Abstract
Bacterial glycosyltransferases play important roles in bacterial fitness and virulence. Oral streptococci have evolved diverse strategies to survive and thrive in the carbohydrate-rich oral cavity. In this review, we discuss 2 important biological processes mediated by 2 distinct groups of glycosyltransferases in oral streptococci that are important for bacterial colonization and virulence. The first process is the glycosylation of highly conserved serine-rich repeat adhesins by a series of glycosyltransferases. Using Streptococcus parasanguinis as a model, we highlight new features of several glycosyltransferases that sequentially modify the serine-rich glycoprotein Fap1. Distinct features of a novel glycosyltransferase fold from a domain of unknown function 1792 are contrasted with common properties of canonical glycosyltransferases. The second biological process we cover is involved in building sticky glucan matrix to establish cariogenic biofilms by an important opportunistic pathogen Streptococcus mutans through the action of a family of 3 glucosyltransferases. We focus on discussing the structural feature of this family as a glycoside hydrolase family of enzymes. While the 2 processes are distinct, they all produce carbohydrate-coated biomolecules, which enable bacteria to stick better in the complex oral microbiome. Understanding the making of the sweet modification presents a unique opportunity to develop novel antiadhesion and antibiofilm strategies to fight infections by oral streptococci and beyond.
Keywords: serine-rich repeat glycoproteins, glycosylation, biofilm, carbohydrate, 3D molecular structure, glucans
Introduction
Oral streptococci initiate colonization of the oral cavity through their abundant adhesion molecules, and many are carbohydrate coated (Kolenbrander et al. 1993). These early colonizers have been associated with healthy oral cavity and also provide a platform for pathogenic keystone bacteria to attach and form a complex balanced dental plaque (Jenkinson 2011; Thurnheer et al. 2014). Disruption of a balanced bacterial community leads to the development of oral infectious diseases. The initial adherence of early colonizers through glycosylated adhesins such as serine-rich repeat proteins (SRRPs) (Zhou and Wu 2009; Lizcano et al. 2012) and other glycoconjugates plays an important role in the formation of dental plaque (Koo et al. 2010; Falsetta et al. 2014).
Glycosyltransferases are the enzymes that are essential for producing diverse and complex of glycoconjugates (Jeon et al. 2011). The glycosidic bond formation catalyzed by glycosyltransferases occurs between carbohydrate and acceptor. The acceptor is diverse, containing a variety of biomolecules such as serine-rich repeats or partially glycosylated serine-rich repeat proteins (Wu, Zeng, et al. 2007; Bu et al. 2008; Zhou and Wu 2009; Zhou et al. 2010; Zhu et al. 2011; Lizcano et al. 2012), glucan matrix synthesized by cariogenic Streptococcus mutans (Lemos et al. 2013), glycolipids (Ito et al. 2011), and receptor polysaccharide (Xu et al. 2003).
Glycosyltransferases were first classified by Campbell and colleagues (1967) based on similarities of amino acid sequences. This was further developed and has evolved to facilitate structural and mechanistic studies, as well as avoid recurrent issues in the functional prediction of a large number of putative glycosyltransferases (Coutinho et al. 2003). Currently, 97 families of glycosyltransferases (over 160,000) have been classified in the Carbohydrate-Active Enzymes database (CAZy; http://www.cazy.org/Glycosyltransferases) (Lombard et al. 2014). It is extremely challenging to define both acceptor and carbohydrate donors for any glycosyltransferase.
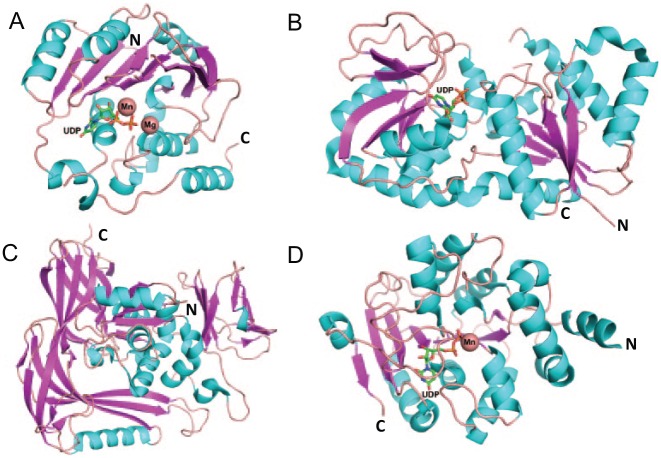
Hundreds of glycosyltransferase structures have been solved in the past 20 years, which presents a quite different picture from what the CAZy classification entails. Most structures of nucleotide-sugar-dependent glycosyltransferases are classified into only 2 glycosyltransferase (GT) folds, GT-A and GT-B (Fig. 1). GT-A–type glycosyltransferases possess a single Rossmann fold (topology β/α/β/α/β) and conserved “DXD” metal binding motif (Charnock and Davies 1999). GT-B–type glycosyltransferases possess a twin Rossmann fold that faces each other and is linked flexibly by an active site within the resulting cleft (Zhu et al. 2011). There is another group of glycosyltransferases with a not well-defined structure fold, called GT-C type (Fig. 1C). GT-C fold possesses 8 to 13 predicted transmembrane segments and displays a DXD motif in the first extracellular/lumenal loop (Igura et al. 2008). Recently, a new glycosyltransferase (DUF1792) has been reported to have a distinct GT structural fold named “GT-D” (Fig. 1D). This family of glycosyltransferases binds to uridine diphosphate (UDP) and requires divalent metal ions to transfer carbohydrate but does not share sequence and structural similarity to any known glycosyltransferases (Zhang et al. 2014).
