Abstract
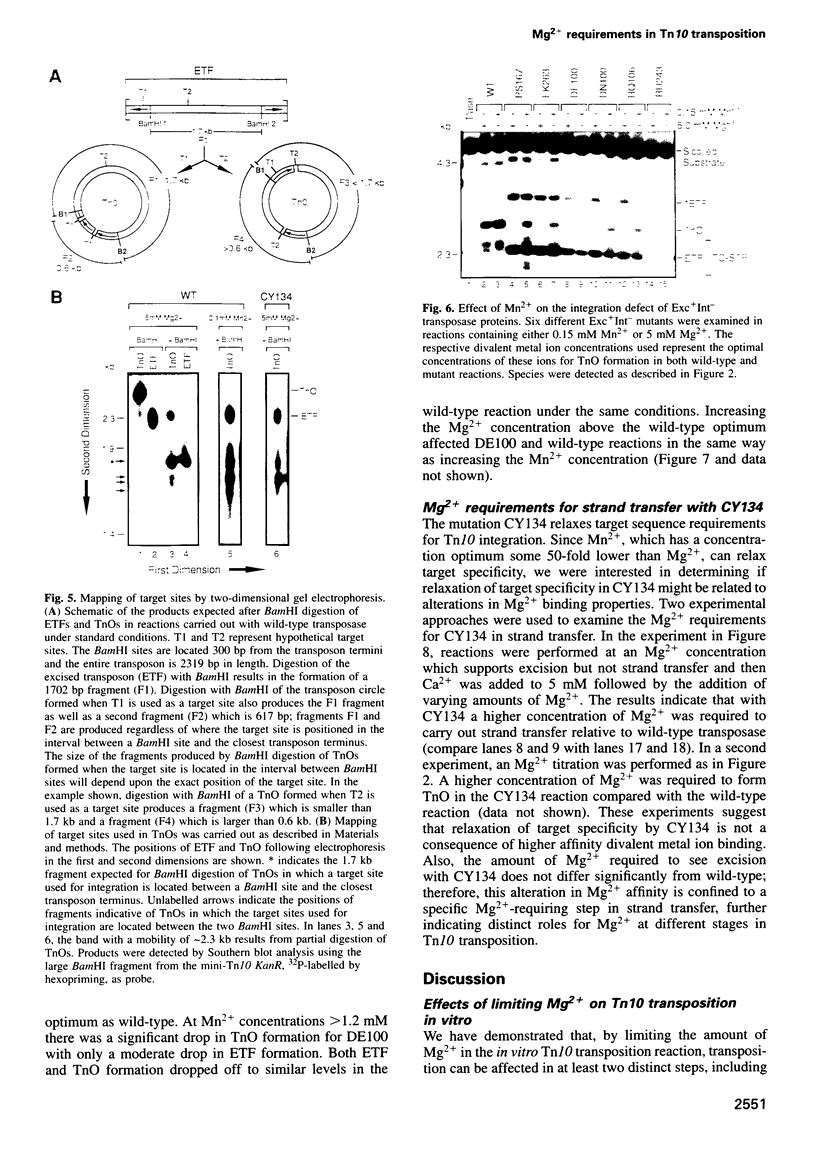
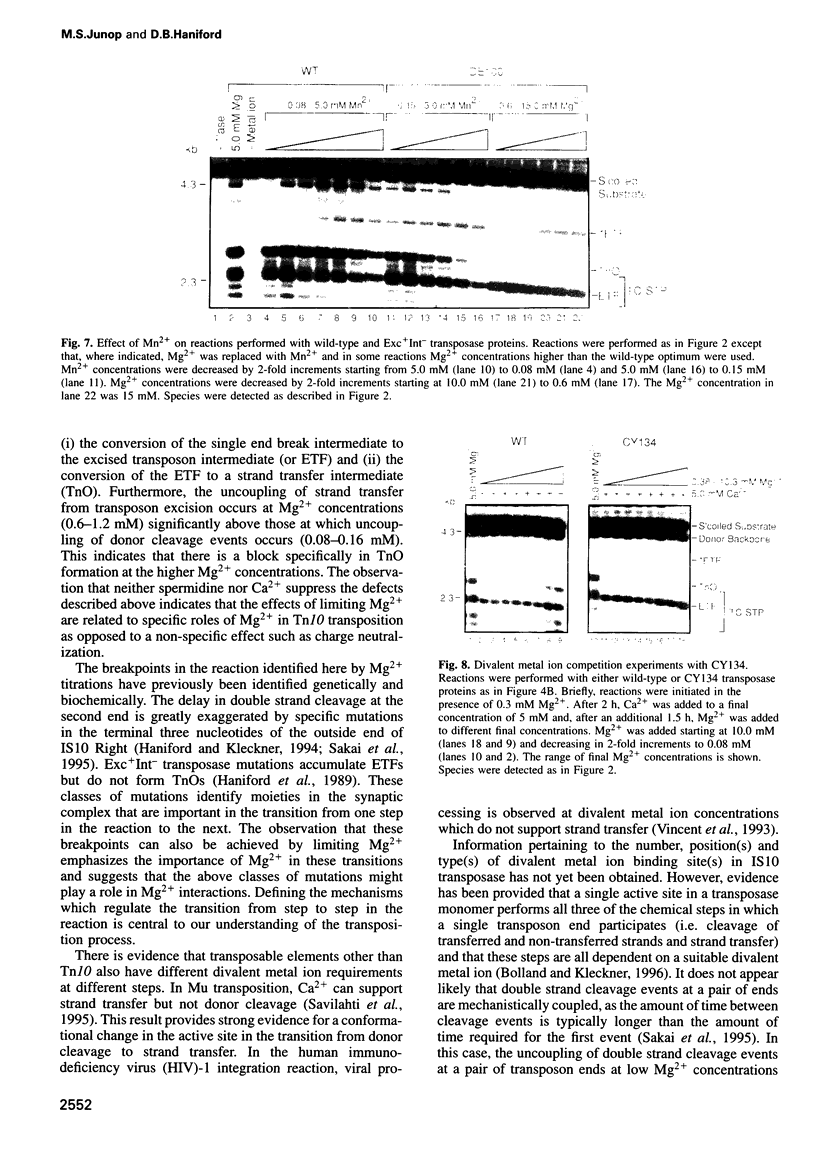
Tn10 transposition takes place by a non-replicative mechanism in which the transposon is excised from donor DNA and integrated into a target site. Mg2+ is an essential cofactor in this reaction. We have examined the Mg2+ requirements at various steps in Tn10 transposition. Results presented here demonstrate that Tn10 excision can occur efficiently at a 16-fold lower Mg2+ concentration than strand transfer and that, at Mg2+ concentrations in the range of 60-fold below the wildt-ype optimum, double strand cleavage events at the two transposon ends are completely uncoupled. These experiments identify specific breakpoints in Tn10 transposition which are sensitive to Mg2+ concentration. Whereas the uncoupling of double strand cleavage events at the two transposon ends most likely reflects the inability of two separate IS10 transposase monomers in the synaptic complex to bind Mg2+, the uncoupling of transposon excision from strand transfer is expected to reflect either a conformational change in the active site or the existence of an Mg2+ binding site which functions specifically in target interactions. We also show that Mn2+ relaxes target specificity in Tn10 transposition and suppresses a class of mutants which are blocked specifically for integration. These observations can be explained by a model in which sequence-specific target site binding is tightly coupled to a conformational change in the synaptic complex which is required for catalysis of strand transfer.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker T. A., Luo L. Identification of residues in the Mu transposase essential for catalysis. Proc Natl Acad Sci U S A. 1994 Jul 5;91(14):6654–6658. doi: 10.1073/pnas.91.14.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J., Kleckner N. Genetic evidence that Tn10 transposes by a nonreplicative mechanism. Cell. 1986 Jun 20;45(6):801–815. doi: 10.1016/0092-8674(86)90555-6. [DOI] [PubMed] [Google Scholar]
- Bender J., Kleckner N. IS10 transposase mutations that specifically alter target site recognition. EMBO J. 1992 Feb;11(2):741–750. doi: 10.1002/j.1460-2075.1992.tb05107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H. W., Kleckner N. Excision of Tn10 from the donor site during transposition occurs by flush double-strand cleavages at the transposon termini. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4648–4652. doi: 10.1073/pnas.89.10.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H. W., Kleckner N. Intramolecular transposition by Tn10. Cell. 1989 Oct 20;59(2):373–383. doi: 10.1016/0092-8674(89)90298-5. [DOI] [PubMed] [Google Scholar]
