with regulation of the immune system. Several studies have reported significant correlation between Pb exposure and modulation of immune response. For example, Kile et al., reported that prenatal exposure to Arsenic cause a shift in immune cell population specifically T-cell population in the cord blood sample [57]. We saw a male-specific decrease in methylation in a CpG island located near the distal transcription start site of a gene encoding an important hematopoiesis controlling transcription factor RUNX1 (Table 2 & Supplementary Figure 1A). Webber et al. showed that the hypomethylation of the distal promoter of RUNX1 is associated with a shift from primitive to definitive hematopoisis and is further promoted by HOXB4 overexpression [58]. We saw a Pb-dependent increase in DNA methylation for another member of Runt domain associated transcription factor, RUNX3 (Table 2 & Supplementary Figure 1B) only in females. RUNX3 deficiency is shown to be associated with myeloproliferative disorder in mouse [59]. Wolff et al. reported that the DNA methylation of RUNX3 increases with age and the process is further accelerated by smoking [60]. In combination with previous data, our data suggest that epigenetic regulation of genes associated with controlling hematopoiesis (e.g., RUNX1 and RUNX3) can play a role in mediating the effect of Pb exposure on immune response in a sex-specific fashion.
The ‘normal’ epigenetic profile of the genome is usually established and stabilized during early developmental years. Exposure to environmental pollutants during this period might lead to permanent maladaptive or adaptive changes in the common progenitor cells. These changes might be detectable across multiple easy-to-collect biospecimens such as DBS. Therefore, it has been suggested that whole blood might serve a viable surrogate to study exposure associated changes in neuronal genes especially in cases of maternal or childhood exposure. We looked at the DNA methylation profiles of genes with known neuronal function in our dataset. We saw an approximately 8% decrease in DNA methylation in a cluster of 5 CpG sites in the promoter region of a single neuronal signaling associated gene, GABRG1, only in males after Pb exposure (Table 2 & Figure 3B). In females, we saw a Pb-associated increase in methylation of several genes related to biological processes such as neuronal maturation (GO:0042551), visual learning (GO:0008542) and regulation of neurotransmitter levels (GO:0001505). We believe that the abundance of DMCs in neuronal genes in females may be an adaptive change which mitigates the effect of chronic Pb exposure on neuronal growth and differentiation in early developmental years in females. Additionally, some of these adaptive DNA methylation changes were also observed in genes associated with response to oxidative stress. For example, we saw an increase in gene body DNA methylation of LONP1 (Table 2 & Figure 4C). LONP1 has been implicated in breakdown of 5-aminolevulinic acid synthase enzyme in the mitochondrial outer membrane [61]. This is an interesting observation, as Pb is a potent inhibitor of ALAD and increases the concentration of 5-aminolevulinic acid (ALA) which can produce ROS [4]. As increase in gene body methylation is associated with increased expression of a gene [62], it is possible that this is indirect evidence of an adaptive methylation change, working to reduce the synthesis of ALA and consequent generation of ROS only in females, and might be a mechanism for resistance to Pb exposure in females.
It is important to note this study has a few limitations. First, this is a pilot study to define gene regions which might be potential epigenetic biomarkers of early Pb exposure and can be used for future studies. Second, for the gene ontology studies we used only known genes in the human genome. This does have a significant effect on the P value of the inferred GO association. So to make sure that reported gene ontology associations are biologically relevant we manually curated the literature for correlation between individual gene function and reported gene-ontology categories. We found that for our dataset, GOstats is accurately able to assign genes to a broad category of gene ontologies which qualitatively defines the possible biological processes being affected by early life Pb exposure.
Conclusion
In this study, we show that modeling Pb exposure associated changes in DNA methylation as coregulated [30], gender-specific methylation clusters provide interesting correlations which warrant further investigation. We also show that these epigenetic changes are observable in DNA extracted from minimally invasive blood collections such as DBS and can serve as an excellent tool for epidemiological studies.
We believe that early life Pb exposure can alter the process of dynamic regulation of DNA methylation via affecting the DNA demethylation pathway. The standard application of the HM450K array is unable to differentiate between 5 mC and the major DNA demethylation by-product 5-hydroxymethylcytosine (5 hmC) [63]. Therefore, for future studies, we will utilize the identified gender-specfic and gene-independent candidate regions from this study and test the ratio of 5 mC to 5 hmC, using established PCR-based detection methods for a much larger cohort [64,65].
