Abstract
Macroautophagy is an intracellular catabolic process involved in the formation of multiple membrane structures ranging from phagophores to autophagosomes and autolysosomes. Dysfunction of macroautophagy is implicated in both physiological and pathological conditions. To date, 38 autophagy-related (ATG) genes have been identified as controlling these complicated membrane dynamics during macroautophagy in yeast; approximately half of these genes are clearly conserved up to human, and there are additional genes whose products function in autophagy in higher eukaryotes that are not found in yeast. The function of the ATG proteins, in particular their ability to interact with a number of macroautophagic regulators, is modulated by posttranslational modifications (PTMs) such as phosphorylation, glycosylation, ubiquitination, acetylation, lipidation, and proteolysis. In this review, we summarize our current knowledge of the role of ATG protein PTMs and their functional relevance in macroautophagy. Unraveling how these PTMs regulate ATG protein function during macroautophagy will not only reveal fundamental mechanistic insights into the regulatory process, but also provide new therapeutic targets for the treatment of autophagy-associated diseases.
Keywords: autophagy, autophagy-related proteins, posttranslational modification
Abbreviations: AMPK, AMP-activated protein kinase; ATG, autophagy-related; MTORC1, mechanistic target of rapamycin complex 1; PE, phosphatidylethanolamine; PTM, posttranslational modification; Ub, ubiquitin; Ubl, ubiquitin like
Introduction
Autophagy, discovered more than half a century ago via electron microscopy,1-3 has been implicated in several physiological and pathological cellular processes including development, differentiation, metabolism, inflammation, myopathies, immunity, and cell death.4-7 Autophagy occurs at a constitutive basal level, but it is upregulated in response to various types of stress.8 Autophagy plays a critical role in maintaining cellular homeostasis and it is considered to be a double-edged sword, primarily acting in a cytoprotective manner, for example in the prevention of certain neurodegenerative diseases, but as a result also promoting survival in some types of cancer.9-11 In addition, although autophagy is critical to maintaining normal cell physiology, dysregulated autophagy can result in compromised cell function and even death.
At least 3 recognized types of autophagy have been identified: chaperone-mediated autophagy, which uses HSPA8/HSC70 (heat shock 70kDa protein 8) to sequester cargo proteins containing a KFERQ motif; microautophagy, which is a direct lysosomal uptake and degradative process; and the best studied, macroautophagy (hereafter referred to as autophagy). As a housekeeping process, autophagy starts with the engulfment of intracellular cargo by the phagophore that then expands through the acquisition of lipids, and ultimately seals to generate a completed double-membrane structure termed the autophagosome. The autophagosome subsequently fuses with the lysosome to form the autolysosome, which has the ability to degrade substrates to produce amino acids and presumably other metabolites such as fatty acids for recycling and reuse (Fig. 1).12 Substrates of autophagy are both selective and nonselective and include unused macromolecules (e.g., nucleic acids, proteins, carbohydrates, and lipids), damaged or superfluous organelles (e.g., endoplasmic reticulum, peroxisomes, ribosomes, mitochondria, and the nucleus), and invasive microbes.13 Multiple membrane sources (including the plasma membrane, endoplasmic reticulum, endosome, mitochondria, and Golgi apparatus) are identified as contributing to the supply of membranes needed for phagophore nucleation and/or expansion, and autophagosome formation,14 indicating that the regulation of the autophagic network is complex.15
Figure 1.
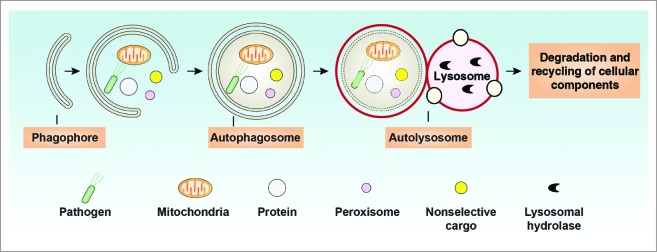
Overview of the autophagy process. Autophagy is a lysosome-mediated degradation and recycling pathway that involves the formation of multiple membrane structures ranging from phagophores to autophagosomes and autolysosomes.
Autophagy is a highly conserved process across eukaryotes.16 The study of the molecular basis of autophagy started with the discovery of Apg5 (now Atg5),16b Aut1 (now Atg3),17 Apg13 (now Atg13),18 and Apg1 (now Atg1; whose mammalian orthologs are ULK1 and ULK2 [unc-51 like autophagy activating kinases 1 and 2])19 in Saccharomyces cerevisiae in 1996-7. Currently, 38 ATG genes have been identified as controlling the induction and complicated membrane dynamics that occur during autophagy in yeasts;20 approximately half of these proteins have clear homologs, orthologs, or paralogs in mammalian cells and plants.15,16 The Atg proteins can form several functional complexes associated with other regulators involved in autophagy initiation and execution at different stages of the process as outlined below.
Induction. In yeast, induction of autophagosome formation is regulated by the Atg1-Atg13-Atg17-Atg31-Atg29 kinase complex.16 In mammals, this complex contains the core proteins ULK1 (or ULK2; hereafter we only refer to ULK1), ATG13, and RB1CC1/FIP200 (RB1-inducible coiled-coil 1; the functional ortholog of yeast Atg17), which is stable and required for induction of autophagosome formation.21,22 In mammals, ATG101 is also part of the complex; this protein, which binds to ATG13, has no known yeast ortholog,23,24 but has an ortholog in C. elegans, EPG-9.25 The mechanistic target of rapamycin complex 1 (MTORC1) and AMP-activated protein kinase (AMPK) are 2 key regulators of the ULK1 kinase complex and will be discussed in detail below. AMPK is active when cells suffer from energy starvation, thus representing a mechanism to switch on autophagy to catabolize cellular components to generate ATP. When nutrients are insufficient, MTORC1 is turned off and autophagy is again turned on, in this case providing a mechanism to restore amino acid levels through the breakdown of proteins. Thus, the regulation of autophagy induction depends, in part, on the balance between external energy and nutrient supply. Other types of stress including hypoxia, and excess reactive oxygen species can also induce autophagy.
Nucleation. During the nucleation of the phagophore, proteins and lipids are recruited for the process that ultimately leads to autophagosome formation. In both yeast and mammals, the class III phosphatidylinositol 3-kinase (PtdIns3K) complex plays an important role in the nucleation of the phagophore. The PtdIns3K complex contains the core proteins PIK3C3/VPS34 (phosphatidylinositol 3-kinase, catalytic subunit type 3), PIK3R4/p150 (phosphoinositide-3-kinase, regulatory subunit 4; the yeast ortholog is Vps15), BECN1 (Beclin 1, autophagy-related; the yeast ortholog is Vps30/Atg6) and ATG14 (also Atg14 in yeast).26-29 One of the key functions of the PtdIns3K complex is the generation of phosphatidylinositol-3-phosphate (PtdIns3P), a phosphoinositide that serves as a landmark on the membrane to recruit other factors involved in the process of autophagosome formation. Regulation of the PtdIns3K complex occurs largely through host cofactors (e.g., ATG14, UVRAG, SH3GLB1/BIF-1, KIAA0226/RUBICON, AMBRA1, HMGB1, and PINK1) that interact with BECN1,30 which is important for the crosstalk between autophagy and apoptosis.31
Elongation. Subsequent to nucleation, the phagophore expands by membrane addition, which is accomplished by 2 ubiquitin-like (Ubl) conjugation systems.32 In yeast, the first of these systems covalently conjugates the Ubl Atg12 to Atg5 by the E2-like enzyme Atg10, whereas the second system conjugates the Ubl Atg8 to phosphatidylethanolamine (PE) by the E2-like enzyme Atg3, after Atg8 has been processed by the cysteine protease Atg4.33,34 Both the Atg8 and Atg12 proteins are activated by the E1-like enzyme Atg7.35 The Atg12–Atg5 conjugate forms a complex with Atg16, which in turn promotes Atg8–PE conjugation in an E3-like manner,36 although it is not essential for this process to occur.37 Mammalian orthologs and paralogs of this system have been identified and function as in yeast. The ATG12–ATG5 conjugation system containing the core proteins ATG5, ATG12, ATG7, ATG10, and ATG16L1 (the ortholog of yeast Atg16); and the Ubl LC3 conjugation system containing the core proteins MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3, a major ortholog family, among the orthologs and paralogs of yeast Atg8), ATG3, ATG4, and ATG7. Unlike yeast, mammals have 4 isoforms of ATG4 (ATG4A to ATG4D) and 3 of LC3 (LC3A to LC3C). In addition to the LC3 subfamily, the GABARAP proteins (including GABARAP, GABARAPL1, and GABARAPL2/GATE-16), another subfamily of Atg8 paralogs in mammals, may regulate a later step of autophagosome maturation.38 As with Atg8 in yeast, LC3 lipidation requires an initial cleavage at the extreme C terminus of LC3 by the ATG4B protease. Notably, the lipidated LC3 and GABARAP paralog protein families of yeast Atg8 specifically associate with phagophore membranes, and the Atg8/LC3/GABARAP proteins determine the size of the autophagosome;39,40 this protein also participates in cargo recruitment and membrane protein complex assembly/disassembly.
In addition to Ubl conjugation systems, Atg9/ATG9-mediated cycling systems containing the core proteins Atg9/ATG9, Atg2/ATG2, and Atg18/WIPI1/2 contribute to the elongation of the phagophore. Atg9/ATG9 is thought to participate in membrane delivery from donor sources to the expanding phagophore in both yeast and mammals, although the precise role of this protein is not known.41,42 In yeast, the movement of Atg9 during phagophore elongation depends on the Atg11, Atg23, and Atg27 proteins, and multimerization of Atg9, whereas the release of Atg9 from the phagophore assembly site (PAS; the site of phagophore nucleation) involves Atg2-Atg18 and Atg1-Atg13.42,43 In mammals, the movement of ATG9 from the trans-Golgi network or late endosomes to the phagophore is regulated by the activity of ULK1, PtdIns3K, and MAPK14/p38 (mitogen-activated protein kinase 14).41,44
4. Completion and fusion. Once autophagosome formation is complete, mammalian autophagosomes move along microtubules in a dynein motor-dependent manner and cluster close to the microtubule-organizing center near the nucleus, where they fuse with lysosomes.45 In axons, this movement requires MAPK8IP1/JIP1 binding to LC3.46 In addition to microtubules, HSPB1 (heat shock 27kDa protein 1)-mediated actin filament dynamics also facilitate the movement of autophagosomes to lysosomes.47 Recent studies have suggested that the SNARE (soluble NSF [N-ethyl-maleimide-sensitive fusion protein] attachment protein receptor) family proteins (e.g., VAMP7, STX17/syntaxin 17, Tlg2, Vam3, and Ykt6) are required for autophagosome fusion with endosomes or lysosomes/vacuoles in mammals and yeast.48-50 The SNARE family consists of more than 30 members in mammalian cells and plays a central role in intracellular membrane trafficking events. Thus, the cytoskeleton along with SNARE proteins participates in autophagosome completion and fusion, reflecting the complexity of the interplay between autophagic and endocytic vesicle trafficking. However, the role of Atg/ATG proteins at this stage of the process remains unknown.
5. Degradation and efflux. The vacuole/lysosome is a degradative organelle, and in general the autophagic process is not complete until the sequestered cargo is broken down and the resulting macromolecules are released back into the cytosol. This part of autophagy relies on the many hydrolases present within the lumen, and on integral membrane transporters that are needed for efflux.51
Collectively, autophagy consists of several sequential steps and is a very complex process that involves Atg/ATG proteins and other components to assemble the required machinery.52 Accumulating evidence indicates that posttranslational modifications of Atg/ATG proteins afford significant flexibility for the regulation of autophagic and nonautophagic pathways.53,54 Here, we highlight emerging evidence for the PTM-mediated regulation of Atg/ATG proteins and its functional relevance in autophagy.
