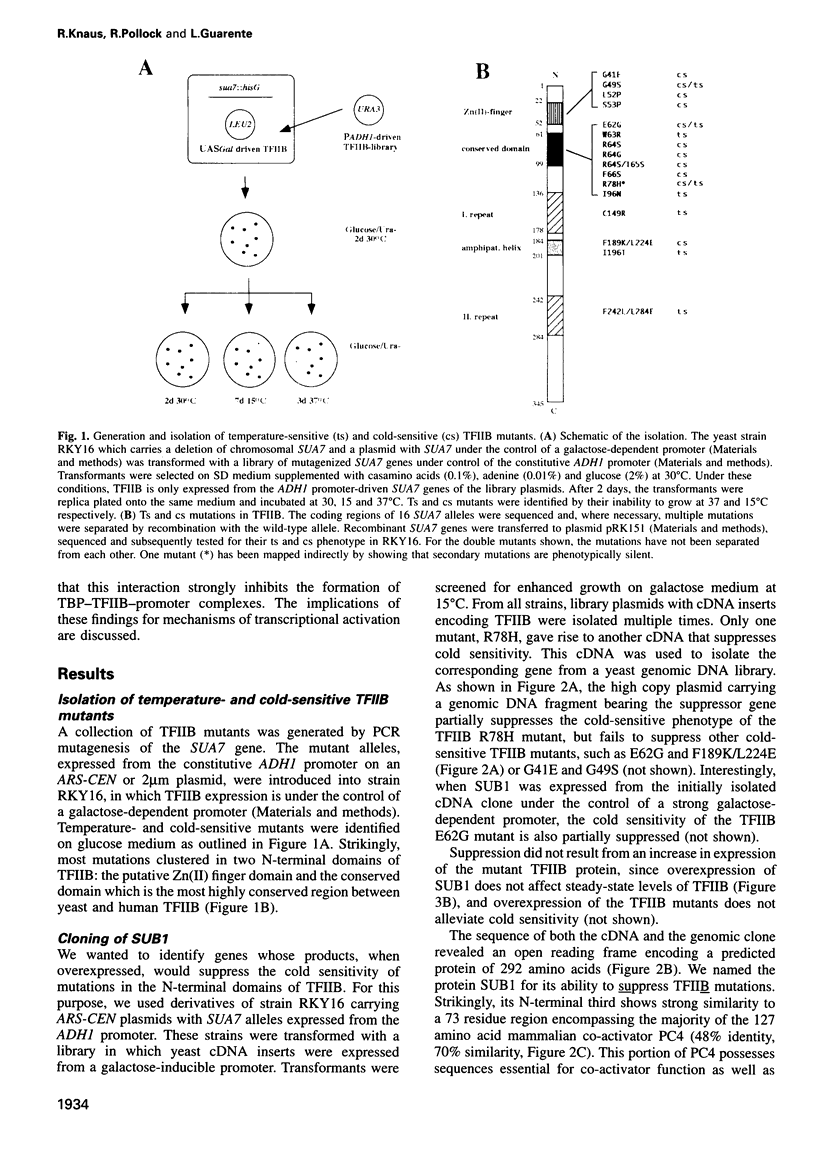
Abstract
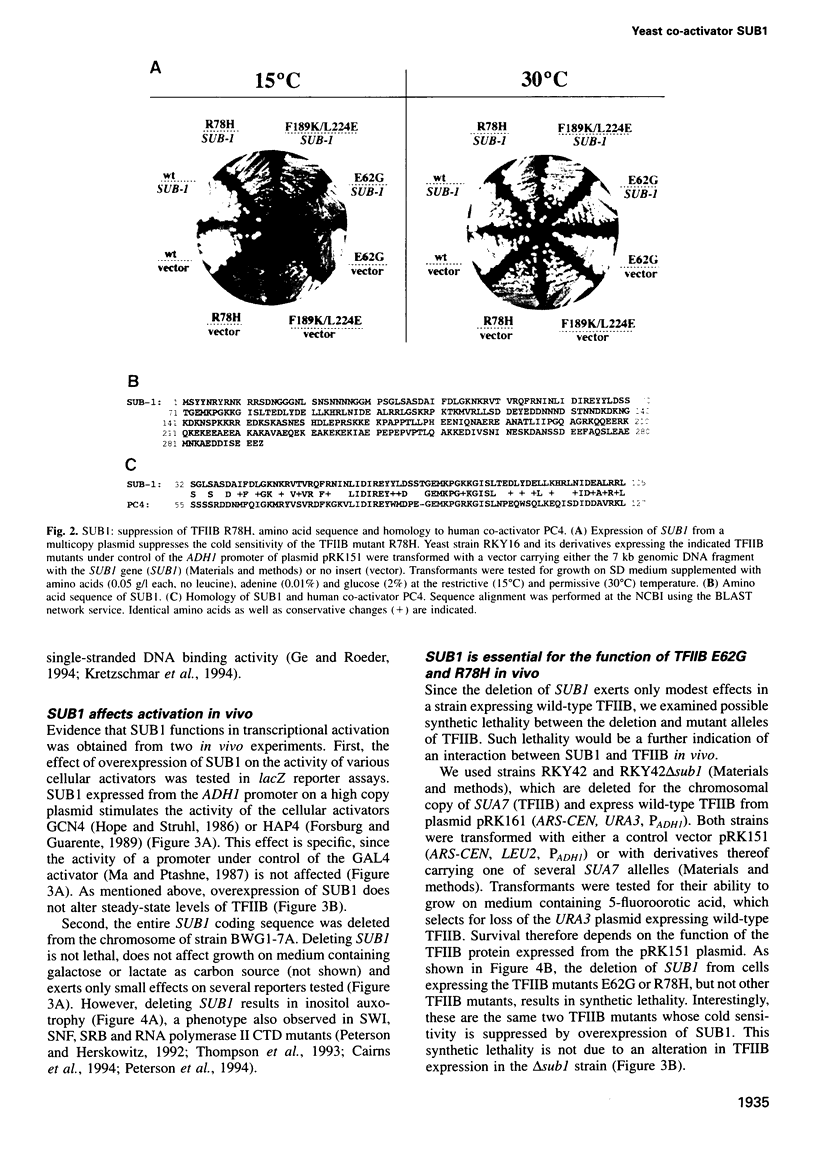
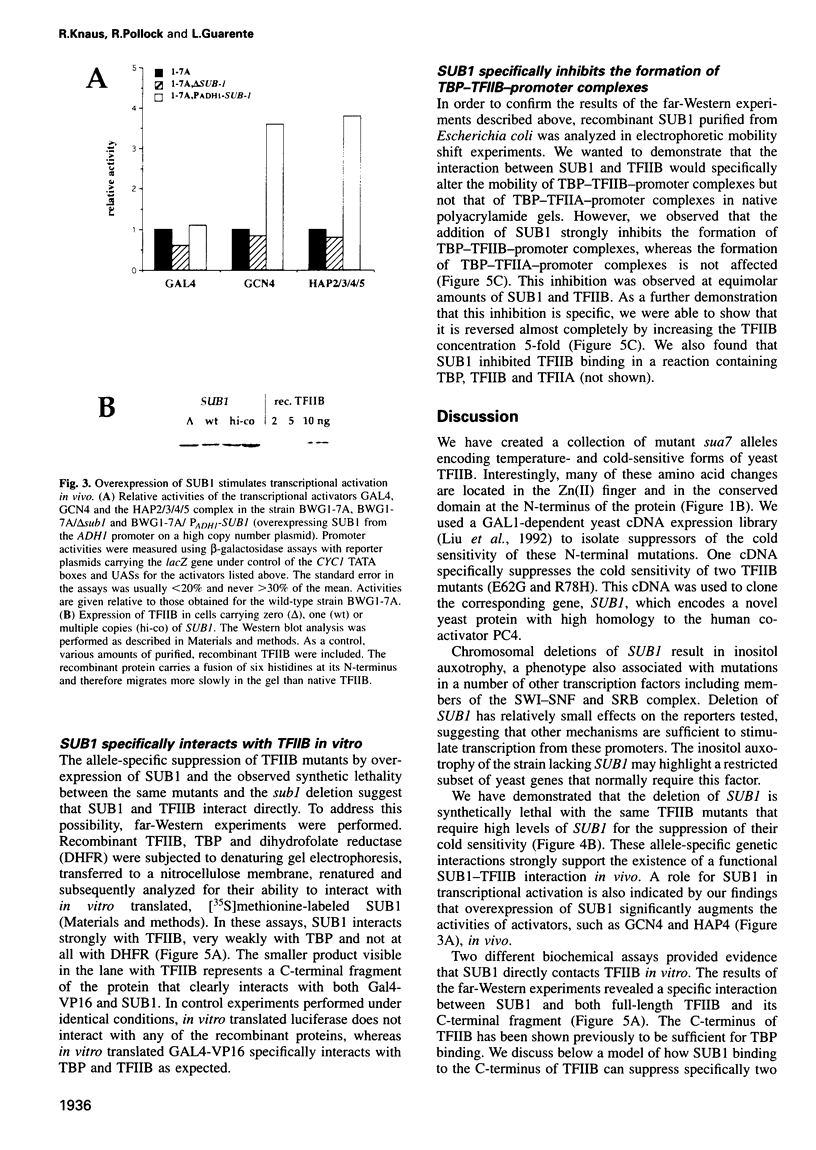
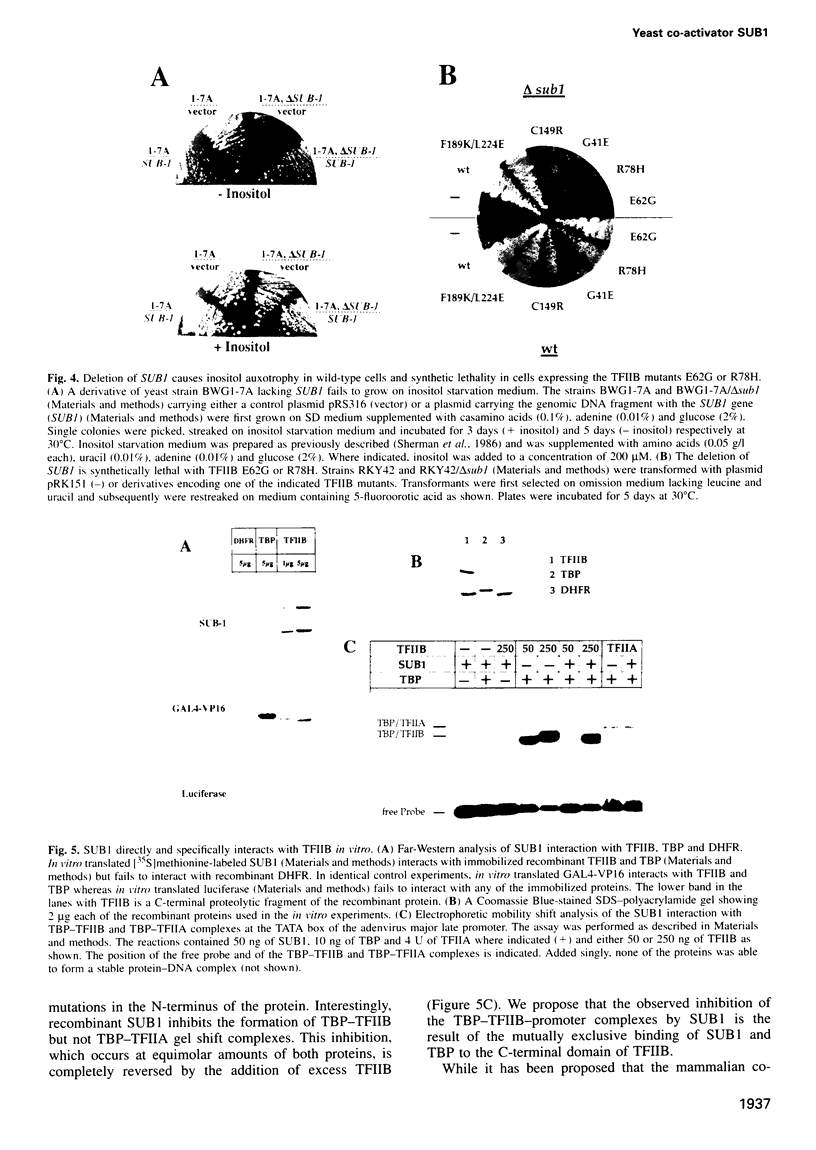
Activation of transcription in eukaryotes depends upon the interplay between transcriptional activators and general transcription factors. While direct contacts between activators and general factors have been demonstrated in vitro, an additional class of proteins, termed co-activators, is also required of transcriptional activation. Here we describe a yeast protein, SUB1, that was isolated as a suppressor of the cold-sensitive TFIIB R78H mutant. The N-terminal third of SUB1 is highly similar to the mammalian co-activator PC4. We show that increased expression of SUB1 suppresses two alleles of TFIIB (E62G, R78H) specifically and that the deletion of SUB1 is lethal in combination with these same two alleles. We show that SUB1 binds to TFIIB in vitro and that it specifically inhibits the formation of TBP-TFIIB-promoter complexes. Furthermore we show that increasing the copy number of SUB1 stimulates transcriptional activation in vivo. Based on our results and recent observations of others, we propose that SUB1 plays a role in the release of TFIIB from the transcription complex during transcription initiation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias J., Alberts A. S., Brindle P., Claret F. X., Smeal T., Karin M., Feramisco J., Montminy M. Activation of cAMP and mitogen responsive genes relies on a common nuclear factor. Nature. 1994 Jul 21;370(6486):226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- Auble D. T., Hansen K. E., Mueller C. G., Lane W. S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994 Aug 15;8(16):1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Becker D. M., Fikes J. D., Guarente L. A cDNA encoding a human CCAAT-binding protein cloned by functional complementation in yeast. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1968–1972. doi: 10.1073/pnas.88.5.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S. L., Cress W. D., Cress A., Triezenberg S. J., Guarente L. Selective inhibition of activated but not basal transcription by the acidic activation domain of VP16: evidence for transcriptional adaptors. Cell. 1990 Jun 29;61(7):1199–1208. doi: 10.1016/0092-8674(90)90684-7. [DOI] [PubMed] [Google Scholar]