Future perspective
Currently, the majority of genome-wide epigenetic studies are heavily reliant on next-generation DNA-sequencing-based techniques. However, in recent years, there has been a concerted effort in the development of third-generation DNA-sequencing-based platforms which use nanopores and ion-current changes for sequencing the DNA (e.g., the Oxford Nanopore Minion™ [Oxford, UK]). Third-generation DNA sequences approaches should be considerably less expensive and will make epigenetic studies in larger human cohorts more affordable and accessible.
Supplementary Material
Executive summary.
Differences in the DNA methylation profile during early human development can be measured in dried blood spots (DBS) collected from the children by finger stabs.
Single nucleotide differences in DNA methylation in autosomal genes between males and females might cause difference in epigenetic susceptibility to lead (Pb) exposure.
Pb exposure-associated changes in clusters of correlated and adjacent 5 mC sites are sexually dimorphic and can be divided into three subtypes: conserved, male-specific and female-specific.
Blood spots from females show significantly higher numbers of differentially methylated clusters (DMCs) than males.
Pb-dependent DNA methylation changes in females appear to be adaptive and protective in nature.
Conserved regions show increase in DNA methylation around the transcription start site of Leptin (LEP), which is associated with the control of body mass.
Increase in gene body DNA methylation of Lon-Peptidase 1 (LONP1) is an adaptive modification seen in females but not in males.
Candidate genes from this study will be explored further as biomarkers of early Pb exposure in future studies in larger cohorts.
Acknowledgements
All of the Infinium data are available upon request to DMR and is available in the GEO database (accession number GSE60598). We thank the Applied Genomics Facility Core at Wayne State University for conducting the HM450K assays.
This research was supported by R01 ES012933 and R21 ES021893 to DM Ruden, the WSU-NIEHS Center (P30 ES020957), a pilot grant from the Institute for Population Sciences, Health Assessment, Administration, Services and Economics (INPHAASE) from Wayne State University and Henry Ford Medical Center to DM Ruden and MO Dereski, and a National Health and Nutrition Examination Survey), BLEEP (The Michigan Bloodspot Environmental Epidemiology Project) pilot grant from the University Research Corridor (URC) to DM Ruden. ICP-MS analyses were supported by NSF grant DUE-9952410 to K Hollacher.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
Internal Review Board (IRB) approval for this study was granted by Wayne State University, the Michigan Department of Community Health and the Michigan Neonatal Bio-bank IRBs prior to enrollment.The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Glorennec P, Bemrah N, Tard A, Robin A, Le Bot B, Bard D. Probabilistic modeling of young children’s overall lead exposure in France: integrated approach for various exposure media. Environ. Int. 2007;33(7):937–945. doi: 10.1016/j.envint.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Jakubowski M. Low-level environmental lead exposure and intellectual impairment in children – the current concepts of risk assessment. Int. J. Occup. Med. Environ. health. 2011;24(1):1–7. doi: 10.2478/s13382-011-0009-z. [DOI] [PubMed] [Google Scholar]
- 3.Christie NT, Costa M. In vitro assessment of the toxicity of metal compounds: IV. disposition of metals in cells: interactions with membranes, glutathione, metallothionein, and DNA. Biol. Trace Elem. Res. 1984;6(2):139–158. doi: 10.1007/BF02916931. [DOI] [PubMed] [Google Scholar]
- 4.Farant JP, Wigfield DC. Biomonitoring lead exposure with delta-aminolevulinate dehydratase (ALA-D) activity ratios. Int. Arch. Occup. Environ. Health. 1982;51(1):15–24. doi: 10.1007/BF00378406. [DOI] [PubMed] [Google Scholar]
- 5.Bakulski KM, Rozek LS, Dolinoy DC, Paulson HL, Hu H. Alzheimer’s disease and environmental exposure to lead: the epidemiologic evidence and potential role of epigenetics. Curr. Alzheimer Res. 2012;9(5):563–573. doi: 10.2174/156720512800617991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altmann L, Gutowski M, Wiegand H. Effects of maternal lead exposure on functional plasticity in the visual cortex and hippocampus of immature rats. Brain Res. Dev. Brain Res. 1994;81(1):50–56. doi: 10.1016/0165-3806(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 7.Wolff MS, Britton JA, Boguski L, et al. Environmental exposures and puberty in inner-city girls. Environ. Res. 2008;107(3):393–400. doi: 10.1016/j.envres.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence DA, mCcabe MJ., Jr Immunomodulation by metals. Int. Immunopharmacol. 2002;2(2–3):293–302. doi: 10.1016/s1567-5769(01)00180-1. [DOI] [PubMed] [Google Scholar]
