Abstract
Contactin-associated protein 4 (Caspr4), also known as contactin-associated protein-like protein (CNTNAP4), is expressed in various regions of the brain. Recent reports suggest that CNTNAP4 is a susceptibility gene of autism spectrum disorders (ASDs). However, the molecular function of Caspr4 in the brain has yet to be identified. In this study, we show an essential role of Caspr4 in neural progenitor cells (NPCs). Caspr4 is expressed in NPCs in the subventricular zone (SVZ), a neurogenic region in the developing cortex. Knocking down of Caspr4 enhances the proliferation of NPCs derived from the SVZ of embryonic day 14 mouse. Neuronal differentiation is increased by overexpression of Caspr4, but decreased by knocking down of Caspr4 in cultured mouse NPCs. Transfection of the intracellular domain of Caspr4 (C4ICD) rescues the abnormal decreased neuronal differentiation of Caspr4-knocking down NPCs. Ligand of Numb protein X2 (LNX2), a binding partner of Numb, interacts with Caspr4 in a PDZ domain-dependent manner and plays a similar role to Caspr4 in NPCs. Moreover, transfection of LNX2 rescues the decreased neuronal differentiation in Caspr4-knocking down NPCs. In contrast, transfection of C4ICD fails to do so in LNX2-knocking down NPCs. These results indicate that Caspr4 inhibits neuronal differentiation in a LNX-dependent manner. Therefore, this study reveals a novel role of Caspr4 through LNX2 in NPCs, which may link to the pathogenesis of ASDs.
Introduction
The functional complexity of the mammalian central nervous system is predicated on the ability to devise cell types, including neurons, astrocytes, and oligodendrocytes during development. Neural progenitor cells (NPCs) are a population of cells, which could self-renew and differentiate into neurons and glial cells [1]. These NPCs proliferate, differentiate, migrate, and eventually integrate into the neural network. The abnormalities in any of these processes will cause dysfunctions of the brain and leads to neurological diseases, such as brain tumors [2,3], schizophrenia [4], depression [5,6], and Alzheimer's disease [7,8].
Autism spectrum disorders (ASDs), which are characterized by impairments in social reciprocity and language development and highly restrictive interests and/or repetitive behaviors, exhibited developmental abnormalities in the hippocampus, the amygdala, and the cerebral cortex of the patients [9]. However, the etiology of ASDs remains unknown. Recent studies indicate that some autism risk genes, such as contactin-associated protein 2 (Caspr2) [10], myocyte enhancer factor 2C [11], and phosphatase and tensin homolog on chromosome 10 [12], modulates the proliferation, differentiation, or migration of NPCs. These studies indicate that NPCs play essential roles in the pathogenesis of ASDs.
Contactin-associated protein 4 (Caspr4), also known as contactin-associated protein-like protein (CNTNAP4), is a transmembrane protein member of the neurexin superfamily involved in neuron–glia interaction and the clustering of K+ channels in myelinated axons [13–17]. CNTNAP4 gene has recently been identified as a novel susceptibility gene of ASDs [18,19]. Caspr4-deficient mice exhibited hypersensitivity in sensory and overgrooming behaviors [20], the phenotypes often observed in mouse models of autism [10,21]. Expression of Caspr4 has been detected in the olfactory bulb, hippocampus, deep cerebellar nuclei, and the substantia nigra [22]. These studies suggest that Caspr4 may play an important role in the brain development. However, the functions of Caspr4 in the brain remain unknown.
Ligand Numb-protein X2 (LNX2), also known as PDZRN1, is one of the members of the LNX family, which also includes LNX1, LNX3, and LNX4. The LNX family of proteins is of special interest as it has been suggested that they serve as molecular scaffolds that localize PDZ containing proteins, including Numb, a cell fate determinant, to specific subcellular sites [23]. LNX1 protein functions as a RING type E3 ubiquitin ligase and promotes degradation of Numb protein [23,24]. Likewise, LNX2 interacts with Numb and Numblike through a mechanism that involves the phosphotyrosine-binding (PTB) domains of Numb and Numblike and the tetrapeptide, NPAF in LNX2 [23]. Moreover, high levels of expression of LNX2 were reported from embryonic day (E) 12.5 in the brain and were evident in the cortical plate at E15.5 [23]. However, the cellular functions of LNX2 in the brain development are unknown.
In this study, we show that both Caspr4 and LNX2 are expressed in NPCs of the subventricular zone (SVZ), a neurogenic region in the embryonic brain. Moreover, we describe that LNX2 binds to Caspr4 in a PDZ domain-dependent manner. We demonstrate that both Caspr4 and LNX2 promote neuronal differentiation while inhibiting the proliferation of NPCs in vitro. We further identify that Caspr4 enhances neuronal differentiation of NPCs in a LNX2-dependent manner. Therefore, this study reveals a novel function of Caspr4 in modulating the proliferation and differentiation of NPCs through LNX2. This study suggests a role of Caspr4 in cortical development, which may link to the pathogenesis of ASDs.
Materials and Methods
Antibodies
Anti-LNX2 (RP670 from Dr. Kerstin, Ludwig Institute for Cancer Research Stockholm Branch, Karolinska Institute), anti-HA (Upstate), anti-myc (9E10), anti-GAPDH (6C5), anti-γ-tubulin (Sigma), anti-MAP2 (Sigma), anti-βIII tubulin (Chemicon; TUJ1), anti-Nestin (Dako), and anti-CaN1 (Abcam). Polyclonal antibodies against Caspr4 were generated by immunizing rabbits with glutathione S-transferase (GST)-fusion protein containing the cytoplasmic domain of human Caspr4.
Plasmids and siRNAs
Yeast expression plasmids: For bait vector construction, the 157 bp cDNA corresponding to the cytoplasmic region of human Caspr4 (hC4ICD) was amplified from human spinal cord cDNA by polymerase chain reaction (PCR) using a forward primer 5′-CGGAATTCCGCATTTA TCAGCAGAAAAG-3′ with an EcoR1 site (in bold) and reverse primer 5′-CGGGATCCT CAGAAGAAGTACTC-3′ with a BamH1 site (in bold) and subsequently cloned into the pGBKT7 vector (Clontech). Similarly, human LNX2 (hLNX2) was cloned from a human brain cDNA library (Clontech) by PCR using a forward primer 5′-CGGAATTCATGGGA ACAACAAGTGATGAGATGGTGTC-3′ with an EcoR1 site and reverse primer 5′-CGGGATCCCTATACAAGGCTGCCAGGCC AAC-3′ with a BamH1 site. The PCR product was subcloned into pGADT7 (Clontech). Mouse postnatal day 10 brain cDNA was isolated. Reverse transcription (RT) reaction was performed using the Superscript First-Strand System (Invitrogen) according to the manufacturer's protocol. Full-length Caspr4 was amplified by PCR using 5′-GGGGTACCGCCACCATGAATATGGGATCTGTC-3′ with a KpnI site and reverse primer 5′-GGATAGCCCTCGAGGAGGAT CATTGCCAATCAGAAGAAATACTC-3′ with a Xho1 site and subsequently cloned into the pcDNA4/V5-His-A vector (Invitrogen). For transfection of NPCs, mouse Caspr4 and LNX2 were subcloned into the pCDF1-MCS1-EF1-copGFP vector (System Biosciences). phLNX2/pGADT7 was digested by EcoR1/BamH1, and the 2.1 kb fragment was cloned into the pCMV-HA vector by EcoR1/BglII with an N-terminal HA tag (Clontech). The mouse LNX2 plasmid (mLNX2/pcDNA3.1/Zeo) and p80-LNX (full-length LNX1 cDNA) were provided by Dr. Kerstin [23]. For myc-tagged expression vector construction, the coding regions of mLNX2 were cloned into pCMV-myc vector by EcoR1/KpnI with an N-terminal myc tag (Clontech). All mLNX2 deletions were amplified by PCR and cloned into pCMV-myc vector by EcoR1/KpnI with an N-terminal myc tag. Small hairpin RNAs (shRNAs) targeting the coding regions of Caspr4 and LNX2 were designed using the Ambion website. Hairpins were inserted into the backbone pFIV-H1-copGFP shRNA cloning vector (System Biosciences). Validation of shRNA constructs was performed by transfecting HEK293T/17 cells with pCMV-myc-mLNX2 or pcDNA4-mCaspr4 and the shRNA plasmid of interest at a 1:2 ratio. Immunoblotting was performed using anti-myc and anti-Caspr4 antibodies to determine the extent of knockdown. The sequences of the shRNAs were as follows: Caspr4 shRNA#1 (194–214) 5′-GGCTGAATCGAAGAGACGG-3′, Caspr4 scrambled (Control) shRNA 5′-TGTATCGCTCGTTCCTACG-3′, LNX2 shRNA#2 (468–488) 5′-GCGGCTTCATTT CAAGCTG-3′, and LNX2 scrambled (Control) shRNA 5′-CGAGCTCTTCAAGAACCAG-3′. siRNAs were purchased from Genepharm Technology. The sequences of siRNA are as follows: mouse Caspr4: 5′-GGAGCACCUUUCAUUGUAATT-3′; mouse LNX2: 5′-GUGACGUGUUGCUGAACAUTT-3′.