- Bolland S., Kleckner N. The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell. 1996 Jan 26;84(2):223–233. doi: 10.1016/s0092-8674(00)80977-0. [DOI] [PubMed] [Google Scholar]
- Bolland S., Kleckner N. The two single-strand cleavages at each end of Tn10 occur in a specific order during transposition. Proc Natl Acad Sci U S A. 1995 Aug 15;92(17):7814–7818. doi: 10.1073/pnas.92.17.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers R. M., Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J Biol Chem. 1994 Mar 18;269(11):8029–8035. [PubMed] [Google Scholar]
- Craig N. L. Unity in transposition reactions. Science. 1995 Oct 13;270(5234):253–254. doi: 10.1126/science.270.5234.253. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi K. Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell. 1987 Nov 6;51(3):493–501. doi: 10.1016/0092-8674(87)90645-3. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Hostomska Z., Hostomsky Z., Jordan S. R., Matthews D. A. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991 Apr 5;252(5002):88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- Dorner L. F., Schildkraut I. Direct selection of binding proficient/catalytic deficient variants of BamHI endonuclease. Nucleic Acids Res. 1994 Mar 25;22(6):1068–1074. doi: 10.1093/nar/22.6.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drelich M., Wilhelm R., Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992 Jun;188(2):459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- Halling S. M., Kleckner N. A symmetrical six-base-pair target site sequence determines Tn10 insertion specificity. Cell. 1982 Jan;28(1):155–163. doi: 10.1016/0092-8674(82)90385-3. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Benjamin H. W., Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991 Jan 11;64(1):171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Chaconas G. Mechanistic aspects of DNA transposition. Curr Opin Genet Dev. 1992 Oct;2(5):698–704. doi: 10.1016/s0959-437x(05)80129-7. [DOI] [PubMed] [Google Scholar]
- Haniford D. B., Chelouche A. R., Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989 Oct 20;59(2):385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- Heitman J. How the EcoRI endonuclease recognizes and cleaves DNA. Bioessays. 1992 Jul;14(7):445–454. doi: 10.1002/bies.950140704. [DOI] [PubMed] [Google Scholar]
- Hsu M., Berg P. Altering the specificity of restriction endonuclease: effect of replacing Mg2+ with Mn2+. Biochemistry. 1978 Jan 10;17(1):131–138. doi: 10.1021/bi00594a019. [DOI] [PubMed] [Google Scholar]