- 9.Hirsch HV, Mercer J, Sambaziotis H, et al. Behavioral effects of chronic exposure to low levels of lead in Drosophila melanogaster. Neurotoxicology. 2003;24(3):435–442. doi: 10.1016/S0161-813X(03)00021-4. [DOI] [PubMed] [Google Scholar]
- 10.Schneider JS, Talsania K, Mettil W, Anderson DW. Genetic diversity influences the response of the brain to developmental lead exposure. Toxicol. Sci. 2014 doi: 10.1093/toxsci/kfu101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobin C, Parisi N, Schaub T, Gutierrez M, Ortega AX. delta-Aminolevulinic acid dehydratase single nucleotide polymorphism 2 and peptide transporter 2*2 haplotype may differentially mediate lead exposure in male children. Arch. Environ. Contam. Toxicol. 2011;61(3):521–529. doi: 10.1007/s00244-011-9645-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulis M, Queiros AC, Beekman R, Martin-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim. Biophys. Acta. 2013;1829(11):1161–1174. doi: 10.1016/j.bbagrm.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Tate PH, Bird AP. Effects of DNA methylation on DNA-binding proteins and gene expression. Curr. Opin. Genet. Dev. 1993;3(2):226–231. doi: 10.1016/0959-437x(93)90027-m. [DOI] [PubMed] [Google Scholar]
- 14.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33(Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 15.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18(11):6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hendrich B, Tweedie S. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 2003;19(5):269–277. doi: 10.1016/S0168-9525(03)00080-5. [DOI] [PubMed] [Google Scholar]
- 17.Arand J, Spieler D, Karius T, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS Genet. 2012;8(6):e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 19.Cingolani P, Cao X, Khetani R, et al. Intronic non-CG DNA hydroxymethylation and alternative mRNA splicing in honey bees. BMC Genomics. 2013;14(1):666. doi: 10.1186/1471-2164-14-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13(8):335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 21.Kile ml, Baccarelli A, Hoffman E, et al. Prenatal arsenic exposure and DNA methylation in maternal and umbilical cord blood leukocytes. Environ. Health Perspect. 2012;120(7):1061–1066. doi: 10.1289/ehp.1104173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ. Health Perspect. 2010;118(6):790–795. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dosunmu R, Alashwal H, Zawia NH. Genome-wide expression and methylation profiling in the aged rodent brain due to early-life Pb exposure and its relevance to aging. Mech. Ageing Dev. 2012;133(6):435–443. doi: 10.1016/j.mad.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ. Health Perspect. 2013;121(8):971–977. doi: 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna CW, Bloom MS, Robinson WP, et al. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum. Reprod. 2012;27(5):1401–1410. doi: 10.1093/humrep/des038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brubaker CJ, Dietrich KN, Lanphear BP, Cecil KM. The influence of age of lead exposure on adult gray matter volume. Neurotoxicology. 2010;31(3):259–266. doi: 10.1016/j.neuro.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jedrychowski W, Perera F, Jankowski J, et al. Gender specific differences in neurodevelopmental effects of prenatal exposure to very low-lead levels: the prospective cohort study in three-year olds. Early Hum. Dev. 2009;85(8):503–510. doi: 10.1016/j.earlhumdev.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 29.Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem. Sci. 2003;28(3):152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- 30.Sofer T, Schifano ED, Hoppin JA, Hou L, Baccarelli AA. A-clustering: a novel method for the detection of co-regulated methylation regions, and regions associated with exposure. Bioinformatics. 2013;29(22):2884–2891. doi: 10.1093/bioinformatics/btt498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petronis KR, Anthony JC. Social epidemiology, intra-neighbourhood correlation, and generalised estimating equations. J. Epidemiol. Community Health. 2003;57(11):914. doi: 10.1136/jech.57.11.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12(2):206–222. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 33.Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Du P, Zhang X, Huang CC, et al. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teschendorff AE, Marabita F, Lechner M, et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29(2):189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4(3):325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 37.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 38.Barfield RT, Kilaru V, Smith AK, Conneely KN. CpGassoc: an R function for analysis of DNA methylation microarray data. Bioinformatics. 2012;28(9):1280–1281. doi: 10.1093/bioinformatics/bts124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wessely F, Emes RD. Identification of DNA methylation biomarkers from Infinium arrays. Front. Genet. 2012;3:161. doi: 10.3389/fgene.2012.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Felsenfeld G. A brief history of epigenetics. Cold Spring Harbor Perspect. Biol. 2014;6(1) doi: 10.1101/cshperspect.a018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker DJ, Eriksson JG, Forsen T, Osmond C. Fetal origins of adult disease: strength of effects and biological basis. Int. J. Epidemiol. 2002;31(6):1235–1239. doi: 10.1093/ije/31.6.1235. [DOI] [PubMed] [Google Scholar]