Yeast two-hybrid screening
The yeast two-hybrid analysis was performed according to the manufacturer's instructions (Clontech; Matchmaker Two-Hybrid System 3). A human brain cDNA library was purchased from Clontech. In total, 3.5×106 transformants (AH109; Clontech) were screened. Thirty-three clones were obtained on synthetic dropout (SD) selection plates lacking the amino acids Ade, His, Trp, and Leu, containing X-a-Gal. These positive colonies were blue, indicating that the MEL1 reporter, a-galactosidase, was activated. These clones were introduced into Escherichia coli. And TOP10 and the protein coding sequence of prey plasmids were analyzed by sequencing and searching the BLAST database.
Mammalian cell lines, culture conditions, and transfection
HEK293T/17, COS7, and CHO cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and 100 U/mL penicillin and streptomycin (all from Invitrogen). Cells were cultured at 37°C in a CO2-humidified incubator. LipofectAMINE 2000 reagent (Invitrogen) and Effectene reagent (QIAGEN) were used for plasmid transfection into cells using conditions that were recommended by the manufacturer.
Mice
C57BL/6J mice were bred in the specific pathogen free condition. All mice used in this study were handled according to the protocols approved by the Institutional Animal Care and Use Committee of the Soochow University.
NPCs culturing, transfection, proliferation, and differentiation
NPCs were isolated from the embryonic day 14 (E14) C57BL/6J mouse SVZ, as described previously [8]. Briefly, NPCs were cultured in the DMEM-F12 (Gibco) containing N2-supplements (GIBCO), 20 ng/mL basic fibroblast growth factor (bFGF) (Sigma), and 20 ng/mL epidermal growth factor (EGF; Sigma). Nucleofector II (Amaxa Biosystems) was used for plasmid transfection into NPCs. For in vitro differentiation assays, 2 days after transfection, the medium was changed to 0.5% bovine serum-containing medium without EGF and bFGF. Under these conditions, the cells differentiated into neurons or glial cells over 3–5 days. For proliferation assay, NPCs were cultured for 4–5 h in the NPC culture medium containing 10 μM 5-bromo-2-deoxyuridine (BrdU). All NPCs for differentiation and proliferation assay were isolated from the E14 mouse SVZ.
Reverse transcription–polymerase chain reaction, quantitative PCR, and western blot analysis
Total cellular and tissue RNAs were extracted using the RNeasy Mini Kit (QIAGEN). RT reactions were performed with the Superscript First-Strand System (Invitrogen) according to the manufacturer's protocol. PCR reactions were performed with Taq DNA polymerase (i-DNA Biotechnology). The primers for reverse transcription–polymerase chain reaction (RT-PCR) and quantitative PCR were designed on the basis of mouse sequences. Hes1: 5′-AAAGCCTATCATGGAGAAGAGGCG-3′ (Forward), 5′-GGAATGCCGGGAGCTATCTTTCTT-3′ (Reverse); Hes5: 5′-CGCATCAACAGCAGCATAGAG-3′ (Forward), 5′-TGGAAGTGGTAAAGCAGCTTC-3′ (Reverse); Caspr4: 5′-GAGCACCTTTCATTGTAACTG-3′ (Forward), 5′-CATTGGTCCATTCGTTCA-3′ (Reverse); LNX2: 5′-CTGCTGGACAAACTGCTGG-3′ (Forward), 5′-CGGGTTAGAGCGGTGGAT-3′ (Reverse).
For western blotting, sample proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene fluoride membrane. The membranes were then blocked with 5% dry milk in TBST plus 0.05% Tween 20; the blots were incubated with the primary antibodies in TBST containing 5% dry milk overnight at 4°C and for 1 h at room temperature. The membranes were washed with TBST and incubated with horseradish peroxidase-conjugated sheep anti-mouse immunoglobulin G (IgG) or donkey anti-rabbit IgG (Amersham Biosciences) for 2 h at room temperature. ECL Plus or ECL Advance Western Blotting Detection Reagents (Amersham Biosciences) were used to visualize the immunoreactive proteins. The densities of the bands were analyzed by Image Lab.
Expression of GST fusion proteins
To generate a GST fusion protein containing the cytoplasmic tail of human Caspr4 (pGST/C4ICD), pGADT7/hC4ICD was digested with EcoR1/Sal1 and cloned into pGEX-4T1 (Amersham Biosciences). In addition, to generate Caspr4ICD with the PDZ binding motif (amino acids, EYFF) deleted, pGST/C4ICDΔEYFF was constructed by the standard PCR amplification. GST fusion constructs were expressed in E. coli BL21 (DE3) pLysS competent cells using conditions recommended by the manufacturer (Amersham Biosciences). Equal amounts of purified GST fusion proteins were separated on a 10% SDS-PAGE gel and visualized by staining with Coomassie brilliant blue.
GST-pull down assay
Human LNX2-HA fusion protein was incubated with C4ICD fused with GST-protein at 4°C overnight and then added to glutathione–Sepharose 4B at 4°C for 2 h. After several washes in lysis buffer, the protein was eluted under glutathione. The samples were separated on a 10% SDS-PAGE gel and subjected to western analysis with a monoclonal anti-HA antibody.
Immunoprecipitation analysis
For immunoprecipitation (IP) analysis, cell lysates were extracted by incubating cells in 1 mL of Tris-NaCl-EDTA (TNE) lysis buffer [10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 1% Nonident P, 5 mM ethylenediaminetetraaceticacid (EDTA), complete protease inhibitor] on ice for 15 min. Cell lysates were centrifuged for 20 min at 4°C. The lysates were precleaned with protein A beads (Roche Diagnostics) for 30 min and incubated with anti-myc antibody overnight, followed by further incubation with protein A for 3 h. After four washes in TNE buffer, IP samples were analyzed by SDS-PAGE and immunoblotting with anti-Caspr4 antibody.
IP analysis from cells and brain lysates
NPCs and brain tissues were lysed in RIPA buffer (50 mM Tris–HCl, pH 9.0, 1% sodium deoxycholate, 2.5 mM EDTA-free complete protease inhibitor). After centrifugation at 16,000 g for 1 h at 4°C, the supernatant was used as the cell lysate. The lysate was precleared with protein A beads for 30 min. After centrifugation at 16,000 g, the supernatant was collected and this lysate was used for the IP analysis. Polyclonal anti-Caspr4 antibody was added to the mouse brain lysate and rotated overnight at 4°C. Protein A beads were added to the mixture and rotated for 3 h. After the precipitates had been washed five times in RIPA buffer, the IP samples were analyzed by SDS-PAGE and immunoblotting.
Immunofluoresence staining
C57BL/6 mice pregnant with E14 fetuses were perfused with phosphate-buffered saline (PBS) under anesthesia, followed by 4% paraformaldehyde in 0.1 M phosphate buffer. The brains of fetuses were removed, postfixed in the same fixative, and then immersed in 30% sucrose at 4°C. Serial 16-μm-thick coronal sections were cut on a freezing microtome. Sections were blocked with 10% normal goat serum in PBS and then incubated with primary antibody in the blocking solution overnight at 4°C. Sections were rinsed three times in PBS with 0.3% Triton X-100 and incubated with appropriate secondary Alexa Fluor-conjugated antibodies for 1 h at room temperature. The sections were then mounted with a mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories) after being rinsed three times with PBS with 0.3% Triton X-100.