- Huisman O., Errada P. R., Signon L., Kleckner N. Mutational analysis of IS10's outside end. EMBO J. 1989 Jul;8(7):2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A., Maschke H., Selent U., Wenz C., Köhler E., Connolly B. A., Thorogood H., Pingoud A. DNA binding specificity of the EcoRV restriction endonuclease is increased by Mg2+ binding to a metal ion binding site distinct from the catalytic center of the enzyme. Biochemistry. 1995 May 9;34(18):6239–6246. doi: 10.1021/bi00018a028. [DOI] [PubMed] [Google Scholar]
- Junop M. S., Hockman D., Haniford D. B. Intragenic suppression of integration-defective IS10 transposase mutants. Genetics. 1994 Jun;137(2):343–352. doi: 10.1093/genetics/137.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzman M., Katz R. A., Skalka A. M., Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989 Dec;63(12):5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy A. K., Haniford D. B. Isolation and characterization of IS10 transposase separation of function mutants: identification of amino acid residues in transposase that are important for active site function and the stability of transposition intermediates. J Mol Biol. 1996 Mar 1;256(3):533–547. doi: 10.1006/jmbi.1996.0106. [DOI] [PubMed] [Google Scholar]
- Kim K., Namgoong S. Y., Jayaram M., Harshey R. M. Step-arrest mutants of phage Mu transposase. Implications in DNA-protein assembly, Mu end cleavage, and strand transfer. J Biol Chem. 1995 Jan 20;270(3):1472–1479. doi: 10.1074/jbc.270.3.1472. [DOI] [PubMed] [Google Scholar]
- Kim Y. C., Grable J. C., Love R., Greene P. J., Rosenberg J. M. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990 Sep 14;249(4974):1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Chalmers R. M., Kwon D., Sakai J., Bolland S. Tn10 and IS10 transposition and chromosome rearrangements: mechanism and regulation in vivo and in vitro. Curr Top Microbiol Immunol. 1996;204:49–82. doi: 10.1007/978-3-642-79795-8_3. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., Winkler F. K. Mg2+ binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 A resolution. Biochemistry. 1995 Jan 17;34(2):683–696. doi: 10.1021/bi00002a036. [DOI] [PubMed] [Google Scholar]
- Kulkosky J., Jones K. S., Katz R. A., Mack J. P., Skalka A. M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992 May;12(5):2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K. Polynucleotidyl transfer reactions in transpositional DNA recombination. J Biol Chem. 1992 Oct 25;267(30):21273–21276. [PubMed] [Google Scholar]
- Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker T. A., Mizuuchi K. Assembly of the active form of the transposase-Mu DNA complex: a critical control point in Mu transposition. Cell. 1992 Jul 24;70(2):303–311. doi: 10.1016/0092-8674(92)90104-k. [DOI] [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Polard P., Chandler M. Bacterial transposases and retroviral integrases. Mol Microbiol. 1995 Jan;15(1):13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- Rice P., Mizuuchi K. Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell. 1995 Jul 28;82(2):209–220. doi: 10.1016/0092-8674(95)90308-9. [DOI] [PubMed] [Google Scholar]
- Sakai J., Chalmers R. M., Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995 Sep 1;14(17):4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savilahti H., Rice P. A., Mizuuchi K. The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J. 1995 Oct 2;14(19):4893–4903. doi: 10.1002/j.1460-2075.1995.tb00170.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selent U., Rüter T., Köhler E., Liedtke M., Thielking V., Alves J., Oelgeschläger T., Wolfes H., Peters F., Pingoud A. A site-directed mutagenesis study to identify amino acid residues involved in the catalytic function of the restriction endonuclease EcoRV. Biochemistry. 1992 May 26;31(20):4808–4815. doi: 10.1021/bi00135a010. [DOI] [PubMed] [Google Scholar]
- Surette M. G., Buch S. J., Chaconas G. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell. 1987 Apr 24;49(2):253–262. doi: 10.1016/0092-8674(87)90566-6. [DOI] [PubMed] [Google Scholar]
- Vermote C. L., Halford S. E. EcoRV restriction endonuclease: communication between catalytic metal ions and DNA recognition. Biochemistry. 1992 Jul 7;31(26):6082–6089. doi: 10.1021/bi00141a018. [DOI] [PubMed] [Google Scholar]
- Vincent K. A., Ellison V., Chow S. A., Brown P. O. Characterization of human immunodeficiency virus type 1 integrase expressed in Escherichia coli and analysis of variants with amino-terminal mutations. J Virol. 1993 Jan;67(1):425–437. doi: 10.1128/jvi.67.1.425-437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vipond I. B., Baldwin G. S., Halford S. E. Divalent metal ions at the active sites of the EcoRV and EcoRI restriction endonucleases. Biochemistry. 1995 Jan 17;34(2):697–704. doi: 10.1021/bi00002a037. [DOI] [PubMed] [Google Scholar]
- Winkler F. K., Banner D. W., Oefner C., Tsernoglou D., Brown R. S., Heathman S. P., Bryan R. K., Martin P. D., Petratos K., Wilson K. S. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993 May;12(5):1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D. C., Groeneger A. A., Plasterk R. H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]