Abstract
Dehydration-Responsive Element Binding proteins (DREB)/C-repeat (CRT) Binding Factors (CBF) have been identified as transcriptional activators during plant responses to cold stress. The objective of this study was to determine the physiological roles of a CBF gene isolated from a cold-tolerant perennial grass species, Kentucky bluegrass (Poa pratensis L.), which designated as PpCBF3, in regulating plant tolerance to freezing stress. Transient transformation of Arabidopsis thaliana mesophyll protoplast with PpCBF3-eGFP fused protein showed that PpCBF3 was localized to the nucleus. RT-PCR analysis showed that PpCBF3 was specifically induced by cold stress (4°C) but not by drought stress [induced by 20% polyethylene glycol 6000 solution (PEG-6000)] or salt stress (150 mM NaCl). Transgenic Arabidopsis overexpressing PpCBF3 showed significant improvement in freezing (-20°C) tolerance demonstrated by a lower percentage of chlorotic leaves, lower cellular electrolyte leakage (EL) and H2O2 and O2 .- content, and higher chlorophyll content and photochemical efficiency compared to the wild type. Relative mRNA expression level analysis by qRT-PCR indicated that the improved freezing tolerance of transgenic Arabidopsis plants overexpressing PpCBF3 was conferred by sustained activation of downstream cold responsive (COR) genes. Other interesting phenotypic changes in the PpCBF3-transgenic Arabidopsis plants included late flowering and slow growth or ‘dwarfism’, both of which are desirable phenotypic traits for perennial turfgrasses. Therefore, PpCBF3 has potential to be used in genetic engineering for improvement of turfgrass freezing tolerance and other desirable traits.
Introduction
Low-temperature stresses such as chilling (0–20°C) and freezing stress (< 0°C) are the most common environmental factors limiting plant growth and productivity in cool climatic regions [1,2]. During chilling stress and the thawing phase following freezing stress, cells loss water due to the rupture of cellular membranes, leading to dehydration and therefore, protecting cellular dehydration is critically important for plant survival of chilling or freezing stress [2]. Plant adaptation to chilling or freezing stress involves various molecular changes, including the induction of many low-temperature response genes and transcription factors, such as dehydration-responsive-element-binding protein (DREB1)/C-repeat binding factors (CBFs) [3,4]. DREB1/CBFs are known to function in regulating plant responses to low temperatures which may induce cellular dehydration [5,6].
DREB1/CBFs are a family of genes conserved in flowering plants and CBF homolog genes have been identified and analyzed in many agronomic crops, such as maize (Zea mays), rice (Oryza sativa), wheat (Triticum aestivum) and soybean (Glycine max) [7–10]. DREB1/CBFs regulate the downstream expression of cold-inducible genes, including cold-regulated genes (COR) [4,11,12]. Enhanced cold tolerance was achieved with DREB1/CBF genes constitutively expressed at high levels in agriculturally important species (i.e. rice and wheat) and model species [i.e. Arabidopsis and tobacco (Nicotiana benthamiana)] [13–18]. For example, transgenic rice plants overexpressing OsDREB1 showed higher survival rate to chilling (2°C) stress compared to the wild type [13]. Transgenic barley (Hordeum vulgare) plants overexpressing TaDREB2 and TaDREB3 showed increased survival rate under frost stress (-6°C) [15]. Heterologenic expression of a CBF3 gene isolated from sweet pepper (Capsicum annuum) conferred enhanced tolerance to chilling stress (4°C) in transgenic tobacco plants through higher accumulations of osmolytes (proline and soluble sugars), higher levels of unsaturated fatty acids, and lower accumulations of H2O2 and O2.-, as well as lower electrolyte leakage and photochemical efficiency [18]. Overexpression of CBF1 enhanced Arabidopsis tolerance to freezing stress (-8°C) as by lower electrolyte leakage and improved whole plant survival [19]. Similarly, overexpressing CBF3 lead to enhanced tolerance to freezing (-6°C) stress by activating downstream cold responsive genes [20]. However, constitutive overexpression of LeCBF1 or AtCBF3 genes had no effects for improving freezing tolerance in freezing-sensitive tomato (Lycopersicon esculentum) due to different CBF regulon [21].In general, it is obvious that the extent of DREB1/CBF affecting plant tolerance to cold stress varies with the level of temperature stress, specific type or group of the gene family, and plant species where the genes are isolated from [8,18,21–23]. Furthermore, most of previous studies examined plant responses to chilling stress or sub-freezing temperatures. Increasing frequency of extremely low temperature below 0°C occurs in cool-climatic regions associated with global climate changes [1]. Nevertheless, the knowledge of DREB1/CBFs transcriptional control of plant tolerance to severe freezing temperatures is limited.