Figure 1.
Structural folds of glycosyltransferases (GTs). Structural ribbon diagrams are colored: purple for β-sheet, cyan for helix, and pink for loops. GT-A fold structure, SpsA (PDB entry, 1QGQ) (A); GT-B fold structure, Gtf3 (PDB entry, 3QKW) (B); GT-C fold structure, STT3 (PDB entry, 2ZAG) (C); GT-D fold structure, DUF1792 (PDB entry 4PHR) (D). Uridine diphosphate is found in 3 glycosyltransferase folds, GT-A, B, and D. Mn and Mg are shown in the sphere.
Glycosyltransferases are diverse, and the number of glycosyltransferases is enormous. The function of numerous glycosyltransferases in oral bacteria still remains to be determined (Appendix Table). In this review, we highlight a series of well-studied glycosyltransferases involved in the glycosylation of SRRPs and another distinct group of glucosyltransferases that are crucial for the synthesis of glucan matrix important for biofilm formation of a cariogenic bacterium S. mutans.
Glycosyltransferases Important for the Glycosylation of SRRPs
Streptococcus parasanguinis is one of the initial colonizers in the oral cavity (Wu et al. 1998). It provides a platform for other oral bacteria to attach and form a complex oral microbiome (Kolenbrander et al. 2006; Jenkinson 2011). Fap1, the first SRRP identified, functions as an important bacterial adhesin (Stephenson et al. 2002). The glycosylation of Fap1 is critical for biofilm formation and bacterial colonization (Zhou and Wu 2009).
The mature Fap1 protein possesses several monosaccharide residues, including glucose, N-acetylglucosamine (GlcNAc), and rhamnose. The glycan sequence of the Fap1 peptide backbone has been determined as Rha1-3Glc1-(Glc1-3Glc NAc1-) 2, 6Glc1-6GlcNAc (Zhang et al. 2014). The glycosyl composition of other SRRPs has been determined for GspB of Streptococcus gordonii (Takamatsu et al. 2004) and Srr1 of Streptococcus agalactiae (Chaze et al. 2014), but the sequential order of the glycans are unknown.
Glycosylation of Fap1 is catalyzed by a series of GTs, sequentially adding glycosyl residues to Fap1. An 11-gene cluster has been identified and implicated in the biogenesis of Fap1 (Zhou and Wu 2009). Among them, 6 genes (gtf1, gtf2, gly, gtf3, galT1, and galT2) encode putative glycosyltransferases, which mediate the glycosylation of Fap1 (Wu, Bu, et al. 2007; Wu, Zeng, et al. 2007; Bu et al. 2008; Zhou and Wu 2009; Wu et al. 2010; Zhou et al. 2010; Zhu et al. 2011; Zhang et al. 2013; Zhang et al. 2014) (Fig. 2). Here we discuss the findings on 3 GTs determined to be important for the glycosylation of Fap1 or other SRRPs.
Figure 2.
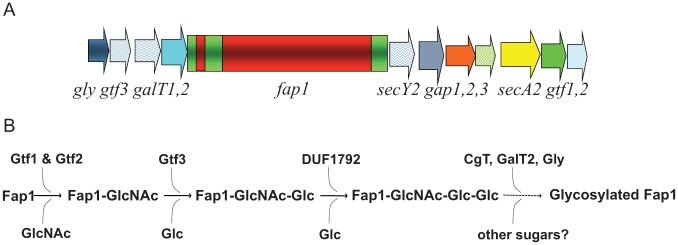
Diagrams of a gene cluster with fap1 and associated glycosyltransferase genes. The fap1 locus has 11 flanking genes. gtf1, gtf2, gtf3, galT1, galT2, and gly are glycosyltransferase genes; secY2, secA2, and gap123 are genes involved in secretion (A). Gtf1 and Gtf2 form a protein complex to initialize the first step of Fap1 glycosylation; Gtf3 mediates the second step; DUF1792 is the third glycosyltransferase to modify Fap1. C terminal GalT1 (CgT), GalT1, and Gly mediate the remaining steps of Fap1 glycosylation to fully modify Fap1 (B).
Gtf1 and Gtf2 initiate the first step of the glycosylation by transferring a GlcNAc reside to Fap1 (Wu, Bu, et al. 2007; Bu et al. 2008). Gtf1 is a typical glycosyltransferase that belongs to the GT-B family of GTs. However, Gtf1 alone only exhibits minimal enzymatic activity, while Gtf2 alone does not have any activity (Wu et al. 2010). Gtf2 enhances the enzymatic activity of Gtf1 by its chaperone-like property (Bu et al. 2008; Wu et al. 2010). Gtf1 and Gtf2 work in concert to form a fully activated protein complex. This novel glycosyltransferase protein complex is highly conserved in other gram-positive bacteria, including S. gordonii, Streptococcus sanguinis, and Streptococcus salivarius as GtfA and GtfB. The GtfA/B complex initiates the glycosylation and transfers GlcNAc to respective SRRPs (Takamatsu et al. 2004; Zhou and Wu 2009; Li et al. 2014; Shi et al. 2014).