Posttranslational Modifications and Atg/ATG Proteins
PTMs play a critical role in the regulation of protein activity involved in almost all aspects of cell structure and function.55 They generally are biochemical processes involving chemical modification of a protein after its translation, resulting in inhibition or enhancement of the protein's activity.56 The major types of PTMs include phosphorylation, glycosylation, ubiquitination, methylation, acetylation, lipidation, and proteolysis. A protein can be modified multiple times by different PTMs or by the same PTM at different residues. PTMs can also occur in a separate or sequential manner on a single protein site.55 In addition, PTMs can be either reversible or irreversible depending on the nature of the modification. Below, we discuss several common types of PTMs that occur on Atg/ATG proteins as well as non-ATG proteins involved in the regulation of autophagy.57-59
Phosphorylation of Atg/ATG Proteins
Phosphorylation is a ubiquitous regulatory mechanism in protein modifications, during which a phosphate (or more than one in many cases) is added. Protein kinases and phosphatases are responsible for catalyzing the addition or removal of a phosphate from their substrates, respectively. Phosphorylation can occur on several amino acids such as serine (“Ser”), threonine (“Thr”), tyrosine (“Tyr”), histidine, and aspartate (“Asp”). In eukaryotes, phosphorylation on serine is the most common, followed by threonine and tyrosine. Protein phosphorylation could act by introducing a negative charge at a specific site that influences a protein's structural conformation, enzymatic activities, molecular association, and/or subcellular localization in signaling pathways.
Atg1/ULK1 complex
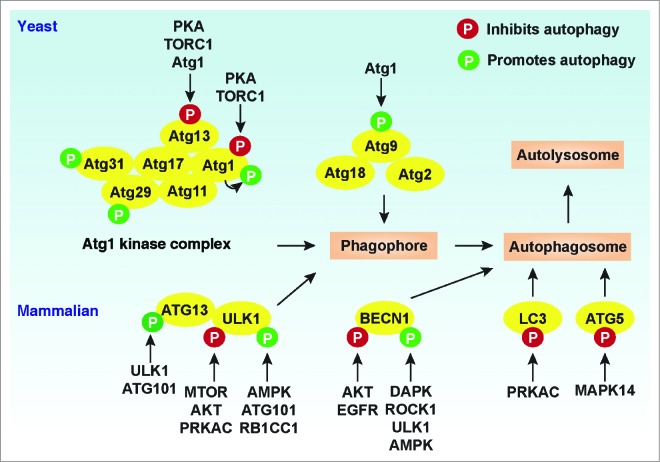
Phosphorylation events play vital roles in the initiation phase of autophagy via regulating Atg1/ULK1 complex assembly and activity in yeast and mammalian cells (Fig. 2). Different laboratories have observed that various protein kinases are responsible for phosphorylation of Atg1/ULK1 at different amino acid sites during autophagy (Table 1). In yeast, the Atg1 complex is composed of several proteins such as Atg1 and Atg13 that are essential for PAS formation as well as retrieval of Atg9 from the PAS to the cytoplasmic pool.42,60 In yeast, the Atg1-Atg13 complex is thought to act downstream of TORC1, a protein complex that functions as a nutrient sensor and controls protein synthesis.61 Atg1 and Atg13 are normally phosphorylated through association with TORC1 under nutrient-rich conditions.62-64 Under starvation conditions or treatment with the TORC1 inhibitor rapamycin, inhibition of TORC1 activities can lead to rapid dephosphorylation of Atg13 at Ser348, Ser437, Ser438, Ser496, Ser535, Ser541, Ser646, and Ser649 and concomitant dephosphorylation of Atg1, which leads to the induction of autophagy.64,65 PKA (protein kinase A) is a second messenger-dependent enzyme that is involved in the regulation of various cellular processes, including autophagy.66,67 Inactivation of the PKA signaling pathway is sufficient to induce autophagy in yeast through dephosphorylation of Atg13 at Ser344, Ser347, and Ser581, and Atg1 at Ser508 and Ser515, which regulates their localization at the PAS.65,68 Furthermore, dephosphorylation of Atg13 may facilitate its interaction with Atg1 or Atg17 and the induction of autophagy. 61,65
Figure 2.
Regulation of Atg/ATG proteins by phosphorylation. In yeast, the initiation of autophagy involves the inhibition of TORC1- and PKA-mediated phosphorylation of Atg1 and Atg13 under starvation conditions. Atg1 is then activated via autophosphorylation, which is responsible for Atg9 localization to the phagophore assembly site. In addition, phosphorylation of Atg29 stimulates autophagy by activating the Atg17-Atg31-Atg29 complex, which is required for subsequent interaction with Atg11. In mammalian cells, phosphorylation of ULK1 and BECN1 by the indicated kinases has dual roles in the regulation of the autophagic response. Phosphorylation of LC3 and ATG5 by PRKAC and MAPK14, respectively, inhibits autophagy. In addition, ULK1- and ATG101-mediated ATG13 phosphorylation promotes selective autophagy.
Table 1.
Regulation of Atg/ATG proteins by PTMs in autophagy
| Yeast | Mammal | PTM | Site | Activated by | PTM Functions | References |
|---|---|---|---|---|---|---|
| Atg1 | ULK1 | Phosphorylation | Thr180 (human) | Autophosphorylation | Maintains ULK1 kinase activity | 76 |
| Phosphorylation | Thr226 (yeast) | Autophosphorylation | Maintains Atg1 kinase activity | 70 | ||
| Phosphorylation | Ser230 (yeast) | Autophosphorylation | Maintains Atg1 kinase activity | 62 | ||
| Phosphorylation | Ser317 (human) Ser777 (human) |
AMPK | Inhibits MTORC1 and promotes autophagy under glucose starvation | 80 | ||
| Phosphorylation | Ser467 (mouse) Ser555 (mouse) Thr574 (mouse) Ser637 (mouse) |
AMPK | Promotes mitophagy | 195 | ||
| Phosphorylation | Ser508 (yeast) Ser515 (yeast) |
PKA | Prevents its association with the PAS and inhibits autophagy |
68 | ||
| Phosphorylation | Ser555 (human) | AMPK | Promotes ULK1 binding to YWHA/14-3-3 | 76 | ||
| Phosphorylation | Ser555 (human) Ser637 (human) Thr659 (human) |
AMPK | Promotes localization of ATG9 to perinuclear clusters |
84 | ||
| Phosphorylation | Ser638 (human) Ser758 (human) |
MTORC1 | Inhibits autophagy upon starvation | 79 | ||
| Phosphorylation | Ser774 (human) | AKT | Inhibits autophagy in response to insulin | 76 | ||
| Phosphorylation | Ser757 (mouse) | MTORC1 | Inhibits ULK1 binding to AMPK and subsequent autophagy | 80 | ||
| Phosphorylation | Ser1043 (mouse) | PRKAC/PKA | Inhibits autophagy | 75 | ||
| Phosphorylation | Ser1047 (mouse) | Autophosphorylation | Promotes the closed clamp conformation of the ULK1 complex |
75 | ||
| Phosphorylation | NA (human) | ATG101 | ATG101 lacks kinase activity. Binds to ATG13 and promotes autophagy |
23 | ||
| Phosphorylation | NA (mouse) | RB1CC1/FIP200 | RB1CC1 lacks kinase activity. Increases ULK1 stability and autophagy | 78 | ||
| Acetylation | Lys162 (human) Lys606 (human) |
KAT5/TIP60 | Stimulates starvation-induced autophagy | 165 | ||
| Ubiquitination | ND (human) | TRAF6 AMBRA1 |
Promotes ULK1 self-association and activity during autophagy | 134 | ||
| Atg3 | ATG3 | Acetylation | Lys19 (yeast) Lys48 (yeast) |
Esa1 | Increases interaction between Atg3 and Atg8 | 161 |
| Acetylation | Lys183 (yeast) | Esa1 | Increases Atg3-mediated Atg8 lipidation | 161 | ||
| Deacetylation | Lys19 (yeast) Lys48 (yeast) |
Rpd3 | Inhibits autophagy | 161 | ||
| Proteolysis | Asp169 (human) | CASP8 | Inhibits autophagy and promotes apoptosis | 185 | ||
| Sulfation | Cysteine (yeast) | Sulfate ion | Increases Atg3-mediated Atg8 lipidation | 193 | ||
| Atg4 | ATG4A/B/C/D | Proteolysis | C-terminal Asp63 (human) | CASP3 | Increases both autophagy and apoptosis | 187 |
| Atg5 | ATG5 | Phosphorylation | Thr75 (mouse) | MAPK14/p38 | Inhibits autophagy | 121 |
| Acetylation | ND (human) | EP300/p300 | Inhibits autophagy | 163 | ||
| Deacetylation | ND (human and mouse) | SIRT1 | Stimulates starvation-induced autophagy | 164 | ||
| Proteolysis | ND (human) | CASP3, 6, 8, 9, 10 | Increases apoptosis and inhibits autophagy | 188 | ||
| Proteolysis | C-terminal Thr193 (human) | CAPN/calpain | Increases apoptosis and inhibits autophagy | 183 | ||
| Atg6 | BECN1 | Phosphorylation | Ser14 (human) | ULK | Increases PtdIns3K complex activity in response to amino acid withdrawal | 92 |
| Phosphorylation | Ser91 (human) Ser94 (human) |
AMPK | Increases PtdIns3K complex activity and membrane association in autophagy or mitophagy | 104,115 | ||
| Phosphorylation | Thr119 (human) | DAPK or ROCK1 |
Dissociates BECN1 from BCL2 | 108,109 | ||
| Phosphorylation | Tyr229 (human) Tyr233 (human) Tyr352 (human) |
EGFR | Inhibits PtdIns3K complex activity | 114 | ||
| Phosphorylation | Ser234 (human) Ser295 (human) |
AKT | Inhibits PtdIns3K complex activity | 113 | ||
| Ubiquitination | Lys117 (mouse) | TRAF6 | Dissociates BECN1 from BCL2 and promotes autophagy | 130 | ||
| Ubiquitination | Lys349 (human) | NEDD4 | Promotes BECN1 degradation | 132 | ||
| Ubiquitination | Lys437 (human) | AMBRA1 | Increases PtdIns3K complex activity and autophagy | 131 | ||
| Deubiquitination | ND (human and mouse) | USP10 USP13 |
Increases PtdIns3K complex activity | 133 | ||
| Proteolysis | C-terminal Asp149 (mouse and human) Asp146 (human) Asp133 (mouse and human) |
CASP3, 6, 8, 9, 10 | Increases apoptosis and inhibits autophagy | 184,188-192 | ||
| Atg7 | ATG7 | Acetylation | ND (human) | EP300/p300 | Inhibits autophagy | 163 |
| Deacetylation | ND (human and mouse) | SIRT1 | Stimulates starvation-induced autophagy | 164 | ||
| Atg8 | LC3A/B/C, GABARAP, GABARAPL1/2 | Phosphorylation | Thr6 Thr29 |
PRKC/PKC | Has no impact on autophagy | 123 |
| Phosphorylation | Ser12 (human LC3) | PRKAC/PKA | Inhibits LC3 recruitment to autophagosome | 122 | ||
| Lipidation | Gly116 (yeast), C-terminal Gly in human and mouse LC3, GABARAP, GABARAPL1/2) | Atg3/ATG3 Atg7/ATG7 |
Converts to phagophore-associating form, LC3-II | 34,196-198 | ||
| Proteolysis | Lys65- Gln77 (human LC3) | 20S proteasome | Degrades LC3 | 181 | ||
| Proteolysis | Arg68 (human LC3) | ATG4B | Affects C-terminal cleavage of LC3B | 176 | ||
| Proteolysis | Gly116 (yeast), C-terminal Gly in human and mouse, LC3, GABARAP, GABARAPL1/2) | Atg4/ATG4A/B | Promotes processing and deconjugation of Atg8/LC3/ GABARAP | 40,167,177 | ||
| Proteolysis | ND (mouse LC3) | CTSE/cathepsin E | Degrades autophagosomal membrane components | 180 | ||
| Acetylation | ND (human LC3) | EP300/p300 | Inhibits autophagy | 163 | ||
| Deacetylation | ND (human and mouse LC3) | SIRT1 | Stimulates starvation-induced autophagy | 164 | ||
| Atg9 | ATG9A/B | Phosphorylation | Multiple Ser residues (yeast) | Atg1 | Recruits Atg8 and Atg18 to the site of autophagosome formation and expands the phagophore membrane | 89 |
| Glycosylation | Asn99 (human ATG9A) | ND | Coordinates membrane transport from donor sources to site of autophagosome formation | 41 | ||
| Atg11 | NA | Phosphorylation | ND (yeast) | ND | Scaffold protein in selecting autophagy for PAS organization |
72 |
| Atg12 | ATG12 | Acetylation (human) | ND | EP300/p300 | Inhibits autophagy | 163 |
| Deacetylation (human and mouse) | ND | SIRT1 | Stimulates starvation-induced autophagy | 164 | ||
| Atg13 | ATG13 | Phosphorylation | Ser48 (human) Thr170 (human) Thr331 (human) Thr428 (human) Thr478 (human) |
ULK1 | Has no impact on starvation-induced autophagy | 91 |
| Phosphorylation | Ser318 (human) | ULK1 | Promotes mitophagy | 90 | ||
| Phosphorylation | Ser344 (yeast) Ser437 (yeast) Ser581 (yeast) |
PKA | Regulates the association of Atg13 with the PAS, and inhibits autophagy | 65 | ||
| Phosphorylation | S348 (yeast) S437 (yeast) S438 (yeast) S496(yeast) S535(yeast), S541(yeast) S646 (yeast) and S649 (yeast) | TORC1 | Regulates the interaction between Atg13 and Atg1, and inhibits autophagy | 64 | ||
| Phosphorylation | ND (Drosophila) |
Atg1 | Promotes Atg13 activity and autophagy | 69 | ||
| Phosphorylation | N/A (human) | ATG101 | Binds to ATG13 and promotes autophagy | 23 | ||
| Atg16 | ATG16L1 | Proteolysis | C-terminal Thr300 (human) | CASP3 | Inhibits autophagy and promotes apoptosis | 186 |
| Atg18 | WIPI1/2 | Phosphorylation | ND (yeast) | PKA | Regulates the lipid binding capacity | 68 |
| Atg29 | N/A | Phosphorylation | Multiple Ser and Thr residues such as Ser197, Ser199, and Ser201 (yeast) | ND | Binds to Atg11 and initiates PAS assembly | 71 |
| Atg30 | N/A | Phosphorylation | Ser112 (yeast) Ser71 (yeast) |
ND | Binds to Atg11 or Atg8, and promotes pexophagy | 116,117 |
| Atg32 | N/A | Phosphorylation | Ser114 (yeast) | MAPK | Binds to Atg11 or Atg8, and promotes mitophagy | 116,118-120 |
| Atg36 | N/A | Phosphorylation | Ser31 (yeast) Ser97 (yeast) |
ND | Binds to Atg11 or Atg8, and promotes pexophagy | 116 |
NA, not applicable; ND, not determined.