Immunocytochemistry on cultured cells and immunostaining on tissue sections were performed, as previously described [8]. For quantification of immunofluorescence, images of fields of cultured cells were captured by digital photomicrography under a×20 objective over the entire surface of each coverslip. All labeled cells were then counted in each photomicrograph. The proportion of neurons was quantified as the numbers of Tuj1+EGFP+ cells divided by the total number of EGFP+ cells in the same fields.
Statistical analyses
Each experiment was repeated more than three times. The statistical analyses were performed by one-way analysis of variance or Student's t-test using SPSS software, as appropriate. In all the graphs, the error bars indicate the standard error of the mean. Significance was accepted at P<0.05 (*P<0.05, **P<0.01, ***P<0.001).
Results
Expression of Caspr4 in the neurogenic ventricular zone and NPCs
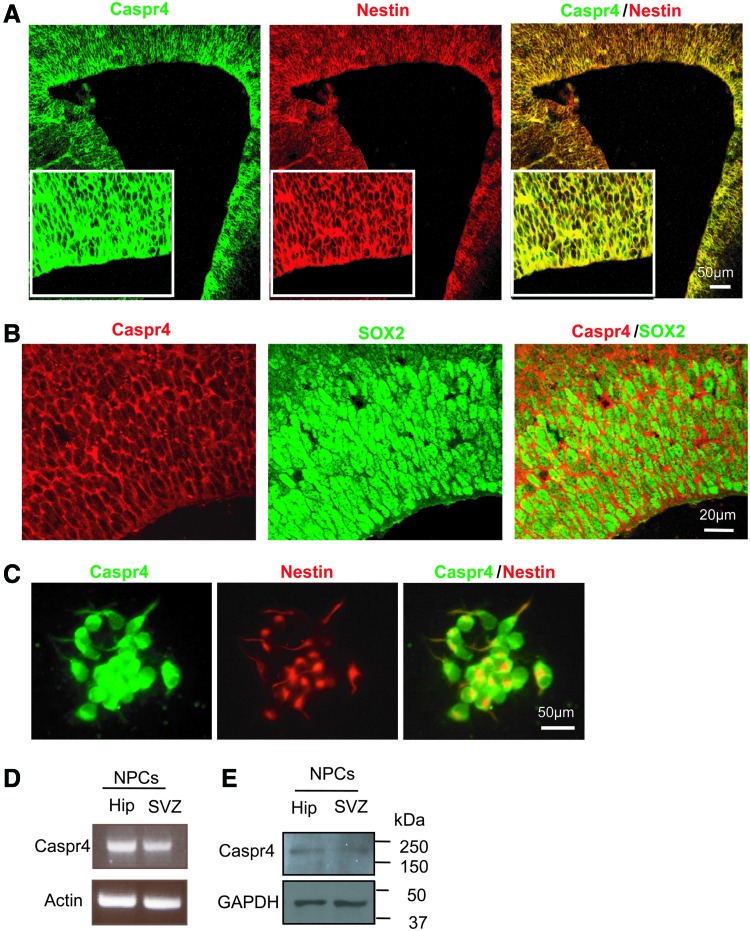
We first investigated the expression profile of Caspr4 in the cortex during development. Western blot analysis showed that expression of Caspr4 in the cortex started at E12 and increased at E14, E16, E18, and peaked at P0 relatively, during the cortex development (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/scd). A similar expression profile of Caspr4 was detected in the SVZ of E12, E14, and E18 mouse brains (Supplementary Fig. S1B). These results suggest that Caspr4 might play a role in the cortex development. NPCs in the SVZ play essential roles in the cortex development [25]. We thus performed immunofluoresence staining with antibodies against Caspr4 and Nestin (Fig. 1A) or Sox2 (Fig. 1B), two markers of NPCs, in the SVZ of E14 (Fig. 1A, B), P0 (Supplementary Fig. S2A), and adult mice (Supplementary Fig. S2C). The results showed that Caspr4 was expressed in Nestin+ (Fig. 1A) or Sox2+ cells (Fig. 1B) in the SVZ. Next, we performed immunofluoresence staining using antibodies against Caspr4 and Nestin in cultured NPCs derived from E14 mouse SVZ. Caspr4 immunoreactivity was detected in cultured NPCs, which exhibited Nestin+ staining (Fig. 1C). In addition, immunoreactivity of Caspr4 was also detected in βIII-tubulin+ (Tuj1, a maker of neuron) cells in the cerebral cortex (Supplementary Fig. S3A) and glial fibrillary acidic protein+ (GFAP, a maker of astrocyte) cells in the corpus callosum (Supplementary Fig. S3E), indicating that Caspr4 is expressed in cortical neurons and astrocytes. The specificity of Caspr4 antibody was validated by coexpression of Caspr4 and myc in HEK293 cells transfected with the plasmid expressing Caspr4 fused with myc at its carboxyl terminus (Supplementary Fig. S4A) and by the lack of immunoreactivity of Caspr4 in the brain sections stained with non-immunoglobulin (Supplementary Fig. S4B). Moreover, Caspr mRNA (Fig. 1D) and protein (Fig. 1E) were detected in cultured NPCs derived from E14 mouse SVZ and hippocampus by RT-PCR and western blot, respectively. These results demonstrate that Caspr4 is expressed by NPCs in the SVZ of the embryonic mouse brain.
FIG. 1.
Contactin-associated protein 4 (Caspr4) is expressed in embryonic neural progenitor cells (NPCs). (A) Double immunostaining for Nestin and Caspr4 in the subventricular zone (SVZ) of embryonic day 14 (E14) mouse. Scale bar: 50 μm. (B) Double immunostaining for Caspr4 and Sox2 in the SVZ of E14 mouse. Scale bar: 20 μm. (C) NPCs isolated from the E14 mouse SVZ were double-stained for Caspr4 and Nestin. Scale bar: 50 μm. (D) Reverse transcription–polymerase chain reaction (RT-PCR) analysis of the expression of Caspr mRNA in the cultured NPCs derived from E14 SVZ and hippocampus (Hip). Actin was detected as the loading control. (E) Western blot analysis of the expression of Caspr protein in the cultured NPCs derived from E14 SVZ and hippocampus (Hip). GAPDH was detected as the loading control. Color images available online at www.liebertpub.com/scd
Caspr4 inhibits the proliferation of NPCs
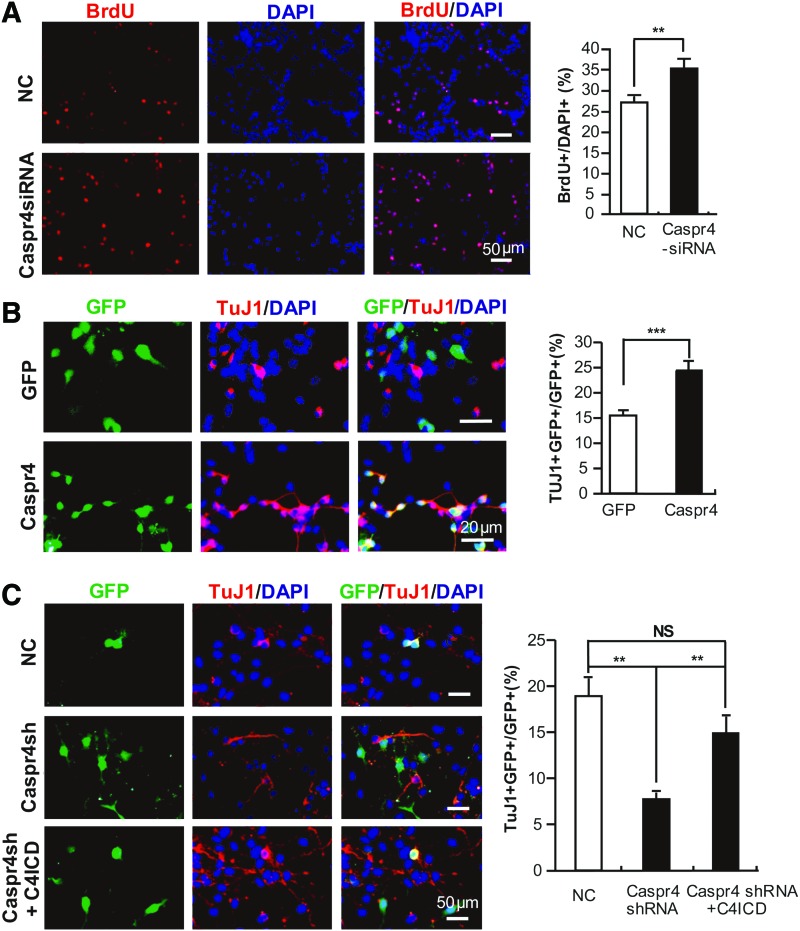
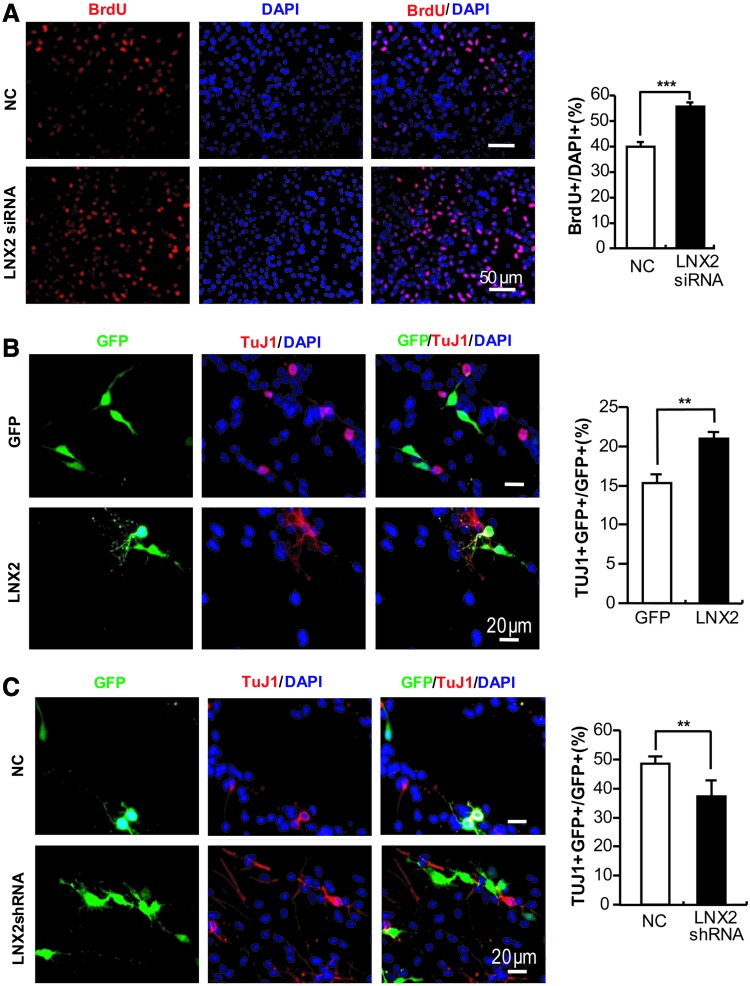