Cool-season perennial grasses used as forage or managed turf are typically exposed to sub-zero freezing temperatures during winter months though limited information is available as to how changes in DREB1/CBF genes may confer cold tolerance and improved survivability. DREB1/CBF genes were identified in perennial ryegrass (Lolium perenne) and tall fescue (Festuca arundinacea) [23–25]. LpCBF3 was induced by chilling stress (4°C) and transgenic Arabidopsis plants overexpressing LpCBF3 showed higher survival rate at -6°C [23,25]. However, physiological roles and molecular mechanisms of DREB1/CBF genes isolated from cold-tolerant perennial grasses have not been well documented and is a topic which deserves further investigation.
The DREB1/CBF gene (PpCBF3) was isolated from Kentucky bluegrass (P. pratensis), a species which displays exceptional cold tolerance [26], to investigate whether PpCBF3 could positively regulate plant tolerance to severe freezing stress (-20°C). The objectives of this study were to determine the specificity of PpCBF3 from Kentucky bluegrass for cold stress responses by characterizing the expression patterns of PpCBF3 in response to different abiotic stresses and to examine physiological functions and downstream genes regulated by PpCBF3 conferring plant tolerance to freezing stress (-20°C) through overexpressing it in Arabidopsis.
Materials and Methods
Growth conditions for Kentucky bluegrass seedlings
Seeds of Kentucky bluegrass cultivar (Poa pratensis ‘Midnight’) were germinated on Petri dishes in darkness in a growth chamber (MT8070iE, Xubang, Henan, China) set to day/night temperature of 25/15°C and humidity of 70%. 7 d old seedlings were transferred to plastic pots filled with a mixture of moss peat (Pindstrup Sphagnum, Ryomgaard, Denmark) and vermiculite (3:1 by volume) and maintained in a greenhouse with natural sunlight and an average day/night temperature of 23/18°C. Plants were irrigated every other day and fertilized weekly with half-strength Hoagland’s nutrient solution [27], and trimmed regularly to keep the plant height at about 5 cm.
Isolation PpCBF3 gene from Kentucky bluegrass
For gene isolation, 4-week-old Kentucky bluegrass seedlings were subjected to 4°C in an incubator with cool-white fluorescent lights for 1 h. Total RNA was isolated from leaves using Rnapure fast isolation Kit (YuanPingHao, Tianjin, China) and cDNA was synthesized by Transcriptor First Strand cDNA Synthesis Kit (Roche, Rotkreuz, Switzerland). RACE (rapid amplification of cDNA ends) PCR was carried out to extend the 3’ region of the PpCBF3 by using gene specific primer 1 (GSP1F1) (S1 Table) for the first cycle and GSP2F2 for the second cycle with each paired to an adaptor-specific primer (Adaptor-R). Degenerate primer from start codon ATG was designed according to CBF3 sequence from other monocot plants and the 5’-end sequence of PpCBF3 was obtained by using forward primer degF and reverse primer degR. Full-length CDS of PpCBF3 was amplified using conCBF3F and conCBF3R primers. The PCR amplification procedure was first initial denaturation at 94°C for 5 min, with 35 cycles (94°C for 30 s, 60°C for 30 s, 72°C for 1 min) followed, and a final extension cycle of 72°C for 7 min. PCR fragment obtained was cloned into pMD19-T Vector (Takara, Kyoto, Japan). Positive clones confirmed by colony PCR were sequenced (Springen Co., Ltd, Nanjing, China).
RT-PCR and qRT-PCR analysis of PpCBF3 gene expression in response to abiotic stresses
In order to determine whether PpCBF3 is specifically responsive to cold stress or broadly responsive to different abiotic stress, PpCBF3 gene expression patterns were analyzed for plants exposed to cold, drought and salt stress. For drought and salt treatment, tillers of Kentucky bluegrass were transferred into half-strength Hoagland’s nutrient solution containing PEG-6000 (20% w/v) to induce drought stress or NaCl (150 mM) to induce salt stress under normal growth temperature (a day/night temperature of 25/15°C). For cold treatment, Kentucky bluegrass plants grown in plastic pots were treatted in an incubator at 4°C. To examine gene responses to different stresses, leaf samples were collected at 0, 15 min, 30 min, 1 h, 3 h, 6 h, 12 h and 24 h of stress treatment and frozen in liquid nitrogen.
For qRT-PCR analysis, total RNA was extracted using a Tripure Isolation Reagent Kit (Roche Diagnostic, Basel, Switzerland) according to the manufacturer’s protocol. The first strand cDNA was synthesized with 1 μg RNA using the PrimeScript RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara, Otsu, Japan). qRT-PCR was performed on Roche LightCycler480 II machine (Roche Diagnostic, Rotkreuz, Switzerland) with SYBR Green I Master reaction system (Roche Diagnostic, Rotkreuz, Switzerland). The PCR condition was as follows: initial denaturation step of 10 min at 95°C, 40 cycles of PCR (95°C for 15 s, 60°C for 15 s, and 72°C for 20 s). Data was collected at 65°C in each cycle. The qRT-PCR analysis used three biological replicates for every treatment, each with three technical replicates. Relative expression level of genes was determined by 2-ΔΔCT method using P. pratensis elongation factor 1α (PpEF1α) as the reference gene.
For RT-PCR analysis, total RNA extraction and first strand cDNA synthesis was done as previously described and primers for reference gene PpEF1α were the same in qRT-PCR.