In addition to yeast and mammals, Atg13 phosphorylation levels are influenced by TORC1 and Atg1 kinases in Drosophila.60,69 Atg1 facilitates hyperphosphorylation of Atg13 and autophagy induction in response to starvation in Drosophila.69 Collectively, these findings suggest that dephosphorylation/phosphorylation of Atg13 acts as an initiating trigger for autophagy through inactivation of TORC1 and PKA (PRKAC in mammals) signals or activation of Atg1/ULK1 kinase depending on the context and species.
Autophosphorylation at Thr226 and Ser230 within the Atg1 kinase-activation loop is required for sustaining Atg1 kinase activity during autophagy induction following treatment with the TORC1 inhibitor rapamycin in yeast.62,70 The presence of Atg13 and Atg17 is required for Atg1 autophosphorylation, although the mechanism remains unknown.70 Besides Atg1 and Atg13, the Atg1 kinase complex in yeast also contains a complex composed of Atg17-Atg31-Atg29. This complex is constitutively formed and activated by the phosphorylation of Atg29 at multiple residues such as Ser197, Ser199, and Ser201 in the C terminus (amino acids 101 to 213) under nitrogen starvation conditions, but the kinases that are responsible remain unknown.71 Phosphorylation of Atg29 likely promotes a conformational change, which is required for the induction of autophagy and its interaction with phosphorylated Atg11, a scaffold protein for binding multiple Atg proteins (e.g., Atg9, Atg19, Atg20, Atg24, Pichia pastoris Atg30, Atg32, and Atg36) in currently identified selective types of autophagy including the cytoplasm-to-vacuole targeting (Cvt) pathway, mitophagy, and pexophagy.71,72 The series of phosphorylation events controlling these interactions can be quite complex. For example, phosphorylation of Atg19 by Hrr25 and of Atg32 by casein kinase 2 is required for their interaction with Atg11. 73,74
Phosphorylation plays dual roles in regulating the scaffolding and kinase functions of ULK1 to coordinate the autophagic response in mammalian cells.75 In human embryonic kidney (HEK)-293 cells, autophosphorylation at Thr180 within the ULK1 kinase-activation loop is required for sustaining ULK1 kinase activity and its intracellular localization to phagophores upon starvation.76 In mammalian cells, MTORC1 contains 5 components: MTOR, the catalytic subunit of the complex; RPTOR/Raptor (regulatory associated protein of MTOR, complex 1); MLST8/GβL (MTOR associated protein, LST8 homolog [S. cerevisiae]); AKT1S1/PRAS40 (AKT1 substrate [proline-rich]); and DEPTOR (DEP domain containing MTOR-interacting protein).77 In human HEK-293 cells, MTORC1 complex components (e.g., RPTOR and MTOR) interact with ULK1/2, which causes ULK1/2 phosphorylation and subsequent ATG13 phosphorylation under nutrient-rich conditions.21,22 Moreover, MTORC1-mediated phosphorylation of ATG13 and ULK1/2 has negative effects on ULK activity.21 ULK1/2 can directly phosphorylate ATG13 and RB1CC1 to regulate autophagosome formation.21 Interestingly, RB1CC1 and ULK1 are functionally connected, and RB1CC1 is also required for the stability and phosphorylation of ULK1 in mouse cells.78
The interactions between MTORC1 and ULK1/2 significantly decrease during starvation, which leads to dephosphorylation of ULK1/2 and ATG13, increased kinase activity of ULK, and subsequent induction of autophagy.21,22 Several MTORC1-mediated phosphorylation sites of ULK1 have been recently mapped. For example, MTORC1 is required for phosphorylation of ULK1 at Ser638 and Ser758 in human cancer cells (e.g., HeLa and U2OS) under nutrient-rich conditions and these sites are dramatically dephosphorylated upon starvation.79 Phosphorylation of ULK1 at Ser757 by MTORC1 inhibits the ULK1-AMPK interaction and subsequent autophagy in mouse cells.80 In contrast, MTORC1-mediated phosphorylation of ULK1 at Ser758 in human U2OS cell results in a reassociation between ULK1 and AMPK and subsequent inhibition of autophagy.79 These studies have revealed a model for ULK1 regulation and autophagy induction involving MTORC1-dependent signaling by which MTORC1 can downregulate autophagy through inactivation and destabilization of ULK1.
The serine/threonine family of kinases termed AKT (v-akt murine thymoma viral and oncogene homolog) regulates multiple biological processes including cell survival, proliferation, and autophagy. AKT is required for phosphorylation of ULK1 at Ser774 in human HEK-293 cells, which limits the autophagic response following insulin treatment.76 In addition to MTORC1 and AKT, PRKAC/PKA (protein kinase A, cAMP-activated) negatively regulates autophagy by direct phosphorylation of ULK1 at Ser1043 in mouse cells, preventing ULK1 activation; ULK1 autophosphorylation at Ser1047 promotes phosphorylation at the Ser1043 site.75
In contrast to the aforementioned negative regulators, AMPK is generally a positive regulator of ULK1 activity. When cellular energy level drops, AMPK is activated and MTORC1 activity becomes inhibited by AMPK-mediated phosphorylation of TSC2 (tuberous sclerosis 2) at Thr1271 and Ser1387, and by phosphorylation of RPTOR at Ser722 and Ser792, which leads to the induction of autophagy.81 In addition to inhibition of MTORC1 activity by AMPK, ULK1 is conversely activated by phosphorylation at multiple sites by AMPK in response to glucose starvation82 and cyclic dinucleotides;83 phosphorylation at Ser317 and Ser777 initiates autophagy in human HEK-293 cells.80 Another study indicates that ULK1 contains 4 AMPK substrate motif sites at Ser467, Ser555, Thr574, and Ser637. These sites are conserved in higher eukaryotes, and as far back as Caenorhabditis elegans (Ser555 and Ser574).82 Importantly, AMPK-mediated ULK1 phosphorylation at Ser467, Ser555, Thr574, and Ser637 is required all or in part for mitophagy induction,82 ATG9 localization,84 TMEM173/STING (transmembrane protein 173)-mediated type I interferon immune response pathway activation,83 and ULK1 interaction with YWHA/14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein).76,84,85
Furthermore, ATG101 was discovered as a binding protein for ATG13 that is important for the stability and basal phosphorylation of both ATG13 and ULK1.23,24 However, it is unlikely that ATG13 and ULK1 are direct subtracts of ATG101, because currently ATG101 is not known to have kinase activity. ULK1 can phosphorylate AMPK and MTORC1 subunits such as RPTOR.86-88 Further studies show that the components of the autophagy machinery such as yeast Atg9 (multiple Ser residues),89 ATG13 (Ser318, Ser48, and multiple Thr residues),90,91 RB1CC1, AMBRA1 (autophagy/beclin-1 regulator 1), BECN1 (Ser14), and DAPK3/ZIPK (death-associated protein kinase 3) are direct phosphorylation targets of Atg1/ULK1 kinase, and these modifications result in complicated negative or positive feedback loop effects.80,89,92-95 Together, these findings suggest that the Atg1/ULK1 serine-threonine kinase can serve as a convergence point for multiple signals that control autophagy and other cellular processes through its alterations in phosphorylation status as well as kinase activity.
Vps34/PIK3C3 and the PtdIns3K complex
Autophagosome formation relies on the generation of PtdIns3P at the PAS and/or on the phagophore membrane by a protein complex containing the PtdIns3K, whose catalytic subunit is termed Vps34/PIK3C3. In yeast, Vps34 forms 2 distinct PtdIns3K complexes; complex I contains Vps34-Vps15-Vps30/Atg6-Atg14-Atg38 and complex II is comprised of Vps34-Vps15-Vps30/Atg6-Vps38. Complex I is involved in generating autophagic membranes, whereas complex II is involved in the vacuolar protein sorting pathway. Vps15 is a serine/threonine protein kinase that mediates phosphorylation of Vps34, a PTM that is required to form the PtdIns3K complexes.96 Vps34 is the sole enzyme in yeast that can phosphorylate phosphatidylinositol to form PtdIns3P. The cellular level and activity of PtdIns3P is also controlled by PtdIns3P phosphatases that belong to the myotubularin family.97 In yeast, the PtdIns3P phosphatase Ymr1 is recruited to the PAS at an early stage of autophagy; subsequent dephosphorylation of PtdIns3P by Ymr1 is required for Atg protein disassembly during autophagosome completion.98 In contrast, the PtdIns3P phosphatase MTMR14/Jumpy (myotubularin related protein 14), and PTPRD/PTPσ (protein tyrosine phosphatase, receptor type, D) act as negative regulators of autophagy by dephosphorylating PtdIns3P or inhibiting PtdIns3P expression in C. elegans and/or mammalian cells.99-102 In addition, depletion of PtdIns3P phosphatases MTMR6 and MTMR7 increase LC3-II in mammalian cells suggesting they are also negative regulators of autophagy, although the mechanism is unknown.100 These findings suggest that phosphorylation/dephosphorylation of PtdIns3P can either positively or negatively regulate autophagic initiation depending on the PtdIns3P kinases and phosphatases involved.