We next investigated whether Caspr4 might play a role in modulating the proliferation of NPCs. NPCs derived from E14 mouse SVZ were transfected with Caspr4 small interference RNA (Caspr4 siRNA), which could downregulate the endogenous expression of Caspr4 in NPCs (Supplementary Fig. S5A) or the control scrambled siRNA (NC, Fig. 2A), respectively. The transfected cells were subjected to BrdU incorporation analysis. Quantification showed that the number of BrdU+ cells was increased in Caspr4 siRNA-transfected NPCs compared to the control NPCs (Fig. 2A). In contrast, apoptosis in Caspr4 siRNA-transfected NPCs showed no difference from the control cells (data not shown). These results demonstrate that Caspr4 inhibits the proliferation of NPCs in vitro.
FIG. 2.
Caspr4 inhibits the proliferation while promoting neuronal differentiation of NPCs. (A) NPCs were transfected with Caspr4 small interference RNA (siRNA) and control siRNA (NC), respectively. Transfected cells were stained for 5-bromo-2-deoxyuridine (BrdU) and 4′,6-diamidino-2-phenylindole (DAPI) after being cultured in the medium containing BrdU for 3 h. The numbers of BrdU+ cells were counted and expressed as the percentage of the number of DAPI+ cells (NC: 26.89±1.26; Caspr4-siRNA: 35.11±2.06). (B) NPCs were transfected with Caspr4 plasmid in a pCDF1-MCS1-EF1-copGFP vector that expresses coGFP (Caspr4) or in an empty vector containing only copGFP (GFP). After 3–4 days of differentiation in vitro, cells were immunostained for βIII tubulin (TUJ1), GFP, and DAPI. The numbers of GFP+TUJ1+ cells were counted and expressed as the percentage of the number of GFP+ cells (GFP: 15.33±1.01; Caspr4: 24.33±1.70). (C) NPCs were transfected with Caspr4 small hairpin RNA (shRNA) or Caspr4 shRNA plus intracellular domain of Caspr4 (C4ICD) as well as a scrambled shRNA (NC) as the control. After 3–4 days of differentiation in vitro, cells were immunostained for βIII tubulin (TUJ1), GFP, and DAPI. The numbers of GFP+TUJ1+ cells were counted and expressed as the percentage of the number of GFP+ cells (NC: 18.59±1.97; Caspr4-shRNA: 7.69±0.80; Caspr4-shRNA plus C4ICD: 14.89±1.82). Scale bars: 50 μm (A–C); 20 μm (B). Values are represented as mean±standard error of the mean (SEM), Student's t-test (A, B); one-way analysis of variance (C). **P<0.01. ***P<0.001. NS, not significant. Color images available online at www.liebertpub.com/scd
Caspr4 promotes neuronal differentiation of NPCs
We further investigated whether Caspr4 might play a role in regulating neuronal differentiation of NPCs. Caspr4 cDNA was transfected together with copepod green fluorescent protein (copGFP) in the same vector but under control of another promoter into cultured NPCs (Caspr4, Fig. 2B). The control cells were transfected with the empty vector expressing copGFP alone (GFP, Fig. 2B). The transfected cells were immunostained for βIII-tubulin, a marker of neurons, and GFP after being cultured under differentiation condition for 3 days. Quantification analysis showed an increased number of βIII-tubulin+ cells in Caspr4-transfected cells compared to the control cells (Fig. 2B). These data suggest that Caspr4 promotes neuronal differentiation of NPCs in vitro. To further test this notion, we transfected short hairpin RNA (shRNA) of Caspr4 in a copGFP expressing vector, which could downregulate the expression of Caspr4 in transfected HEK293T cells (Supplementary Fig. S5B), to cultured NPCs and immunostained them for βIII-tubulin and GFP after 3 days in vitro differentiation (Fig. 2C). The number of βIII-tubulin+ cells was significantly decreased in the NPCs transfected with the Caspr4 shRNA compared with that of the control group transfected with scrambled shRNA (Fig. 2C). Caspr4 is a type I transmembrane protein that contains a large extracellular and a short intracellular domain [26]. The intracellular domain of Caspr4 (C4ICD) contains a PDZ binding motif, through which it may interact with different PDZ domain-containing proteins known to participate in membrane protein trafficking [27], the establishment of specialized membrane domains [28], clustering of transmembrane receptors [29], and the generation of multiprotein signaling complexes [30]. We therefore asked whether Caspr4 promotes neuronal differentiation of NPCs through C4ICD. C4ICD plasmid and Caspr4 shRNA were cotransfected to NPCs. The transfected cells were immunostained for βIII-tubulin and GFP at 3 days after being cultured under differentiation condition. Interestingly, the abnormally reduced neuronal differentiation of Caspr4 shRNA-transfected NPCs was rescued by coexpression of C4ICD (Fig. 2C). These results indicate that Caspr4 promotes neuronal differentiation of NPCs through its intracellular domain.