Isolation and transient transformation of Arabidopsis mesophyll protoplast
For gene transformation and a phenotypic control, Arabidopsis thaliana accession Columbia (col-0) was used. Seeds of Arabidopsis were treated with 1.3% NaClO for 15 min and washed thoroughly with sterile water six times. Seeds were sowed on MS medium [28] with 0.2% low melting-point agarose for uniform separation, vernalized in darkness for 72 h at 4°C, and grown on MS medium at 23°C with a 16 h photoperiod under 120 μmol m-2 s-1 light density (designated as normal condition) in a controlled growth chamber (Haier, Qingdao, China).
Arabidopsis protoplasts were isolated according to Wu et al. (2009) [29]. The upper and lower epidermal surface of arabidopsis leaves was affixed to Time tape (Time Med, Burr Ridge, IL) and Magic tape (3 M, St. Paul, MN), respectively. The lower side of epidermal cell layer was peeled by pulling off the Magic tape. The peeled leaves were immersed in 10 ml enzyme solution (with lower surface in the sulution) [1% cellulase ‘Onozuka’ R10 (Yakult, Tokyo, Japan), 0.25% macerozyme ‘Onozuka’ R10 (Yakult), 0.4 M mannitol, 10 mM CaCl2, 20mM KCl, 0.1% BSA and 20 mM MES, PH 5.7] for 1 h in light, the protoplasts released into solution were collected by centrifuged at 100 ×g for 3 min and washed twice with prechilled W5 solution [154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 5 mM glucose, and 2mM MES, PH 5.7] and incubated on ice for 30 min. Protoplasts were finally resuspended in modified MMg solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM MES, PH 5.7) with a concentration of 5 ×105 cells mL-1.
To investigate the subcellular localization of PpCBF3 in living cells, the ORF (open reading frame) of PpCBF3 without a stop codon in a pENTR1A Dual Selection Vector (Invitrogen, Carlsbad, CA, USA) was LR-recombinated with a small binary vector P2GWF7.0 (~6.7 Kb). Thus PpCBF3 was fused to the N-terminal of eGFP reporter gene under the cauliflower mosaic virus 35S (CaMV35S) promoter. About 30 μg PpCBF3-eGFP recombinant plasmids were transiently transformed into Arabidopsis protoplasts by the PEG4000 (Fluka, USA)-mediated method [29]. After incubating in 6-well plates at 25°C for 16 h in light, photographs were captured by Confocal Laser Scanning Microscope (Carl Zeiss, Jena, Germany).
Construction of plant expression vector and transformation of Arabidopsis thaliana
The ORF of PpCBF3 with EcoRI and EcoRV sites was first cloned into a pENTR1A Dual Selection Vector and transformed into pEarleyGate 103 destination plasmid using LR Clonase II enzyme mix (Invitrogen, Carlsbad, CA, USA) [30] with CaMV35S as promoter. The CaMV35S::PpCBF3 constructs were introduced into Agrobacterium tumefaciens strain EHA105. Floral dip method [31] was adopted to obtain transgenic Arabidopsis. Seeds (T1) from transgenic plants were selected on MS medium [with 20 μg ml-1 glufosinate ammonium (Sigma, USA) supplemented]. The glufosinate ammonium-resistant T1 seedlings were tested by PCR analysis using the primers for PpCBF3 (conCBF3R) and pEarleyGate103 expression vector (103F). T2 seeds produced from the T1 plants expressing PpCBF3 gene were collected for further analysis.
Phenotypic analysis of transgenic Arabidopsis under normal temperature and freezing stress
T2 seeds of transgenic lines were germinated on MS medium (supplemented with 20 μg ml-1 glufosinate ammonium) to select for positive transgenic plants and WT seeds were germinated on MS medium concurrently. One-week-old seedlings of transgenic plants and WT were then transplanted to new MS medium in Petri dishes and grown at normal condition (23°C with a 16 h photoperiod under 120 μmol m-2 s-1 light densities). Freezing tests were performed following the protocol by Li et al. (2014) [32] with some modifications. 4-week-old transgenic lines and WT plants were acclimated at 4°C in an incubator with cool-white fluorescent lights for 3 d, and subsequently exposed to freezing temperatures gradually decreasing from -2 to -20°C within 2 h and finally at -20°C for 3 h in a freeze chamber (Beckman Coulter, Brea, CA). Plants were then moved to 4°C for 24 h and recovered at 23°C for 7 d.
The typical symptoms of freezing injury include loss of chlorophyll and membrane stability [33]. Therefore, several commonly used stress indicators, the percentage of chlorotic leaves per plant, leaf chlorophyll content (Chl), leaf photochemical efficiency, and membrane stability, were measured after plants were thawed following the freezing treatment to evaluate whether over-expressing PpCBF3 would alleviate freezing damages.
The percentage of leaf chlorosis was calculated based on the number of leaves displaying yellow divided by the total number of leaves per plant. Photographs of plants were taken using single lens reflex camera (Nikon D5100, Thailand).
