Abstract
Maintaining genome integrity and transmission of intact genomes is critical for cellular, organismal, and species survival. Cells can detect damaged DNA, activate checkpoints, and either enable DNA repair or trigger apoptosis to eliminate the damaged cell. Aberrations in these mechanisms lead to somatic mutations and genetic instability, which are hallmarks of cancer. Considering the long history of host-microbe coevolution, an impact of microbial infection on host genome integrity is not unexpected, and emerging links between microbial infections and oncogenesis further reinforce this idea. In this review, we compare strategies employed by viruses, bacteria, and parasites to alter, subvert, or otherwise manipulate host DNA damage and repair pathways. We highlight how microbes contribute to tumorigenesis by directly inducing DNA damage, inactivating checkpoint controls, or manipulating repair processes. We also discuss indirect effects resulting from inflammatory responses, changes in cellular metabolism, nuclear architecture, and epigenome integrity, and the associated evolutionary tradeoffs.
Introduction
Microbial infections mount hostile attacks on host signaling pathways and cellular integrity. Perhaps nothing is as challenging to host cells as pathogen attacks on the host genome and processes that protect genome integrity. It is inevitable that in some contexts pathogen-induced genomic damage contributes to tumorigenesis, and as such, 20% of all human cancers are causally related to pathogenic agents (de Martel et al., 2012; Zur Hausen, 2009). Pathogens employ a plethora of strategies to harness or inactivate the DNA damage response (DDR), a host mechanism that protects genome integrity, and circumvent barriers imposed by DNA damage checkpoints (reviewed by Guerra et al., 2011; Turnell and Grand, 2012; Weitzman et al., 2010).
In this review, we compare the strategies employed by viruses, bacteria, and parasites to affect the host’s DNA damage and repair pathways. We highlight specific direct and indirect mechanisms involved as well as the genotoxic outcomes of these host-pathogen interactions (see Figure 1). The evolutionary tradeoffs resulting from the impact of host-microbe interactions on genome integrity are also discussed. Studies of viruses, bacteria, and parasites highlight the importance of host cellular pathways that maintain genome and epigenome integrity and provide insights into therapeutic approaches to pathogen-induced tumorigenesis.
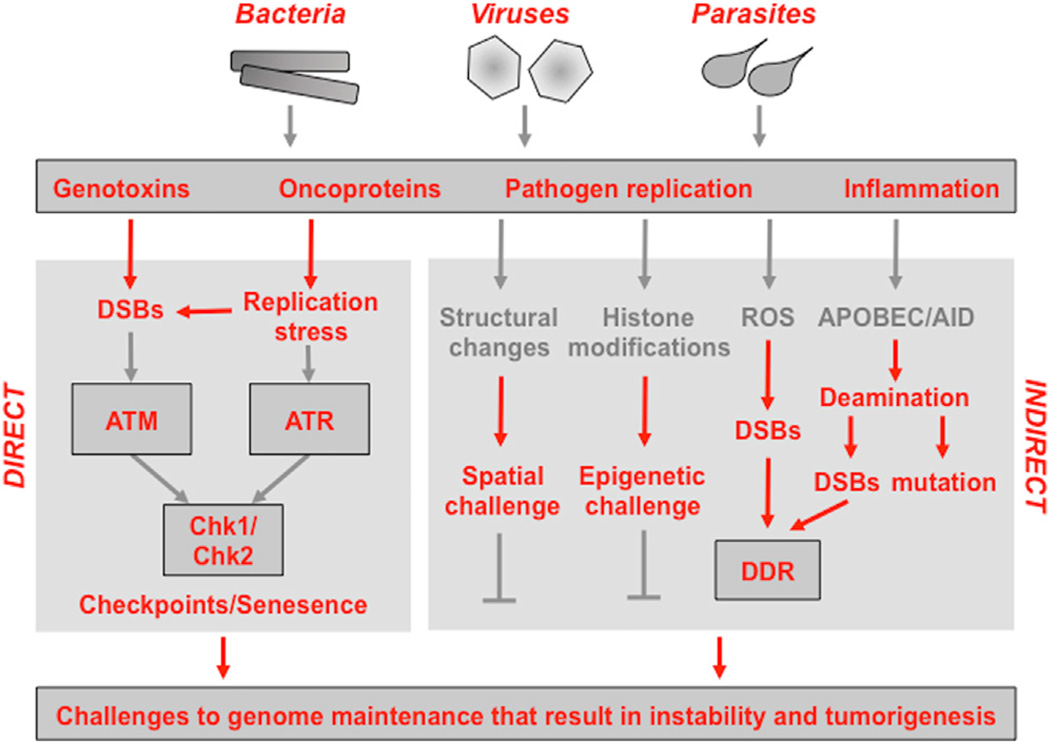
Figure 1. Interactions between Microbial Pathogens and Pathways that Maintain Host Genome Integrity.
The schematic illustrates how infectious agents (viruses, prokaryotic bacteria, or eukaryotic parasites) associated with tumorigenesis converge on common strategies that challenge DNA integrity and genome stability. These include direct effects of pathogen-encoded genotoxins and oncoproteins as well as indirect outcomes of microbial infection and inflammatory responses. Please see the main text for detailed explanations of the host pathways that are impacted by the direct and indirect activities of microbial pathogens.
Genome Integrity and Tumorigenesis
While it is generally accepted that most cancers are genetically unstable, the origins of this instability and the molecular mechanisms responsible for inducing tumorigenic mutations and rearrangements are numerous and unclear in several cases. The resolution and scale of genetic instability varies considerably, from subtle sequence changes involving base substitutions, deletions, or insertions of a few nucleotides to aneuploidy and gross alterations in chromosome structure (Lengauer et al., 1998). Picking apart the role of specific initiators and drivers critical for tumor initiation remains a nontrivial challenge. Microbial infection can influence cellular functions that represent classical hallmarks of cancer, including stimulating proliferative growth, evading growth suppression, and preventing apoptosis, as well as emerging hallmarks, such as altered cellular energetics and avoidance of immune destruction (Hanahan and Weinberg, 2011; see also review by Mesri et al., 2014 in this issue). Infectious agents can act as direct carcinogens or can indirectly contribute to tumorigenesis through induction of chronic inflammation, leading to either localized mutational changes and/or global chromosomal defects, which are features of the cancer genomic landscape.
DNA Damage and Repair
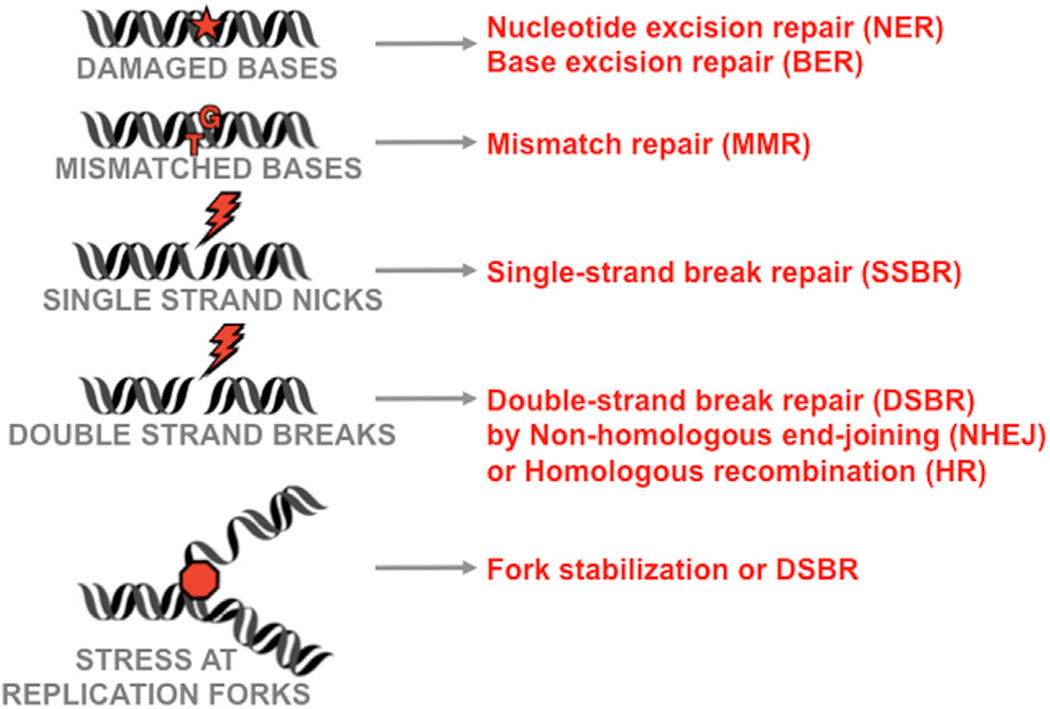
Repair pathways recognize and restore a range of DNA anomalies, including mismatches, abnormal bases, stalled replication forks, single-stranded DNA nicks, and double-stranded DNA breaks (DSBs) (Friedberg et al., 2006) (see Figure 2). The mismatch repair (MMR), base excision repair (BER), and nucleotide excision repair (NER) pathways respond to specific lesions in DNA residues. DSBs present a particularly dangerous lesion and are repaired by two principal pathways: the error-prone nonhomologous end-joining (NHEJ) pathway functions in all phases of the cell cycle, while the high-fidelity homologous recombination (HR) pathway requires a template for repair and utilizes available sister chromatids during the S and G2 phases of the cell cycle (see Figure 2). In all cases, sensor proteins recognize the damage and activate signaling cascades to recruit mediators for repair and induction of proliferation checkpoints (reviewed by Ciccia and Elledge, 2010). DNA damage activates protein kinases, including the ataxia telangiectasia mutated protein kinase (ATM), the ATM and Rad3-related kinase (ATR), the DNA-dependent protein kinase (DNA-PK), and the downstream checkpoint effector kinases Chk1 and Chk2 (Ciccia and Elledge, 2010) (see Figure 3). In addition to phosphorylation, other posttranslational modifications (PTMs) are critical for the cellular DNA damage and repair machinery (reviewed by Polo and Jackson, 2011). Covalent, but reversible, modification by ubiquitin and SUMO recently emerged as crucial components of the cellular DDR (Jackson and Durocher, 2013). The DDR network is established through recognition of these PTMs by high-affinity binding modules, such as BRCT and FHA domains that bind phosphorylated epitopes (reviewed by Reinhardt and Yaffe, 2013), Tudor domains that bind methylated sites, ubiquitin-binding domains (UBDs), and SUMO-interacting motifs (SIMs) (Polo and Jackson, 2011). PTMs affect localization, protein-protein interactions, and protein activity to regulate damage recognition and repair processes.