LNX2 interacts with Caspr4
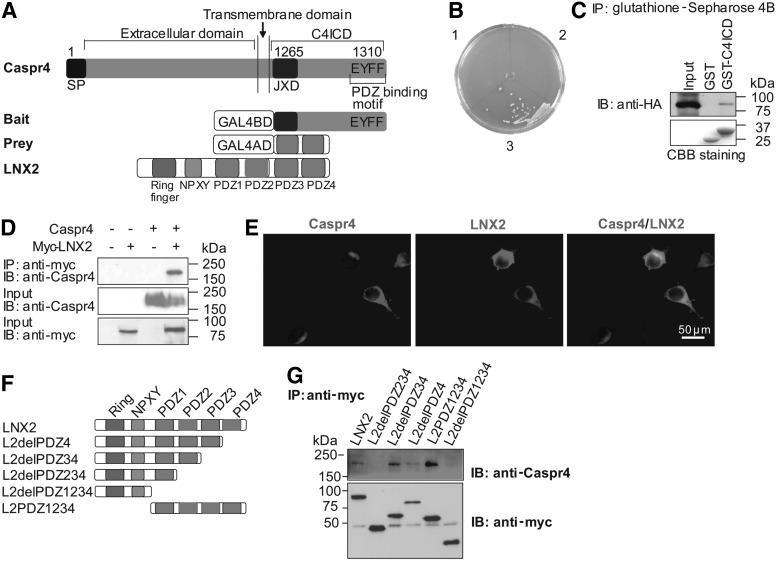
Given that Caspr4 promotes neuronal differentiation of NPCs through C4ICD, we were interested in identifying binding proteins interacting with C4ICD by yeast two-hybrid screening. Using bait that consisted of C4ICD (amino acid residues 1,265–1,310), we identified prey that coded amino acids 433–690 of LNX2, an N-terminal truncated form of LNX2 (Fig. 3A). To confirm this interaction, we transformed yeast cells with the following three groups of plasmids: (1) Caspr4ICD/pGBKT7 and pGADT7; (2) pGBKT7 and LNX2/pGADT7 (full length of LNX2 fused with the GAL4 activation domain); and (3) Caspr4ICD/pGBKT7 and LNX2/pGADT7 (Fig. 3B). On SD plates that lack amino acids Ade, His, Leu, and Trp and contain X-α-Gal, blue colonies were only apparent when the bait plasmid, C4ICD/pGBKT7, and the prey plasmid, LNX2/pGADT7, were cotransformed into the yeast cells (Fig. 3B). The interaction between Caspr4 and LNX2 was further confirmed by the GST pull-down assay and IP. C4ICD-GST, but not GST, could pull-down LNX2-HA protein from LNX2-HA-transfected-COS7 cells (Fig. 3C). IP analysis showed that anti-myc antibody could only precipitate Caspr4 from Caspr4- and LNX2-myc-cotransfected HEK293T cells (Fig. 3D). Furthermore, immunostaining using antibodies against Caspr4 and LNX2 showed that Caspr4 and LNX2 were colocalized along cell membranes of Caspr4- and LNX2-myc-cotransfected HEK293T cells (Fig. 3E). These results indicate that Caspr4 interacts with LNX2 in both yeast and mammalian cells.
FIG. 3.
Ligand of Numb protein X2 (LNX2) interacts with Caspr4. (A) Schematic representations of full-length Caspr4, the bait protein that has the last 47 amino acids of the Caspr4 cytoplasmic domain (C4ICD, amino acids 1,265–1,310), the prey clone that codes the amino acids 433–690 of LNX2, and full-length LNX2 that contains one Ring finger, one NPXY, and four PDZ domains. (B) Yeast cells were transformed with the following plasmids: Caspr4ICD/pGBKT7 and pGADT7 (1), pGBKT7 and LNX2/pGADT7 (2), and Caspr4ICD/pGBKT7 and LNX2/pGADT7 (3). Transformants were spread onto SD/ −Ade/ −His/ −Leu/ −Trp medium plate containing X-α-Gal. Colonies were only observed in yeast cells coexpressing C4ICD and LNX2. (C) Protein extracts from LNX2-HA-transfected COS7 cells were incubated and pulled-down with GST and GST-C4ICD proteins. Bound proteins were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and immunoblotted with anti-HA antibody (upper panel) or stained with Coomassie brilliant blue (CBB staining; lower panel). (D) Caspr4 was cotransfected with LNX2-myc into HEK293T cells. Immunoprecipitation was performed with an anti-myc antibody and probed with an anti-Caspr4 antibody. The inputs were immunoblotted with both antibodies against Caspr4 and myc as indicated. (E) HEK293T cells were cotransfected with Caspr4 and LNX2-myc. After 24 h post-transfection, double immunofluorescence staining was performed on these cells by using antibodies against Caspr4 and myc. Scale bar: 50 μm. (F, G) Schematic representation of truncated LNX2 constructs, which were tagged with a myc at their N-terminus (F). Caspr4 was cotransfected with a myc-tagged full-length (FL-LNX2) and different truncated LNX2 (L2delPDZ234, L2delPDZ34, L2delPDZ4, L2PDZ1234, and L2delPDZ1234 into HEK293T cells. Coimmunoprecipitation was performed using the anti-myc antibody and probed with antibodies against Caspr4 (upper panel) and myc (lower panel) (G).
LNX2 binding to Caspr4 requires the second PDZ domain
LNX2 consists of an N-terminal ring-finger domain and four PDZ domains. The ring-finger domain functions as an E2-dependent, E3 ubiquitin ligase [23,24]. The PDZ domains of LNX2 contain the binding sites for one of the proteins own PDZ domains and so LNX proteins can form PDZ-dependent oligomeric as well as heteromeric complexes with each other [23]. To identify the binding sites of LNX2 to Caspr4, we constructed various truncated LNX2, which were tagged with myc (Fig. 3F), and cotransfected them respectively with Caspr4 cDNA into HEK293T cells. IP analysis showed that Caspr4 could be precipitated by full-length LNX2, truncated LNX2 deleted PDZ3 and PDZ4 domains (L2delPDZ34), truncated LNX2 deleted PDZ4 domain (L2delPDZ4), and the PDZ (1–4) domains of LNX2 (L2PDZ1234) (Fig. 3G). In contrast, truncated LNX2 deleted PDZ2, PDZ3, and PDZ4 (L2delPDZ234) and truncated LNX2 deleted all four PDZ domains (L2delPDZ1234) failed to precipitate Caspr4 from the cotransfected cells (Fig. 3G). These results suggest that the binding of LNX2 to Caspr4 requires its second PDZ domain. Thus, LNX2 binds to Caspr4 in a PDZ domain-dependent manner.
LNX2 coexpression with Caspr4 in the SVZ and NPCs
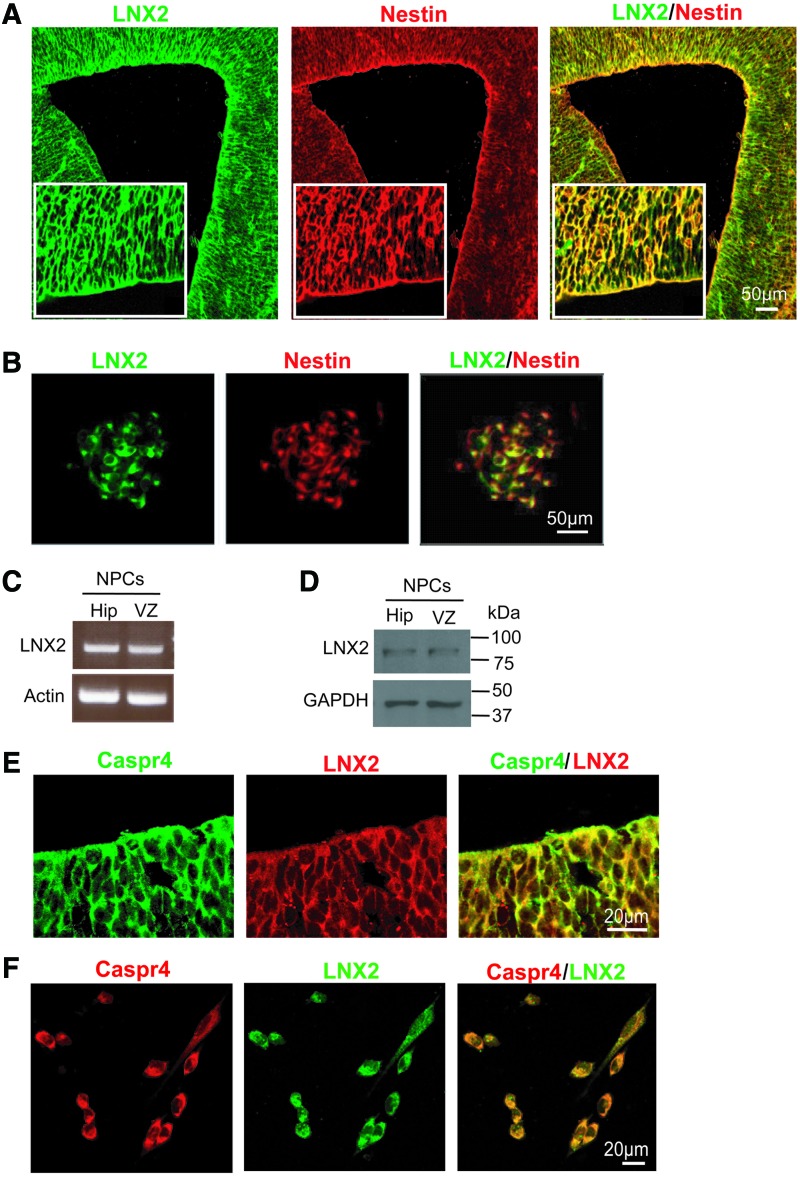
Caspr4 is expressed in NPCs in the SVZ and modulates the proliferation and neuronal differentiation of NPCs. We then asked whether LNX2, as a novel binding partner of Caspr4, might play a role in NPCs as well. The expression of LNX2 in the cortex was detected from E10, increased at E12, E14, E16, E18, and peaked at P0 (Supplementary Fig. 6A), which showed a similar expression profile to Caspr4. Similar to Caspr4, LNX2 was detected in the SVZ during the embryonic stage (Supplementary Fig. 6B). Immunostaining by using antibodies against LNX2 and Nestin in E14 (Fig. 4A), P0 (Supplementary Fig. S2B), and adult (Supplementary Fig. 2D) mouse SVZ or in the cultured NPCs derived from E14 mouse SVZ (Fig. 4B) showed that LNX2 was expressed in Nestin+ cells. Similar to Caspr4, LNX2 mRNA (Fig. 4C) and protein (Fig. 4D) were detected in cultured NPCs derived from E14 mouse SVZ and hippocampus by RT-PCR and western blot, respectively. In addition, similar to Caspr4, LNX2 immunoreactivity was detected in βIII-tubulin+ cortical neurons in the cerebral cortex (Supplementary Fig. S3C) and GFAP+ astrocytes in the corpus callosum (Supplementary Fig. S3F). Moreover, immunostaining by using antibodies against LNX2 and Caspr4 in the E14 SVZ or in the cultured NPCs showed that LNX2 colocalized with Caspr4 in the cells along the SVZ of E14 mice (Fig. 4E) and the cultured NPCs derived from the SVZ (Fig. 4F). These results indicate that LNX2 is coexpressed with Caspr4 by NPCs in the SVZ of embryonic mouse.
