Abstract
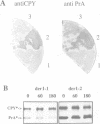
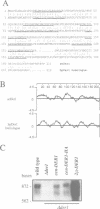
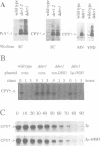
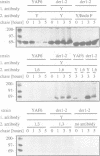
The endoplasmic reticulum (ER) of the yeast Saccharomyces cerevisiae contains of proteolytic system able to selectively degrade misfolded lumenal secretory proteins. For examination of the components involved in this degradation process, mutants were isolated. They could be divided into four complementation groups. The mutations led to stabilization of two different substrates for this process. The mutant classes were called 'der' for 'degradation in the ER'. DER1 was cloned by complementation of the der1-2 mutation. The DER1 gene codes for a novel, hydrophobic protein, that is localized to the ER. Deletion of DER1 abolished degradation of the substrate proteins. The function of the Der1 protein seems to be specifically required for the degradation process associated with the ER. The depletion of Der1 from cells causes neither detectable growth phenotypes nor a general accumulation of unfolded proteins in the ER. In DER1-deleted cells, a substrate protein for ER degradation is retained in the ER by the same mechanism which also retains lumenal ER residents. This suggests that DER1 acts in a process that directly removes protein from the folding environment of the ER.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antebi A., Fink G. R. The yeast Ca(2+)-ATPase homologue, PMR1, is required for normal Golgi function and localizes in a novel Golgi-like distribution. Mol Biol Cell. 1992 Jun;3(6):633–654. doi: 10.1091/mbc.3.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranski T. J., Koelsch G., Hartsuck J. A., Kornfeld S. Mapping and molecular modeling of a recognition domain for lysosomal enzyme targeting. J Biol Chem. 1991 Dec 5;266(34):23365–23372. [PubMed] [Google Scholar]
- Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993 Nov 19;75(4):717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Lippincott-Schwartz J. Degradation of proteins within the endoplasmic reticulum. Curr Opin Cell Biol. 1991 Aug;3(4):592–600. doi: 10.1016/0955-0674(91)90028-w. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Suzuki C. K., Klausner R. D. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science. 1990 Jan 5;247(4938):79–82. doi: 10.1126/science.2294595. [DOI] [PubMed] [Google Scholar]
- Chiang H. L., Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991 Mar 28;350(6316):313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- Christianson T. W., Sikorski R. S., Dante M., Shero J. H., Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992 Jan 2;110(1):119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Cvrcková F., Nasmyth K. Yeast G1 cyclins CLN1 and CLN2 and a GAP-like protein have a role in bud formation. EMBO J. 1993 Dec 15;12(13):5277–5286. doi: 10.1002/j.1460-2075.1993.tb06223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean N., Pelham H. R. Recycling of proteins from the Golgi compartment to the ER in yeast. J Cell Biol. 1990 Aug;111(2):369–377. doi: 10.1083/jcb.111.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doms R. W., Lamb R. A., Rose J. K., Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993 Apr;193(2):545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- Dorner A. J., Wasley L. C., Kaufman R. J. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J. 1992 Apr;11(4):1563–1571. doi: 10.1002/j.1460-2075.1992.tb05201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field J., Nikawa J., Broek D., MacDonald B., Rodgers L., Wilson I. A., Lerner R. A., Wigler M. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol Cell Biol. 1988 May;8(5):2159–2165. doi: 10.1128/mcb.8.5.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger A., Knop M., Wolf D. H. Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur J Biochem. 1993 Dec 1;218(2):565–574. doi: 10.1111/j.1432-1033.1993.tb18410.x. [DOI] [PubMed] [Google Scholar]