For phenotype analysis under normal condition, 14-day-old Arabidopsis seedlings were transplanted into plastic pots and maintained at normal growth condition for up to 6 weeks. The number of rosette leaves per plants was counted and photographs of plants were taken using the camera described above.
Leaf Chl was calculated as described in Arnon (1949) [34]. First fully-expanded leaves (0.1 g) from plant top were detached from plants and soaked in 95% dimethylsulfoxide in darkness for about 72 h in order to extract chlorophyll completely. The absorbance of Chl extract was measured at 663 and 645 nm using a spectrophotometer (GE Healthcare Life Sciences, Cambridge, UK).
Leaf photochemical efficiency expressed as the ratio of variable to maximum fluorescence (Fv/Fm) was determined in both transgenic lines and WT exposed to freezing stress. Leaf Fv/Fm was measured with a fluorescence induction monitor (OPTI-Sciences, Hudson, USA) following 30 min dark adaptation.
Cellular membrane stability was measured as leaf electrolyte leakage (EL). Fully-expanded leaves (about 0.1 g) of WT and transgenic lines were detached and incubated in 15 ml distilled deionized water. The initial level of EL (Ci) was measured using a conductance meter (Thermo Scientific, Baverly, USA) after shaken for 24 h at room temperature. Leaf tissue was killed in an autoclave at 121°C for 30 min. The conductance of the incubation solution was measured after 24 h incubation on a shaker. Relative EL was calculated as EL = (Ci/Cmax) × 100 [35].
Histochemical detection of H2O2 and O2 .- in transgenic plants exposed to freezing stress
The production of hydrogen peroxide (H2O2) in leaves exposed to -20°C for 1 h (firstly acclimated at 4°C for 24 h) and thawed at 4°C for 24 h was detected by 3, 3’-diaminobenzidine (DAB) staining as described by Lee et al. (2002) [36]. Three-week-old seedlings of transgenic and WT lines were vacuum-incubated with 0.1 mg ml-1 DAB (sigma, USA) in 50 mM Tris-acetate buffer (pH 5.0). Solutions with seedlings were incubated at 25°C in the darkness for 24 h. Chlorophyll was removed by incubation in 70% ethanol at room temperature for 24 h.
Detection of superoxide free radicals were performed as described previously [36]. All seedlings acclimated at 4°C for 24 h, then exposed to freezing stress (-20°C for 1 h) and thawed at 4°C for 24 h were harvested, and vacuum-infiltrated [0.1 mg ml-1 nitroblue tetrozolium (NBT) in 25 mM HEPES buffer (pH7.6)]. Samples were subsequently incubated at 25°C for 2 h in the darkness. Stained samples were bleached in 70% ethanol and incubated at 25°C for 24 h to thoroughly remove the chlorophyll. Photographs were taken using a stereomicroscope (Olympus, Tokyo, Japan).
qRT-PCR analysis of PpCBF3 gene and downstream genes in transgenic plants and WT in response to cold stress
Leaves of 3-week-old transgenic and WT plants exposed to 4°C in a freezer with cool-white fluorescent lights for 4 h were harvested and frozen in liquid nitrogen for further analysis. For qRT-PCR analysis in Arabidopsis, methods were the same as mentioned above (S1 Table). 2-ΔΔCT method was adopted to calculate the expression levels of genes with AtActin2 as the reference gene. The qRT-PCR analysis had three biological replicates for every treatment, each with three technical replicates.
Western blot analysis of CBF protein in PpCBF3-transgenic and WT Arabidopsis
Crude proteins of transgenic and WT lines were extracted with phosphate buffer (pH 7.0), incubated on ice for 0.5 h and centrifuged at 12,000 g for 30 min. Protein samples were separated by 12% SDS-PAGE. The proteins were semi-dry blotted (Amersham Biosciences, Piscataway, USA) onto a PVDF membrane (Roche, Basel, Switzerland). For detection, the membranes were incubated for 2 h with the Anti-GFP mouse monoclonal antibody (1:5000) (CMCTAG, Milwaukee, USA) after incubation overnight at 4°C in blocking buffer (5% non-fat dried milk in PBST). Membranes were then rinsed 4 times in PBST and incubated with the secondary antibody [horseradish-peroxidase conjugated goat anti-mouse antibody (Abgent, San Diego, America) 1: 5 000 in blocking buffer] for 1 h. Finally, the membranes were incubated with Pierce ECL Western Blotting Substrate (Thermo Scientific, Rockford, USA) and detected by a CCD Video Camera Imaging System (Vilber Lourmat, Marne la Vallée, France). The contrast and brightness of resulting pictures were processed with BIO-1D software (Vilber Lourmat, Marne la Vallée, France).
Statistical analysis of phenotypic and gene expression data
Physiological data and qRT-PCR data were subjected to the analysis of variance (ANOVA) analysis of variance according to the general linear model procedure of SAS (SAS 9.0, Cary, NC). Differences between the means were distinguished by Fisher’s protected least significance difference (LSD) test at the 0.05 probability level.