Both Gtf1 and Gtf2 possess a conserved domain of unknown function (DUF) 1975 at their N-terminus. DUF1975 mediates the interaction between Gtf1 and Gtf2 (Bu et al. 2008; Wu and Wu 2011). The crystal structure of GtfA from Streptococcus pneumoniae (Shi et al. 2014) revealed GtfA has a typical GT-B fold at the C terminus and a new “add-on” domain at the N terminus by DUF1975. Add-on domains are found in many GTs, including eukaryotic O-GlcNAc transferase (Clarke et al. 2008; Martinez-Fleites et al. 2008; Lazarus et al. 2011). Like many other add-on domains, DUF 1975 is involved in substrate binding in addition to its binding to its interacting partner, GtfB. Unlike other add-on domains, DUF1975 adopts a novel β-meander structure of a twisted 10-stranded antiparallel β-sheet, and the insertion of DUF 1975 makes GtfA a novel O-GlcNAc transferase. DUF1975 is critical for the recognition of both coactivator GtfB and acceptor substrate pneumococcal serine-rich repeat protein (PsrP) (Shi et al. 2014). However, it is not clear how DUF1975 mediates interaction with GtfB and how it recognizes its acceptor substrate. Future studies will focus on solving the complex structure of GtfA/B with its substrate, which may shed new mechanistic insights. Despite the lack of the complex structure, the unique protein-protein interaction module DUF1975 found in the enzymatic complex can be explored to design novel inhibitors that disrupt the interaction, thereby achieving the inhibition of bacterial adhesions and infection. This approach is promising as DUF1975 is highly conserved in gram-positive bacteria and structurally distinct from other add-on domains found in other GTs in eukaryotic cells.
Glycosylation of Fap1 is a multistep process (Zhou and Wu 2009). The initial glycosylation is mediated by the Gtf1/2 protein complex. Gtf3, a glucosyltransferase, transfers Glc to the GlcNAc-modified Fap1 to complete the second step of the glycosylation (Zhou et al. 2010). Of note, Gtf3 was initially annotated as a nucleotide sugar synthetase by its sequence homology. Biochemical and structural studies unequivocally demonstrated it is a bona fide glucosyltransferase. This indicates sequence homology-based predication has its limitations and sometimes can be misleading. Structural studies have revealed that Gtf3 is a GT-B–type GT with a typical twin Rossmann fold (Zhu et al. 2011). Unlike Gtf1, Gtf3 does not require a molecular chaperone, and it is stable and fully functional by itself. Interestingly, Gtf3 forms a homotetramer to achieve the maximum enzymatic activity (Zhu et al. 2011). The transition between homo- and heteromers of glycosyltransferases can modulate formation of optimal active glycosyltransferases complexes (Hassinen et al. 2011). Whether this second step of the Fap1 glycosylation is tightly coordinated with the first step via this organizational interplay is unknown and awaits further investigation. Other Gtf3-like proteins are also conserved in many gram-positive bacteria that produce SRRPs, including Streptococcus mitis, S. sanguinis, S. gordonii, Streptococcus australis, and S. salivarius (Zhu et al. 2015). However, the precise function of most Gtf3-like proteins in these bacteria has not been investigated.
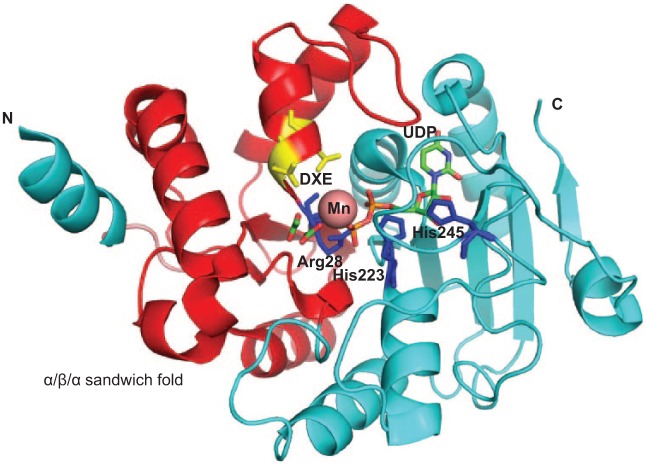
GalT1 is involved in the subsequent glycosylation step. It is coded by a putative glycosyltransferase gene that is located at the same locus with gtf3. GalT1 possesses 2 domains: DUF1792 at the N terminus and a typical GT-A glycosyltransferase domain at the C terminus. We have demonstrated that DUF1792 alone is sufficient to catalyze the third step of the Fap1 glycosylation by transferring another Glc residue to GlcNAc-Glc–modified Fap1 (Zhang et al. 2014). The C terminus of GalT1 is dispensable for this activity, suggesting it might mediate the glycosylation of Fap1 by adding a different glycosyl residue after the DUF1792 modification (Zhang et al. 2014). The crystal structure of DUF1792 has revealed a novel glycosyltransferase fold that is composed of 3 regions: an N-terminal region that has 2 α/β/α sandwich domains consisting of an atypical DXE metal binding motif, a central region that contains a Rossmann-like fold, and a C-terminal region that is composed of a long coil region, a short helical turn, and a short β-strand (Fig. 3). Since DUF1792 is structurally distinct from all known GT folds of glycosyltransferases, we have assigned DUF1792 as a new GT structural fold: GT-D (Zhang et al. 2014). DUF1792 is highly conserved in streptococci, lactobacilli, and even gram-negative bacteria and may represent a universal glycosyltransferase fold (Zhang et al. 2014). Since the distribution of DUF1792 is limited to bacterial species, it can be explored as an antibiotic target to develop antibacterial therapy.
