Abstract
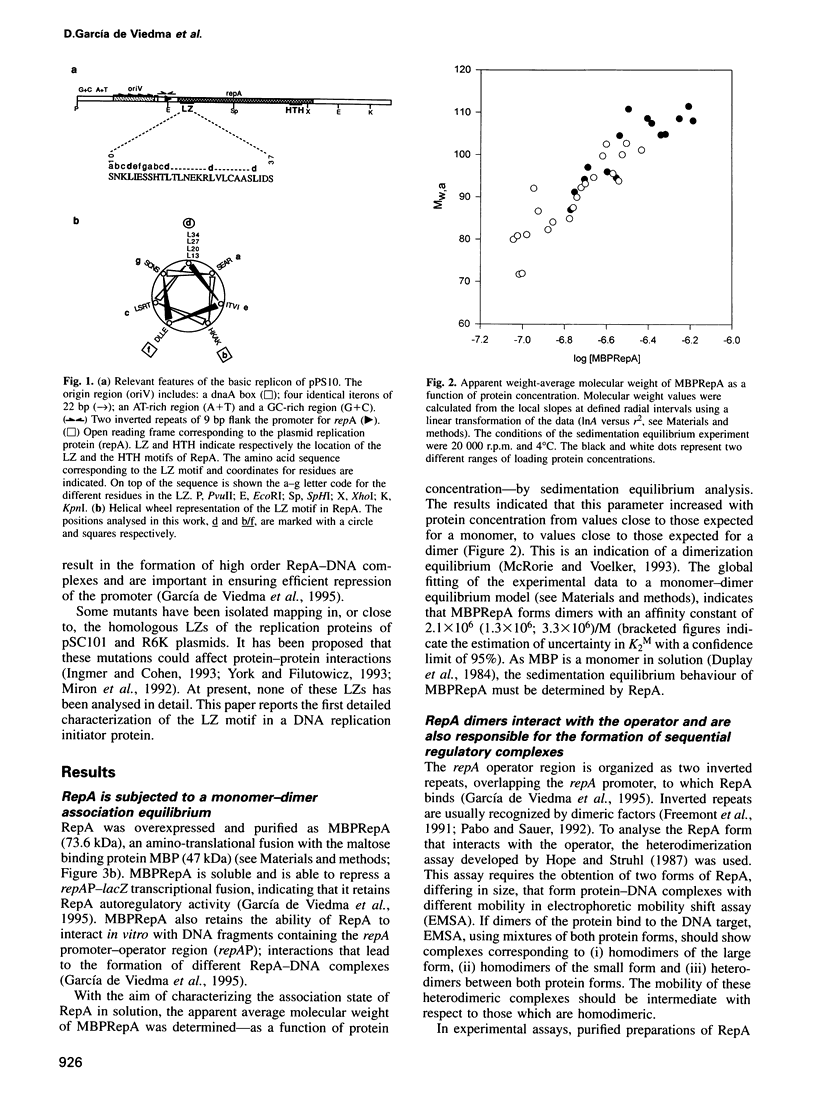
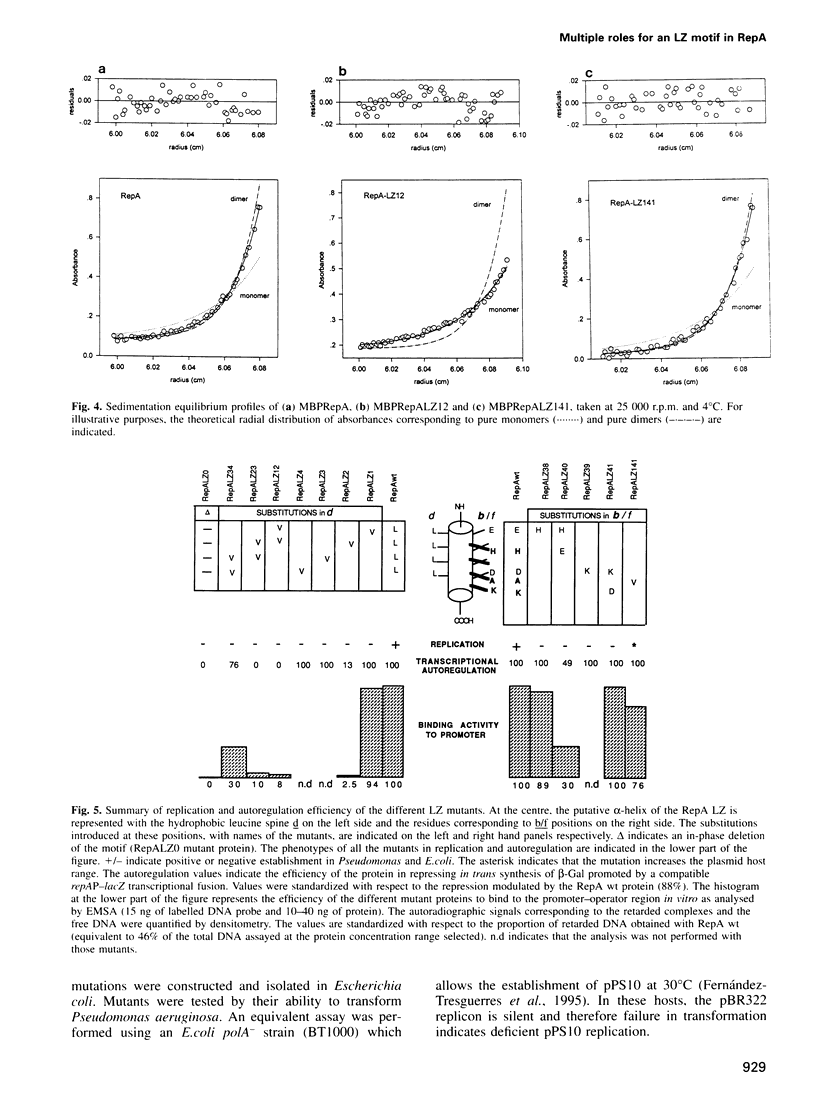
RepA is the replication initiator protein of the Pseudomonas plasmid pPS10 and is also able to autoregulate its own synthesis. Here we report a genetic and functional analysis of a leucine zipper-like (LZ) motif located at the N-terminus of RepA. It is shown that the LZ motif modulates the equilibrium between monomeric and dimeric forms of the protein and that monomers of RepA interact with sequences at the origin of replication, oriV, while dimers are required for interactions of RepA at the repA promoter. Further, different residues of the LZ motif are seen to have different functional roles. Leucines at the d positions of the putative alpha-helix are relevant in the formation of RepA dimers required for transcriptional autoregulation. They also modulate other RepA-RepA interactions that result in cooperative binding of protein monomers to the origin of replication. The residues at the b/f positions of the putative helix play no relevant role in RepA-RepA interactions. These residues do not affect RepA autoregulation but do influence replication, as demonstrated by mutants that, without affecting binding to oriV, either increase the host range of the plasmid or are inactive in replication. It is proposed that residues in b/f positions play a relevant role in interactions between RepA and host replication factors.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andera L., Schneider G. J., Geiduschek E. P. An analysis of subunit exchange in the dimeric DNA-binding and DNA-bending protein, TF1. Biochimie. 1994;76(10-11):933–940. doi: 10.1016/0300-9084(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra S. P., Kumar A., Lewis M. S., Widen S. G., Abbotts J., Karawya E. M., Hughes S. H., Shiloach J., Wilson S. H., Lewis M. S. Protein-protein interactions of HIV-1 reverse transcriptase: implication of central and C-terminal regions in subunit binding. Biochemistry. 1991 Dec 17;30(50):11707–11719. doi: 10.1021/bi00114a015. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Cole J. L., Gehman J. D., Shafer J. A., Kuo L. C. Solution oligomerization of the rev protein of HIV-1: implications for function. Biochemistry. 1993 Nov 9;32(44):11769–11775. doi: 10.1021/bi00095a004. [DOI] [PubMed] [Google Scholar]
- DasGupta S., Mukhopadhyay G., Papp P. P., Lewis M. S., Chattoraj D. K. Activation of DNA binding by the monomeric form of the P1 replication initiator RepA by heat shock proteins DnaJ and DnaK. J Mol Biol. 1993 Jul 5;232(1):23–34. doi: 10.1006/jmbi.1993.1367. [DOI] [PubMed] [Google Scholar]
- Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984 Aug 25;259(16):10606–10613. [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., Harrison S. C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992 Dec 24;71(7):1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Fernández-Tresguerres M. E., Martín M., García de Viedma D., Giraldo R., Díaz-Orejas R. Host growth temperature and a conservative amino acid substitution in the replication protein of pPS10 influence plasmid host range. J Bacteriol. 1995 Aug;177(15):4377–4384. doi: 10.1128/jb.177.15.4377-4384.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freemont P. S., Lane A. N., Sanderson M. R. Structural aspects of protein-DNA recognition. Biochem J. 1991 Aug 15;278(Pt 1):1–23. doi: 10.1042/bj2780001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Frank R., Blöcker H., Scholz P., Bagdasarian M., Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48(1):119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- García de Viedma D., Giraldo R., Ruiz-Echevarría M. J., Lurz R., Díaz-Orejas R. Transcription of repA, the gene of the initiation protein of the Pseudomonas plasmid pPS10, is autoregulated by interactions of the RepA protein at a symmetrical operator. J Mol Biol. 1995 Mar 24;247(2):211–223. doi: 10.1006/jmbi.1994.0134. [DOI] [PubMed] [Google Scholar]
- Giraldo R., Nieto C., Fernandez-Tresguerres M. E., Diaz R. Bacterial zipper. Nature. 1989 Dec 21;342(6252):866–866. doi: 10.1038/342866a0. [DOI] [PubMed] [Google Scholar]
- Glover J. N., Harrison S. C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature. 1995 Jan 19;373(6511):257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Kim P. S., Alber T. Crystal structure of an isoleucine-zipper trimer. Nature. 1994 Sep 1;371(6492):80–83. doi: 10.1038/371080a0. [DOI] [PubMed] [Google Scholar]
- Harbury P. B., Zhang T., Kim P. S., Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993 Nov 26;262(5138):1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hodges R. S., Semchuk P. D., Taneja A. K., Kay C. M., Parker J. M., Mant C. T. Protein design using model synthetic peptides. Pept Res. 1988 Sep-Oct;1(1):19–30. [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. C., Newell N. E., Tidor B., Sauer R. T. Probing the roles of residues at the e and g positions of the GCN4 leucine zipper by combinatorial mutagenesis. Protein Sci. 1993 Jul;2(7):1072–1084. doi: 10.1002/pro.5560020701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. C., O'Shea E. K., Kim P. S., Sauer R. T. Sequence requirements for coiled-coils: analysis with lambda repressor-GCN4 leucine zipper fusions. Science. 1990 Dec 7;250(4986):1400–1403. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