Results
Isolation of PpCBF3 from Kentucky bluegrass and subcellular localization of PpCBF3-eGFP fused protein
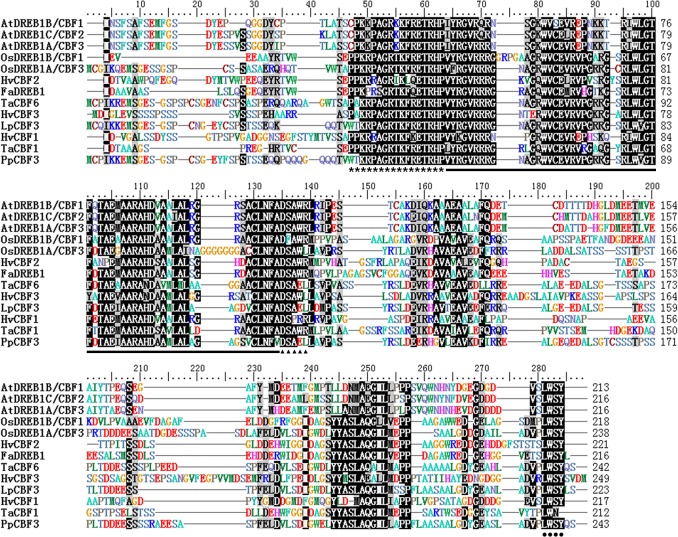
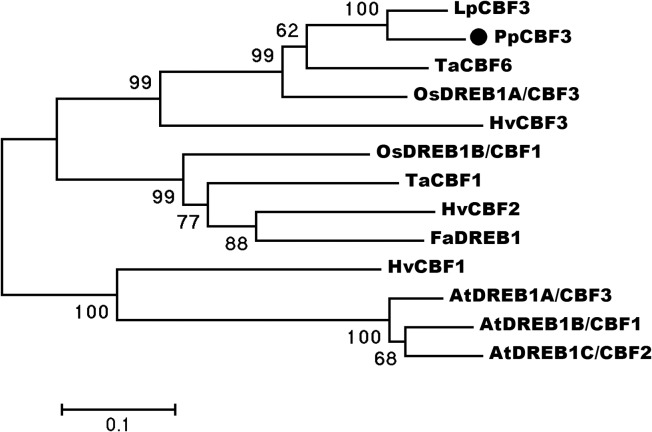
A 569 bp nucleotide fragment obtained by 3’ RACE and 5’-end of cDNA was extended by homology-based cloning. Finally, a full-length sequence consisted of 860 bp nucleotides with a 732 bp ORF encoding a 243-residue polypeptide was obtained. Sequence analysis at http://blast.ncbi.nlm.nih.gov/Blast.cgi by Blastx programme showed that the gene had high similarity to DREB1/CBF in other plant species (Fig 1); about 81% similarity to LpCBF3 in perennial ryegrass and 64% to OsDREB1A/CBF3 in rice. A phylogenetic tree revealed that the gene is a putative ortholog of LpCBF3 (Fig 2). We designated the gene as PpCBF3 and the accession number in NCBI database was KP258182. In addition to an AP2 conserved domain serving a DNA-binding role, two signature sequences (PKK/RPAGRxKFxETRHP and DSAEL) also presented in PpCBF3. The C-terminal LWSY motif was also conserved in PpCBF3 compared with other CBF-like proteins (Fig 1). The PpCBF3 isolated from Kentucky bluegrass encoded an ortholog of CBF3-like transcription factor.
Fig 1. Sequence homology analysis of PpCBF3 with DREB/CBFs from Arabidopsis, rice, wheat, barley, tall fescue and perennial ryegrass.
Conserved AP2 DNA-binding domain is indicated by black line, two signature sequence of CBF are marked by asterisk and filled triangle. ‘LWSY’ domain at the end of C terminal is indicated by black dots. Accession numbers of the proteins are as follows: AtDREB1B/CBF1 (AAC49662), AtDREB1C/CBF2 (AAD15976), AtDREB1A/CBF3 (AAD15977), OsDREB1B/CBF1 (AAN02488), OsDREB1A/CBF3 (AAN02486), HvCBF2 (AAM13419), FaDREB1 (AAQ98965), TaCBF6 (AAX28964), HvCBF3 (AAG59618), LpCBF3 (AAX57275), HvCBF1 (AAL84170), TaCBF1 (AAL37944), PpCBF3 (KP258182). Sequence alignment was done by Cluster W.
Fig 2. Phylogenetic tree of PpCBF3 and other DREB/CBF proteins.
The tree is constructed by MEGA 4.0 software (Tamura et al. 2007) based on alignment of complete protein sequences. Black dot indicates PpCBF3 protein. Accession number of protein sequences used here are the same as in Fig 1.
As shown in Fig 3, the GFP signal was confined predominantly to the nucleus and was merged with DAPI in the individual cells transferred with the PpCBF3-eGFP recombinant plasmids. PpCBF3 was present as a nuclear protein.
Fig 3. Subcellular localization of PpCBF3 protein.