Figure 3.
Structure of DUF1792. The α/β/α sandwich fold is colored in red. Key residues Arg28, His223, and His 245 are colored in blue. The DXE motif is highlighted in yellow. Uridine diphosphate (UDP) is shown as a stick. Mn is shown in a sphere.
Another 2 genes, galT2 and gly, located at the same gene cluster, are predicted to encode 2 putative glycosyltransferases, which mediate the last 2 steps of the Fap1 glycosylation. The glycosyl specificity of GalT2 and Gly has not been defined yet. Whether they are coordinated with GalT1 and other GTs to complete the modification of Fap1 is unknown.
Genetic, biochemical, and structural biology studies have allowed us to propose a sequential Fap1 glycosylation pathway. Of note, the organization and number of annotated glycosyltransferases in the gene cluster are quite diverse (Zhou and Wu 2009). In S. parasanguinis, 4 glycosyltransferase genes (gly, gtf3, galT1, and galT2) are located upstream of fap1 (Bu et al. 2008). In other gram-positive bacteria that possess SRRP genes, putative glycosyltransferase genes are clustered downstream of srrp genes. In a search of sequenced bacterial genomes, we found that S. pneumoniae possesses a very large number of annotated glycosyltransferases, ranging from 4 to 11, which may indicate differential modification of PsrP by various S. pneumoniae strains (Zhou and Wu 2009). Both organization and number of annotated glycosyltransferases vary even within the same bacterial species such as S. agalactiae (Seifert et al. 2006). SRRPs and protein glycosylation have been implicated in bacterial pathogenesis (Zhou and Wu 2009; Lizcano et al. 2012), and it is tempting to hypothesize that the diversity of GTs in various bacterial species and strains may allow the bacteria to make differentially modified adhesion molecules tailored to their unique host environments.
Structural Features of 3 GTs Involved in Sequential Glycosylation of SRRPs
UDP Binding
All GTs identified in the pathway bind to nucleotide substrates. For instance, the structures of GtfA, Gtf3, and DUF1792 all contain UDP (Zhu et al. 2011; Shi et al. 2014; Zhang et al. 2014). UDP binding residues are quite conserved. Arg328 from GtfA, Arg 179 from Gtf3, and Arg28 from DUF1792 interact with UDP (Zhu et al. 2011; Shi et al. 2014; Zhang et al. 2014). Biochemical experiments confirmed that the interaction between Arg and phosphate atom of UDP is important for enzymatic activity (Shi et al. 2014; Zhang et al. 2014; Zhu et al. 2011). This interaction is also found in other GTs (Pedersen et al. 2000) and is considered a conserved feature of the UDP-dependent glycosyltransferase. Likewise, Lys residue is also critical for the enzymatic activity. Lys333 from GtfA, Lys246 from Gtf3, and Lys205 from DUF1792 are all observed close enough to possibly interact with UDP (Zhu et al. 2011; Shi et al. 2014; Zhang et al. 2014). These findings suggest that both residues mediate neutralization of the negative charge on phosphoryl groups of UDP during catalysis and also may help UDP to leave the active site after catalysis.
In addition, there is a phenyl ring conserved to interact with the uracil ring of UDP. Mutation of Tyr19 in GtfA, Tyr211 in Gtf3, and either His 223 or His 245 in DUF1792 has significantly inhibited enzymatic activity (Zhu et al. 2011; Shi et al. 2014; Zhang et al. 2014). These results suggest that Arg, Tyr, or His in the UDP binding site is critical for the enzymatic activity. These studies may assist the rational design of small molecules that target these enzymes at the precise positions.
Metal Binding
The DXD metal binding motif is considered a signature for GT-A fold glycosyltransferase as the motif plays crucial role in binding to sugar-nucleoside diphosphate and divalent metal ion (Busch et al. 1998). However, concerns arise as this motif may not be as conserved as previously thought (Coutinho et al. 2003; Lairson et al. 2008). Nevertheless, the DXD motif is crucial for the activity of most GT-A types of glycosyltransferases. Recently, DUF1792 has been reported as the first glycosyltransferase in the GT-D fold family. It has a DXE metal binding motif, which is invariable in all DUF1792 homologs (Zhang et al. 2014), indicating the motif represents a bona fide property of this glycosyltransferase family.
Glucosyltransferases That Catalyze the Synthesis of Glucans in Oral Streptococci
Glucosyltransferases (GTFs) play an important role in the sucrose-dependent accumulation of mutans streptococci on the tooth surface. Extensive biochemical studies have been conducted on streptococcal GTFs, such as GtfG from S. gordonii (Vickerman et al. 1996; Vickerman and Clewell 1997), Gtf from S. sanguis (Buchan and Jenkinson 1990), Gtfs from Streptococcus sobrinus (Colby and Russell 1997), and GtfBCD from S. mutans (Bowen and Koo 2011). In this review, we mainly focus on GtfBCD from S. mutans, a major opportunistic pathogen for dental caries.