Figure 2. Common DNA Damage Intermediates and Repair Pathways.
Although much of this review is focused on double-strand break repair pathways, many of these other repair processes can be impacted by microbial infections.
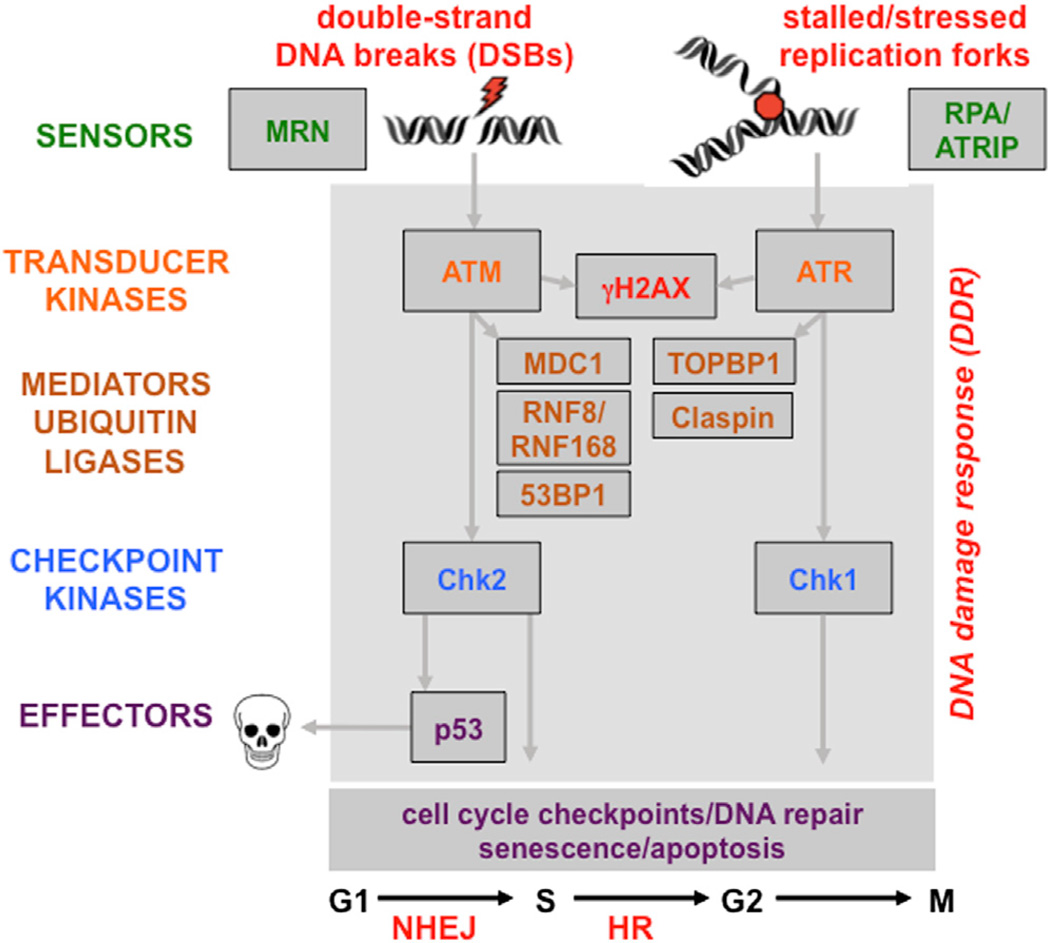
Figure 3. The Network of DDR Signaling Pathways.
Sensors recognize structures at sites of DNA damage induced by DNA breaks and replication stress. Signal is transmitted by transducer kinases, and mediator proteins are recruited to amplify the signal through ubiquitin modifications and checkpoint kinases. The effectors of the pathway mediate cellular processes required for the maintenance of genome integrity.
Temporal regulation of damage recognition and repair is crucial to maintain genome integrity, and PTMs serve to recruit repair factors to damage sites. The ATM kinase is activated by DSBs, whereas ATR responds to single-strand DNA (ssDNA) resulting from resected DNA at breaks or stalled replication forks (Maréchal and Zou, 2013). Each DNA damage lesion has a specific damage sensor. Specifically, the Mre11/Rad50/Nbs1 (MRN) complex senses DSBs, whereas ATR is recruited to ssDNA bound by RPA through its cofactor ATRIP and is activated by binding proteins such as TOPBP1 (see Figure 3). Phosphorylation of the histone variant H2AX at DSB sites forms γH2AX, a marker of breaks. Phosphorylated γH2AX is bound by the mediator of checkpoints, MDC1, which is itself phosphorylated and then recognized by the FHA domain of the E3 ligase RNF8. Protein ubiquitination at DSBs signals the recruitment of RNF168, an E3 ligase with ubiquitin-binding domains. RNF8/ RNF168-dependent ubiquitination promotes recruitment and retention of repair factors, and deubiquitinating proteins dynamically regulate ubiquitin chains at damage sites. Numerous DNA repair factors are SUMOylated in the DDR, and recognition by SIM binding promotes interactions at repair sites or can lead to turnover by SUMO-targeted ubiquitin ligases (Polo and Jackson, 2011). Acetylation also plays an important role in DSB repair, and the acetylase Tip60 is recruited to modify histones and DDR proteins (Price and D’Andrea, 2013). All these critical orchestrating events for the DDR can be perturbed by microbial infections, with detrimental consequences for host genomic integrity.
Pathogen Contributions to Tumorigenesis
It is recognized that infection is a major contributor to cancer, with certain infectious agents classified as direct human carcinogens (de Martel et al., 2012). Some pathogens encode proteins required to maintain their own genetic integrity (e.g., bacteria), whereas others rely extensively on the host machinery (e.g., viruses). Pathogens impact host cell genomes directly through genotoxins and oncoproteins that induce changes to cellular DNA and by impairing DNA repair mechanisms (see Figure 1). Genomic integrity is also impacted indirectly as a result of pathogen replication and induced inflammation. Microbial pathogens manipulate cellular environments to create conditions conducive to their own replication; they alter cellular structures, divert signaling pathways, modify host epigenetic programs, and influence metabolism. The host DDR cascades activated by pathogens serve to combat infection and can therefore be viewed as a branch of cellular defense. These are counteracted by numerous microbial strategies, with devastating consequences for host cells and the infected organism. Mining host and pathogen genomes provides insights into the intricate mechanisms that contribute to this dynamic interface and coevolution of defense and counterattack strategies in tumorigenesis.
Viruses
Viruses are the major infectious contributors to cancers. The history of tumor virology and in-depth analysis of viral oncoproteins has been covered in previous reviews (Howley and Livingston, 2009; Moore and Chang, 2010; Zur Hausen, 2009). A number of viruses are associated with human cancer: (1) Epstein-Barr virus (EBV) is associated with Burkitt lymphoma and Hodgkin’s lymphoma and is detected in nasopharyngeal carcinoma; (2) hepatitis B virus (HBV) and hepatitis C virus (HCV) are major etiological factors for hepatocellular carcinoma and have been associated with non-Hodgkin lymphoma; (3) human T-lymphotrophic virus 1 (HTLV-1) is involved in adult T cell leukemia; (4) high-risk human papillomaviruses (HPVs) cause cervical carcinomas and are prevalent in other anogenital cancers and head and neck cancers; (5) Kaposi’s sarcoma-associated herpesvirus (KSHV) is the causative agent of Kaposi’s sarcoma, primary effusion lymphoma, and multicentric Castleman’s disease; and (6) Merkel cell polyomavirus (MCV) is implicated with Merkel cell carcinoma. These are considered directly carcinogenic in humans (except for MCV) but are unlikely to be the only viral agents that contribute toward tumorigenesis. The oncogenic potential of these viruses is unleashed over long periods, due to accumulated mutations and genome instability. Some virus genomes can persist in tumors in a latent state, where there is no active virus replication or virion production. For some tumor viruses, the viral genome becomes integrated into the host genome (e.g., HBV, HTLV-1, HPV, and MCV), whereas the herpesvirus genomes (EBV and KSHV) are typically maintained as latent episomal genomes. Viral proteins act as direct oncogenes required to maintain the transformed phenotype as well as indirect effectors that promote cancer through infection and inflammation. Recent genomic sequencing approaches to cancer have identified viruses associated with human tumors (Feng et al., 2008), located integration sites, and defined viral impacts on cellular gene expression profiles (Tang et al., 2013). Some viruses that are not directly implicated as causative agents in human cancer also target common molecular pathways relevant to transformation, and transient inactivation of essential tumor suppressors could theoretically facilitate the accumulation of transforming genetic lesions (Niller et al., 2011). Viral infection can cause transformation of nonpermissive cell types and induce tumors in experimental animal models. The links between viruses and DNA damage pathways in cancer highlighted how viruses contribute to the complex multistep process of transformation (McFadden and Luftig, 2013; Turnell and Grand, 2012; Wallace and Galloway, 2014). In addition to the direct effects of transforming viral oncoproteins, infection can also indirectly affect tumor suppression and alter epigenetic regulators of cellular function. Oncogene expression (both viral and cellular) induces replicative stress and DDR activation, which lead to activation of cell-cycle checkpoints, p53-mediated apoptosis, and senescence; these barriers must be overcome for tumorigenesis (McFadden and Luftig, 2013).