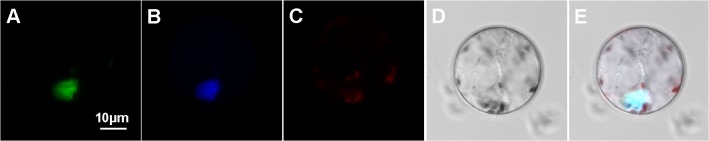
(A) Fluorescence image of Arabidopsis mesophyll protoplast expressing the PpCBF3-eGFP fusion protein. (B) Fluorescence image of nucleus in protoplast stained with DAPI. (C) Fluorescence image of mesophyll in Arabidopsis protoplast. (D) Image of Arabidopsis mesophyll protoplast under bright field. (E) Merged fluorescence image of Arabidopsis protoplast expressing the PpCBF3-eGFP fusion protein and stained with DAPI. (A)-(C), dark field. (D), (E), bright field. All bar = 10 μm.
PpCBF3 expression in response to abiotic stresses
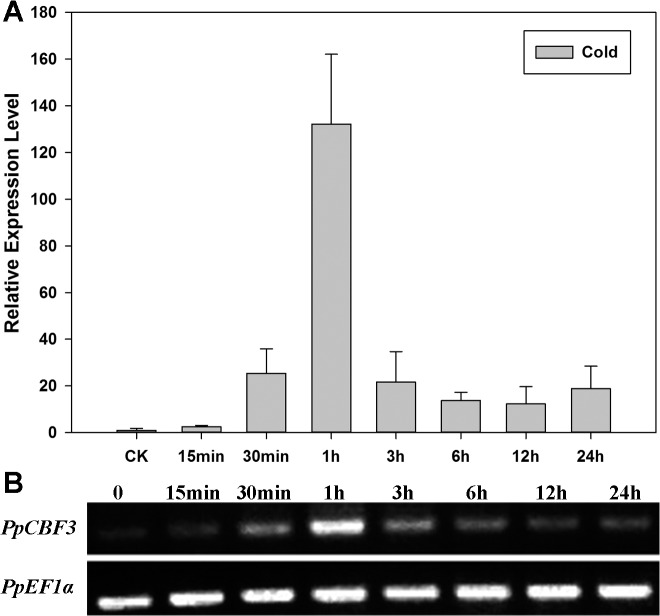
qRT-PCR analysis demonstrated that PpCBF3 exhibited a rapid response to cold stress (4°C), as the expression level increased within 15 min, reached to the highest level by 1 h of treatment, and maintained high expression even after 24 h of cold treatment compared to the initial non-stress level (Fig 4A). Semi-quantitative RT-PCR results also showed the similar expression pattern (Fig 4B). However, PpCBF3 expression did not respond to drought stress induced by PEG-6000 or salt stress in Kentucky bluegrass (S1 Fig).
Fig 4. Relative mRNA expression level of PpCBF3 in leaves of P.
pratensis at different time scale under 4 o C by RT-PCR and qRT-PCR. (A) qRT-PCR analysis of PpCBF3 expression level. Three independent experiments show the similar results and here shows one of the results. Values are means ± SD of three technical repetitions. (B) RT-PCR analysis of PpCBF3 expression level.
Growth and physiological characterization of freezing tolerance of PpCBF3-transgenic Arabidopsis
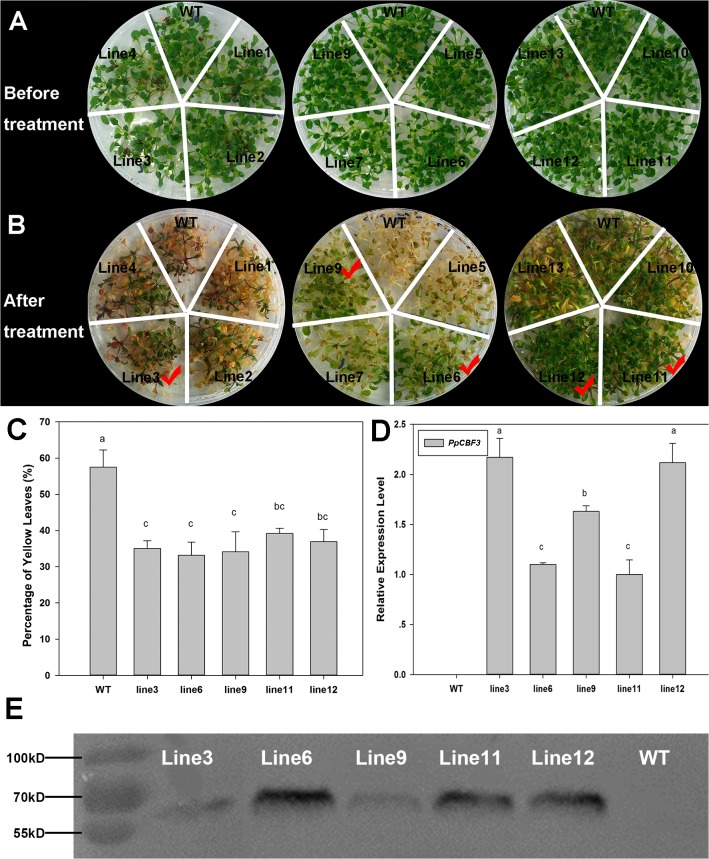
To characterize the physiological functions of PpCBF3 involved in freezing tolerance, 16 transgenic lines of Arabidopsis were obtained (confirmed by PCR, S2 Fig) and 5 transgenic lines (Fig 5A–5C) exhibiting desirable phenotypes and improved freezing tolerance were selected for further experiments. qRT-PCR and Western blot analysis showed that PpCBF3 was highly expressed in all 5 transgenic lines but not in WT (Fig 5D and 5E). The transgenic lines resumed growth following 7 d of recovery from 3 h exposure to -20°C (Fig 5A and 5B). The percentage of chlorotic leaves was significantly lower in the transgenic lines than in WT following freezing stress (Fig 5C).