The human core complex of the PtdIns3K complex consists of 3 major components including PIK3C3/VPS34, PIK3R4/p150, and BECN1. Additional components contribute to the formation of at least 3 distinct PtdIns3K complexes that are either stimulatory or inhibitory for autophagy. Human ATG14/ATG14L/BARKOR and UVRAG are likely functional orthologs of yeast Atg14 and Vps38, respectively, and along with AMBRA1 function in stimulatory complexes; KIAA0226/Rubicon is a component of the inhibitory complex. Phosphorylation of PIK3C3 at Thr159 and Thr668 by CDK1 (cyclin-dependent kinase 1), CDK5, or AMPK inhibits autophagy,103,104 whereas phosphorylation of PIK3C3 at multiple sites by PRKD/PKD (protein kinase D) promotes autophagy.105 BECN1, a mammalian ortholog of Vps30/Atg6, is considered to function in part as a scaffold/platform, which allows multiple signals to converge.31
Following starvation or MTOR inhibition, activated ULK1 directly phosphorylates BECN1 on Ser14 to increase PtdIns3K complex activation, and participates in autophagy initiation by activation of ATG14-bound PIK3C3 in human HEK-293 cells.92 There are 2 general classes of BCL2 (B-cell CLL/lymphoma 2) proteins; antiapoptotic proteins, which include BCL2, BCL2L1/BCL-xL, BCL2L2/BCL-W, and MCL1 that inhibit autophagy, and proapoptotic BH3-only proteins such as BNIP3, BAD, BIK, PMAIP1/NOXA, BBC3/PUMA, and BCL2L11/BIMEL that induce autophagy.106 Interaction of BCL2 with BECN1 prevents the association of BECN1 with PIK3C3 and this process is negatively regulated by phosphorylation.107 Phosphorylation of BECN1 at Thr119 by DAPK or ROCK1 (Rho-associated, coiled-coil containing protein kinase 1) triggers BCL2L1 or BCL2 dissociation from BECN1 in human HEK-293 or HeLa cells, therefore activating the PtdIns3K complex and autophagy upon nutrient deprivation.108,109 Moreover, MAPK8/JNK1 (mitogen-activated protein kinase 8) and MAPK1/ERK2 mediate phosphorylation of BCL2 and promote autophagy by facilitating the association of BECN1 with PIK3C3 or HMGB1 (high mobility group box 1), respectively.110,111 HMGB1 is a nuclear protein that translocates to the cytoplasm to induce autophagy through binding BECN1111 although HMGB1-independent autophagy exists.112 In contrast, AKT-mediated BECN1 phosphorylation at Ser295 and Ser234 limits autophagy in human cancer cells through promoting YWHA/14-3-3-dependent BECN1 sequestration to the cytoskeletal protein VIM/vimentin, which contributes to a number of cancer hallmarks ranging from cell proliferation and survival to angiogenesis and increased metabolism.113 The oncogenic receptor tyrosine kinase EGFR (epidermal growth factor receptor)-mediated BECN1 phosphorylation at Tyr229, Tyr233 and Tyr352 decreases PtdIns3K complex activation and autophagy in lung cancer cells, which may contribute to tumor progression and chemoresistance.114 Moreover, AMPK activates autophagy and mitophagy in response to starvation or valinomycin by phosphorylation of BECN1 at Ser91 and Ser94, and this process depends on ATG14.104,115 Thus, Vps34/PIK3C3-PtdIns3K complex activity in autophagy is tightly controlled, either negatively or positively, by phosphorylation of its components.
Other
Besides Atg1/ULK1 and the Vps34/PIK3C3-containing PtdIns3K complex, phosphorylation can fine-tune the function of other Atg proteins (e.g., Atg30, Atg32 and Atg36) involved in pexophagy and mitophagy in yeast116-120 and the activity of Ubl systems in mammalian cells (Table 1). In mammalian cells, ATG5 is a component of the ATG12–ATG5 conjugation system, acting as an E3-like ubiquitin ligase for LC3 conversion to the PE-conjugated LC3-II form as indicated above. This conversion is critical for autophagosome formation. Phosphorylation of ATG5 at Thr75 by MAPK14 inhibits starvation- and lipopolysaccharide-induced autophagy, which indicates a negative regulatory role of MAPK14 in autophagosome maturation.121 LC3 conversion to the lipidated form and its localization to the phagophore or autophagosome membrane serve as a marker for monitoring autophagy.40 PRKAC/PKA-mediated LC3 phosphorylation at Ser12 in the human SH-SY5Y neuroblastoma cell line prevents LC3 turnover and recruitment to the phagophore in response to rapamycin and parkinsonian neurotoxin (e.g., 1-methyl-4-phenylpyridinium).122 In contrast, PRKC/PKC inhibits autophagy in an LC3 phosphorylation-independent manner, although PRKC mediates LC3 phosphorylation at Thr6 and Thr29 following phorbo-12-myristate-13-acetate/PMA or calyculin A treatment in HEK-293 cells.123 Further systematic analyses will facilitate our understanding of the relationship between LC3 phosphorylation and autophagy induction in response to different stressors.
Glycosylation of ATG Proteins
Glycosylation is one of the major PTMs that have a significant effect in the biosynthetic-secretory pathways involving the endoplasmic reticulum and Golgi apparatus. Approximately half of all proteins undergo this modification in which sugar moieties are added to specific amino acids; glycosylation can be critical to achieve proper protein folding, distribution, stability, and activity.
ATG9/Atg9 is a multispanning membrane protein that functions in the delivery of lipid that is used for the expansion of the phagophore at the early stage of autophagy.42,124,125 Self-interaction and aggregates of Atg9 are required for its trafficking and function at the step of phagophore expansion in yeast.43 There are 2 isoforms of ATG9 in mammalian cells; ATG9A is ubiquitously detected in adult tissues, while ATG9B is highly expressed in the placenta (trophoblast cells) and pituitary gland.126 ATG9 is the only transmembrane ATG protein that is absolutely required for autophagosome formation. ATG9A is normally localized in the trans-Golgi network and late endosomes, whereas treatment with the MTORC1 inhibitor rapamycin or amino acid starvation can cause its redistribution to LC3-positive autophagic vesicles.41 ATG9A has 4 possible N-glycosylation sites (N99, N129, N224, and N507), and only Asn99 is known to be glycosylated in human HEK-293 cells.41 In part due to its glycosylation, ATG9A appears to function in coordinating membrane transport from donor sources to the autophagosome formation site.41,127
Ubiquitination of ATG Proteins
Ubiquitination is one of the crucial mechanisms for the destruction of cellular proteins after their synthesis to implement a continual state of protein flux. Proteins that are tagged by attachment of the 8-kDa, 76 amino acid polypeptide ubiquitin (Ub) are then recognized by the 26S proteasome to catalyze the degradation of the ubiquitinated protein. The completion of the reaction cascade requires the Ub activating enzyme (E1), a Ub conjugating enzyme (E2), a Ub ligase (E3), and, in some cases, a Ub chain elongation factor (E4).128 Ubiquitination of targeted proteins leads to either their degradation or alteration of signaling capabilities.
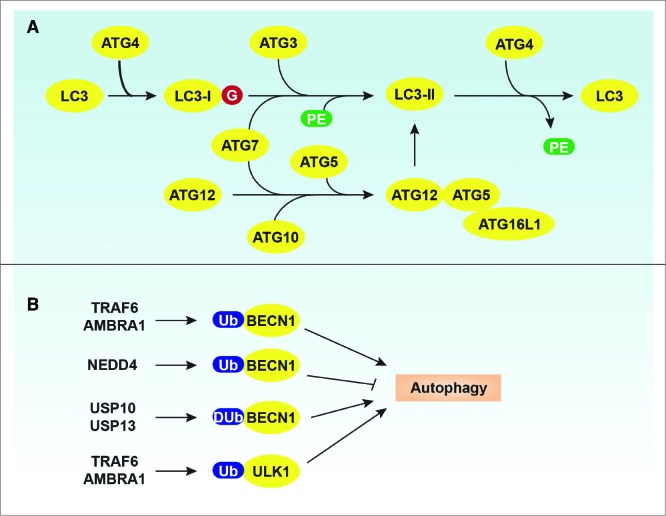
Ubiquitination plays multiple roles in the regulation of autophagy in organisms ranging from yeast to mammals, although the mechanism may be distinct from the ubiquitin proteasome pathway.16,129 Ubiquitination events are considered to be involved in complex assembly and translocation through the regulation of ATG protein levels as well as their interactions with other proteins. Aside from the 2 Ubl conjugation systems (ATG12–ATG5 and LC3-II) that are responsible for phagophore expansion during autophagy as discussed in the Introduction (Fig. 3A),33,34 actual ubiquitination occurs on ATG proteins such as BECN1 and ULK1 (Fig. 3B). BCL2 binding to BECN1 prevents PtdIns3K complex activity, suggesting that dissociation between BECN1 and BCL2 is required for autophagy induction.107 In addition to BECN1 phosphorylation,108 Lys63-linked ubiquitination of BECN1 at Lys117 by TRAF6 (TNF receptor-associated factor 6, E3 ubiquitin protein ligase)130 or at Lys437 by AMBRA1131 regulate its binding to BCL2 as well as subsequent PtdIns3K complex activity. TRAF6-mediaed ubiquitination of BECN1 amplifies lipopolysaccharide-, IFNG/interferon γ- and amino acid starvation-induced autophagy in mouse macrophages.130 WASH (WAS protein family homolog/Wiskott–Aldrich syndrome protein [WASP] and SCAR homolog), a member of the WASP family with a role in endosomal sorting, inhibits AMBRA1-mediated ubiquitination of BECN1 and starvation-induced autophagy in human HeLa cancer cells.131 Lys11- and Lys63-linked ubiquitination of BECN1 by NEDD4 (neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase) promote BECN1 proteasomal degradation in the absence of PIK3C3 in human HeLa cancer cells.132 Moreover, deubiquitinating enzymes such as USP10 (ubiquitin specific peptidase 10) and USP13 (ubiquitin specific peptidase 13 [isopeptidase T-3]) are involved in the regulation of BECN1 ubiquitination in human and mouse cells, which promotes autophagy.133 In contrast, USP10 and USP13 are inhibited by spautin-1, a novel inhibitor of autophagy, which causes increased ubiquitination and degradation of BECN1 in cancer cells.133 Additionally, AMBRA1 interacting with TRAF6 promotes Lys63-linked ubiquitination of ULK1 and its subsequent stabilization and activity during autophagy induction, whereas MTORC1-mediated AMBRA1 phosphorylation at Ser52 inhibits its role in ULK1 modification in human HEK-293 cells.134 Interestingly, a study on the deubiquitinating enzyme interactome indicates that ULK1 is a potential target of USP10 in human HEK-293 cells, although the functional significance of this finding remains unknown.135
Figure 3.
Regulation of ATG proteins by ubiquitination. (A) Two Ubl conjugation systems in mammalian cells. (B) Ubiquitination or deubiquitination of BECN1 and ULK1 regulate the autophagic response.
Ubiquitination is also essential for cargo recognition and selective autophagy processes.136-140 Several targets of selective autophagy have been described, including mitochondria (mitophagy),141 peroxisomes (pexophagy),142 lysosomes (lysophagy),143 lipid droplets (lipophagy),144 secretory granules (zymophagy),145 the endoplasmic reticulum (reticulophagy),146 nucleus (nucleophagy),147 RNA (RNautophagy),148 pathogens (xenophagy),149 ribosomes (ribophagy),150 aggregate-prone proteins (aggrephagy),151 and ferritin (ferritinophagy).152 In addition to cargo degradation, selective autophagy also contributes to the secretion of immune molecules such as cytokines (e.g., IL1) and damage-associated molecular pattern molecules (e.g., HMGB1).153 The selective recruitment of these targets requires selective receptors (e.g., SQSTM1/p62, BNIP3L, NBR1, CALCOCO2, and OPTN) and scaffold proteins, which usually contain a Ub-binding domain and Atg8/LC3/GABARAP-interacting region. Thus, selective autophagy uses Ub as a degradation signal and Ub-binding proteins as specific receptors to transport cargo to the lysosome for degradation or secretion.136-140
Acetylation of Atg/ATG Proteins
Acetylation is described as a reaction that transfers an acetyl group onto a chemical compound. The posttranslational acetylation of histone and nonhistone proteins has been reported to play an important role in cell biology and associated regulatory processes.
In addition to histones154-156 and transcription factors (e.g., FOXO1 [forkhead box O1] and FOXO3),157,158 ATG proteins are regulated through acetylation/deacetylation modifications that influence the outcome of autophagy (Fig. 4).58,59 The NAD-dependent deacetylase SIRT1 (sirtuin 1)-mediated deacetylation of FOXO3 is essential for autophagy induction and cell survival, and the mechanism by which acetylated FOXO3 promotes the transcription of Lc3 and Bnip3 in mouse tissues.157,159 Different from FOXO3-mediated transcriptional control of ATG gene expression, FOXO1 promotes oxidative stress- or starvation-induced autophagy independent of its transcriptional activity.158 FOXO1 shuttles between the cytoplasm and the nucleus. In response to stress, FOXO1 is acetylated after dissociation from SIRT2 (sirtuin 2), which leads to the translocation of FOXO1 from the nucleus to the cytoplasm, where acetylated FOXO1 binds to ATG7 to promote autophagy in human cancer cells.158 FOXO1 can also be deacetylated by SIRT1, which leads to upregulation of RAB7, a small GTPase that controls membrane trafficking and the maturation of phagosomes and autophagic vacuoles.160 This process increases starvation-induced autophagic flux in cardiac myocytes.160
Figure 4.