Bacteria
The best-characterized example of bacterial contribution to cancer is Heliobacter pylori infection, which is well established as a major risk factor for gastritis leading to gastric cancer (reviewed by Touati, 2010; see review by Hatakeyama, 2014 in this issue). Other bacterial infections have not been classified as direct human carcinogens but may result in genomic alterations and transformation associated with human cancer (Touati, 2010; Vogelmann and Amieva, 2007). Infections can be mutagenic as a direct result of cytotoxins or can cause mutations indirectly as a consequence of chronic inflammation. Furthermore, recent studies in germfree mice have demonstrated the tumor-promoting effects of the microbiota on cancer progression in a range of organs, suggesting that antibiotics can reduce tumor burden in human patients. Changes in the complex community of microbes that inhabit the gastrointestinal tract (the gut microbiome) have been shown to contribute to tumorigenesis in inflammation-driven colon cancer (Zackular et al., 2013; see also review by Sears and Garrett, 2014 in this issue). Another example is gallbladder cancer, which is associated with chronic Salmonella typhi infections (Samaras et al., 2010). Mucosa-associated lymphoid tissue (MALT) lymphomas can be caused by uncontrolled adaptive immune responses upon chronic bacterial infections, including H. pylori and Campylobacter jejuni (Guidoboni et al., 2006). Bacterial infections may contribute to cancer progression in a number of complex ways, such as challenges to the integrity of epithelium and the intestinal barrier, effects on host-microbiota interactions and bacterial communities, and chronic inflammation.
Parasites
The association between parasite infection and cancer is less clear, although there is some emerging clinical and epidemiological evidence. The flagellate Trichomonas vaginalis has been linked to cervical and prostate cancer (Sutcliffe, 2010). Malaria was linked to development of endemic Burkitt lymphoma (eBL) by Denis Burkitt in Sub-Saharan Africa in the 1960’s, and malaria antibodies have been detected in cancer patients. The oncogenic herpes virus EBV contributes to the pathogenesis of eBL, and it was suggested that P. falciparum can disturb the immune system in young children by expanding the B cell pool and reactivating latent EBV (Bouvard et al., 2012). The related apicomplexan Toxoplasma gondii has been associated with some adenomas and lymphomas (Zhang et al., 2002). The clearest direct links between parasites and tumorigenesis are the apicomplexan parasites Theileria annulata, Theileria parva, and Cryptosporidium parvum. C. parvum infection causes diarrhea and has been implicated in gastrointestinal carcinoma in an experimental mouse model (Certad et al., 2010). Although the mechanism is unknown, recent studies demonstrated gastrointestinal neoplasia and adenoma formation in the gut in immunocompromised mice infected with C. parvum (Benamrouz et al., 2012). Theileria spp. infect leukocytes in bovine hosts and induce a transformed lymphoproliferative disease with features of cancer. Many studies have described host cell signaling pathways that contribute to Theileria-induced transformation, including activation of host c-Jun and Myc oncogenes (Chaussepied and Langsley, 1996; Shiels et al., 2006), and the importance of these pathways has been demonstrated in immunocompromised mouse models (Lizundia et al., 2006).
Pathogen Oncoproteins and Genotoxins and Their Intersection with DDR Mechanisms
Manipulation of DDR Signaling by Viral Infections
Viruses activate DDR signaling pathways, as well as systematically dismantle aspects of the DDR network, through multiple strategies (reviewed by Turnell and Grand, 2012) (Figure 4). Virus infection activates DDR signaling through both ATM and ATR kinases. The extent to which damage-signaling cascades activated by viruses overlap with those observed in response to DSBs or replication stress, or whether there are unique pathogen-directed substrates, remains to be determined. One feature that distinguishes viruses from other microbial infections is that their genetic material is directly released into the infected cell, resulting in a conflict with the cellular genome, especially for DNA viruses that replicate in the nucleus (Weitzman et al., 2010). These foreign genomes could be recognized as DNA damage, although in most viral systems it has not been conclusively determined whether the viral genome itself is sufficient to trigger the cellular responses. Viral replication intermediates can activate the DDR, and viral replication proteins can also directly induce damage to host genomes. In the case of HPV, replication initiated from integrated HPV origins induces a damage response that may promote genome instability (Kadaja et al., 2009), and the HPV-E1 replication protein alone activates the DDR (Fradet-Turcotte et al., 2011; Sakakibara et al., 2011). The DDR can also be activated by virus infection as a result of the aberrant entry into S phase, by replicative stress, directly by viral oncoproteins, and indirectly through reactive oxygen species (ROS) production. Viruses often overcome the inhibitory effects of DDR and the checkpoints that they have activated. This is seen for HPV, where the E6 and E7 oncoproteins that are the primary viral factors responsible for initiation and progression of cervical cancer contribute to many functions that combine to subvert cell-cycle checkpoints, induce genomic instability, and inhibit apoptosis (reviewed by Moody and Laimins, 2010; Wallace and Galloway, 2014).
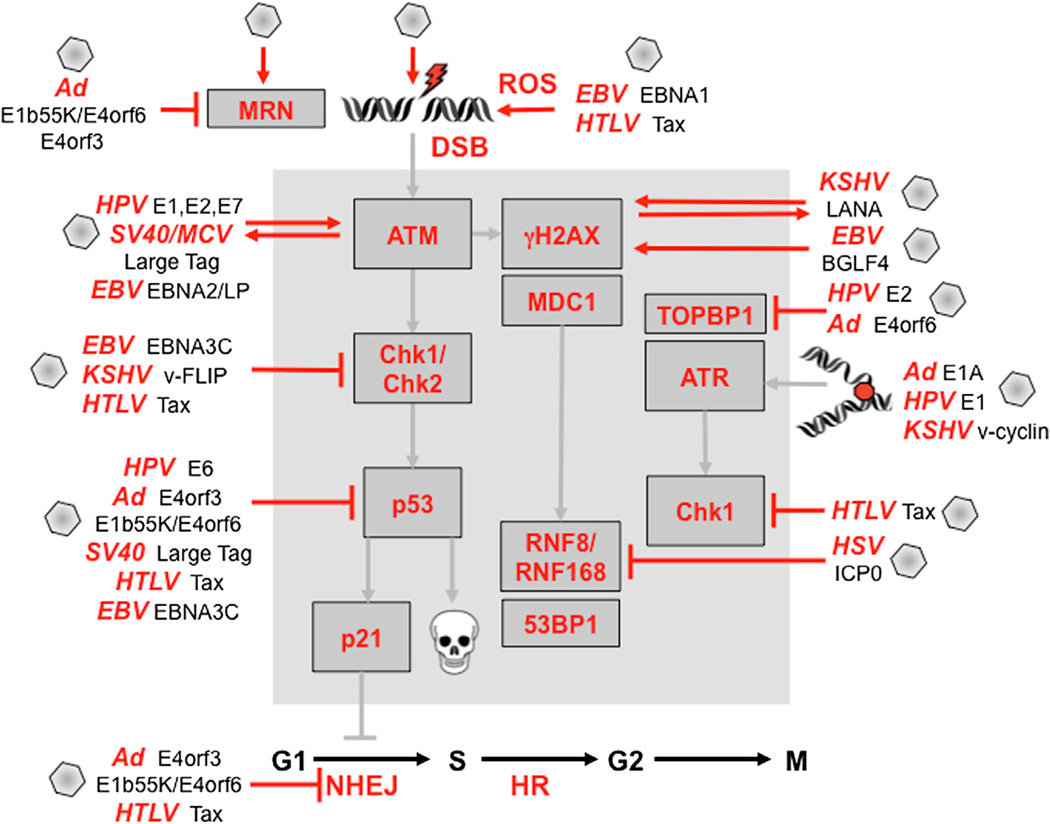
Figure 4. Viruses and Viral Products that Target the DNA Damage and Repair Pathways.
Viral infection and viral gene products (shown outside the gray boxes) activate DDR signaling (shown within the gray boxes) directly or indirectly through induction of ROS and replication stress. Viral oncoproteins activate and are acted upon by damage kinases, as indicated by two-way arrows. Viruses act at every stage of the DDR pathway to disrupt signaling checkpoints and repair processes, as shown. Many of these have been shown to contribute to the process of cellular transformation (see the main text for details and references).
Human herpesviruses activate aspects of cellular damage signaling that can promote efficient lytic replication (Lilley et al., 2005; Wilkinson and Weller, 2004). Herpesviruses are also characterized by their ability to establish latency, where viral gene expression from episomal genomes is silenced by cellular chromatinization. Viral oncoproteins expressed by the latency program of some transforming herpesviruses can also induce a DDR. For example, EBV infection of B cells in culture induces transient activation of ATM signaling as a result of cellular hyper-proliferation driven by early latent oncoproteins (Nikitin et al., 2010). The related KSHV also elicits oncogene-induced senescence via a virus-encoded cyclin (Koopal et al., 2007). Growth suppressive consequences of DDR activation by viruses can be limiting for viruses. During latent infection by herpesviruses, outgrowth of immortalized cells is only achieved in rare circumstances where senescence is overcome via countermea-sures by virus-encoded latent proteins (Leidal et al., 2012b; Nikitin and Luftig, 2012). For KSHV, it is observed that viral oncogene expression during early stages of infection activates ATM signaling, but the DDR is downregulated in advanced Kaposi’s sarcoma tumors (Koopal et al., 2007). Oncogene-induced senescence is overcome by EBV proteins EBNA3C (Nikitin et al., 2010) and LMP1 (Yang et al., 2000) and by KSHV protein v-FLIP (Leidal et al., 2012a). The efficiency of B cell immortalization by EBV infection can be enhanced by inhibition of the ATM and Chk2 kinases (Nikitin et al., 2010).
Viruses manipulate the DDR network at multiple points, blocking signaling and checkpoints at the step of DNA damage sensors, the damage kinases, the checkpoint kinases, and the effector molecules (Figure 4). Early adenovirus (Ad) proteins prevent activation of ATM and ATR by degrading and mislocalizing the MRN complex (Carson et al., 2003; 2009). Downstream checkpoint kinases are targeted by the HTLV-Tax oncoprotein that binds and inactivates both Chk1 and Chk2 (Boxus et al., 2008) and by the EBV-EBNA3C protein that inactivates the Chk2 effector kinase (Choudhuri et al., 2007). During herpes simplex virus (HSV) infection, recruitment of repair factor to sites of incoming virus genomes is blocked by degradation of RNF8 and RNF168 by the viral ICP0 ubiquitin ligase (Lilley et al., 2011; Lilley et al., 2010). Viral targeting of the DNA-PK will have a direct effect on the ability of the cell to repair breaks through NHEJ. DNA-PK is targeted by Ad-E4 proteins (Boyer et al., 1999) and the Tax oncoprotein of HTLV-1 (Durkin et al., 2008; Ramadan et al., 2008). Ad oncoproteins prevent both DDR signaling and processing of the virus genome by degrading the MRN complex (Carson et al., 2003; Stracker et al., 2002), as well as other repair factors such as TOPBP1, and DNA ligase IV (Turnell and Grand, 2012).