- Ingmer H., Cohen S. N. Excess intracellular concentration of the pSC101 RepA protein interferes with both plasmid DNA replication and partitioning. J Bacteriol. 1993 Dec;175(24):7834–7841. doi: 10.1128/jb.175.24.7834-7841.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiai M., Wada C., Kawasaki Y., Yura T. Replication initiator protein RepE of mini-F plasmid: functional differentiation between monomers (initiator) and dimers (autogenous repressor). Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3839–3843. doi: 10.1073/pnas.91.9.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John M., Briand J. P., Granger-Schnarr M., Schnarr M. Two pairs of oppositely charged amino acids from Jun and Fos confer heterodimerization to GCN4 leucine zipper. J Biol Chem. 1994 Jun 10;269(23):16247–16253. [PubMed] [Google Scholar]
- Johnson M. L., Correia J. J., Yphantis D. A., Halvorson H. R. Analysis of data from the analytical ultracentrifuge by nonlinear least-squares techniques. Biophys J. 1981 Dec;36(3):575–588. doi: 10.1016/S0006-3495(81)84753-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. The role of the leucine zipper in the fos-jun interaction. Nature. 1988 Dec 15;336(6200):646–651. doi: 10.1038/336646a0. [DOI] [PubMed] [Google Scholar]
- König P., Richmond T. J. The X-ray structure of the GCN4-bZIP bound to ATF/CREB site DNA shows the complex depends on DNA flexibility. J Mol Biol. 1993 Sep 5;233(1):139–154. doi: 10.1006/jmbi.1993.1490. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Manen D., Upegui-Gonzalez L. C., Caro L. Monomers and dimers of the RepA protein in plasmid pSC101 replication: domains in RepA. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):8923–8927. doi: 10.1073/pnas.89.19.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A., Mukherjee S., Bastia D. Activation of distant replication origins in vivo by DNA looping as revealed by a novel mutant form of an initiator protein defective in cooperativity at a distance. EMBO J. 1992 Mar;11(3):1205–1216. doi: 10.1002/j.1460-2075.1992.tb05161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C., Giraldo R., Fernández-Tresguerres E., Díaz R. Genetic and functional analysis of the basic replicon of pPS10, a plasmid specific for Pseudomonas isolated from Pseudomonas syringae patovar savastanoi. J Mol Biol. 1992 Jan 20;223(2):415–426. doi: 10.1016/0022-2836(92)90661-3. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Mechanism of specificity in the Fos-Jun oncoprotein heterodimer. Cell. 1992 Feb 21;68(4):699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Pu W. T., Struhl K. The leucine zipper symmetrically positions the adjacent basic regions for specific DNA binding. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6901–6905. doi: 10.1073/pnas.88.16.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saudek V., Pastore A., Morelli M. A., Frank R., Gausepohl H., Gibson T. The solution structure of a leucine-zipper motif peptide. Protein Eng. 1991 Jun;4(5):519–529. doi: 10.1093/protein/4.5.519. [DOI] [PubMed] [Google Scholar]
- Schuermann M., Hunter J. B., Hennig G., Müller R. Non-leucine residues in the leucine repeats of Fos and Jun contribute to the stability and determine the specificity of dimerization. Nucleic Acids Res. 1991 Feb 25;19(4):739–746. doi: 10.1093/nar/19.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropsha A., Bowen J. P., Brown F. K., Kizer J. S. Do interhelical side chain-backbone hydrogen bonds participate in formation of leucine zipper coiled coils? Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9488–9492. doi: 10.1073/pnas.88.21.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler J. A., Nüsslein V., Otto B., Klein A., Bonhoeffer F., Herrmann R., Gloger L., Schaller H. Isolation and characterization of thermosensitive Escherichia coli mutants defective in deoxyribonucleic acid replication. J Bacteriol. 1973 Mar;113(3):1381–1388. doi: 10.1128/jb.113.3.1381-1388.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner S., Gottesman S., Skowyra D., Hoskins J., McKenney K., Maurizi M. R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York D., Filutowicz M. Autoregulation-deficient mutant of the plasmid R6K-encoded pi protein distinguishes between palindromic and nonpalindromic binding sites. J Biol Chem. 1993 Oct 15;268(29):21854–21861. [PubMed] [Google Scholar]
- Zhou N. E., Kay C. M., Hodges R. S. Synthetic model proteins: the relative contribution of leucine residues at the nonequivalent positions of the 3-4 hydrophobic repeat to the stability of the two-stranded alpha-helical coiled-coil. Biochemistry. 1992 Jun 30;31(25):5739–5746. doi: 10.1021/bi00140a008. [DOI] [PubMed] [Google Scholar]