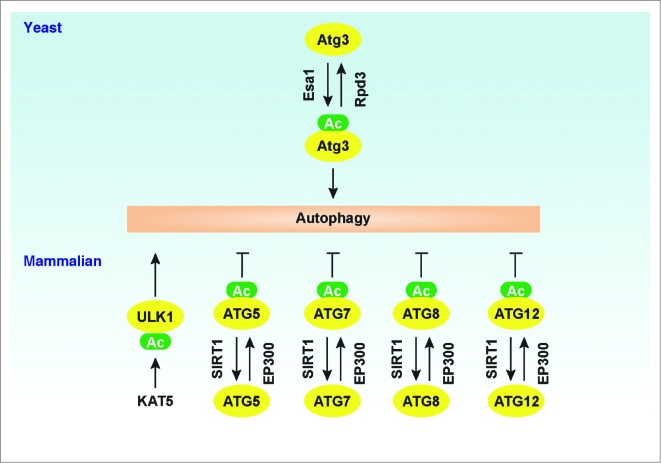
Regulation of Atg/ATG proteins by acetylation. In yeast, acetylation of Atg3 by Esa1 increases autophagy, whereas deacetylation of Atg3 by Rpd3 decreases autophagy. In mammalian cells, acetylation of ATGs (ATG5, ATG7, ATG8, and ATG12) by EP300 decreases autophagy, whereas deacetylation of these ATGs by SIRT1 increases autophagy. In addition, KAT5-mediated ULK1 acetylation promotes autophagy.
As noted above, Atg3 is a ubiquitin-conjugating enzyme (E2) analog that conjugates Atg8 to PE after Atg4-dependent processing and subsequent activation of the C-terminal residue by Atg7 in yeast.34 In yeast, Atg3 acetylation levels regulate autophagic processes through different mechanisms.161 For example, upregulation of Atg3 acetylation at Lys19 and Lys48 by the Esa1 acetyltransferase promotes interaction between Atg3 and Atg8, whereas downregulation of Atg3 acetylation at these sites by the histone deacetylase Rpd3 limits autophagy.161 Moreover, upregulation of Atg3 acetylation at Lys183 by Esa1 increases the Atg3 lipid-conjugating activity.161 In addition to LC3, ATG3 can also be conjugated to ATG12 in mammals or higher eukaryotes, which increases mitochondrial mass, improves bioenergetic function, and inhibits cell death.162 It is unknown whether ATG3 acetylation regulates this process in higher eukaryotes.
Knockdown of the EP300/p300 acetyltransferase inhibits acetylation of ATG5, ATG7, ATG8, and ATG12, which in turn promotes autophagy in human HeLa cells under starvation conditions.163 In contrast, overexpressed SIRT1 in human (e.g., HeLa) and mouse (e.g., embryonic fibroblast) cells can directly deacetylate ATG5, ATG7, LC3, and ATG12, which in turn initiates autophagy during starvation, whereas sirt1−/- cells and mice exhibit the accumulation of damaged organelles and autophagy deficiency.164 These findings suggest a dual role of acetylation in the regulation of ATG function in autophagy induction. Furthermore, interplay between acetylation and other PTMs (e.g., phosphorylation and lipidation) also affect autophagic processes through multi-PTMs of ATG proteins.161,165 For example, under growth factor deprivation conditions, activation of GSK3 (glycogen synthase kinase 3) leads to phosphorylation of KAT5/TIP60 (K [lysine] acetyltransferase 5) at Ser86, which directly acetylates ULK1 at Lys162 and Lys606 in human cancer cells.165 Acetylation of ULK1 by KAT5 increases both kinase activity and the function of ULK1 at the stage of autophagy induction.165 These observations indicate that cooperation between acetyltransferases and deacetylases provides a molecular basis for the fine-tuning of autophagy.
Lipidation of Atg/ATG Proteins
Lipidation is a modification that gives proteins distinct membrane affinities. Membrane localization of these proteins depends on their covalent modifications by specific lipids, resulting in communication with additional proteins associated with the intracellular face of the membrane.
Members of the Atg8/LC3/GABARAP family of proteins are covalently attached to phagophore membranes, functioning in phagophore elongation and cargo recognition, and the lipid conjugation of Atg8/LC3/GABARAP is essential for autophagosome formation.16,34,166 In addition, the lipidated form of Atg8/LC3/GABARAP is associated with the phagophore and autophagosome membrane, and therefore is extensively used to examine autophagic activity.40,167 During the LC3 lipidation cascade in mammalian cells, nascent LC3 is proteolytically processed to expose a C-terminal glycine (equivalent to the removal of the arginine residue of yeast Atg8). The resulting LC3-I is first activated by ATG7 via a thioester bond that is formed between a catalytic cysteine residue of ATG7 and the C-terminal glycine of LC3-I. The LC3–ATG7 intermediate exchanges with ATG3 in forming a second thioester intermediate, LC3–ATG3. Finally, the ATG12–ATG5-ATG16L1 E3 complex functions as a scaffold and promotes the lipid conjugation of LC3 through recruiting the E2 enzyme ATG3 to PE.168 This process converts the soluble form, LC3-I, into the lipidated form, LC3-II, which can attach to the phagophore membrane (Fig. 3). A structural and mutational study found that both ATG12 and ATG5 are directly involved in E3 activity through residues that are assembled into a continuous surface patch upon conjugation.169 In addition to binding to ATG5, ATG12 has another surface (at the side opposite from the interface with ATG5), which functions as the site for interaction with ATG3. This interaction is critical for E2-E3 complex formation.169,170 In addition, the N-terminal 20 amino acids of mouse and human ATG3 form a membrane curvature-sensing amphipathic helix that allows LC3/GABARAP lipidation to proceed efficiently.171 Finally, the N-terminal domain of ATG16L1 is responsible for the localization of the ATG12–ATG5 conjugate to membranes during autophagosome formation.168,170
Proteolysis of Atg/ATG Proteins
Some proteins are activated by proteolysis following translation because they are synthesized as inactive precursors, and proteolysis is involved in several different stages of autophagy. In the initiation phase of autophagy, Atg8 and its orthologs are synthesized as inactive precursors, which are then activated through removal of a C-terminal polypeptide segment. In yeast, Atg8 is cleaved by the cysteine protease Atg4 following translation, removing its C-terminal arginine residue and leaving the glycine 116 residue at the C terminus. This C-terminal glycine is required for interaction with the conjugation system and subsequent Atg8 lipidation.34 Atg4 is also required for a deconjugation step in which PE is removed from Atg8 on the surface of the autophagosome.172 The deconjugation step is required for disassembly of Atg proteins and the completion of autophagosome formation.173 Thus, Atg4 functions in 2 stages of autophagy, phagophore expansion and subsequent autophagosome completion. The single Atg4 in yeast has 4 mammalian orthologs, ATG4A, ATG4B, ATG4C, and ATG4D. Whereas ATG4A and ATG4B have potent enzymatic activities, ATG4C and ATG4D seem to have only minor effects on LC3 processing.174 Like Atg4 in yeast, ATG4B plays a role in the processing and deconjugation of LC3B in mammalian cells.167,175 Arginine 68 in human LC3B is a specific residue facilitating the interaction between LC3B and ATG4B by forming a salt bridge.176 As discussed above, the proteolytically processed LC3 (LC3-I) is subjected to conjugation with PE catalyzed by ATG7 and ATG3, yielding the phagophore membrane-associated form, PE-conjugated LC3 (LC3-II).40,167,177 Consequently, starvation-stimulated formation of reactive oxygen species, specifically H2O2, in mitochondria can lead to oxidation of the catalytic site of ATG4A/B (Cys81).178 Oxidized ATG4A/B loses deconjugation activity resulting in an increase in membrane-associated LC3-II (Fig. 3).178 Thus, ATG4 activity in the autophagic process may be redox-regulated.
In the execution of autophagy, lysosomal hydrolases/ca-thepsins are involved in the degradation of autophagosomal membrane components.179 As the association of LC3 with the autophagosome membrane is a reversible process, the deconjugated LC3 is recycled and participates in a new conjugation reaction (as mentioned above), whereas LC3-II trapped inside the autophagosome is degraded following fusion with the lysosome. Macrophages derived from CTSE/cathepsin E-deficient mice exhibit abnormal membrane trafficking and delayed degradation of autophagosomal membrane proteins.180 The ATP- and ubiquitin-independent 20S proteasome can completely degrade LC3.181 This process requires the N-terminal helices of LC3, and can be inhibited by SQSTM1.181 SQSTM1 can bind LC3-II and bring SQSTM1-containing protein aggregates to the autophagosome for degradation.182 These findings suggest a negative or competitive regulation mechanism between the proteasome and autophagy.181
CAPN/calpains and CASP/caspases can selectively cleave ATGs, which regulates the balance and switch between autophagy to apoptosis in response to various stimuli (Fig. 5).183-192 For example, ATG5 is cleaved at Thr193 by CAPN1 and CAPN2, but not CASP3, CASP6, CASP7, CASP8, or CASP9, in an apoptotic human cervix adenocarcinoma cell line (HeLa) and a human T lymphocyte cell line (Jurkat).183 Truncated ATG5 lacks autophagy-inducing capacity, whereas it can induce apoptosis through binding to BCL2L1 at the mitochondria and promoting the release of mitochondrial prodeath factors such as CYCS/cytochrome c.183 Interestingly, the combination of TNFSF10/TRAIL (tumor necrosis factor [ligand] superfamily, member 10) and arginine deprivation induces ATG5 cleavage through CASP6, CASP9, and CASP10, but not CASP8 activation in human melanoma cell lines (A375 and A2058), which contributes to apoptosis;188 however, the CASP cleavage sites of ATG5 remain unknown.
Figure 5.
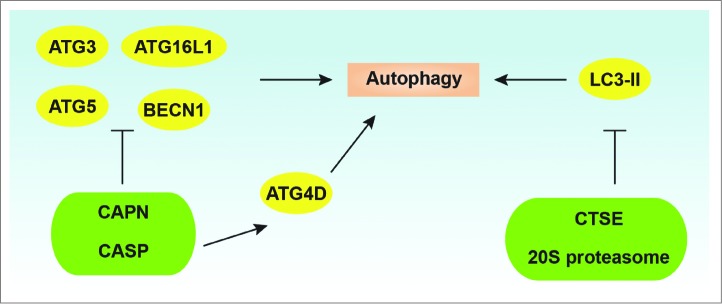
Regulation of ATG proteins by proteolysis. CAPN/calpain and CASP can inhibit autophagy by degrading or cleaving ATGs such as ATG3, ATG5, ATG16L1, and BECN1. In contrast, CASP-mediated ATG4D cleavage promotes autophagy. In addition, CTSE/cathepsin E and the 20S proteasome are involved in the degradation of the phagophore and autophagosome membrane component LC3-II.
Cleavage of BECN1 has been observed in multiple apoptotic cells, and this cleavage is mediated by CASP3, CASP6, CASP8, CASP9, and CASP10.184,188,189,191,192 The cleaved C-terminal BECN1 cannot induce autophagy, but induces apoptosis once it translocates into the mitochondria and cooperates with other proapoptotic BCL2 members.184 Asp149,184,190 Asp133,184,191 and Asp146191 at the C terminus of BECN1 have been reported as CASP cleavage sites in human or mouse BECN1, which depends on the cell type and treatment. In addition to caspases, calpains might be responsible for BECN1 cleavage, although the putative cleavage site(s) remains unidentified.184 Moreover, cleavage of ATG3 at Asp169 by CASP8185 and cleavage of ATG16L1 at Thr300 by CASP3186 impairs autophagy during receptor-activated cell death or bacterial infection. In contrast to the above discussion of cleaved ATG proteins lacking autophagy activity, cleavage of ATG4D at Asp163 by CASP3 enhances the priming and delipidation of GABARAPL1, but not GABARAPL2 and LC3 in human cancer cells.187 These findings represent the molecular crosstalk between apoptosis and autophagy, although it is clearly complex.5
Sulfation of Atg Proteins
Sulfation occurs in the trans-Golgi network and is mediated by a membrane-bound protein with its active site on the lumenal side. A sulfate ion is bound near the catalytic cysteine of Atg3 in yeast.193 The sulfate ion of Atg3 may mimic the phosphate group of PE and bind to the PE-binding site.193 In addition, 3 basic residues (Lys183, His232, Lys235), which surround the sulfate ion, might recognize the phosphate moiety of PE.193 This may provide a molecular basis for understanding the mechanism of the Atg3-mediated lipidation reaction.