S. mutans is capable of rapidly synthesizing large amounts of exopolysaccharide through a family of GTFs and readily forming biofilms. S. mutans has 3 separate GTFs (GtfBCD). This family of GTFs is distinct from GTs we discussed above. GTFs catalyze the cleavage of sucrose into fructose and glucose, as well as use the energy released from the cleavage of the glycosidic bond to drive concurrent transfer of the glucose residue to a growing glucan polymer (van Hijum et al. 2006). GTFs are named after their ability to transfer the glucose residue, but the nomenclature fails to consider the essential activity of GTFs to split sucrose into glucose and fructose.
In fact, GTFs belong to members of glycoside hydrolase (GH) family 70 (Hamada and Slade 1980). Each GTF enzyme has distinctive properties varying in synthesizing a structurally distinct glucan from sucrose (Colby and Russell 1997). GtfB synthesizes water-insoluble glucans rich in α(1-3)-linkage, GtfC produces a mixture of soluble glucans rich in α(1-6)-linkage and insoluble glucans, and GtfD makes primarily soluble glucans (Bowen and Koo 2011).
GTFs produced by S. mutans play a key role in the cariogenic process as the glucan synthesized by GTFs enhances the biofilm formation and colonization of cariogenic bacteria. Furthermore, production of the glucans is also accompanied by the increase in the amount of acids produced, and glucans help localize generated acids to the glucan matrix and enamel interface to form an acidic sealing zone, which facilitates the dissolution of dental enamel and promotes the development of dental caries (Krzysciak et al. 2013). This acidic sealing zone may resemble an analogous structural zone found at the interface between osteoclasts and bone surfaces when osteoclasts polarize to form acidic, hydrolase-rich resorption lacunae to degrade bone (Heckel et al. 2009). GTFs have become an important target for drug discovery in the search of small-molecule compounds that inhibit cariogenic biofilms. The details of the biological function of each GTF have been beautifully reviewed elsewhere (Bowen and Koo 2011), so we focus on discussing the structure of GTFs and the potential application.
GTFs from S. mutans are responsible for catalyzing the formation of glucan with various types of glucosidic linkages, such as α(1-3), α(1-4), or α(1-6) bonds, from sucrose (Ito et al. 2011). GTFs from S. mutans are composed of 3 distinct functional domains: the N-terminal variable junction region, the catalytic region, and the C-terminal glucan binding region (Monchois et al. 1999; Ito et al. 2011) (Fig. 4). Protein sequence alignments of different GTFs indicate that the catalytic domain is highly conserved (Monchois et al. 1999), but the alignment does not explain the particular type of activity conferred by each GTF. Structural studies are in need to solve the puzzle.
Figure 4.

Organization of GtfC domains.
The crystal structure of GtfC from amino acid residues 244 to 1087 has been solved (Ito et al. 2011). In the structure, GtfC possesses 4 domains: A, B, C, and IV (Fig. 5). Each domain, with the exception of domain C, is composed of 2 separate regions of the polypeptide chain. Domain C is composed of a single stretch of amino acid residues (693 to 828). The overall structure of GtfC shares great similarity to GH family 13 amylases, especially in domain A, which forms the core of the catalytic domain of GtfC. Domain A has a common TIM-barrel motif of GH family 13 amylases and also has 2 additional α helices (Ito et al. 2011). Overall, GtfB, GtfC, and GtfD share more than 40% amino acid sequence identity. Many of the amino acids in the catalytic domain are conserved, except Glu431 and Asp593 (Ito et al. 2011). Glu431 is in a position opposite the active center and occupies a part of the metal binding. Asp593 is in the additional α-helix in domain A, which appears to be the critical residue for acceptor sugar orientation (Ito et al. 2011). It has been shown that replacement of Thr at this position in GtfD with Asp or Glu promotes the synthesis of insoluble glucan instead of producing soluble glucan, while replacement of this Asp residue in GtfB promotes synthesis of soluble glucans rather than insoluble glucan (Shimamura et al. 1994). Therefore, Asp593 is considered the only residue that determines the linkage specificity of glucans produced by GtfBCD. In addition, site-directed mutagenesis of a GtfC homolog, GtfR of Streptococcus oralis, reveals that the additional α-helices in domain A and the adjacent loop could interact with different substrates with different length and modification, which would alter the reaction specificity of GTFs (Hellmuth et al. 2008).
Figure 5.
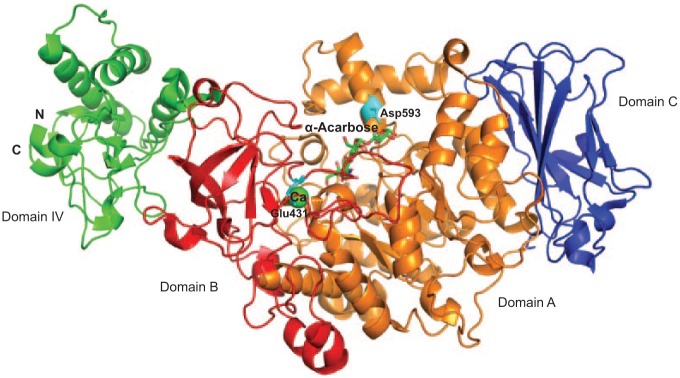
Structure of the catalytic domain of GtfC from Streptococcus mutans. Domain A is colored in orange, domain B is colored in red, domain C is colored in blue, and domain IV is colored in green. Residues Glu431 and Asp593 are colored in cyan. α-Acarbose is shown as a stick. Ca is shown as a sphere and colored in green (PDB entry, 3AIC).