- Berger S. L., Piña B., Silverman N., Marcus G. A., Agapite J., Regier J. L., Triezenberg S. J., Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992 Jul 24;70(2):251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- Cairns B. R., Kim Y. J., Sayre M. H., Laurent B. C., Kornberg R. D. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1950–1954. doi: 10.1073/pnas.91.5.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrivia J. C., Kwok R. P., Lamb N., Hagiwara M., Montminy M. R., Goodman R. H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993 Oct 28;365(6449):855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Colgan J., Ashali H., Manley J. L. A direct interaction between a glutamine-rich activator and the N terminus of TFIIB can mediate transcriptional activation in vivo. Mol Cell Biol. 1995 Apr;15(4):2311–2320. doi: 10.1128/mcb.15.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S., Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992 Feb 7;68(3):597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ewen M. E., Newsome D., Gerdes M., DeCaprio J. A., Lawrence J. B., Livingston D. M. Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev. 1994 Apr 15;8(8):869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- Eisenmann D. M., Dollard C., Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989 Sep 22;58(6):1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L., Guarente L. Identification and characterization of HAP4: a third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989 Aug;3(8):1166–1178. doi: 10.1101/gad.3.8.1166. [DOI] [PubMed] [Google Scholar]
- Fujita T., Nolan G. P., Liou H. C., Scott M. L., Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993 Jul;7(7B):1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- Ge H., Roeder R. G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994 Aug 12;78(3):513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Hoey T., Thut C. J., Admon A., Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993 Nov 5;75(3):519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Goodrich J. A., Tjian R. TBP-TAF complexes: selectivity factors for eukaryotic transcription. Curr Opin Cell Biol. 1994 Jun;6(3):403–409. doi: 10.1016/0955-0674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Guarente L., Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983 Apr;32(4):1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Buratowski S., Sharp P. A., Guarente L. Isolation of the gene encoding the yeast TATA binding protein TFIID: a gene identical to the SPT15 suppressor of Ty element insertions. Cell. 1989 Sep 22;58(6):1173–1181. doi: 10.1016/0092-8674(89)90515-1. [DOI] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., Brown S. A., Clark C. D., Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992 Dec;6(12A):2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Horiuchi J., Silverman N., Marcus G. A., Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995 Mar;15(3):1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbalzano A. N., Kwon H., Green M. R., Kingston R. E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994 Aug 11;370(6489):481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- Kelleher R. J., 3rd, Flanagan P. M., Kornberg R. D. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990 Jun 29;61(7):1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994 May 20;77(4):599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Koleske A. J., Young R. A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994 Mar 31;368(6470):466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Kaiser K., Lottspeich F., Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994 Aug 12;78(3):525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- Kwok R. P., Lundblad J. R., Chrivia J. C., Richards J. P., Bächinger H. P., Brennan R. G., Roberts S. G., Green M. R., Goodman R. H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994 Jul 21;370(6486):223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- Lee J. W., Ryan F., Swaffield J. C., Johnston S. A., Moore D. D. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature. 1995 Mar 2;374(6517):91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Green M. R. Mechanism of action of an acidic transcriptional activator in vitro. Cell. 1991 Mar 8;64(5):971–981. doi: 10.1016/0092-8674(91)90321-o. [DOI] [PubMed] [Google Scholar]
- Lin Y. S., Ha I., Maldonado E., Reinberg D., Green M. R. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991 Oct 10;353(6344):569–571. doi: 10.1038/353569a0. [DOI] [PubMed] [Google Scholar]
- Liu H., Krizek J., Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992 Nov;132(3):665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Marcus G. A., Silverman N., Berger S. L., Horiuchi J., Guarente L. Functional similarity and physical association between GCN5 and ADA2: putative transcriptional adaptors. EMBO J. 1994 Oct 17;13(20):4807–4815. doi: 10.1002/j.1460-2075.1994.tb06806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Dingwall A., Scott M. P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):2905–2908. doi: 10.1073/pnas.91.8.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992 Feb 7;68(3):573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- Pinto I., Ware D. E., Hampsey M. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell. 1992 Mar 6;68(5):977–988. doi: 10.1016/0092-8674(92)90040-j. [DOI] [PubMed] [Google Scholar]
- Pinto I., Wu W. H., Na J. G., Hampsey M. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem. 1994 Dec 2;269(48):30569–30573. [PubMed] [Google Scholar]
- Piña B., Berger S., Marcus G. A., Silverman N., Agapite J., Guarente L. ADA3: a gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993 Oct;13(10):5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon D., Weil P. A. Immunopurification of yeast TATA-binding protein and associated factors. Presence of transcription factor IIIB transcriptional activity. J Biol Chem. 1993 Jul 25;268(21):15325–15328. [PubMed] [Google Scholar]