Conclusions
In the 1960s, Christian de Duve, a Nobel Prize-winning Belgian cytologist and biochemist, coined and described “autophagy,” which literally means “self-eating.” During the past 15 years, significant progress has been made toward understanding the morphology, mechanism, and regulation of autophagy.52,194 Currently, autophagy has become a hot field in translational medicine. The most important regulators for autophagy are autophagy-related genes/proteins and the first members were identified in yeast in 1996-7. As we have discussed in this review, multiple PTMs exist on ATG proteins during the complicated membrane reshuffling processes in yeast and mammalian cells that are part of autophagy (Table 1). We are surely at a very stimulating and exciting stage in the study of ATG protein modifications. Usually, the patterns of modification across a single ATG protein are combinatorial. Different PTMs can exhibit distinct downstream responses in autophagy. Although these PTMs play a critical role in the regulation of ATG protein functions in different stage of autophagy, many specific questions remain to be answered: How do different PTM “codes” communicate to fine-tune the process of autophagy? What are the essential enzymes responsible for specific PTMs in different ATG proteins? What are the effects of PTMs on structural modifications of ATG proteins? How do PTMs qualitatively and quantitatively sense different forms of autophagic stimulus in different species? How should PTMs in autophagy be systematically measured and monitored? Of course, the mechanistic basis of PTM encoding presents an urgent scientific and technological challenge. The complexity of crosstalk and cellular signaling may help explain the PTM “code.” More work is needed for a complete description of the process, mechanism, and function of PTMs in the regulation of autophagy. This will not only reveal fundamental mechanistic insights into the regulatory process, but also provide new therapeutic targets for the treatment of autophagy-associated diseases. In particular, dissection of the upstream pathways that regulate the PTM will be crucial for developing pharmaceutics to target autophagy.
Acknowledgments
We apologize to the researchers who were not referenced due to space limitations. We thank Christine Heiner (Department of Surgery, University of Pittsburgh) for her critical reading of the manuscript.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health (R01CA160417 to D.T., and R01GM053396 to D.J.K.), a 2013 Pancreatic Cancer Action Network-AACR Career Development Award (Grant Number 13-20-25-TANG), The National Natural Science Foundation-Guangdong Joint Fund (U1132005 to X.S.), and Science and Information Technology of Guangzhou Key Project (2011Y1-00038 to X.S.). Work done in support of findings reviewed in this manuscript was aided by the Core Support of the University of Pittsburgh Cancer Institute (P30CA047904).
References
- 1. Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/ 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. . Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/ 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature 2011; 469:323-35; PMID:21248839; http://dx.doi.org/ 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741-52; PMID:17717517; http://dx.doi.org/ 10.1038/nrm2239 [DOI] [PubMed] [Google Scholar]
- 6. Rabinowitz JD, White E. Autophagy and metabolism. Science 2010; 330:1344-8; PMID:21127245; http://dx.doi.org/ 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol 2010; 12:823-30; PMID:20811354; http://dx.doi.org/ 10.1038/ncb0910-823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280-93; PMID:20965422; http://dx.doi.org/ 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med 2013; 368:651-62; PMID:23406030; http://dx.doi.org/ 10.1056/NEJMra1205406 [DOI] [PubMed] [Google Scholar]
- 10. Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27-42; PMID:18191218; http://dx.doi.org/ 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004; 306:990-5; PMID:15528435; http://dx.doi.org/ 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; http://dx.doi.org/ 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hou W, Zhang Q, Yan Z, Chen R, Zeh Iii HJ, Kang R, Lotze MT, Tang D. Strange attractors: DAMPs and autophagy link tumor cell death and immunity. Cell Death Dis 2013; 4:e966; PMID:24336086; http://dx.doi.org/ 10.1038/cddis.2013.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol 2010; 12:831-5; PMID:20811355; http://dx.doi.org/ 10.1038/ncb0910-831 [DOI] [PubMed] [Google Scholar]
- 15. Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a. Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res 2014; 24:24-41; PMID:24366339; http://dx.doi.org/ 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16b.Kametaka S, Matsuura A, Wada Y, Ohsumi Y. Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene 1996; 178:139-43. [DOI] [PubMed] [Google Scholar]
- 17. Schlumpberger M, Schaeffeler E, Straub M, Bredschneider M, Wolf DH, Thumm M. AUT1, a gene essential for autophagocytosis in the yeast Saccharo-myces cerevisiae. J Bacteriol 1997; 179:1068-76; PMID:9023185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene 1997; 192:207-13; PMID:9224892; http://dx.doi.org/ 10.1016/S0378-1119(97)00031-0 [DOI] [PubMed] [Google Scholar]
- 19. Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997; 192:245-50; PMID:9224897; http://dx.doi.org/ 10.1016/S0378-1119(97)00084-X [DOI] [PubMed] [Google Scholar]
- 20. Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol 2013; 203:299-313; PMID:24165940; http://dx.doi.org/ 10.1083/jcb.201304123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, Cao J, Kundu M, Kim DH. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell 2009; 20:1992-2003; PMID:19225151; http://dx.doi.org/ 10.1091/mbc.E08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, Miura Y, Iemura S, Natsume T, Takehana K, Yamada N, et al. . Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell 2009; 20:1981-91; PMID:19211835; http://dx.doi.org/ 10.1091/mbc.E08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009; 5:973-9; PMID:19597335; http://dx.doi.org/ 10.4161/auto.5.7.9296 [DOI] [PubMed] [Google Scholar]
- 24. Mercer CA, Kaliappan A, Dennis PB. A novel, human Atg13 binding protein, Atg101, interacts with ULK1 and is essential for macroautophagy. Autophagy 2009; 5:649-62; PMID:19287211; http://dx.doi.org/ 10.4161/auto.5.5.8249 [DOI] [PubMed] [Google Scholar]
- 25. Liang Q, Yang P, Tian E, Han J, Zhang H. The C. elegans ATG101 homolog EPG-9 directly interacts with EPG-1/Atg13 and is essential for autophagy. Autophagy 2012; 8:1426-33; PMID:22885670; http://dx.doi.org/ 10.4161/auto.21163 [DOI] [PubMed] [Google Scholar]
- 26. Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360-72; PMID:18843052; http://dx.doi.org/ 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152:519-30; PMID:11157979; http://dx.doi.org/ 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672-6; PMID:10604474; http://dx.doi.org/ 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- 29. Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J 2009; 417:747-55; PMID:18957027; http://dx.doi.org/ 10.1042/BJ20081865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 2008; 27 Suppl 1:S137-48; PMID:19641499; http://dx.doi.org/ 10.1038/onc.2009.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18:571-80; PMID:21311563; http://dx.doi.org/ 10.1038/cdd.2010.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2:211-6; PMID:11265251; http://dx.doi.org/ 10.1038/35056522 [DOI] [PubMed] [Google Scholar]
- 33. Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature 1998; 395:395-8; PMID:9759731; http://dx.doi.org/ 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- 34. Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. . A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488-92; PMID:11100732; http://dx.doi.org/ 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- 35. Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7pCvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell 1999; 10:1337-51; PMID:10233148; http://dx.doi.org/ 10.1091/mbc.10.5.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5·Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem 2002; 277:18619-25; PMID:11897782; http://dx.doi.org/ 10.1074/jbc.M111889200 [DOI] [PubMed] [Google Scholar]
- 37. Cao Y, Cheong H, Song H, Klionsky DJ. In vivo reconstitution of autophagy in Saccharomyces cerevi-siae. J Cell Biol 2008; 182:703-13; PMID:18725539; http://dx.doi.org/ 10.1083/jcb.200801035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J 2010; 29:1792-802; PMID:20418806; http://dx.doi.org/ 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290-8; PMID:18508918; http://dx.doi.org/ 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID:11060023; http://dx.doi.org/ 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888-900; PMID:16940348; http://dx.doi.org/ 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- 42. Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79-90; PMID:14723849; http://dx.doi.org/ 10.1016/S1534-5807(03)00402-7 [DOI] [PubMed] [Google Scholar]
- 43. He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell 2008; 19:5506-16; PMID:18829864; http://dx.doi.org/ 10.1091/mbc.E08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Webber JL, Tooze SA. Coordinated regulation of autophagy by p38α MAPK through mAtg9 and p38IP. EMBO J 2010; 29:27-40; PMID:19893488; http://dx.doi.org/ 10.1038/emboj.2009.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Monastyrska I, Rieter E, Klionsky DJ, Reggiori F. Multiple roles of the cytoskeleton in autophagy. Biol Rev Camb Philos Soc 2009; 84:431-48; PMID:19659885; http://dx.doi.org/ 10.1111/j.1469-185X.2009.00082.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu MM, Nirschl JJ, Holzbaur EL. LC3 binding to the scaffolding protein JIP1 regulates processive dynein-driven transport of autophagosomes. Dev Cell 2014; 29:577-90; PMID:24914561; http://dx.doi.org/ 10.1016/j.devcel.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ, III, Lotze MT. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab 2011; 13:701-11; PMID:21641551; http://dx.doi.org/ 10.1016/j.cmet.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell 2011; 146:303-17; PMID:21784250; http://dx.doi.org/ 10.1016/j.cell.2011.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. SNARE proteins are required for macroautophagy. Cell 2011; 146:290-302; PMID:21784249; http://dx.doi.org/ 10.1016/j.cell.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomeslysosomes. Cell 2012; 151:1256-69; PMID:23217709; http://dx.doi.org/ 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 51. Yang Z, Huang J, Geng J, Nair U, Klionsky DJ. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol Biol Cell 2006; 17:5094-104; PMID:17021250; http://dx.doi.org/ 10.1091/mbc.E06-06-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 2014; 20:460-73; PMID:23725295; http://dx.doi.org/ 10.1089/ars.2013.5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713-20; PMID:23817233; http://dx.doi.org/ 10.1038/ncb2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO Rep 2013; 14:143-51; PMID:23337627; http://dx.doi.org/ 10.1038/embor.2012.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deribe YL, Pawson T, Dikic I. Post-translational modifications in signal integration. Nat Struct Mol Biol 2010; 17:666-72; PMID:20495563; http://dx.doi.org/ 10.1038/nsmb.1842 [DOI] [PubMed] [Google Scholar]
- 56. Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol 2003; 21:255-61; PMID:12610572; http://dx.doi.org/ 10.1038/nbt0303-255 [DOI] [PubMed] [Google Scholar]
- 57. McEwan DG, Dikic I. The three musketeers of autophagy: phosphorylation, ubiquitylation and acetylation. Trends Cell Biol 2011; 21:195-201; PMID:21277210; http://dx.doi.org/ 10.1016/j.tcb.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Banreti A, Sass M, Graba Y. The emerging role of acetylation in the regulation of autophagy. Autophagy 2013; 9:819-29; PMID:23466676; http://dx.doi.org/ 10.4161/auto.23908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hamai A, Codogno P. New targets for acetylation in autophagy. Sci Signal 2012; 5:pe29; PMID:22763338 [DOI] [PubMed] [Google Scholar]
- 60. Mizushima N. The role of the Atg1ULK1 complex in autophagy regulation. Curr Opin Cell Biol 2010; 22:132-9; PMID:20056399; http://dx.doi.org/ 10.1016/j.ceb.2009.12.004 [DOI] [PubMed] [Google Scholar]
- 61. Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150:1507-13; PMID:10995454; http://dx.doi.org/ 10.1083/jcb.150.6.1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kijanska M, Dohnal I, Reiter W, Kaspar S, Stoffel I, Ammerer G, Kraft C, Peter M. Activation of Atg1 kinase in autophagy by regulated phosphorylation. Autophagy 2010; 6:1168-78; PMID:20953146; http://dx.doi.org/ 10.4161/auto.6.8.13849 [DOI] [PubMed] [Google Scholar]
- 63. Scott SV, Nice DC, III, Nau JJ, Weisman LS, Kamada Y, Keizer-Gunnink I, Funakoshi T, Veenhuis M, Ohsumi Y, Klionsky DJ. Apg13p and Vac8p are part of a complex of phosphoproteins that are required for cytoplasm to vacuole targeting. J Biol Chem 2000; 275:25840-9; PMID:10837477; http://dx.doi.org/ 10.1074/jbc.M002813200 [DOI] [PubMed] [Google Scholar]