Manipulation of DDR Signaling by Bacterial Infections
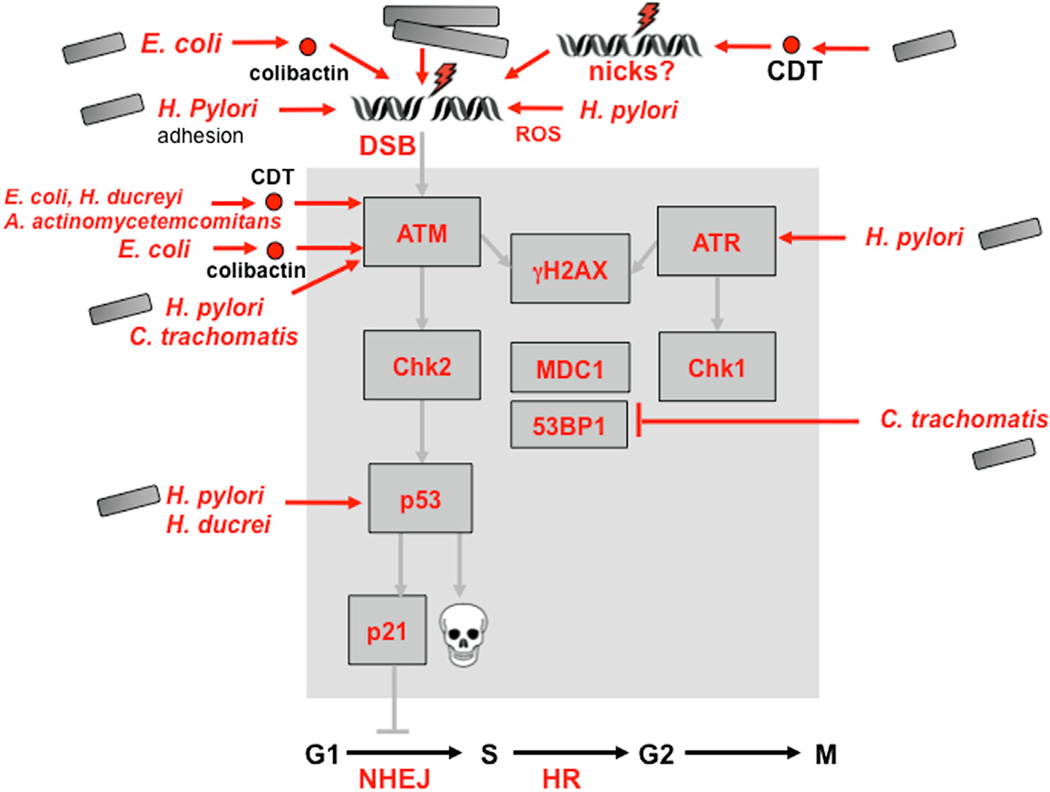
Several pathogenic bacterial strains have been demonstrated to induce DSBs and the DDR (Figure 5). H. pylori infection of human gastric epithelial cells induces DNA damage accompanied by ATM and ATR signaling to γH2AX, Chk1, and Chk2, checkpoint activation, and apoptosis (Jang et al., 2012; Toller et al., 2011). Recently, Chlamydia infections were also reported to induce DSB hallmarks, including γH2AX (Chumduri et al., 2013). In addition to activating aspects of the DDR, bacteria can also block further downstream steps, as seen with Chlamydia. DSBs induced by Chlamydia trachomatis infection fail to recruit repair proteins such as 53BP1, and the Chk1/ Chk2 kinases are not activated (Chumduri et al., 2013). Bacterial infections can also impact repair pathways: H. pylori infection leads to inhibition of nucleotide repair by downregulation at both the gene and protein level of factors involved in MMR and BER (Machado et al., 2010).
Figure 5. Bacterial Infections and Secreted Products Impact Many Aspects of the Eukaryotic Damage Response.
Damage is induced by bacterial cell adhesion, genotoxins, and production of ROS. There is also a limited impact on downstream signaling, which affects recruitment of damage factors and, potentially, repair processes (see the main text for details and references).
The effects of chronic inflammation may also be augmented by bacterial toxins that directly induce DNA damage (reviewed by Guerra et al., 2011). Cytolethal distending toxins (CDTs) are genotoxins produced by many clinically important bacterial pathogens (Jinadasa et al., 2011) that can induce cell-cycle arrest and apoptosis in a broad range of mammalian cell lines (Guerra et al., 2011). The active subunit of CDT is a phosphodi-esterase that can initiate DSBs and a DDR similar to that elicited by ionizing radiation (Alaoui-El-Azher et al., 2010). This was demonstrated for CDT from Aggregatibacter actinomycetemcomitans (Alaoui-El-Azher et al., 2010), Haemophilus ducreyi (Frisan et al., 2003), and Escherichia coli (Fedor et al., 2013). The active subunit CdtB translocates to the host nucleus, and its DNase activity may contribute to DNA damage and carcinogenic potential (Elwell and Dreyfus, 2000; Nesić et al., 2004). DSBs in human cells transiently infected with commensal and extraintestinal pathogenic E. coli may be due to colibactin, another toxin encoded by a widely distributed genomic island that activates the ATM-Chk2 signaling pathway and the G2/M checkpoint (Nougayrède et al., 2006). CDT and colibactin can be considered bacterial genotoxins that directly induce DNA damage and genomic instability (Cuevas-Ramos et al., 2010; Nesić et al., 2004). Chronic exposure to sublethal doses of CDT increases mutations, chromosomal aberrations, and anchorage-independent growth while decreasing DDR activation (Guidi et al., 2013). The H. hepaticus CDT promotes dysplasia in a mouse model of hepatocarcinogenesis (Ge et al., 2007), and E. coli colibactin was functionally linked to colorectal cancer development in mice (Arthur et al., 2012). Most of these effects have been reported using high doses of CDT, but recent experiments with lower doses of E. coli CDT suggested that damage signaling is caused by induction of ssDNA nicks that are subsequently converted to DSBs during S phase (Fedor et al., 2013). DNA breaks and recruitment of repair factors have also been reported for H. pylori in a manner that depends upon direct pathogen contact with human cells but is independent of known virulence factors (Toller et al., 2011). Together, these observations suggest molecular mechanisms for the observed genetic alterations detected in bacterial-induced carcinogenesis.
Manipulation of DDR Signaling by Parasitic Infections
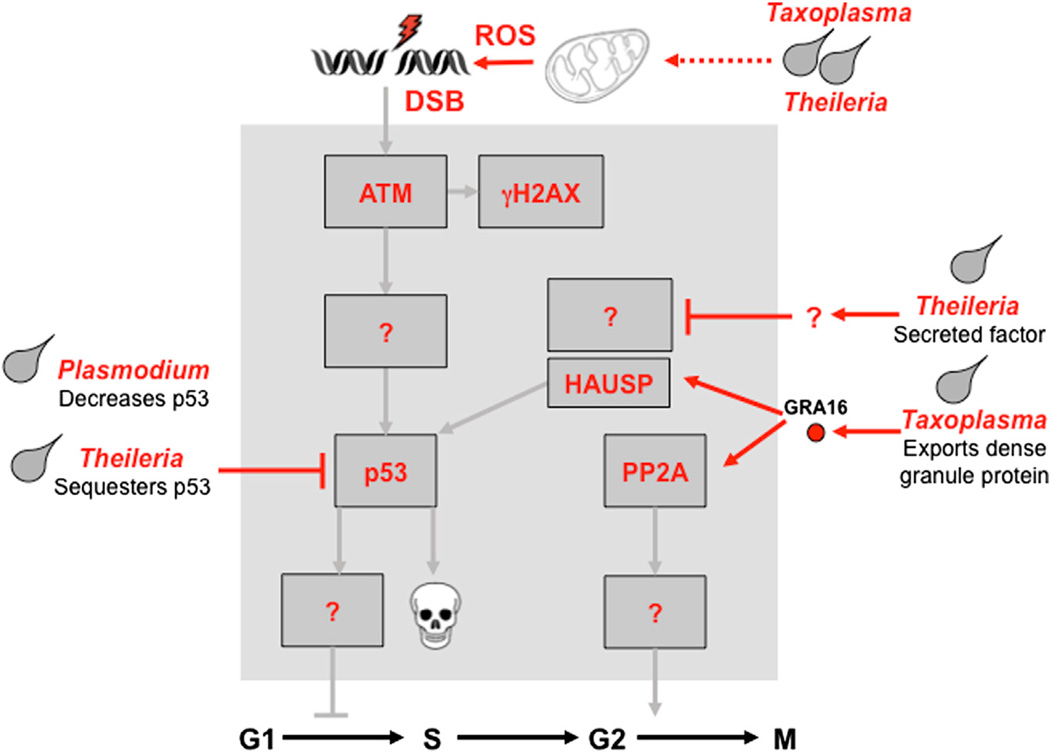
In general, knowledge of the direct mechanisms by which parasites contribute to cancer and manipulate the DDR lags behind studies of viruses and bacteria (Figure 6). There is evidence that apicomplexan parasites directly target the p53 pathway; for example, Theileria annulata infection was linked to cytoplasmic sequestration of p53 and physical association with parasite membranes (Haller et al., 2010). T. parva-infected lymphocytes have increased levels of MDM2, with alternatively spliced isoforms and impaired DNA damage responses (Hayashida et al., 2013). Parasite-encoded proteins responsible for manipulating host pathways have remained relatively elusive. A recent report identified a key dense granule protein, GRA16, that is exported by Toxoplasma and targets host nuclear proteins such as deubiquitinase HAUSP and PP2A phosphatase (Bougdour et al., 2013). This results in deregulation of the p53 checkpoint pathway and is critical for parasite pathogenesis. Theileria infection induces expression of the miR-155 oncomiR, which targets DET1, a component of the ubiquitin ligase complex containing DNA damage-binding protein 1 (DDB1) (Marsolier et al., 2013). These recent studies provide evidence for links between parasite-encoded proteins and host signaling pathways that regulate apoptosis and damage signaling. Although these events were linked to parasite survival, they have not been formally linked to DDR signaling, and we can therefore only speculate about their direct contribution to host cell phenotypes, genome integrity, and eventual tumorigenesis.