The structure of the GtfC has 3 forms: free enzyme form and in complex with an inhibitor acarbose and a carbohydrate acceptor maltose (Ito et al. 2011). The inhibitor of GtfC, acarbose, is a pseudo-tetrasaccharide that contains a nonhydrolyzable nitrogen-linked bond, which blocks the catalytic activity of various GHs. Maltose, a carbohydrate acceptor of GtfC, is not only a known transglycosyl acceptor for GTFs (Van den Steen et al. 1998) but also a weak inhibitor of GTFs (Newbrun et al. 1983). The complex structure of maltose and GtfC suggests that maltose serves as a competitive inhibitor of the second acceptor, sucrose (Ito et al. 2011). All these structural details, especially the unique features of the catalytic domain from GtfC, can be useful in the design of novel inhibitors targeting GTFs. We are actively pursuing structure-based discovery of inhibitory compounds targeting GTFs.
In addition to synthesizing different glucans, GtfBCD also have distinct roles in the building of dental plaque. GtfC is absorbed to enamel, GtfB binds to bacteria or fungi and enhances adhesion to plaque, and GtfD catalyzes the biosynthesis of a soluble polysaccharide, which acts as a primer for GtfB (Bowen and Koo 2011; Gregoire et al. 2011). Since the diverse glycosyl linkage products are determined by limited differences in the conserved catalytic domain, the distinct roles in the formation of dental plaque can be attributed to low homology among putative glucan binding modules found in each GTF. Further characterization will help determine distinct function of each GTF.
Gene Regulation of GTFs
Although the role of a number of GTs and GTFs in their host is well studied, the mechanisms that control the expression of these proteins are poorly understood. So far, only the gene regulation of GTFs from S. mutans and S. gordonii has been documented. In S. mutans, response regulators CovR and VicR differentially regulate the expression of both GtfB and GtfC by binding directly to both the gtfB and gtfC promoters (Senadheera et al. 2005; Biswas and Biswas 2006). It is unknown how the 2 response regulators coordinate this regulation. In S. gordonii, rgg, a gene located upstream of gtfG, was shown to positively regulate the expression of GtfG (Sulavik and Clewell 1996; Vickerman and Clewell 1997). Deciphering the molecular mechanisms and identifying genes that directly regulate GTs awaits further investigation.
Conclusions and Future Perspectives
Oral streptococci live in a carbohydrate-rich environment. They have evolved a variety of strategies to cope with this niche. This review highlighted only 2 aspects of the sweet modification by oral streptococci: making glycoproteins and establishing sticky glucans. As the number of glycosyltransferases is diverse and enormous, it would not be surprising to see more interesting structural folds, specific biochemical activities, and biological functions emerging. Since glycosyltransferases can sequentially modify serine-rich peptide substrates efficiently, one practical utility of studying the glycosylation pathway is to engineer carbohydrate-coated proteins using a rich resource of glycosyltransferases. In addition, biological function of this sweet modification of SRRPs is unknown. As many important pathogens also possess these carbohydrate-coated SRRPs, it will be interesting to learn whether and how the sweet modification can modulate biological function of bacterial-host interactions.
Furthermore, as the production of glucan matrix is crucial for cariogenic biofilm formation of S. mutans, solving crystal structures of each GTF will shed new mechanistic insights into the matrix production. Of note, this family of GTFs is also found in commensal streptococci, and it would be interesting to study how they contribute to build a complex biofilm matrix when coexisting with S. mutans in the oral microbiome.
Author Contributions
F. Zhu, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; H. Zhang, contributed to data acquisition, analysis, and interpretation, drafted the manuscript; H. Wu, contributed to conception and design, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Footnotes
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
The work in the authors’ laboratory was supported by National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR) grants R01DE017954 (HW) and R01DE022350 (HW).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Biswas S, Biswas I. 2006. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans. J Bacteriol. 188(3):988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen WH, Koo H. 2011. Biology of Streptococcus mutans–derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 45(1):69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu S, Li Y, Zhou M, Azadin P, Zeng M, Fives-Taylor P, Wu H. 2008. Interaction between two putative glycosyltransferases is required for glycosylation of a serine-rich streptococcal adhesin. J Bacteriol. 190(4):1256–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan RA, Jenkinson HF. 1990. Glucosyltransferase production by Streptococcus sanguis Challis and comparison with other oral streptococci. Oral Microbiol Immunol. 5(2):63–71. [DOI] [PubMed] [Google Scholar]
- Busch C, Hofmann F, Selzer J, Munro S, Jeckel D, Aktories K. 1998. A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J Biol Chem. 273(31):19566–19572. [DOI] [PubMed] [Google Scholar]