- Flynn G. C., Chappell T. G., Rothman J. E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989 Jul 28;245(4916):385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Fra A. M., Fagioli C., Finazzi D., Sitia R., Alberini C. M. Quality control of ER synthesized proteins: an exposed thiol group as a three-way switch mediating assembly, retention and degradation. EMBO J. 1993 Dec;12(12):4755–4761. doi: 10.1002/j.1460-2075.1993.tb06164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fra A., Sitia R. The endoplasmic reticulum as a site of protein degradation. Subcell Biochem. 1993;21:143–168. doi: 10.1007/978-1-4615-2912-5_7. [DOI] [PubMed] [Google Scholar]
- Franzusoff A., Schekman R. Functional compartments of the yeast Golgi apparatus are defined by the sec7 mutation. EMBO J. 1989 Sep;8(9):2695–2702. doi: 10.1002/j.1460-2075.1989.tb08410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut J. R., Hendershot L. M. The modification and assembly of proteins in the endoplasmic reticulum. Curr Opin Cell Biol. 1993 Aug;5(4):589–595. doi: 10.1016/0955-0674(93)90127-c. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Guide to yeast genetics and molecular biology. Methods Enzymol. 1991;194:1–863. [PubMed] [Google Scholar]
- Hammond C., Helenius A. Folding of VSV G protein: sequential interaction with BiP and calnexin. Science. 1994 Oct 21;266(5184):456–458. doi: 10.1126/science.7939687. [DOI] [PubMed] [Google Scholar]
- Hardwick K. G., Lewis M. J., Semenza J., Dean N., Pelham H. R. ERD1, a yeast gene required for the retention of luminal endoplasmic reticulum proteins, affects glycoprotein processing in the Golgi apparatus. EMBO J. 1990 Mar;9(3):623–630. doi: 10.1002/j.1460-2075.1990.tb08154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Hlodan R., Langer T. Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem Sci. 1994 Jan;19(1):20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Herscovics A., Orlean P. Glycoprotein biosynthesis in yeast. FASEB J. 1993 Apr 1;7(6):540–550. doi: 10.1096/fasebj.7.6.8472892. [DOI] [PubMed] [Google Scholar]
- Hill K. J., Stevens T. H. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol Biol Cell. 1994 Sep;5(9):1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S., Simoni R. D. 3-Hydroxy-3-methylglutaryl-coenzyme A reductase and T cell receptor alpha subunit are differentially degraded in the endoplasmic reticulum. J Biol Chem. 1992 May 5;267(13):9080–9086. [PubMed] [Google Scholar]
- Johnson L. M., Bankaitis V. A., Emr S. D. Distinct sequence determinants direct intracellular sorting and modification of a yeast vacuolar protease. Cell. 1987 Mar 13;48(5):875–885. doi: 10.1016/0092-8674(87)90084-5. [DOI] [PubMed] [Google Scholar]
- Klausner R. D., Sitia R. Protein degradation in the endoplasmic reticulum. Cell. 1990 Aug 24;62(4):611–614. doi: 10.1016/0092-8674(90)90104-m. [DOI] [PubMed] [Google Scholar]
- Knittler M. R., Dirks S., Haas I. G. Molecular chaperones involved in protein degradation in the endoplasmic reticulum: quantitative interaction of the heat shock cognate protein BiP with partially folded immunoglobulin light chains that are degraded in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1995 Feb 28;92(5):1764–1768. doi: 10.1073/pnas.92.5.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittler M. R., Haas I. G. Interaction of BiP with newly synthesized immunoglobulin light chain molecules: cycles of sequential binding and release. EMBO J. 1992 Apr;11(4):1573–1581. doi: 10.1002/j.1460-2075.1992.tb05202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M. J., Mori K. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol Cell Biol. 1993 Feb;13(2):877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Bonifacino J. S., Yuan L. C., Klausner R. D. Degradation from the endoplasmic reticulum: disposing of newly synthesized proteins. Cell. 1988 Jul 15;54(2):209–220. doi: 10.1016/0092-8674(88)90553-3. [DOI] [PubMed] [Google Scholar]
- Mallet L., Bussereau F., Jacquet M. Nucleotide sequence analysis of an 11.7 kb fragment of yeast chromosome II including BEM1, a new gene of the WD-40 repeat family and a new member of the KRE2/MNT1 family. Yeast. 1994 Jun;10(6):819–831. doi: 10.1002/yea.320100612. [DOI] [PubMed] [Google Scholar]