- 42.Haworth KE, Farrell WE, Emes RD, et al. Combined influence of gene-specific cord blood methylation and maternal smoking habit on birth weight. Epigenomics. 2013;5(1):37–49. doi: 10.2217/epi.12.72. [DOI] [PubMed] [Google Scholar]
- 43.Hsiung DT, Marsit CJ, Houseman EA, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2007;16(1):108–114. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Aryee MJ, Padyukov L, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 2013;31(2):142–147. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun YV, Smith AK, Conneely KN, et al. Epigenomic association analysis identifies smoking-related DNA methylation sites in African Americans. Hum. Genet. 2013;132(9):1027–1037. doi: 10.1007/s00439-013-1311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilsner JR, Hall MN, Liu X, et al. Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS ONE. 2012;7(5):e37147. doi: 10.1371/journal.pone.0037147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics. 2013;5(5):487–500. doi: 10.2217/epi.13.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS ONE. 2010;5(4):e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penaloza CG, Estevez B, Han DM, Norouzi M, Lockshin RA, Zakeri Z. Sex-dependent regulation of cytochrome P450 family members Cyp1a1, Cyp2e1, and Cyp7b1 by methylation of DNA. FASEB J. 2014;28(2):966–977. doi: 10.1096/fj.13-233320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barrett JR. Children’s health. Sex-specific cognitive effects of lead. Environ. Health Perspect. 2009;117(9):A393. doi: 10.1289/ehp.117-a393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tretter L, Adam-Vizi V. Alpha-ketoglutarate dehydrogenase: a target and generator of oxidative stress. Philos. Trans. R Soc. Lond. B Biol. Sci. 2005;360(1464):2335–2345. doi: 10.1098/rstb.2005.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Padilla MA, Elobeid M, Ruden DM, Allison DB. An examination of the association of selected toxic metals with total and central obesity indices: NHANES 99–02. Int. J. Environ. Res. Public Health. 2010;7(9):3332–3347. doi: 10.3390/ijerph7093332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selenscig D, Rossi A, Chicco A, Lombardo YB. Increased leptin storage with altered leptin secretion from adipocytes of rats with sucrose-induced dyslipidemia and insulin resistance: effect of dietary fish oil. Metabolism. 2010;59(6):787–795. doi: 10.1016/j.metabol.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 56.Shen W, Wang C, Xia L, et al. Epigenetic modification of the leptin promoter in diet-induced obese mice and the effects of N-3 polyunsaturated fatty acids. Sci. Rep. 2014;4:5282. doi: 10.1038/srep05282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kile ml, Houseman EA, Baccarelli AA, et al. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics. 2014;9(5):774–782. doi: 10.4161/epi.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webber BR, Iacovino M, Choi SH, Tolar J, Kyba M, Blazar BR. DNA methylation of Runx1 regulatory regions correlates with transition from primitive to definitive hematopoietic potential in vitro and in vivo. Blood. 2013;122(17):2978–2986. doi: 10.1182/blood-2013-03-489369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang CQ, Motoda L, Satake M, et al. Runx3 deficiency results in myeloproliferative disorder in aged mice. Blood. 2013;122(4):562–566. doi: 10.1182/blood-2012-10-460618. [DOI] [PubMed] [Google Scholar]
- 60.Wolff EM, Liang G, Cortez CC, et al. RUNX3 methylation reveals that bladder tumors are older in patients with a history of smoking. Cancer Res. 2008;68(15):6208–6214. doi: 10.1158/0008-5472.CAN-07-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tian Q, Li T, Hou W, Zheng J, Schrum LW, Bonkovsky HL. Lon peptidase 1 (LONP1)-dependent breakdown of mitochondrial 5-aminolevulinic acid synthase protein by heme in human liver cells. J. Biol. Chem. 2011;286(30):26424–26430. doi: 10.1074/jbc.M110.215772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chopra P, Papale LA, White AT, et al. Array-based assay detects genome-wide 5-mC and 5-hmC in the brains of humans, non-human primates, and mice. BMC Genomics. 2014;15:131. doi: 10.1186/1471-2164-15-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang Y, Pastor WA, Shen Y, Tahiliani M, Liu DR, Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5(1):e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by mlL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.