- 64. Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, Ohsumi Y. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol 2010; 30:1049-58; PMID:19995911; http://dx.doi.org/ 10.1128/MCB.01344-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A 2009; 106:17049-54; PMID:19805182; http://dx.doi.org/ 10.1073/pnas.0903316106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The RascAMP-dependent protein kinase signaling pathway regulates an early step of the autophagy process in Saccharomyces cerevisiae. J Biol Chem 2004; 279:20663-71; PMID:15016820; http://dx.doi.org/ 10.1074/jbc.M400272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2007; 18:4180-9; PMID:17699586; http://dx.doi.org/ 10.1091/mbc.E07-05-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Budovskaya YV, Stephan JS, Deminoff SJ, Herman PK. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc Natl Acad Sci U S A 2005; 102:13933-8; PMID:16172400; http://dx.doi.org/ 10.1073/pnas.0501046102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nagy P, Karpati M, Varga A, Pircs K, Venkei Z, Takats S, Varga K, Erdi B, Hegedus K, Juhasz G. Atg17FIP200 localizes to perilysosomal Ref(2)P aggregates and promotes autophagy by activation of Atg1 in. Autophagy 2014; 10; 453-67; PMID:24419107; http://dx.doi.org/ 10.4161/auto.27442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yeh YY, Wrasman K, Herman PK. Autophosphorylation within the Atg1 activation loop is required for both kinase activity and the induction of autophagy in Saccharomyces cerevisiae. Genetics 2010; 185:871-82; PMID:20439775; http://dx.doi.org/ 10.1534/genetics.110.116566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, Yip CK, Klionsky DJ. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A 2013; 110:E2875-84; PMID:23858448; http://dx.doi.org/ 10.1073/pnas.1300064110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell 2005; 16:1593-605; PMID:15659643; http://dx.doi.org/ 10.1091/mbc.E04-11-1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, Abert C, Ammerer G, Martens S, Kraft C. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep 2014; 15:862-70; PMID:24968893; http://dx.doi.org/ 10.15252/embr.201438932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kanki T, Kurihara Y, Jin X, Goda T, Ono Y, Aihara M, Hirota Y, Saigusa T, Aoki Y, Uchiumi T, Kang D. Casein kinase 2 is essential for mitophagy. EMBO Rep 2013; 14:788-94; PMID:23897086; http://dx.doi.org/ 10.1038/embor.2013.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dorsey FC, Rose KL, Coenen S, Prater SM, Cavett V, Cleveland JL, Caldwell-Busby J. Mapping the phosphorylation sites of Ulk1. J Proteome Res 2009; 8:5253-63; PMID:19807128; http://dx.doi.org/ 10.1021/pr900583m [DOI] [PubMed] [Google Scholar]
- 76. Bach M, Larance M, James DE, Ramm G. The serinethreonine kinase ULK1 is a target of multiple phosphorylation events. Biochem J 2011; 440:283-91; PMID:21819378; http://dx.doi.org/ 10.1042/BJ20101894 [DOI] [PubMed] [Google Scholar]
- 77. Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009; 122:3589-94; PMID:19812304; http://dx.doi.org/ 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, Mizushima N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181:497-510; PMID:18443221; http://dx.doi.org/ 10.1083/jcb.200712064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shang L, Chen S, Du F, Li S, Zhao L, Wang X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc Natl Acad Sci U S A 2011; 108:4788-93; PMID:21383122; http://dx.doi.org/ 10.1073/pnas.1100844108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell 2008; 30:214-26; PMID:18439900; http://dx.doi.org/ 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Egan DF, Shackelford DB, Mihaylova MM, Gelino SR, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, Asara JM, Fitzpatrick J, Dillin A, Viollet B, Kundu M, Hansen M, Shaw RJ. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456-61; PMID:21205641; http://dx.doi.org/ 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 2013; 155:688-98; PMID:24119841; http://dx.doi.org/ 10.1016/j.cell.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mack HI, Zheng B, Asara JM, Thomas SM. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 2012; 8:1197-214; PMID:22932492; http://dx.doi.org/ 10.4161/auto.20586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PLoS One 2010; 5:e15394; PMID:21072212; http://dx.doi.org/ 10.1371/journal.pone.0015394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Loffler AS, Alers S, Dieterle AM, Keppeler H, Franz-Wachtel M, Kundu M, Campbell DG, Wesselborg S, Alessi DR, Stork B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 2011; 7:696-706; PMID:21460634; http://dx.doi.org/ 10.4161/auto.7.7.15451 [DOI] [PubMed] [Google Scholar]
- 87. Dunlop EA, Hunt DK, Acosta-Jaquez HA, Fingar DC, Tee AR. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 2011; 7:737-47; PMID:21460630; http://dx.doi.org/ 10.4161/auto.7.7.15491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jung CH, Seo M, Otto NM, Kim DH. ULK1 inhibits the kinase activity of mTORC1 and cell proliferation. Autophagy 2011; 7:1212-21; PMID:21795849; http://dx.doi.org/ 10.4161/auto.7.10.16660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, et al. . Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell 2014; 53:471-83; PMID:24440502; http://dx.doi.org/ 10.1016/j.molcel.2013.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, et al. . Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell 2011; 43:572-85; PMID:21855797; http://dx.doi.org/ 10.1016/j.molcel.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Alers S, Loffler AS, Paasch F, Dieterle AM, Keppeler H, Lauber K, Campbell DG, Fehrenbacher B, Schaller M, Wesselborg S, et al. . Atg13 and FIP200 act independently of Ulk1 and Ulk2 in autophagy induction. Autophagy 2011; 7:1423-33; PMID:22024743; http://dx.doi.org/ 10.4161/auto.7.12.18027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741-50; PMID:23685627; http://dx.doi.org/ 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk12 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol 2012; 32:2-11; PMID:22025673; http://dx.doi.org/ 10.1128/MCB.06159-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. . The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191:155-68; PMID:20921139; http://dx.doi.org/ 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tang HW, Wang YB, Wang SL, Wu MH, Lin SY, Chen GC. Atg1-mediated myosin II activation regulates autophagosome formation during starvation-induced autophagy. EMBO J 2011; 30:636-51; PMID:21169990; http://dx.doi.org/ 10.1038/emboj.2010.338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J 1993; 12:2195-204; PMID:8387919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett 2010; 584:1313-8; PMID:20188094; http://dx.doi.org/ 10.1016/j.febslet.2010.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, Reggiori F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol 2012; 22:1545-53; PMID:22771041; http://dx.doi.org/ 10.1016/j.cub.2012.06.029] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yu X, Ma J, Lin F, Zhao W, Fu X, Zhao ZJ. Myotubularin family phosphatase ceMTM3 is required for muscle maintenance by preventing excessive autophagy in Caenorhabditis elegans. BMC Cell Biol 2012; 13:28; PMID:23114011; http://dx.doi.org/ 10.1186/1471-2121-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 2009; 28:2244-58; PMID:19590496; http://dx.doi.org/ 10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic 2010; 11:468-78; PMID:20059746; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01034.x [DOI] [PubMed] [Google Scholar]
- 102. Martin KR, Xu Y, Looyenga BD, Davis RJ, Wu CL, Tremblay ML, Xu HE, MacKeigan JP. Identification of PTPsigma as an autophagic phosphatase. J Cell Sci 2011; 124:812-9; PMID:21303930; http://dx.doi.org/ 10.1242/jcs.080341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, Yuan J. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell 2010; 38:500-11; PMID:20513426; http://dx.doi.org/ 10.1016/j.molcel.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013; 152:290-303; PMID:23332761; http://dx.doi.org/ 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ 2012; 19:788-97; PMID:22095288; http://dx.doi.org/ 10.1038/cdd.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Erlich S, Mizrachy L, Segev O, Lindenboim L, Zmira O, Adi-Harel S, Hirsch JA, Stein R, Pinkas-Kramarski R. Differential interactions between Beclin 1 and Bcl-2 family members. Autophagy 2007; 3:561-8; PMID:17643073; http://dx.doi.org/ 10.4161/auto.4713 [DOI] [PubMed] [Google Scholar]
- 107. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927-39; PMID:16179260; http://dx.doi.org/ 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 108. Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 2009; 10:285-92; PMID:19180116; http://dx.doi.org/ 10.1038/embor.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gurkar AU, Chu K, Raj L, Bouley R, Lee SH, Kim YB, Dunn SE, Mandinova A, Lee SW. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun 2013; 4:2189; PMID:23877263; http://dx.doi.org/ 10.1038/ncomms3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678-88; PMID:18570871; http://dx.doi.org/ 10.1016/j.molcel.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ, 3rd, Lotze MT. Endogenous HMGB1 regulates autophagy. J Cell Biol 2010; 190:881-92; PMID:20819940; http://dx.doi.org/ 10.1083/jcb.200911078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Sun X, Tang D. HMGB1-dependent and -independent autophagy. Autophagy 2014; 10; 1873-6; PMID:25126737; http://dx.doi.org/ 10.4161/auto.32184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012; 338:956-9; PMID:23112296; http://dx.doi.org/ 10.1126/science.1225967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, Grishin NV, Peyton M, Minna J, Bhagat G, Levine B. EGFR-mediated beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013; 154:1269-84; PMID:24034250; http://dx.doi.org/ 10.1016/j.cell.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol 2013; 33:3675-88; PMID:23878393; http://dx.doi.org/ 10.1128/MCB.00079-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep 2013; 14:441-9; PMID:23559066; http://dx.doi.org/ 10.1038/embor.2013.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell 2008; 14:365-76; PMID:18331717; http://dx.doi.org/ 10.1016/j.devcel.2007.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kondo-Okamoto N, Noda NN, Suzuki SW, Nakatogawa H, Takahashi I, Matsunami M, Hashimoto A, Inagaki F, Ohsumi Y, Okamoto K. Autophagy-related protein 32 acts as autophagic degron and directly initiates mitophagy. J Biol Chem 2012; 287:10631-8; PMID:22308029; http://dx.doi.org/ 10.1074/jbc.M111.299917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, Kang D. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell 2011; 22:3206-17; PMID:21757540; http://dx.doi.org/ 10.1091/mbc.E11-02-0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Mao K, Wang K, Zhao M, Xu T, Klionsky DJ. Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J Cell Biol 2011; 193:755-67; PMID:21576396; http://dx.doi.org/ 10.1083/jcb.201102092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Keil E, Hocker R, Schuster M, Essmann F, Ueffing N, Hoffman B, Liebermann DA, Pfeffer K, Schulze-Osthoff K, Schmitz I. Phosphorylation of Atg5 by the Gadd45beta-MEKK4-p38 pathway inhibits autophagy. Cell Death Differ 2013; 20:321-32; PMID:23059785; http://dx.doi.org/ 10.1038/cdd.2012.129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cherra SJ, III, Kulich SM, Uechi G, Balasubramani M, Mountzouris J, Day BW, Chu CT. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol 2010; 190:533-9; PMID:20713600; http://dx.doi.org/ 10.1083/jcb.201002108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Jiang H, Cheng D, Liu W, Peng J, Feng J. Protein kinase C inhibits autophagy and phosphorylates LC3. Biochem Biophys Res Commun 2010; 395:471-6; PMID:20398630; http://dx.doi.org/ 10.1016/j.bbrc.2010.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012; 198:219-33; PMID:22826123; http://dx.doi.org/ 10.1083/jcb.201202061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005; 1:101-9; PMID:16874040; http://dx.doi.org/ 10.4161/auto.1.2.1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric-oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem 2005; 280:18283-90; PMID:15755735; http://dx.doi.org/ 10.1074/jbc.M413957200 [DOI] [PubMed] [Google Scholar]
- 127. Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell 2010; 21:3998-4008; PMID:20861302; http://dx.doi.org/ 10.1091/mbc.E10-05-0457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Wang X, Pattison JS, Su H. Posttranslational modification and quality control. Circ Res 2013; 112:367-81; PMID:23329792; http://dx.doi.org/ 10.1161/CIRCRESAHA.112.268706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Kuang E, Qi J, Ronai Z. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem Sci 2013; 38:453-60; PMID:23870665; http://dx.doi.org/ 10.1016/j.tibs.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 2010; 3:ra42; PMID:20501938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, Hou N, Cheng X, Sun Q, Li L, Yang X, Fan Z. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J 2013; 32:2685-96; PMID:23974797; http://dx.doi.org/ 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J 2012; 441:399-406; PMID:21936852; http://dx.doi.org/ 10.1042/BJ20111424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, Cai Y, Norberg HV, Zhang T, Furuya T, et al. . Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 2011; 147:223-34; PMID:21962518; http://dx.doi.org/ 10.1016/j.cell.2011.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. . mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; PMID:23524951; http://dx.doi.org/ 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- 135. Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009; 138:389-403; PMID:19615732; http://dx.doi.org/ 10.1016/j.cell.2009.04.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol 2014; 16:495-501; PMID:24875736; http://dx.doi.org/ 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- 137. Rogov V, Dotsch V, Johansen T, Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol Cell 2014; 53:167-78; PMID:24462201; http://dx.doi.org/ 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- 138. Jin M, Liu X, Klionsky DJ. SnapShot: selective autophagy. Cell 2013; 152:368- e2; PMID:23332767; http://dx.doi.org/ 10.1016/j.cell.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell 2009; 34:259-69; PMID:19450525; http://dx.doi.org/ 10.1016/j.molcel.2009.04.026 [DOI] [PubMed] [Google Scholar]