FIG. 4.
Expression of LNX2 in the SVZ and NPCs. (A) Double immunostaining for Nestin and LNX2 in the SVZ of E14 mouse brain. Scale bar: 50 μm. (B) Cultured NPCs isolated from the SVZ of E14 mouse brain were immunostained for LNX2 and Nestin. Scale bar: 50 μm. (C) RT-PCR analysis of the expression of LNX2 mRNA in cultured NPCs derived from E14 SVZ and hippocampus (Hip). Actin was detected as loading control. (D) Western blot analysis of the expression of LNX2 in cultured NPCs derived from E14 SVZ and hippocampus (Hip). GAPDH was detected as loading control. (E) Double immunostaining for LNX2 and Caspr4 in the SVZ of E14 mouse brain. Scale bar: 20 μm. (F) NPCs isolated from the SVZ of E14 mouse brain were immunostained for LNX2 and Caspr4. Scale bar: 50 μm. Color images available online at www.liebertpub.com/scd
LNX2 inhibits the proliferation while promoting neuronal differentiation of NPCs
We then investigated whether LNX2 might play a same role as Caspr4 in the proliferation of NPCs. NPCs transfected with LNX2 siRNA, which could downregulate the expression of LNX2 in LNX2-transfected HEK293T cells (694, Supplementary Fig. S5C) or the control scrambled siRNA (NC, Supplementary Fig. S5C), were subjected to BrdU incorporation analysis. Quantification showed that the number of BrdU+ cells was increased in LNX2 siRNA-transfected NPCs (Fig. 5A). Thus, similar to Caspr4, LNX2 inhibits the proliferation of NPCs in vitro.
FIG. 5.
LNX2 inhibits the proliferation but promotes neuronal differentiation of NPCs. (A) NPCs were transfected with LNX2 siRNA and its scrambled siRNA (NC), respectively. Transfected cells were stained for BrdU and DAPI after being cultured in the medium containing BrdU for 3 h. The numbers of BrdU+ cells were counted and expressed as the percentage of the number of DAPI+ cells (NC: 39.68±1.39; LNX2 siRNA: 55.23±1.75). (B, C) NPCs were transfected with LNX2 cDNA in a pCDF1-MCS1-EF1-copGFP vector expressing coGFP [LNX2, (B)] or in an empty vector containing only copGFP [GFP, (B)] and a LNX2 shRNA [LNX2 shRNA, (C)] or a scrambled shRNA as a control [NC, (C)]. After 3–4 days of differentiation in vitro, cells were double-stained for TUJ1, GFP, and DAPI. The numbers of GFP+TUJ1+ cells were counted and expressed as the percentage of the number of GFP+ cells. (B) Control: 15.33±1.01; LNX2: 20.76±0.77; (C) Control: 48.50±2.17; LNX2-shRNA: 37.24±5.56. Scale bars: 50 μm (A), 20 μm (B, C). Values are represented as mean±SEM, Student's t-test. **P<0.01. ***P<0.001. Color images available online at www.liebertpub.com/scd
We next investigated whether LNX2 might play a role in promoting neuronal differentiation of NPCs. Using a similar approach as in Fig. 2B and C, LNX2-transfected NPCs showed an increased number of βIII-tubulin+ cells (Fig. 5B), whereas LNX2-shRNA-transfected NPCs (Supplementary Fig. S5D) showed a decreased number of βIII-tubulin+ cells (Fig. 5C), compared to the control cells (Fig. 5B, C) after differentiation. These results indicate that LNX2, similar to Caspr4, inhibits the proliferation while promoting neuronal differentiation of NPCs.
LNX2 is an essential downstream element of Caspr4 in promoting neuronal differentiation of NPCs
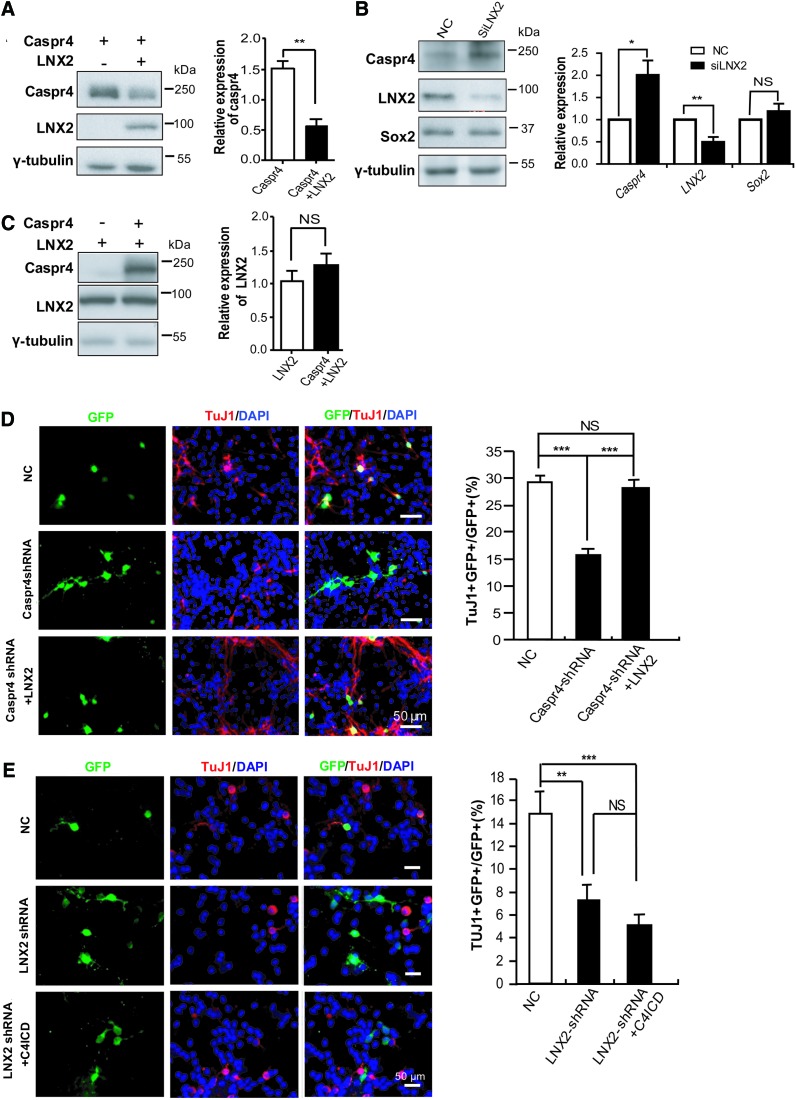
LNX2 functions as a RING-type E3 ubiquitin ligase and promotes degradation of Numb protein [24]. Considering the interaction between LNX2 and Caspr4, we investigated whether LNX2 might regulate the degradation of Caspr4. Consistent with this idea, the levels of Caspr4 were reduced by overexpression of LNX2 in HEK293 cells (Fig. 6A) and were increased by knocking down of LNX2 in cultured NPCs (Fig. 6B). These results indicate that LNX2 play a role in regulation of Caspr4 protein expression. In contrast, the levels of LNX2 remain unchanged by overexpression of Caspr4 (Fig. 6C). Given that LNX2 plays a similar role to Caspr4 in neuronal differentiation of NPCs, it is unlikely that LNX2 functions on NPCs through degradation of Caspr4. Therefore, to further clarify the relationship of LNX2 and Caspr4 in modulating neuronal differentiation of NPCs, we cotransfected LNX2 plasmid and Caspr4 shRNA into cultured NPCs and compared the capability of the transfected cells to differentiate neurons. As previous result, Caspr4 shRNA-transfected NPCs differentiated less βIII-tubulin+ neurons than the control cells transfected with the scrambled shRNA (Fig. 6D). In contrast, cotransfection of LNX2 with Caspr4 shRNA rescued the decreased neuronal differentiation of Caspr4 shRNA-transfected NPCs (Fig. 6D). However, when we cotransfected C4ICD and LNX2 shRNA into cultured NPCs, the result showed that C4ICD failed to rescue the decreased neuronal differentiation of LNX2 shRNA-transfected NPCs (Fig. 6E), although it rescued the decreased βIII-tubulin+ neuron numbers in Caspr4 shRNA-transfected cells (Fig. 2C). These results suggest that C4ICD promotes neuronal differentiation in a LNX2-dependent manner, and LNX2 acts as the downstream molecule of Caspr4 in promoting neuronal differentiation of NPCs.