Fig 5. Expression analysis of PpCBF3 and phenotype of WT and transgenic lines under normal and cold treatment conditions.
(A) Shows 4-week-old seedlings of WT and transgenic lines grow under normal condition. (B) Phenotype of WT and transgenic lines after -20°C freezing stress. Red tick represents the stable transformed lines finally selected for further analysis. (C) Percentage of chlorotic leaves is calculated in (B). More than half yellow color of one leaf is designated as chlorolic leaf. Values are means ± SD of fifteen independent plants. Different letters on the top of bars indicate significant differences (P < 0.05) between WT and transgenic lines under the same growth condition. (D) Detection of the relative mRNA expression level of PpCBF3 in WT and transgenic lines grow under normal condition. (E) PpCBF3-GFP fused protein is detected by western blot in transgenic lines but not in WT.
Under non-stress temperature, no differences in leaf EL were detected between the WT and transgenic lines (Fig 6A). Following freezing treatment, EL was significantly lower (about 35%) in all transgenic lines than in the WT (> 60%) (Fig 6A). Leaf chlorophyll content and Fv/Fm ratio decreased after freezing treatments in the WT and transgenic lines, but the transgenic lines maintained significantly higher chlorophyll content (approximately 0.8 in transgenic lines versus 0.5 in WT) and Fv/Fm (approximately 0.8 in transgenic lines versus 0.6 in WT) than WT (Fig 6B and 6C).
Fig 6. Physiological index including relative electrolyte leakage (EL), total chlorophyll content (Chl) and photochemical efficiency (Fv/Fm) in WT and transgenic plants under normal and freezing treated.
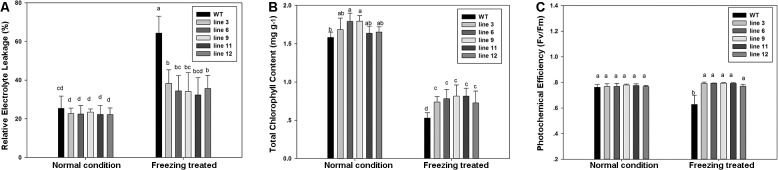
Values are means ± SD of twelve independent plants. The same letters atop bars indicate that there is no significant difference at P < 0.05.
To identify if transgenic plants accumulated less H2O2 and O2 .- under freezing stress at -20°C, leaves were stained with DAB and NBT. Leaves of the WT showed a dark blue color while those of all transgenic plants were stained light blue (Fig 7A and 7B), which indicated less accumulation of O2 .- in transgenic plants compare to the WT. Similarly, less H2O2 production was found in transgenic plants than in the WT, as shown by lesser intensity of brown staining of transgenic leaves (Fig 7C and 7D).
Fig 7. Detection of ROS in WT and transgenic Arabidopsis plants.
(A) NBT staining for superoxide in WT and transgenic Arabidopsis under normal condition and (B) freezing stress. (C) DAB staining for hydrogen peroxide in WT and transgenic Arabidopsis under normal condition and (D) freezing stress.
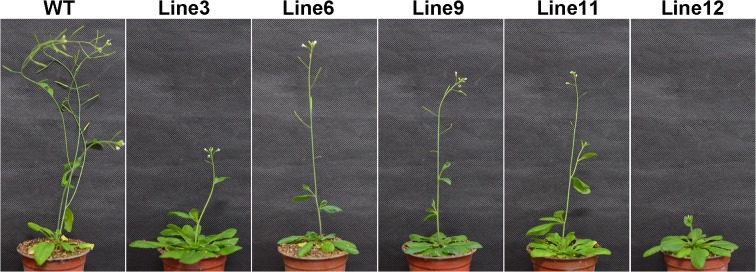
Phenotypic alterations due to overexpression of PpCBF3
In addition to differential phenotypic and physiological changes in response to freezing stress between the WT and transgenic plants, PpCBF3-transgenic plants grew slower than the WT under normal temperatures (Fig 8). The WT started to flower following 29 d of seedling emergence whereas transgenic lines did not flower until 34–43 d following seedling emergence (Table 1). The transgenic plants had an average 21 rosette leaves per plant before flowering compared with 13 rosette leaves in the WT (Table 1). After 6 weeks growth under non-stress temperature conditions, the WT bore many siliques and began to senescence. However, the transgenic plants were just beginning to flower or bore few siliques (Fig 8).