- Pugh B. F., Tjian R. Mechanism of transcriptional activation by Sp1: evidence for coactivators. Cell. 1990 Jun 29;61(7):1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- Reese J. C., Apone L., Walker S. S., Griffin L. A., Green M. R. Yeast TAFIIS in a multisubunit complex required for activated transcription. Nature. 1994 Oct 6;371(6497):523–527. doi: 10.1038/371523a0. [DOI] [PubMed] [Google Scholar]
- Roberts S. G., Green M. R. Activator-induced conformational change in general transcription factor TFIIB. Nature. 1994 Oct 20;371(6499):717–720. doi: 10.1038/371717a0. [DOI] [PubMed] [Google Scholar]
- Roberts S. G., Ha I., Maldonado E., Reinberg D., Green M. R. Interaction between an acidic activator and transcription factor TFIIB is required for transcriptional activation. Nature. 1993 Jun 24;363(6431):741–744. doi: 10.1038/363741a0. [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D. Structure and function of the yeast URA3 gene. Differentially regulated expression of hybrid beta-galactosidase from overlapping coding sequences in yeast. J Mol Biol. 1983 Nov 15;170(4):883–904. doi: 10.1016/s0022-2836(83)80193-4. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N., Agapite J., Guarente L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffield J. C., Melcher K., Johnston S. A. A highly conserved ATPase protein as a mediator between acidic activation domains and the TATA-binding protein. Nature. 1995 Mar 2;374(6517):88–91. doi: 10.1038/374088a0. [DOI] [PubMed] [Google Scholar]
- Thompson C. M., Koleske A. J., Chao D. M., Young R. A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993 Jul 2;73(7):1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Turcotte B., Guarente L. HAP1 positive control mutants specific for one of two binding sites. Genes Dev. 1992 Oct;6(10):2001–2009. doi: 10.1101/gad.6.10.2001. [DOI] [PubMed] [Google Scholar]
- Winston F., Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992 Nov;8(11):387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- Zawel L., Kumar K. P., Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995 Jun 15;9(12):1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]