Figure 6. Parasite Infection and Effect on DDR Pathways.
In contrast to viral and bacterial infections, the mechanistic links between infection by intracellular parasites and DDR signaling pathways remain poorly defined. There are a few examples of parasite-encoded proteins, which are secreted into the host cell and directly target signaling pathways. It is also likely that parasite modulation of mitochondria and ROS levels affects host cell signaling indirectly (see the main text for details and references). The contrast between the details in this figure and those of viral infection highlight future avenues of research.
Indirect Effects of Microbial Infections on Genome Integrity
In addition to the direct effects of specific pathogen-encoded proteins on the DDR as discussed above, there are numerous examples of indirect effects on host genome integrity resulting from pathogen infection and the associated inflammatory responses.
Reactive Oxygen Species
An obvious indirect effect of pathogen infection is the production of ROS. Virus infection and viral oncoproteins can induce ROS that cause DNA damage and DDR activation. ROS-dependent damage signaling is induced by HTLV-Tax (Kinjo et al., 2010) and EBV-EBNA1 (Gruhne et al., 2009). Persistent infection of human cells with the bacterial pathogen Chlamydia trachomatis induces γH2AX, possibly due to increased ROS (Chumduri et al., 2013). ROS production also underlies the transformed phenotype associated with intracellular Theilieria parasites and drives the metabolic reprogramming of host cells (Medjkane et al., 2013). The widespread effects of microbe infection on inflammation and ROS production inevitably contribute to pathogen-induced tumorigenesis but fall beyond the scope of this review.
Spatial Reorganization of Cellular Structures
Since dynamic subnuclear structures facilitate cellular responses to DNA damage, disruption of nuclear architecture by microbes can impact cellular function. Viral infections often disrupt nuclear substructures by targeting specific cellular proteins, and pathogens create new protein complexes to promote replication. Cellular proteins involved in DNA replication and repair accumulate at virus-induced structures and are exploited to aid virus replication (reviewed by Schmid et al., 2014). The small circular genomes of polyomaviruses replicate at centers where large T antigen colocalizes with the MRN complex and ATM (Li et al., 2013a; Zhao et al., 2008). ATM activation by HPV is also accompanied by accumulation of repair factors at replication centers and viral genome amplification in differentiated cells appears dependent on damage signaling (Moody and Laimins, 2009). In some cases, there appears to be selective recruitment of DNA repair proteins to virus replication centers. ATR and its cofactors ATRIP and RPA accumulate at replication centers during Ad infection, although ATR signaling is prevented by strategies that vary between different Ad serotypes (Blackford et al., 2010; Carson et al., 2009). Viral proteins can also be substrates of DNA damage kinases, such as ATM-mediated phosphorylation of the simian virus type 40 (SV40) large T antigen (Shi et al., 2005), which promotes virus replication (Zhao et al., 2008).
Viral genomes released from virus particles in the nucleus can be recognized by cellular proteins that assemble structures at sites associated with the incoming viral genomes. These include DNA repair proteins as well as components of PML nuclear bodies (Everett et al., 2006; Lilley et al., 2011). Other cellular sensors of foreign DNA may also play a role in recognition of viral genomes. In particular, IFI16 is a protein implicated in DNA damage complexes (Ouchi and Ouchi, 2008) that may also act as a nuclear sensor of herpesvirus genomes (Kerur et al., 2011; Li et al., 2013b). The MRN DNA repair complex is inactivated by some Ad oncoproteins, and in the absence of these proteins the MRN complex accumulates at sites of viral DNA replication (Stracker et al., 2002). There appears to be a connection between the sensors of DNA damage that respond to virus genomes and DNA sensors that mediate innate immune responses. Both IFI16 and Mre11 have been implicated in sensing foreign DNA in the nucleus and cytoplasm and play roles in induction of antiviral cytokines (Kondo et al., 2013; Li et al., 2013b). Targeting of these DNA sensors by virus proteins to counteract innate immune processes may therefore also impact their role in DNA repair responses and thus indirectly influence the host’s genome integrity. Viral proteins can also form structures that sequester or inactivate cellular repair proteins. Ad-E1b55K and Ad-E4orf3 oncoproteins accumulate in cytoplasmic aggresomes that sequester both p53 and the MRN complex proteins (Araujo et al., 2005; Liu et al., 2005). The Ad-E4orf3 oncoprotein also polymerizes to form nuclear structures that immobilize the MRN complex and prevent recruitment to DNA damage sites (Carson et al., 2009; Ou et al., 2012). The HTLV-Tax protein is another example of a viral oncoprotein that sequesters MDC1 in pseudo DNA damage sites that compete with the regular cellular DDR (Belgnaoui et al., 2010).
Epigenetic Reprogramming upon Infection
Since DNA damage and DNA repair are influenced by nucleo-some structure and chromatin architecture (Price and D’Andrea, 2013), pathogen influences on host epigenomes can indirectly affect host genome integrity. Histones are subject to a plethora of complex and dynamic PTMs, including phosphorylation, acetylation, methylation, and ubiquitination, that define the degree of DNA accessibility and facilitate protein recruitment. The histone variant H2AX, a key player in the response to DSBs, can undergo multiple PTMs, including phosphorylation that generates γH2AX, acetylation by Tip60, and ubiquitination by RNF8/RNF168 (Price and D’Andrea, 2013). Viruses, bacteria, and parasites all provoke pathways that result in histone modifications and chromatin remodeling of infected cells, providing tactics to manipulate the DDR.
Virus
Phosphorylated H2AX (γH2AX) is observed during infection by many viruses. Staining for γH2AX is detected at replication centers or sites of the large T antigen for the polyomaviruses SV40 (Zhao et al., 2008) and MCV (Li et al., 2013a). Activated ATM induces γH2AX during HPV infection (Moody and Laimins, 2009) and may recruit other DNA repair factors that accumulate at viral centers and at foci induced by expression of replication proteins E1 and E2 (Gillespie et al., 2012; Sakakibara et al., 2011). In addition to cellular kinases, there are herpesvirus kinases that directly phosphorylate H2AX to promote infection (Tarakanova et al., 2007). These herpesvirus kinases are conserved across the family and can also phosphorylate other DNA repair proteins including Tip60 (Li et al.,2011). Since γH2AX accumulates at sites associated with incoming HSV viral genomes and facilitates virus replication (Lilley et al., 2011), it could also play a role in protecting ends of viral genomes or facilitating circularization of viral DNA. In the case of KSHV, infection induces γH2AX (Koopal et al., 2007), and its interaction with the latency-associated nuclear antigen (LANA) may increase binding to viral terminal repeats and promote episome persistence (Jha et al., 2013). It will be important to establish the extent to which γH2AX and other damage-associated histone modifications detected during virus infection occur on viral versus cellular DNA. Viruses may also encode factors that protect viral genomes from recognition by cellular histones and DNA repair sensors. The histone-like protein VII of Ad has been suggested to protect incoming virus genomes from recognition and checkpoint activation (Karen and Hearing, 2011), and it is possible that subsequent release of this protein from viral DNA allows it to become part of cellular chromatin.
Alterations to chromatin marks induced by pathogens may have multiple outcomes, regulating DNA repair responses in addition to gene expression and antiviral defenses. The E1A oncoprotein of Ad prevents monoubiquitination of H2B as a way to block interferon induction (Fonseca et al., 2012). Monoubiquitination of H2B is observed upon DNA damage and is important for repair, implying that viral evasion of innate immunity may also impact DNA repair. Degradation of RNF8/RNF168 ligases by HSV-ICP0 protein results in decreased ubiquitinated H2A that prevents recruitment of downstream cellular repair factors but may also overcome transcriptional silencing (Lilley et al., 2010; 2011). Some viral oncoproteins directly impact epigenetic control of the host genome. The HBV-X protein (HBX) induces aberrant epigenetic modifications at multiple levels, including hypermethylation of genomic DNA due to increases in DNA methyltransferases, hypomethylation within promoters of tumor-promoting genes, and altered histone modifications (Tian et al., 2013).
Bacteria and Parasites
Our understanding of pathogen-associated alterations of histone modifications is still rudimentary, and we do not yet have a sense of the global effects of infection on histone function. Many bacteria and parasites produce secreted factors that enter infected cell nuclei and interfere with chromatin regulation (reviewed by Bierne et al., 2012), but other than the γH2AX mark discussed earlier, the impact of these chromatin regulators on the DDR is unclear. Cellular transformation induced by the parasite Theileria is accompanied by modulation of host histone-modifying enzymes, such as the H3K4 methyltransferase SMYD3, which contributes to the transformed and metastatic phenotypes (Cock-Rada et al., 2012). Secreted bacterial proteins can directly modify the host chromatin landscape and promote efficient intracellular replication by repressing host gene expression (Rolando et al., 2013), that could also impact DNA damage responses. It is intriguing to speculate that microbial infections that affect host epigenetic machineries might leave a mark that would influence subsequent infections and tumor initiation. It is, however, too early to conclude whether these epigenetic modifications affect DNA damage and repair pathways and the extent to which histone PTMs contribute to tumorigenesis associated with bacterial or parasite infections.
Indirect Effects of Evolutionary Tradeoffs
Pathogen and host coevolution in the context of mutual conflict shapes both genomes. Signs of positive selection are characteristic evolutionary signatures of these genetic conflicts. In fact, DNA repair proteins show signs of positive selection (Demogines et al., 2010). Positive selection in genes with fundamental cellular functions, such as DNA repair, can result in alleles that are not fully optimized for their cellular functions due to the pressure to avoid inactivation by pathogenic attacks. Thus, the evolution of host genes that enables them to counteract microbial proteins interacting with repair pathways may make them vulnerable to accumulation of genome instability.