- 140. Klionsky DJ, Schulman BA. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat Struct Mol Biol 2014; 21:336-45; PMID:24699082; http://dx.doi.org/ 10.1038/nsmb.2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12:9-14; PMID:21179058; http://dx.doi.org/ 10.1038/nrm3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy 2005; 1:75-83; PMID:16874024; http://dx.doi.org/ 10.4161/auto.1.2.1737 [DOI] [PubMed] [Google Scholar]
- 143. Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 2013; 4:2111; PMID:23817530; http://dx.doi.org/ 10.1038/ncomms3111 [DOI] [PubMed] [Google Scholar]
- 144. Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature 2009; 458:1131-5; PMID:19339967; http://dx.doi.org/ 10.1038/nature07976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Grasso D, Ropolo A, Lo Re A, Boggio V, Molejon MI, Iovanna JL, Gonzalez CD, Urrutia R, Vaccaro MI. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem 2011; 286:8308-24; PMID:21173155; http://dx.doi.org/ 10.1074/jbc.M110.197301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 2007; 3:285-7; PMID:17351330; http://dx.doi.org/ 10.4161/auto.3930 [DOI] [PubMed] [Google Scholar]
- 147. Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, Nishino I. Autophagic degradation of nuclear components in mammalian cells. Autophagy 2009; 5:795-804; PMID:19550147; http://dx.doi.org/ 10.4161/auto.8901 [DOI] [PubMed] [Google Scholar]
- 148. Fujiwara Y, Furuta A, Kikuchi H, Aizawa S, Hatanaka Y, Konya C, Uchida K, Yoshimura A, Tamai Y, Wada K, Kabuta T. Discovery of a novel type of autophagy targeting RNA. Autophagy 2013; 9:403-9; PMID:23291500; http://dx.doi.org/ 10.4161/auto.23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 2005; 120:159-62; PMID:15680321 [DOI] [PubMed] [Google Scholar]
- 150. Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3pBre5p ubiquitin protease. Nat Cell Biol 2008; 10:602-10; PMID:18391941; http://dx.doi.org/ 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- 151. Overbye A, Fengsrud M, Seglen PO. Proteomic analysis of membrane-associated proteins from rat liver autophagosomes. Autophagy 2007; 3:300-22; PMID:17377489; http://dx.doi.org/ 10.4161/auto.3910 [DOI] [PubMed] [Google Scholar]
- 152. Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014; 509:105-9; PMID:24695223; http://dx.doi.org/ 10.1038/nature13148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Zhang Q, Kang R, Zeh HJ, 3rd, Lotze MT, Tang D. DAMPs and autophagy: cellular adaptation to injury and unscheduled cell death. Autophagy 2013; 9:451-8; PMID:23388380; http://dx.doi.org/ 10.4161/auto.23691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Eisenberg T, Knauer H, Schauer A, Buttner S, Ruckenstuhl C, Carmona-Gutierrez D, Ring J, Schroeder S, Magnes C, Antonacci L, et al. . Induction of autophagy by spermidine promotes longevity. Nat Cell Biol 2009; 11:1305-14; PMID:19801973; http://dx.doi.org/ 10.1038/ncb1975 [DOI] [PubMed] [Google Scholar]
- 155. Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, Cabrera S, Benit P, Rustin P, Criollo A, et al. . Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol 2011; 192:615-29; PMID:21339330; http://dx.doi.org/ 10.1083/jcb.201008167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, Hermanson O, Rosenfeld MG, Klionsky DJ, Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature 2013; 500:468-71; PMID:23863932; http://dx.doi.org/ 10.1038/nature12313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest 2010; 120:1043-55; PMID:20335657; http://dx.doi.org/ 10.1172/JCI41376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Zhao Y, Yang J, Liao W, Liu X, Zhang H, Wang S, Wang D, Feng J, Yu L, Zhu WG. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol 2010; 12:665-75; PMID:20543840; http://dx.doi.org/ 10.1038/ncb2069 [DOI] [PubMed] [Google Scholar]
- 159. Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. . FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab 2007; 6:458-71; PMID:18054315; http://dx.doi.org/ 10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- 160. Hariharan N, Maejima Y, Nakae J, Paik J, Depinho RA, Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res 2010; 107:1470-82; PMID:20947830; http://dx.doi.org/ 10.1161/CIRCRESAHA.110.227371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Yi C, Ma M, Ran L, Zheng J, Tong J, Zhu J, Ma C, Sun Y, Zhang S, Feng W, et al. . Function and molecular mechanism of acetylation in autophagy regulation. Science 2012; 336:474-7; PMID:22539722; http://dx.doi.org/ 10.1126/science.1216990 [DOI] [PubMed] [Google Scholar]
- 162. Radoshevich L, Murrow L, Chen N, Fernandez E, Roy S, Fung C, Debnath J. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell 2010; 142:590-600; PMID:20723759; http://dx.doi.org/ 10.1016/j.cell.2010.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem 2009; 284:6322-8; PMID:19124466; http://dx.doi.org/ 10.1074/jbc.M807135200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, Tsokos M, Alt FW, Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A 2008; 105:3374-9; PMID:18296641; http://dx.doi.org/ 10.1073/pnas.0712145105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Ruan K, Wang Z, et al. . GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 2012; 336:477-81; PMID:22539723; http://dx.doi.org/ 10.1126/science.1217032 [DOI] [PubMed] [Google Scholar]
- 166. Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007; 282:37298-302; PMID:17986448; http://dx.doi.org/ 10.1074/jbc.C700195200 [DOI] [PubMed] [Google Scholar]
- 167. Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004; 117:2805-12; PMID:15169837; http://dx.doi.org/ 10.1242/jcs.01131 [DOI] [PubMed] [Google Scholar]
- 168. Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 2008; 19:2092-100; PMID:18321988; http://dx.doi.org/ 10.1091/mbc.E07-12-1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol 2013; 20:59-66; PMID:23202584; http://dx.doi.org/ 10.1038/nsmb.2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Metlagel Z, Otomo C, Takaesu G, Otomo T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc Natl Acad Sci U S A 2013; 110:18844-9; PMID:24191030; http://dx.doi.org/ 10.1073/pnas.1314755110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. Lipidation of the LC3GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol 2014; 16:415-24; PMID:24747438; http://dx.doi.org/ 10.1038/ncb2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. The reversible modification regulates the membrane-binding state of Apg8Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151:263-76; PMID:11038174; http://dx.doi.org/ 10.1083/jcb.151.2.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Nair U, Yen W-L, Mari M, Cao Y, Xie Z, Baba M, Reggiori F, Klionsky DJ. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 2012; 8:780-93; PMID:22622160; http://dx.doi.org/ 10.4161/auto.19385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Li M, Hou Y, Wang J, Chen X, Shao ZM, Yin XM. Kinetics comparisons of mammalian Atg4 homologues indicate selective preferences toward diverse Atg8 substrates. J Biol Chem 2011; 286:7327-38; PMID:21177865; http://dx.doi.org/ 10.1074/jbc.M110.199059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Satoo K, Noda NN, Kumeta H, Fujioka Y, Mizushima N, Ohsumi Y, Inagaki F. The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J 2009; 28:1341-50; PMID:19322194; http://dx.doi.org/ 10.1038/emboj.2009.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Liu C, Ma H, Wu J, Huang Q, Liu JO, Yu L. Arginine68 is an essential residue for the C-terminal cleavage of human Atg8 family proteins. BMC Cell Biol 2013; 14:27; PMID:23721406; http://dx.doi.org/ 10.1186/1471-2121-14-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Hemelaar J, Lelyveld VS, Kessler BM, Ploegh HL. A single protease, Apg4B, is specific for the autophagy-related ubiquitin-like proteins GATE-16, MAP1-LC3, GABARAP, and Apg8L. J Biol Chem 2003; 278:51841-50; PMID:14530254; http://dx.doi.org/ 10.1074/jbc.M308762200 [DOI] [PubMed] [Google Scholar]
- 178. Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 2007; 26:1749-60; PMID:17347651; http://dx.doi.org/ 10.1038/sj.emboj.7601623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Kaminskyy V, Zhivotovsky B. Proteases in autophagy. Biochim Biophys Acta 2012; 1824:44-50; PMID:21640203; http://dx.doi.org/ 10.1016/j.bbapap.2011.05.013 [DOI] [PubMed] [Google Scholar]
- 180. Tsukuba T, Yanagawa M, Kadowaki T, Takii R, Okamoto Y, Sakai E, Okamoto K, Yamamoto K. Cathepsin E deficiency impairs autophagic proteolysis in macrophages. PLoS One 2013; 8:e82415; PMID:24340026; http://dx.doi.org/ 10.1371/journal.pone.0082415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Gao Z, Gammoh N, Wong PM, Erdjument-Bromage H, Tempst P, Jiang X. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy 2010; 6:126-37; PMID:20061800; http://dx.doi.org/ 10.4161/auto.6.1.10928 [DOI] [PubMed] [Google Scholar]
- 182. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62SQSTM1 binds directly to Atg8LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 183. Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 2006; 8:1124-32; PMID:16998475; http://dx.doi.org/ 10.1038/ncb1482 [DOI] [PubMed] [Google Scholar]
- 184. Wirawan E, Vande Walle L, Kersse K, Cornelis S, Claerhout S, Vanoverberghe I, Roelandt R, De Rycke R, Verspurten J, Declercq W, et al. . Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis 2010; 1:e18; PMID:21364619; http://dx.doi.org/ 10.1038/cddis.2009.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Oral O, Oz-Arslan D, Itah Z, Naghavi A, Deveci R, Karacali S, Gozuacik D. Cleavage of Atg3 protein by caspase-8 regulates autophagy during receptor-activated cell death. Apoptosis 2012; 17:810-20; PMID:22644571; http://dx.doi.org/ 10.1007/s10495-012-0735-0 [DOI] [PubMed] [Google Scholar]
- 186. Murthy A, Li Y, Peng I, Reichelt M, Katakam AK, Noubade R, Roose-Girma M, DeVoss J, Diehl L, Graham RR, et al. . A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature 2014; 506:456-62; PMID:24553140; http://dx.doi.org/ 10.1038/nature13044 [DOI] [PubMed] [Google Scholar]
- 187. Betin VM, Lane JD. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J Cell Sci 2009; 122:2554-66; PMID:19549685; http://dx.doi.org/ 10.1242/jcs.046250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. You M, Savaraj N, Kuo MT, Wangpaichitr M, Varona-Santos J, Wu C, Nguyen DM, Feun L. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol Cell Biochem 2013; 374:181-90; PMID:23180246; http://dx.doi.org/ 10.1007/s11010-012-1518-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010; 29:1717-9; PMID:20101204; http://dx.doi.org/ 10.1038/onc.2009.519 [DOI] [PubMed] [Google Scholar]
- 190. Luo S, Rubinsztein DC. Apoptosis blocks Beclin 1-dependent autophagosome synthesis: an effect rescued by Bcl-xL. Cell Death Differ 2010; 17:268-77; PMID:19713971; http://dx.doi.org/ 10.1038/cdd.2009.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Li H, Wang P, Sun Q, Ding WX, Yin XM, Sobol RW, Stolz DB, Yu J, Zhang L. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res 2011; 71:3625-34; PMID:21444671; http://dx.doi.org/ 10.1158/0008-5472.CAN-10-4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Cho D-H, Jo YK, Hwang JJ, Lee YM, Roh SA, Kim JC. Caspase-mediated cleavage of ATG6/Beclin-1 links apoptosis to autophagy in HeLa cells. Cancer Lett 2009; 274:95-100; PMID:18842334; http://dx.doi.org/ 10.1016/j.canlet.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 193. Yamada Y, Suzuki NN, Hanada T, Ichimura Y, Kumeta H, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J Biol Chem 2007; 282:8036-43; PMID:17227760; http://dx.doi.org/ 10.1074/jbc.M611473200 [DOI] [PubMed] [Google Scholar]
- 194. Ohsumi Y. Historical landmarks of autophagy research. Cell Res 2014; 24:9-23; PMID:24366340; http://dx.doi.org/ 10.1038/cr.2013.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. . Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science 2011; 331:456-61; PMID:21205641; http://dx.doi.org/ 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep 2008; 9:859-64; PMID:18704115; http://dx.doi.org/ 10.1038/embor.2008.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a Protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem 2001; 276:1701-6; PMID:11096062; http://dx.doi.org/ 10.1074/jbc.C000752200 [DOI] [PubMed] [Google Scholar]
- 198. Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3pAut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem 2002; 277:13739-44; PMID:11825910; http://dx.doi.org/ 10.1074/jbc.M200385200 [DOI] [PubMed] [Google Scholar]