- Campbell JA, Davies GJ, Bulone V, Henrissat B. 1997. A classification of nucleotide-diphospho-sugar glycosyltransferases based on amino acid sequence similarities. Biochem J. 326(Pt 3):929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Davies GJ. 1999. Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry. 38(20):6380–6385. [DOI] [PubMed] [Google Scholar]
- Chaze T, Guillot A, Valot B, Langella O, Chamot-Rooke J, Di Guilmi AM, Trieu-Cuot P, Dramsi S, Mistou MY. 2014. O-glycosylation of the N-terminal region of the serine-rich adhesin Srr1 of Streptococcus agalactiae explored by mass spectrometry. Mol Cell Proteomics. 13(9):2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke AJ, Hurtado-Guerrero R, Pathak S, Schüttelkopf AW, Borodkin V, Shepherd SM, Ibrahim AF, van Aalten DM. 2008. Structural insights into mechanism and specificity of O-GlcNAc transferase. Embo J. 27(20):2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby SM, Russell RR. 1997. Sugar metabolism by mutans streptococci. Soc Appl Bacteriol Symp Ser. 26:80S–88S. [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. 2003. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 328(2):307–317. [DOI] [PubMed] [Google Scholar]
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai CH, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 82(5):1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoire S, Xiao J, Silva BB, Gonzalez I, Agidi PS, Klein MI, Ambatipudi KS, Rosalen PL, Bauserman R, Waugh RE, et al. 2011. Role of glucosyltransferase B in interactions of Candida albicans with Streptococcus mutans and with an experimental pellicle on hydroxyapatite surfaces. Appl Environ Microbiol. 77(18):6357–6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada S, Slade HD. 1980. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 44(2):331–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassinen A, Pujol FM, Kokkonen N, Pieters C, Kihlström M, Korhonen K, Kellokumpu S. 2011. Functional organization of Golgi N- and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J Biol Chem. 286(44):38329–38340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel T, Czupalla C, Expirto Santo AI, Anitei M, Arantzazu Sanchez-Fernandez M, Mosch K, Krause E, Hoflack B. 2009. Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc Natl Acad Sci U S A. 106(5):1451–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmuth H, Wittrock S, Kralj S, Dijkhuizen L, Hofer B, Seibel J. 2008. Engineering the glucansucrase GTFR enzyme reaction and glycosidic bond specificity: toward tailor-made polymer and oligosaccharide products. Biochemistry. 47(25):6678–6684. [DOI] [PubMed] [Google Scholar]
- Igura M, Maita N, Kamishikiryo J, Yamada M, Obita T, Maenaka K, Kohda D. 2008. Structure-guided identification of a new catalytic motif of oligosaccharyltransferase. EMBO J. 27(1):234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Ito S, Shimamura T, Weyand S, Kawarasaki Y, Misaka T, Abe K, Kobayashi T, Cameron AD, Iwata S. 2011. Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans. J Mol Biol. 408(2):177–186. [DOI] [PubMed] [Google Scholar]
- Jenkinson HF. 2011. Beyond the oral microbiome. Environ Microbiol. 13(12):3077–3087. [DOI] [PubMed] [Google Scholar]
- Jeon JG, Rosalen PL, Falsetta ML, Koo H. 2011. Natural products in caries research: current (limited) knowledge, challenges and future perspective. Caries Res. 45(3):243–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. 1993. Coaggregation-specific adherence among human oral plaque bacteria. FASEB J. 7(5):406–413. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Rickard AH, Jakubovics NS, Chalmers NI, Diaz PI. 2006. Bacterial interactions and successions during plaque development. Periodontol 2000. 42:47–79. [DOI] [PubMed] [Google Scholar]
- Koo H, Xiao J, Klein MI, Jeon JG. 2010. Exopolysaccharides produced by Streptococcus mutans glucosyltransferases modulate the establishment of microcolonies within multispecies biofilms. J Bacteriol. 192(12):3024–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysciak W, Pluskwa KK, Jurczak A, Koscielniak D. 2013. The pathogenicity of the Streptococcus genus. Eur J Clin Microbiol Infect Dis. 32(11):1361–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lairson LL, Henrissat B, Davies GJ, Withers SG. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 77:521–555. [DOI] [PubMed] [Google Scholar]
- Lazarus MB, Nam Y, Jiang J, Sliz P, Walker S. 2011. Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature. 469(7331):564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Quivey RG, Jr, Koo H, Abranches J. 2013. Streptococcus mutans: a new Gram-positive paradigm? Microbiology. 159(Pt 3):436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Huang X, Li J, Zeng J, Zhu F, Fan W, Hu L. 2014. Both GtfA and GtfB are required for SraP glycosylation in Staphylococcus aureus. Curr Microbiol. 69(2):121–126. [DOI] [PubMed] [Google Scholar]
- Lizcano A, Sanchez CJ, Orihuela CJ. 2012. A role for glycosylated serine-rich repeat proteins in gram-positive bacterial pathogenesis. Mol Oral Microbiol. 27(4):257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42(Database Issue):D490–D495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ. 2008. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 15(7):764–765. [DOI] [PubMed] [Google Scholar]
- Monchois V, Willemot RM, Monsan P. 1999. Glucansucrases: mechanism of action and structure-function relationships. FEMS Microbiol Rev. 23(2):131–151. [DOI] [PubMed] [Google Scholar]
- Newbrun E, Hoover CI, Walker GJ. 1983. Inhibition by acarbose, nojirimycin and 1-deoxynojirimycin of glucosyltransferase produced by oral streptococci. Arch Oral Biol. 28(6):531–536. [DOI] [PubMed] [Google Scholar]