Cellular defenses that have evolved to protect the host from invading pathogens could also inadvertently be contributors to genomic instability and carcinogenesis. The AID/APOBEC cytidine deaminases play important roles in the immune system, acting in both innate and adaptive pathways (reviewed by Pavri and Nussenzweig, 2011; Refsland and Harris, 2013). Activation-induced cytidine deaminase (AID), a key mediator of the adaptive humoral immune response, functions to diversify antigen receptor genes through processes of somatic hypermutation (SHM) and class-switch recombination (CSR). AID may also play a role in innate immune defense by its activity on viral genomes that limits their fitness (Refsland and Harris, 2013) and via upregulation of ligands for activating natural killer cell receptors that marks infected cells for elimination (Bekerman et al., 2013). The human apolipoprotein B mRNA editing enzyme, catalytic poly-peptide-like 3 (APOBEC3) proteins comprise seven host restriction factors (APOBEC3A–APOBEC3H) that constitute an innate barrier to retroviruses, endogenous retroelements, and DNA viruses. These AID/APOBEC enzymes share the ability to catalyze deamination of cytidine to uracil residues, which can be processed by cellular DNA glycosylases to produce abasic sites. If not repaired by the high-fidelity repair systems, the resulting U:G mismatches will be replicated into C-to-T transitions. Processing by BER produces nicks, and if sites are closely located then they can result in DSBs. The antiviral and immune functions of AID/APOBEC proteins therefore rely on their ability to generate mutations, but these come at a high cost if not exquisitely regulated.
It has recently emerged that aberrant activity of AID/APOBEC proteins can generate somatic mutations and breaks in cellular genomes, thus turning them into powerful DNA mutators (Burns et al., 2013; Landry et al., 2011; Okazaki et al., 2007; Robbiani and Nussenzweig, 2013; Suspène et al., 2011). Constitutive and ubiquitous AID expression in a transgenic mouse model led to accumulation of mutations in nonimmunoglobulin (non-Ig) genes and produced T cell lymphomas and epithelial tumors (Okazaki et al., 2003). The murine genome possesses only one APOBEC3 gene, and the impact of human APOBEC3 proteins expressed in transgenic mice has not yet been reported. Although expression and activity of AID and APOBEC enzymes are strongly regulated, pathogens or inflammatory responses could disrupt these control mechanisms and inadvertently promote genome instability (Shimizu et al., 2012). Expression of AID and APOBEC3 proteins can be activated by cytokine signaling resulting from chronic inflammation and microbial infections (Petersen-Mahrt et al., 2009; Shimizu et al., 2012). The activation of NF-κB can produce aberrant AID protein expression in H. pylori gastric cancer tissues (Matsumoto et al., 2007) and by HCV in hepatocytes (Endo et al., 2007). Viral oncoproteins may also lead more directly to increases in AID activity that could contribute to genomic instability, such as the latent membrane protein of EBV, which upregulates AID expression in B cells (Kim et al., 2013). In contrast to these studies that implicate AID/APOBECs as potential contributors to pathogen-induced carcinogenesis, there is also evidence to suggest that DNA damage induced by these enzymes in response to virus infection could also enhance recognition of infected cells (Norman et al., 2011) and potentially limit transformation (Bekerman et al., 2013). Upregulation of AID by infection of human B cells with the oncogenic KSHV may act on viral genomes to limit reactivation and spread, while also acting on host genomes to induce DNA damage may result in upregulation of ligands that stimulate immune cells to remove infected cells (Bekerman et al., 2013). Therefore, pathogens may upset the delicate balance of AID/ APOBECs between efficient immune function and aberrant activity on the host genome. It has been speculated that observed epidemiological association between malaria and EBV could involve parasite induction of AID by malarial antigens and that parasite-induced B cell proliferation in children could increase the EBV-infected compartment or reactivate latent EBV via increased AID activity (Bouvard et al., 2012). There is also some evidence that repair pathways involving BER are downregulated in conditions where bacterial infection is associated with cancer (Touati, 2010). Activation of AID/APOBEC mutator activity as a result of microbial infections may be difficult to ascribe to infection once pathogens are cleared, making determination of causality challenging.
Concluding Remarks
The link between infection and cancer is now uncontested, and yet the molecular mechanisms by which DNA damage contributes to pathogen-induced genomic instability and tumorigenesis by infectious agents remain unclear. It is likely that a combination of genetic and epigenetic aberrations induced by infections combine to generate molecular events underlying pathogen-induced tumorigenesis. Viruses directly assault cells with delivery of foreign genetic material, and every DNA tumor virus so far examined has intersected with the cellular DNA damage apparatus. By contrast, relatively little is known about the role of these pathways in bacterial and parasite infections. Nevertheless, it is clear that microbial pathogens can induce genetic change by increasing DNA damage and decreasing DNA repair activity that transiently produces a mutator phenotype, while simultaneously blocking checkpoints and apoptosis. We predict that future investigation of microbial (both prokaryote and eukaryote)-host interactions will provide further insight into genome-to-genome crosstalk in disease. DNA damage pathways respond to pathogen infections, but in most cases it is still unclear what structures are sensed by the cellular machinery. Dissecting the intersection of microbial pathogens with host DDR machinery will reveal how cellular sensors recognize distinct DNA damage. Emerging proteomic techniques will provide a clearer view of cellular responses to pathogen infection and their intersection with DDR signaling pathways. Furthermore, understanding microbial infections associated with human cancers opens up opportunities for therapeutic intervention and prevention.
ACKNOWLEDGMENTS
We apologize to groups whose primary research papers could not be cited due to space constraints. We thank colleagues who have worked in our respective labs for helpful discussions and contributions to the field. Work on viruses and DNA repair in the lab of M.D.W. has been supported by grants from the National Institutes of Health (AI067952, CA097093, AI074967, and NS082240) and funds from the Children’s Hospital of Philadelphia. Work on parasites and transformation in the lab of J.B.W. was supported by grants from the Fondation de France (FdF #2102), Association pour le Recherche contre le Cancer (ARC #4975 and #7990), the Association for International Cancer Research (AICR, #08-0111), the French National Research Agency (ANR; Blanc SVSE 3 #090601), and the ȌWho Am I?” Laboratory of Excellence #ANR-11-LABX-0071 funded by the French government through its “Investments for the Future” program operated by the ANR under grant #ANR-11-IDEX-0005-01.
REFERENCES
- Alaoui-El-Azher M, Mans JJ, Baker HV, Chen C, Progulske-Fox A, Lamont RJ, Handfield M. Role of the ATM-checkpoint kinase 2 pathway in CDT-mediated apoptosis of gingival epithelial cells. PLoS ONE. 2010;5:e11714. doi: 10.1371/journal.pone.0011714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 2005;79:11382–11391. doi: 10.1128/JVI.79.17.11382-11391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 2012;338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekerman E, Jeon D, Ardolino M, Coscoy L. A role for host activation-induced cytidine deaminase in innate immune defense against KSHV. PLoS Pathog. 2013;9:e1003748. doi: 10.1371/journal.ppat.1003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Fryrear KA, Nyalwidhe JO, Guo X, Semmes OJ. The viral oncoprotein tax sequesters DNA damage response factors by tethering MDC1 to chromatin. J. Biol. Chem. 2010;285:32897–32905. doi: 10.1074/jbc.M110.146373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benamrouz S, Conseil V, Creusy C, Calderon E, Dei-Cas E, Certad G. Parasites and malignancies, a review, with emphasis on digestive cancer induced by Cryptosporidium parvum (Alveolata: Apicomplexa) Parasite. 2012;19:101–115. doi: 10.1051/parasite/2012192101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Hamon M, Cossart P. Epigenetics and bacterial infections. Cold Spring Harb Perspect Med. 2012;2:a010272. doi: 10.1101/cshperspect.a010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackford AN, Patel RN, Forrester NA, Theil K, Groitl P, Stewart GS, Taylor AM, Morgan IM, Dobner T, Grand RJ, Turnell AS. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. USA. 2010;107:12251–12256. doi: 10.1073/pnas.0914605107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougdour A, Durandau E, Brenier-Pinchart MP, Ortet P, Barakat M, Kieffer S, Curt-Varesano A, Curt-Bertini RL, Bastien O, Coute Y, et al. Host cell subversion by Toxoplasma GRA16, an exported dense granule protein that targets the host cell nucleus and alters gene expression. Cell Host Microbe. 2013;13:489–500. doi: 10.1016/j.chom.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan RA, Grosse Y, Lauby-Secretan B, El Ghissassi F, Benbrahim-Tallaa L, Guha N, Straif K WHO International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of malaria and of some polyomaviruses. Lancet Oncol. 2012;13:339–340. doi: 10.1016/s1470-2045(12)70125-0. [DOI] [PubMed] [Google Scholar]
- Boxus M, Twizere JC, Legros S, Dewulf JF, Kettmann R, Willems L. The HTLV-1 Tax interactome. Retrovirology. 2008;5:76. doi: 10.1186/1742-4690-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology. 1999;263:307–312. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Schwartz RA, Stracker TH, Lilley CE, Lee DV, Weitzman MD. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson CT, Orazio NI, Lee DV, Suh J, Bekker-Jensen S, Araujo FD, Lakdawala SS, Lilley CE, Bartek J, Lukas J, Weitzman MD. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 2009;28:652–662. doi: 10.1038/emboj.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Certad G, Creusy C, Guyot K, Mouray A, Chassat T, Delaire B, Pinon A, Sitja-Bobadilla A, Alvarez-Pellitero P, Praet M, et al. Fulminant cryptosporidiosis associated with digestive adenocarcinoma in SCID mice infected with Cryptosporidium parvum TUM1 strain. Int. J. Parasitol. 2010;40:1469–1475. doi: 10.1016/j.ijpara.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Chaussepied M, Langsley G. Theileria transformation of bovine leukocytes: a parasite model for the study of lymphoproliferation. Res. Immunol. 1996;147:127–138. doi: 10.1016/0923-2494(96)83165-8. [DOI] [PubMed] [Google Scholar]