FIG. 6.
LNX2 is a downstream element of Caspr4 in promoting neuronal differentiation of NPCs. (A) Caspr4 were cotransfected with LNX2 or the empty vector into HEK293T cells. The cells were harvested at 48 h after transfection and subjected to western blot analysis using antibodies against Caspr4 and LNX2. γ-Tubulin (tubulin) was detected as a loading control. Quantification analysis of the blots was shown. (B) NPCs were transfected with LNX2 siRNA or a scrambled siRNA (NC) as a control. The cells were harvested at 48 h after transfection and subjected to western blot analysis using antibodies against Caspr4, LNX2, and Sox2. γ-Tubulin (tubulin) was detected as a loading control. Quantification analysis of the blots was shown. (C) LNX2 were cotransfected with Caspr4 or the empty vector into HEK293T cells. The cells were harvested at 48 h after transfection and subjected to western blot analysis using antibodies against Caspr4 and LNX2. γ-Tubulin (tubulin) was detected as a loading control. Quantification analysis of the blots was shown. (D) NPCs were transfected with Caspr4 shRNA, Caspr4 shRNA plus LNX2, or a scrambled shRNA (NC) as a control. Cells were immunostained for βIII tubulin (TUJ1), GFP, and DAPI after 3–4 days of differentiation in vitro. The numbers of GFP+TUJ1+ neurons were counted and expressed as the percentage of the number of total GFP+ cells (NC: 29.23±1.37; Caspr4-shRNA:15.46±0.78; Caspr4-shRNA plus LNX2: 28.20±1.48). (E) NPCs transfected with LNX2 shRNA or LNX2 shRNA plus C4ICD or a scrambled shRNA (NC) were immunostained for βIII tubulin (TUJ1), GFP, and DAPI after 3–4 days of differentiation in vitro. The numbers of GFP+TUJ1+ neurons were counted and expressed as the percentage of the number of total GFP+ cells (NC: 14.84±1.79; LNX2 shRNA: 7.24±1.31; LNX2 shRNA plus C4ICD: 5.04±0.85). Scale bars: 50 μm. Values are represented as mean±SEM. *P<0.05; **P<0.01; ***P<0.001; NS, non-significant. Color images available online at www.liebertpub.com/scd
Discussion
The Caspr family contains five members. Caspr1 and Caspr2 play essential roles in the establishment of the polarized domains in myelinated axons [13–17]. Caspr3 is expressed by basket cells in the cerebellum, by oligodendrocytes, and is distributed along axons in the corpus callosum, spinal cord, and peripheral nerves [26]. Caspr5 expression is detected in both the fetal development and adult brain [22]. Caspr4 is expressed in specific neuronal subpopulations in the olfactory bulb, hippocampus, deep cerebellar nuclei, and the substantia nigra [22]. However, the functions of Caspr3, Caspr4, and Caspr5 in the brain remain unknown. Both Caspr2 and Caspr4 have recently been identified as risk genes of ASDs [10,18]. Caspr2-deficient mice show deficits in social interaction behavior and abnormality in neuronal migration during the cortex development [10], suggesting a role of Caspr2 in the cortex development. In this study, we show that Caspr4 is expressed in NPCs. We further have confirmed that Caspr4 promotes neuronal differentiation of NPCs. Our study indicates Caspr4 may play important roles in the cortex development. This point is consistent with the recent genetic studies, which have shown that deletion of Caspr4 exhibits strong association with children developmental delay and ASDs [18–20].
Similar to other members of Caspr family, Caspr4 consists of a large extracellular domain, a single membrane-spanning domain, and a short intracellular domain [26]. The intracellular domain of Caspr4 (C4ICD) has a PDZ binding motif, which may interact with the PDZ domain-containing proteins involved in multiple cellular and biological processes [26]. It has reported that C4ICD associates with CASK and Mint1 [26], two PDZ domain-containing proteins. In this study, we report that C4ICD interacts with LNX2. LNX2 is an E3 ubiquitin ligase, which contains a RING finger and four PDZ domains [23,24]. LNX2 interacts with the cell fate determinant Numb through the first PDZ domain and a Numb PTB domain binding site [23,24]. However, the binding of LNX2 to Caspr4 requires the second PDZ domain, suggesting that LNX2 may recognize different substrates through distinct PDZ domains. Moreover, we show that C4ICD rescues the decreased neuronal differentiation of NPCs by knocking down of Caspr4, indicating C4ICD is required for the function of Caspr4 in NPCs. This is distinct from Caspr, which inhibits neurite outgrowth through its extracellular domain [31].
Consistent with that LNX2 functions as E3 ubiquitin ligase [24], a decrease in the levels of Caspr4 by overexpression of LNX2 has been observed. However, the fact that LNX2 plays a similar role to Caspr4 in modulating the proliferation and differentiation of NPCs excludes the possibility that LNX2 promotes neuronal differentiation through the degradation of Caspr4. If LNX2 functions in NPCs through decreased Caspr4 expression, knocking down of Caspr4 should have the same effect as overexpression of LNX2 in NPCs. We further demonstrate that C4ICD rescues the decreased neuronal differentiation in Caspr4-knocking down NPCs, but it failed to do so in LNX2-knocking down NPCs. In contrast, LNX2 rescues the decreased neuronal differentiation in Caspr4-knocking down NPCs. These observations suggest that LNX2 may function as the downstream of Caspr4 in NPC differentiation. LNX2 interacts and degrades Numb and thus enhances Notch signaling [23,24], which represses neuronal differentiation during the cortical development [32]. LNX2 is recently found to enhance WNT signaling as well [33], which promotes neuronal differentiation during the development [34]. Moreover, as a PDZ domain containing protein, LNX2 interacts with various proteins such as junctional adhesion molecule 4, Coxsackievirus and adenovirus receptor, and CD8α [35–37]. Thus, identification of the downstream targets of LNX2 may help to explain how Caspr4 modulates NPCs through LNX2.