- Pedersen LC, Tsuchida K, Kitagawa H, Sugahara K, Darden TA, Negishi M. 2000. Heparan/chondroitin sulfate biosynthesis structure and mechanism of human glucuronyltransferase I. J Biol Chem. 275(44):34580–34585. [DOI] [PubMed] [Google Scholar]
- Seifert KN, Adderson EE, Whiting AA, Bohnsack JF, Crowley PJ, Brady LJ. 2006. A unique serine-rich repeat protein (Srr-2) and novel surface antigen (epsilon) associated with a virulent lineage of serotype III Streptococcus agalactiae. Microbiology. 152(Pt 4):1029–1040. [DOI] [PubMed] [Google Scholar]
- Senadheera MD, Guggenheim B, Spatafora GA, Huang YC, Choi J, Hung DC, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG. 2005. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 187(12):4064–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi WW, Jiang YL, Zhu F, Yang YH, Shao QY, Yang HB, Ren YM, Wu H, Chen Y, Zhou CZ. 2014. Structure of a novel O-linked N-acetyl-d-glucosamine (O-GlcNAc) transferase, GtfA, reveals insights into the glycosylation of pneumococcal serine-rich repeat adhesins. J Biol Chem. 289(30):20898–20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura A, Nakano YJ, Mukasa H, Kuramitsu HK. 1994. Identification of amino acid residues in Streptococcus mutans glucosyltransferases influencing the structure of the glucan product. J Bacteriol. 176(16):4845–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson AE, Wu H, Novak J, Tomana M, Mintz K, Fives-Taylor P. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol Microbiol. 43(1):147–157. [DOI] [PubMed] [Google Scholar]
- Sulavik MC, Clewell DB. 1996. Rgg is a positive transcriptional regulator of the Streptococcus gordonii gtfG gene. J Bacteriol. 178(19):5826–5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Bensing BA, Sullam PM. 2004. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol 52(1):189–203. [DOI] [PubMed] [Google Scholar]
- Thurnheer T, Belibasakis GN, Bostanci N. 2014. Colonisation of gingival epithelia by subgingival biofilms in vitro: role of “red complex” bacteria. Arch Oral Biol. 59(9):977–986. [DOI] [PubMed] [Google Scholar]
- Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. 1998. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 33(3):151–208. [DOI] [PubMed] [Google Scholar]
- van Hijum SA, Kralj S, Ozimek LK, Dijkhuizen L, van Geel-Schutten IG. 2006. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol Mol Biol Rev. 70(1):157–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman MM, Clewell DB. 1997. Regulation of Streptococcus gordonii glucosyltransferase. Adv Exp Med Biol. 418:661–664. [DOI] [PubMed] [Google Scholar]
- Vickerman MM, Sulavik MC, Minick PE, Clewell DB. 1996. Changes in the carboxyl-terminal repeat region affect extracellular activity and glucan products of Streptococcus gordonii glucosyltransferase. Infect Immun. 64(12):5117–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. 2007a. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J Bacteriol. 189(4):1390–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mintz KP, Ladha M, Fives-Taylor PM. 1998. Isolation and characterization of Fap1, a fimbriae-associated adhesin of Streptococcus parasanguis FW213. Mol Microbiol. 28(3):487–500. [DOI] [PubMed] [Google Scholar]
- Wu H, Zeng M, Fives-Taylor P. 2007b. The glycan moieties and the N-terminal polypeptide backbone of a fimbria-associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis. Infect Immun. 75(5):2181–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Wu H. 2011. A molecular chaperone mediates a two-protein enzyme complex and glycosylation of serine-rich streptococcal adhesins. J Biol Chem. 286(40):34923–34931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Zhou M, Wu H. 2010. Purification and characterization of an active N-acetylglucosaminyltransferase enzyme complex from Streptococci. Appl Environ Microbiol. 76(24):7966–7971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu DQ, Thompson J, Cisar JO. 2003. Genetic loci for coaggregation receptor polysaccharide biosynthesis in Streptococcus gordonii 38. J Bacteriol. 185(18):5419–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu F, Ding L, Zhou M, Wu R, Wu H. 2013. Preliminary X-ray crystallographic studies of an N-terminal domain of unknown function from a putative glycosyltransferase from Streptococcus parasanguinis. Acta Crystallogr Sect F Struct Biol Cryst Commun. 69(Pt 5):520–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhu F, Yang T, Ding L, Zhou M, Li J, Haslam SM, Dell A, Erlandsen H, Wu H. 2014. The highly conserved domain of unknown function 1792 has a distinct glycosyltransferase fold. Nat Commun. 5:4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wu H. 2009. Glycosylation and biogenesis of a family of serine-rich bacterial adhesins. Microbiology. 155(Pt 2):317–327. [DOI] [PubMed] [Google Scholar]
- Zhou M, Zhu F, Dong S, Pritchard DG, Wu H. 2010. A novel glucosyltransferase is required for glycosylation of a serine-rich adhesin and biofilm formation by Streptococcus parasanguinis. J Biol Chem. 285(16):12140–12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Erlandsen H, Ding L, Li J, Huang Y, Zhou M, Liang X, Ma J, Wu H. 2011. Structural and functional analysis of a new subfamily of glycosyltransferases required for glycosylation of serine-rich streptococcal adhesins. J Biol Chem. 286(30):27048–27057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Zhang H, Wu H. 2014. A conserved domain is crucial for acceptor substrate binding in a family of glucosyltransferases. J Bacteriol. 197(3):510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.