- Choudhuri T, Verma SC, Lan K, Murakami M, Robertson ES. The ATM/ATR signaling effector Chk2 is targeted by Epstein-Barr virus nuclear antigen 3C to release the G2/M cell cycle block. J. Virol. 2007;81:6718–6730. doi: 10.1128/JVI.00053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chumduri C, Gurumurthy RK, Zadora PK, Mi Y, Meyer TF. Chlamydia infection promotes host DNA damage and proliferation but impairs the DNA damage response. Cell Host Microbe. 2013;13:746–758. doi: 10.1016/j.chom.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, Chluba J, Langsley G, Weitzman JB. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Cancer Res. 2012;72:810–820. doi: 10.1158/0008-5472.CAN-11-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Ramos G, Petit CR, Marcq I, Boury M, Oswald E, Nougayr- ède JP. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martel C, Ferlay J, Franceschi S, Vignat J, Bray F, Forman D, Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Demogines A, East AM, Lee JH, Grossman SR, Sabeti PC, Paull TT, Sawyer SL. Ancient and recent adaptive evolution of primate non-homologous end joining genes. PLoS Genet. 2010;6:e1001169. doi: 10.1371/journal.pgen.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin SS, Guo X, Fryrear KA, Mihaylova VT, Gupta SK, Belgnaoui SM, Haoudi A, Kupfer GM, Semmes OJ. HTLV-1 Tax oncoprotein subverts the cellular DNA damage response via binding to DNA-dependent protein kinase. J. Biol. Chem. 2008;283:36311–36320. doi: 10.1074/jbc.M804931200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elwell CA, Dreyfus LA. DNase I homologous residues in CdtB are critical for cytolethal distending toxin-mediated cell cycle arrest. Mol. Microbiol. 2000;37:952–963. doi: 10.1046/j.1365-2958.2000.02070.x. [DOI] [PubMed] [Google Scholar]
- Endo Y, Marusawa H, Kinoshita K, Morisawa T, Sakurai T, Okazaki IM, Watashi K, Shimotohno K, Honjo T, Chiba T. Expression of activation-induced cytidine deaminase in human hepatocytes via NF-kappaB signaling. Oncogene. 2007;26:5587–5595. doi: 10.1038/sj.onc.1210344. [DOI] [PubMed] [Google Scholar]
- Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor Y, Vignard J, Nicolau-Travers ML, Boutet-Robinet E, Watrin C, Salles B, Mirey G. From single-strand breaks to double-strand breaks during S-phase: a new mode of action of the Escherichia coli Cytolethal Distending Toxin. Cell. Microbiol. 2013;15:1–15. doi: 10.1111/cmi.12028. [DOI] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, Moore PS. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J, Mymryk JS. Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte A, Bergeron-Labrecque F, Moody CA, Lehoux M, Laimins LA, Archambault J. Nuclear accumulation of the papillomavirus E1 helicase blocks S-phase progression and triggers an ATM-dependent DNA damage response. J. Virol. 2011;85:8996–9012. doi: 10.1128/JVI.00542-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Second Edition. New York: ASM Press; 2006. [Google Scholar]
- Frisan T, Cortes-Bratti X, Chaves-Olarte E, Stenerlöw B, Thelestam M. The Haemophilus ducreyi cytolethal distending toxin induces DNA double-strand breaks and promotes ATM-dependent activation of RhoA. Cell. Microbiol. 2003;5:695–707. doi: 10.1046/j.1462-5822.2003.00311.x. [DOI] [PubMed] [Google Scholar]
- Ge Z, Rogers AB, Feng Y, Lee A, Xu S, Taylor NS, Fox JG. Bacterial cytolethal distending toxin promotes the development of dysplasia in a model of microbially induced hepatocarcinogenesis. Cell. Microbiol. 2007;9:2070–2080. doi: 10.1111/j.1462-5822.2007.00939.x. [DOI] [PubMed] [Google Scholar]
- Gillespie KA, Mehta KP, Laimins LA, Moody CA. Human papillomaviruses recruit cellular DNA repair and homologous recombination factors to viral replication centers. J. Virol. 2012;86:9520–9526. doi: 10.1128/JVI.00247-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhne B, Sompallae R, Marescotti D, Kamranvar SA, Gastaldello S, Masucci MG. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl. Acad. Sci. USA. 2009;106:2313–2318. doi: 10.1073/pnas.0810619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra L, Guidi R, Frisan T. Do bacterial genotoxins contribute to chronic inflammation, genomic instability and tumor progression? FEBS J. 2011;278:4577–4588. doi: 10.1111/j.1742-4658.2011.08125.x. [DOI] [PubMed] [Google Scholar]
- Guidi R, Guerra L, Levi L, Stenerlöw B, Fox JG, Josenhans C, Masucci MG, Frisan T. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell. Microbiol. 2013;15:98–113. doi: 10.1111/cmi.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidoboni M, Ferreri AJ, Ponzoni M, Doglioni C, Dolcetti R. Infectious agents in mucosa-associated lymphoid tissue-type lymphomas: pathogenic role and therapeutic perspectives. Clin. Lymphoma Myeloma. 2006;6:289–300. doi: 10.3816/CLM.2006.n.003. [DOI] [PubMed] [Google Scholar]
- Haller D, Mackiewicz M, Gerber S, Beyer D, Kullmann B, Schneider I, Ahmed JS, Seitzer U. Cytoplasmic sequestration of p53 promotes survival in leukocytes transformed by Theileria. Oncogene. 2010;29:3079–3086. doi: 10.1038/onc.2010.61. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hatakeyama M. Helicobacter pylori CagA and Gastric Cancer: A Paradigm for Hit-and-Run Carcinogenesis. Cell Host Microbe. 2014;15(this issue):306–316. doi: 10.1016/j.chom.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Hayashida K, Kajino K, Hattori M, Wallace M, Morrison I, Greene MI, Sugimoto C. MDM2 regulates a novel form of incomplete neoplastic transformation of Theileria parva infected lymphocytes. Exp. Mol. Pathol. 2013;94:228–238. doi: 10.1016/j.yexmp.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Howley PM, Livingston DM. Small DNA tumor viruses: large contributors to biomedical sciences. Virology. 2009;384:256–259. doi: 10.1016/j.virol.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Durocher D. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 2013;49:795–807. doi: 10.1016/j.molcel.2013.01.017. [DOI] [PubMed] [Google Scholar]
- Jang SH, Lim JW, Morio T, Kim H. Lycopene inhibits Helico-bacter pylori-induced ATM/ATR-dependent DNA damage response in gastric epithelial AGS cells. Free Radic. Biol. Med. 2012;52:607–615. doi: 10.1016/j.freeradbiomed.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Jha HC, Upadhyay SK, A J Prasad M, Lu J, Cai Q, Saha A, Robertson ES. H2AX phosphorylation is important for LANA-mediated Kaposi’s sarcoma-associated herpesvirus episome persistence. J. Virol. 2013;87:5255–5269. doi: 10.1128/JVI.03575-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinadasa RN, Bloom SE, Weiss RS, Duhamel GE. Cytolethal distending toxin: a conserved bacterial genotoxin that blocks cell cycle progression, leading to apoptosis of a broad range of mammalian cell lineages. Microbiology. 2011;157:1851–1875. doi: 10.1099/mic.0.049536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 2009;5:e1000397. doi: 10.1371/journal.ppat.1000397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karen KA, Hearing P. Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J. Virol. 2011;85:4135–4142. doi: 10.1128/JVI.02540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, Otageri P, Chandran B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9:363–375. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim WS, Park C. Epstein-Barr virus latent membrane protein 1 increases genomic instability through Egr-1-mediated up-regulation of activation-induced cytidine deaminase in B-cell lymphoma. Leuk. Lymphoma. 2013;54:2035–2040. doi: 10.3109/10428194.2013.769218. [DOI] [PubMed] [Google Scholar]
- Kinjo T, Ham-Terhune J, Peloponese JM, Jr, Jeang KT. Induction of reactive oxygen species by human T-cell leukemia virus type 1 tax correlates with DNA damage and expression of cellular senescence marker. J. Virol. 2010;84:5431–5437. doi: 10.1128/JVI.02460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Kobayashi J, Saitoh T, Maruyama K, Ishii KJ, Barber GN, Komatsu K, Akira S, Kawai T. DNA damage sensor MRE11 recognizes cytosolic double-stranded DNA and induces type I interferon by regulating STING trafficking. Proc. Natl. Acad. Sci. USA. 2013;110:2969–2974. doi: 10.1073/pnas.1222694110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopal S, Furuhjelm JH, Järviluoma A, Jäämaa S, Pyakurel P, Pussi-nen C, Wirzenius M, Biberfeld P, Alitalo K, Laiho M, Ojala PM. Viral oncogene-induced DNA damage response is activated in Kaposi sarcoma tumorigenesis. PLoS Pathog. 2007;3:1348–1360. doi: 10.1371/journal.ppat.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–450. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidal AM, Cyr DP, Hill RJ, Lee PW, McCormick C. Subversion of autophagy by Kaposi’s sarcoma-associated herpesvirus impairs oncogene-induced senescence. Cell Host Microbe. 2012a;11:167–180. doi: 10.1016/j.chom.2012.01.005. [DOI] [PubMed] [Google Scholar]
- Leidal AM, Pringle ES, McCormick C. Evasion of oncogene-induced senescence by gammaherpesviruses. Curr Opin Virol. 2012b;2:748–754. doi: 10.1016/j.coviro.2012.09.009. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Li R, Zhu J, Xie Z, Liao G, Liu J, Chen MR, Hu S, Woodard C, Lin J, Taverna SD, et al. Conserved herpesvirus kinases target the DNA damage response pathway and TIP60 histone acetyltransferase to promote virus replication. Cell Host Microbe. 2011;10:390–400. doi: 10.1016/j.chom.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Diaz J, Tsang SH, Buck CB, You J. Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation. J. Virol. 2013a;87:9173–9188. doi: 10.1128/JVI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013b;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. USA. 2005;102:5844–5849. doi: 10.1073/pnas.0501916102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010;29:943–955. doi: 10.1038/emboj.2009.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley CE, Chaurushiya MS, Boutell C, Everett RD, Weitzman MD. The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog. 2011;7:e1002084. doi: 10.1371/journal.ppat.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Shevchenko A, Shevchenko A, Berk AJ. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 2005;79:14004–14016. doi: 10.1128/JVI.79.22.14004-14016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizundia R, Chaussepied M, Huerre M, Werling D, Di Santo JP, Langsley G. c-Jun NH2-terminal kinase/c-Jun signaling promotes survival and metastasis of B lymphocytes transformed by Theileria. Cancer Res. 2006;66:6105–6110. doi: 10.1158/0008-5472.CAN-05-3861. [DOI] [PubMed] [Google Scholar]