- Melnick J., Dul J. L., Argon Y. Sequential interaction of the chaperones BiP and GRP94 with immunoglobulin chains in the endoplasmic reticulum. Nature. 1994 Aug 4;370(6488):373–375. doi: 10.1038/370373a0. [DOI] [PubMed] [Google Scholar]
- Metcalf P., Fusek M. Two crystal structures for cathepsin D: the lysosomal targeting signal and active site. EMBO J. 1993 Apr;12(4):1293–1302. doi: 10.1002/j.1460-2075.1993.tb05774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Sant A., Kohno K., Normington K., Gething M. J., Sambrook J. F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (BiP) gene by unfolded proteins. EMBO J. 1992 Jul;11(7):2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullis K., Faloona F., Scharf S., Saiki R., Horn G., Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol. 1986;51(Pt 1):263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- Ng D. T., Watowich S. S., Lamb R. A. Analysis in vivo of GRP78-BiP/substrate interactions and their role in induction of the GRP78-BiP gene. Mol Biol Cell. 1992 Feb;3(2):143–155. doi: 10.1091/mbc.3.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T., Jackson M., Peterson P. A. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989 Aug 25;58(4):707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Hardwick K. G., Lewis M. J. Sorting of soluble ER proteins in yeast. EMBO J. 1988 Jun;7(6):1757–1762. doi: 10.1002/j.1460-2075.1988.tb03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Kaiser C. A., Orlean P., Albright C., Rose M. D., Robbins P. W., Botstein D. Structure of the yeast endoplasmic reticulum: localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991 Dec;7(9):891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Pryer N. K., Wuestehube L. J., Schekman R. Vesicle-mediated protein sorting. Annu Rev Biochem. 1992;61:471–516. doi: 10.1146/annurev.bi.61.070192.002351. [DOI] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B., Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994 May;19(1):55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Rothman J. E. Polypeptide chain binding proteins: catalysts of protein folding and related processes in cells. Cell. 1989 Nov 17;59(4):591–601. doi: 10.1016/0092-8674(89)90005-6. [DOI] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A., Maintz M., Kehle T., Herzog V. In vivo iodination of a misfolded proinsulin reveals co-localized signals for Bip binding and for degradation in the ER. EMBO J. 1995 Mar 15;14(6):1091–1098. doi: 10.1002/j.1460-2075.1995.tb07092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano R. H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol. 1988;157:533–544. doi: 10.1016/0076-6879(88)57102-1. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R., Meldolesi J. Endoplasmic reticulum: a dynamic patchwork of specialized subregions. Mol Biol Cell. 1992 Oct;3(10):1067–1072. doi: 10.1091/mbc.3.10.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana C., Stevens T. H. The yeast EUG1 gene encodes an endoplasmic reticulum protein that is functionally related to protein disulfide isomerase. Mol Cell Biol. 1992 Oct;12(10):4601–4611. doi: 10.1128/mcb.12.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsley F. M., Pelham H. R. The KKXX signal mediates retrieval of membrane proteins from the Golgi to the ER in yeast. Eur J Cell Biol. 1994 Jun;64(1):211–216. [PubMed] [Google Scholar]
- Wileman T., Kane L. P., Carson G. R., Terhorst C. Depletion of cellular calcium accelerates protein degradation in the endoplasmic reticulum. J Biol Chem. 1991 Mar 5;266(7):4500–4507. [PubMed] [Google Scholar]
- Wilson I. A., Niman H. L., Houghten R. A., Cherenson A. R., Connolly M. L., Lerner R. A. The structure of an antigenic determinant in a protein. Cell. 1984 Jul;37(3):767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Yaffe M. P., Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J., Kane L. P., Exley M., Wileman T. Regulation of selective protein degradation in the endoplasmic reticulum by redox potential. J Biol Chem. 1993 Sep 15;268(26):19810–19818. [PubMed] [Google Scholar]