Supplementary Material
Acknowledgments
This work was supported by grants to Q.-H.M. from the National Program on Key Basic Research Project (2013CB945602), National Natural Science Foundation of China (31171313), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Suzhou Science and Technology Development Program (SZS201205), Soochow University Satrtup Foundation (Q421500110), to Z.-C.X. from the Talent Program, Yunnan Province, China and Monash Professorial Fellowship, Monash University, Australia; Y.T. as Grant-in-Aid for General Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (15500275, 20590149). We thank Dr. Kerstin for providing LNX2 and LNX1 cDNA and LNX2 antibody, Dr. Kazutada Watanabe for his support and advice, and Ms. Sha sha Lu for organizing the figures.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Temple S. (2001). The development of neural stem cells. Nature 414:112–117 [DOI] [PubMed] [Google Scholar]
- 2.Liu HK, Wang Y, Belz T, Bock D, Takacs A, Radlwimmer B, Barbus S, Reifenberger G, Lichter P. and Schutz G. (2010). The nuclear receptor tailless induces long-term neural stem cell expansion and brain tumor initiation. Genes Dev 24:683–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, Grimmer MR, Lau J, Chesler L, et al. (2012). Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 21:601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reif A, Fritzen S, Finger M, Strobel A, Lauer M, Schmitt A. and Lesch KP. (2006). Neural stem cell proliferation is decreased in schizophrenia, but not in depression. Mol Psychiatry 11:514–522 [DOI] [PubMed] [Google Scholar]
- 5.Eisch AJ. and Petrik D. (2012). Depression and hippocampal neurogenesis: a road to remission?. Science 338:72–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snyder JS, Soumier A, Brewer M, Pickel J. and Cameron HA. (2011). Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature 476:458–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taupin P. (2009). Adult neurogenesis, neural stem cells and Alzheimer's disease: developments, limitations, problems and promises. Curr Alzheimer Res 6:461–470 [DOI] [PubMed] [Google Scholar]
- 8.Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, et al. (2008). A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat Cell Biol 10:283–294 [DOI] [PubMed] [Google Scholar]
- 9.State MW. and Sestan N. (2012). Neuroscience. The emerging biology of autism spectrum disorders. Science 337:1301–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, et al. (2011). Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell 147:235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang YJ, et al. (2008). Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proc Natl Acad Sci U S A 105:9397–9402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amiri A, Cho W, Zhou J, Birnbaum SG, Sinton CM, McKay RM. and Parada LF. (2012). Pten deletion in adult hippocampal neural stem/progenitor cells causes cellular abnormalities and alters neurogenesis. J Neurosci 32:5880–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Einheber S, Zanazzi G, Ching W, Scherer S, Milner TA, Peles E. and Salzer JL. (1997). The axonal membrane protein Caspr, a homologue of neurexin IV, is a component of the septate-like paranodal junctions that assemble during myelination. J Cell Biol 139:1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charles P, Tait S, Faivre-Sarrailh C, Barbin G, Gunn-Moore F, Denisenko-Nehrbass N, Guennoc AM, Girault JA, Brophy PJ. and Lubetzki C. (2002). Neurofascin is a glial receptor for the paranodin/Caspr-contactin axonal complex at the axoglial junction. Curr Biol 12:217–220 [DOI] [PubMed] [Google Scholar]
- 15.Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, Trimmer JS, Shrager P. and Peles E. (1999). Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron 24:1037–1047 [DOI] [PubMed] [Google Scholar]
- 16.Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, et al. (2003). Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol 162:1149–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Traka M, Goutebroze L, Denisenko N, Bessa M, Nifli A, Havaki S, Iwakura Y, Fukamauchi F, Watanabe K, et al. (2003). Association of TAG-1 with Caspr2 is essential for the molecular organization of juxtaparanodal regions of myelinated fibers. J Cell Biol 162:1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, et al. (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485:246–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang LS, Hranilovic D, Wang K, Lindquist IE, Yurcaba L, Petkovic ZB, Gidaya N, Jernej B, Hakonarson H. and Bucan M. (2010). Population-based study of genetic variation in individuals with autism spectrum disorders from Croatia. BMC Med Genet 11:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karayannis T, Au E, Patel JC, Kruglikov I, Markx S, Delorme R, Heron D, Salomon D, Glessner J, et al. (2014). Cntnap4 differentially contributes to GABAergic and dopaminergic synaptic transmission. Nature 511:236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han S, Tai C, Westenbroek RE, Yu FH, Cheah CS, Potter GB, Rubenstein JL, Scheuer T, de la Iglesia HO. and Catterall WA. (2012). Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature 489:385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traut W, Weichenhan D, Himmelbauer H. and Winking H. (2006). New members of the neurexin superfamily: multiple rodent homologues of the human CASPR5 gene. Mamm Genome 17:723–731 [DOI] [PubMed] [Google Scholar]
- 23.Rice DS, Northcutt GM. and Kurschner C. (2001). The Lnx family proteins function as molecular scaffolds for Numb family proteins. Mol Cell Neurosci 18:525–540 [DOI] [PubMed] [Google Scholar]
- 24.Nie J, McGill MA, Dermer M, Dho SE, Wolting CD. and McGlade CJ. (2002). LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J 21:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbin JG, Gaiano N, Juliano SL, Poluch S, Stancik E. and Haydar TF. (2008). Regulation of neural progenitor cell development in the nervous system. J Neurochem 106:2272–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiegel I, Salomon D, Erne B, Schaeren-Wiemers N. and Peles E. (2002). Caspr3 and caspr4, two novel members of the caspr family are expressed in the nervous system and interact with PDZ domains. Mol Cell Neurosci 20:283–297 [DOI] [PubMed] [Google Scholar]
- 27.Garner CC, Nash J. and Huganir RL. (2000). PDZ domains in synapse assembly and signalling. Trends Cell Biol 10:274–280 [DOI] [PubMed] [Google Scholar]
- 28.Gonzalez-Mariscal L, Betanzos A. and Avila-Flores A. (2000). MAGUK proteins: structure and role in the tight junction. Semin Cell Dev Biol 11:315–324 [DOI] [PubMed] [Google Scholar]
- 29.Sheng M. and Pak DT. (2000). Ligand-gated ion channel interactions with cytoskeletal and signaling proteins. Annu Rev Physiol 62:755–778 [DOI] [PubMed] [Google Scholar]
- 30.Scott K. and Zuker CS. (1998). Assembly of the Drosophila phototransduction cascade into a signalling complex shapes elementary responses. Nature 395:805–808 [DOI] [PubMed] [Google Scholar]
- 31.Devanathan V, Jakovcevski I, Santuccione A, Li S, Lee HJ, Peles E, Leshchyns'ka I, Sytnyk V. and Schachner M. (2010). Cellular form of prion protein inhibits Reelin-mediated shedding of Caspr from the neuronal cell surface to potentiate Caspr-mediated inhibition of neurite outgrowth. J Neurosci 30:9292–9305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J. and Conlon RA. (1997). Conservation of the Notch signalling pathway in mammalian neurogenesis. Development 124:1139–1148 [DOI] [PubMed] [Google Scholar]
- 33.Camps J, Pitt JJ, Emons G, Hummon AB, Case CM, Grade M, Jones TL, Nguyen QT, Ghadimi BM, et al. (2013). Genetic amplification of the NOTCH modulator LNX2 upregulates the WNT/beta-catenin pathway in colorectal cancer. Cancer Res 73:2003–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N. and Gotoh Y. (2004). The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 131:2791–2801 [DOI] [PubMed] [Google Scholar]
- 35.Kansaku A, Hirabayashi S, Mori H, Fujiwara N, Kawata A, Ikeda M, Rokukawa C, Kurihara H. and Hata Y. (2006). Ligand-of-Numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene 25:5071–5084 [DOI] [PubMed] [Google Scholar]
- 36.Mirza M, Hreinsson J, Strand ML, Hovatta O, Soder O, Philipson L, Pettersson RF. and Sollerbrant K. (2006). Coxsackievirus and adenovirus receptor (CAR) is expressed in male germ cells and forms a complex with the differentiation factor JAM-C in mouse testis. Exp Cell Res 312:817–830 [DOI] [PubMed] [Google Scholar]
- 37.D'Agostino M, Tornillo G, Caporaso MG, Barone MV, Ghigo E, Bonatti S. and Mottola G. (2011). Ligand of Numb proteins LNX1p80 and LNX2 interact with the human glycoprotein CD8alpha and promote its ubiquitylation and endocytosis. J Cell Sci 124:3545–3556 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.