- Machado AM, Figueiredo C, Seruca R, Rasmussen LJ. Helicobacter pylori infection generates genetic instability in gastric cells. Biochim. Biophys. Acta. 2010;1806:58–65. doi: 10.1016/j.bbcan.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Maréchal A, Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013;5:5. doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolier J, Pineau S, Medjkane S, Perichon M, Yin Q, Flemington E, Weitzman MD, Weitzman JB. OncomiR addiction is generated by a miR-155 feedback loop in Theileria-transformed leukocytes. PLoS Pathog. 2013;9:e1003222. doi: 10.1371/journal.ppat.1003222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Marusawa H, Kinoshita K, Endo Y, Kou T, Morisawa T, Azuma T, Okazaki IM, Honjo T, Chiba T. Helicobacter pylori infection triggers aberrant expression of activation-induced cytidine deaminase in gastric epithelium. Nat. Med. 2007;13:470–476. doi: 10.1038/nm1566. [DOI] [PubMed] [Google Scholar]
- McFadden K, Luftig MA. Interplay between DNA tumor viruses and the host DNA damage response. Curr. Top. Microbiol. Immunol. 2013;371:229–257. doi: 10.1007/978-3-642-37765-5_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medjkane S, Perichon M, Marsolier J, Dairou J, Weitzman JB. Theileria induces oxidative stress and HIF1α activation that are essential for host leukocyte transformation. Oncogene. 2013 doi: 10.1038/onc.2013.134. Published online May 13, 2013. http://dx.doi.org/10.1038/onc.2013.134. [DOI] [PubMed] [Google Scholar]
- Mesri EA, Feitelson M, Munger K. Human Viral Oncogenesis: A Cancer Hallmarks Analysis. Cell Host Microbe. 2014;15(this issue):266–282. doi: 10.1016/j.chom.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 2009;5:e1000605. doi: 10.1371/journal.ppat.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- Moore PS, Chang Y. Why do viruses cause cancer? Highlights of the first century of human tumour virology. Nat. Rev. Cancer. 2010;10:878–889. doi: 10.1038/nrc2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesić D, Hsu Y, Stebbins CE. Assembly and function of a bacterial genotoxin. Nature. 2004;429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- Nikitin PA, Luftig MA. The DNA damage response in viral-induced cellular transformation. Br. J. Cancer. 2012;106:429–435. doi: 10.1038/bjc.2011.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, et al. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8:510–522. doi: 10.1016/j.chom.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niller HH, Wolf H, Minarovits J. Viral hit and run-oncogenesis: genetic and epigenetic scenarios. Cancer Lett. 2011;305:200–217. doi: 10.1016/j.canlet.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Norman JM, Mashiba M, McNamara LA, Onafuwa-Nuga A, Chiari-Fort E, Shen W, Collins KL. The antiviral factor APOBEC3G enhances the recognition of HIV-infected primary T cells by natural killer cells. Nat. Immunol. 2011;12:975–983. doi: 10.1038/ni.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gott-schalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- Okazaki IM, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T. Constitutive expression of AID leads to tumorigenesis. J. Exp. Med. 2003;197:1173–1181. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki IM, Kotani A, Honjo T. Role of AID in tumorigenesis. Adv. Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X, et al. A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell. 2012;151:304–319. doi: 10.1016/j.cell.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouchi M, Ouchi T. Role of IFI16 in DNA damage and checkpoint. Front. Biosci. 2008;13:236–239. doi: 10.2741/2673. [DOI] [PubMed] [Google Scholar]
- Pavri R, Nussenzweig MC. AID targeting in antibody diversity. Adv. Immunol. 2011;110:1–26. doi: 10.1016/B978-0-12-387663-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Petersen-Mahrt SK, Coker HA, Pauklin S. DNA deaminases: AIDing hormones in immunity and cancer. J. Mol. Med. 2009;87:893–897. doi: 10.1007/s00109-009-0496-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Jackson SP. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 2011;25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell. 2013;152:1344–1354. doi: 10.1016/j.cell.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan E, Ward M, Guo X, Durkin SS, Sawyer A, Vilela M, Osgood C, Pothen A, Semmes OJ. Physical and in silico approaches identify DNA-PK in a Tax DNA-damage response interactome. Retrovirology. 2008;5:92. doi: 10.1186/1742-4690-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr. Top. Microbiol. Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 2013;14:563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- Rolando M, Sanulli S, Rusniok C, Gomez-Valero L, Bertholet C, Sahr T, Margueron R, Buchrieser C. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe. 2013;13:395–405. doi: 10.1016/j.chom.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Sakakibara N, Mitra R, McBride AA. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 2011;85:8981–8995. doi: 10.1128/JVI.00541-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaras V, Rafailidis PI, Mourtzoukou EG, Peppas G, Falagas ME. Chronic bacterial and parasitic infections and cancer: a review. J. Infect. Dev. Ctries. 2010;4:267–281. doi: 10.3855/jidc.819. [DOI] [PubMed] [Google Scholar]
- Schmid M, Speiseder T, Dobner T, Gonzalez RA. DNA virus replication compartments. J. Virol. 2014;88:1404–1420. doi: 10.1128/JVI.02046-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Garrett WS. Microbes, Microbiota, and Colon Cancer. Cell Host Microbe. 2014;15(this issue):317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Dodson GE, Shaikh S, Rundell K, Tibbetts RS. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J. Biol. Chem. 2005;280:40195–40200. doi: 10.1074/jbc.C500400200. [DOI] [PubMed] [Google Scholar]
- Shiels B, Langsley G, Weir W, Pain A, McKellar S, Dobbelaere D. Alteration of host cell phenotype by Theileria annulata and Theileria parva: mining for manipulators in the parasite genomes. Int. J. Parasitol. 2006;36:9–21. doi: 10.1016/j.ijpara.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Marusawa H, Endo Y, Chiba T. Inflammation-mediated genomic instability: roles of activation-induced cytidine deaminase in carcinogenesis. Cancer Sci. 2012;103:1201–1206. doi: 10.1111/j.1349-7006.2012.02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–352. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- Suspène R, Aynaud MM, Guétard D, Henry M, Eckhoff G, Marchio A, Pineau P, Dejean A, Vartanian JP, Wain-Hobson S. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc. Natl. Acad. Sci. USA. 2011;108:4858–4863. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe S. Sexually transmitted infections and risk of prostate cancer: review of historical and emerging hypotheses. Future Oncol. 2010;6:1289–1311. doi: 10.2217/fon.10.95. [DOI] [PubMed] [Google Scholar]
- Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakanova VL, Leung-Pineda V, Hwang S, Yang CW, Matatall K, Basson M, Sun R, Piwnica-Worms H, Sleckman BP, Virgin HW., 4th Gamma-herpesvirus kinase actively initiates a DNA damage response by inducing phosphorylation of H2AX to foster viral replication. Cell Host Microbe. 2007;1:275–286. doi: 10.1016/j.chom.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Yang W, Song J, Wu Y, Ni B. Hepatitis B virus X protein-induced aberrant epigenetic modifications contributing to human hepatocellular carcinoma pathogenesis. Mol. Cell. Biol. 2013;33:2810–2816. doi: 10.1128/MCB.00205-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toller IM, Neelsen KJ, Steger M, Hartung ML, Hottiger MO, Stucki M, Kalali B, Gerhard M, Sartori AA, Lopes M, Müller A. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA. 2011;108:14944–14949. doi: 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati E. When bacteria become mutagenic and carcinogenic: lessons from H. pylori. Mutat. Res. 2010;703:66–70. doi: 10.1016/j.mrgentox.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Turnell AS, Grand RJ. DNA viruses and the cellular DNA-damage response. J. Gen. Virol. 2012;93:2076–2097. doi: 10.1099/vir.0.044412-0. [DOI] [PubMed] [Google Scholar]
- Vogelmann R, Amieva MR. The role of bacterial pathogens in cancer. Curr. Opin. Microbiol. 2007;10:76–81. doi: 10.1016/j.mib.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Wallace NA, Galloway DA. Manipulation of cellular DNA damage repair machinery facilitates propagation of human papillomaviruses. Semin. Cancer Biol. 2014 doi: 10.1016/j.semcancer.2013.12.003. Published online January. 8, 2014. http://dx.doi.org/10.1016/j.semcancer.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Micro-biol. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- Wilkinson DE, Weller SK. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 2004;78:4783–4796. doi: 10.1128/JVI.78.9.4783-4796.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, He Z, Xin B, Cao L. LMP1 of Epstein-Barr virus suppresses cellular senescence associated with the inhibition of p16INK4a expression. Oncogene. 2000;19:2002–2013. doi: 10.1038/sj.onc.1203515. [DOI] [PubMed] [Google Scholar]
- Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, Schloss PD. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4:e00692–e13. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li Q, Hu P, Cheng H, Huang G. Two case reports of pituitary adenoma associated with Toxoplasma gondii infection. J. Clin. Pathol. 2002;55:965–966. doi: 10.1136/jcp.55.12.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Madden-Fuentes RJ, Lou BX, Pipas JM, Gerhardt J, Rigell CJ, Fanning E. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in Simian virus 40-infected primate cells. J. Virol. 2008;82:5316–5328. doi: 10.1128/JVI.02677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zur Hausen H. The search for infectious causes of human cancers: where and why. Virology. 2009;392:1–10. doi: 10.1016/j.virol.2009.06.001. [DOI] [PubMed] [Google Scholar]