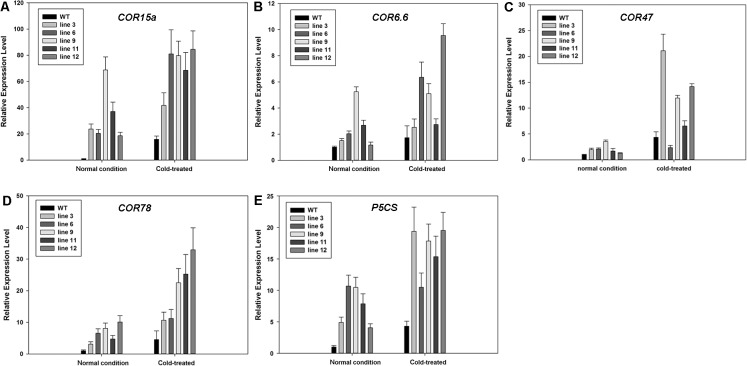
Fig 8. Relative mRNA expression level of downstream genes of DREB/CBF in WT and transgenic plants under normal and cold treatment (4°C).
(A)-(D) Relative mRNA expression level of cold responsive gene (COR) in WT and transgenic plants under normal and cold treatment (4°C). (E) Relative mRNA expression level of P5CS controlling the rate- limiting step of glutamate- derived proline biosynthesis in WT and transgenic plants under normal and cold treatment (4°C).
Table 1. Effects of PpCBF3 expression on flowering time and rosette leaf number.
| Plants | Time to flowering (days)* | Rosette leaves per plant (n) # |
|---|---|---|
| WT | 29 | 13.0±1.93 (14)b |
| Line 3 | 43 | 21.8±1.98 (13)a |
| Line 6 | 34 | 20.7±2.26 (10)a |
| Line 9 | 36 | 21.5±3.14 (11)a |
| Line 11 | 37 | 21.6±1.67 (10)a |
| Line 12 | 43 | 21.5±1.97 (10)a |
a, bMeans followed by the same letter are not significantly different at α = 0.05 based on single degree of freedom contrasts from within an analysis of variance.
*Period of time between plants germination and appearance of first flower buds in a population of “n” plants.
#Means representing the rosette leaves number of WT and 5 transgenic lines divided by the number “n”.
Expression of DREB/CBF down-stream target genes induced by PpCBF3
To determine whether overexpressing PpCBF3 may activate DREB1/CBF downstream target genes such as COR15a, COR6.6, COR47 and COR78, the relative expression level of target genes was determined by qRT-PCR analysis in transgenic plants and WT under normal temperature and 4°C treatments. The results showed that under normal temperature, these genes were expressed in transgenic plants but exhibited lower level in WT (Fig 9A–9D). After 4°C treatment, they were up-regulated in both the WT and transgenic plants but these target genes showed higher expression levels in transgenic plants compared to WT. In addition, relative expression level of Δ1-pyrroline-5-carboxylate synthase (P5CS) was detected which also showed higher transcript levels in transgenic plants than the WT both under normal and cold stress conditions (Fig 9E).
Fig 9. Growth characteristic of 6-week-old WT and transgenic Arabidopsis under normal growth condition.
All transgenic plants show late flowering phenotype compared with WT. The growth chamber condition is set at 23 oC, 16h/8h light/dark, 70% humidity, 120 μmol m-2 s-1 light density.
Discussion
PpCBF3 belongs to DREB/CBF family and specifically responsive to cold stress
The PpCBF3 isolated from Kentucky bluegrass contains a conserved AP2 DNA binding domain and belongs to the DREB1/CBFs subfamily of AP2 superfamily. Previous study showed that the DREB subfamily was further divided into six small groups from A-1 to A-6 [37]. The presence of two signature sequences, one was ‘DSAW’ motif at the end of AP2 domain and the other was ‘LWSY’ motif at the end of C-terminal region, testified that PpCBF3 belonged to the A-1 group. Phylogenetic analysis showed that PpCBF3 was clustered with LpCBF3 and OsDREB1A. PpCBF3 contains a putative nuclear localization signal (NLS) upstream of the AP2 domain and was localized in the cell nucleus. This was consistent with the transcription activator function of CBF genes as found by others [38]. These results suggested that PpCBF3 is a transcription factor.
The CBF genes are known to be responsive to various abiotic stresses [5,39,40]. In this study, PpCBF3 expression responded rapidly to chilling stress (4°C) but did not change in response to drought stress induced by PEG-6000 or salt stress (S1 Fig). It has been reported that not all of the six CBF homologs in Arabidopsis are functionally related to cold tolerance. For example, CBF4 is mainly participated in drought resistance whereas the other two homologs, DDF1 and DDF2, take part in the regulation of gibberellins biosynthesis and salinity tolerance [22,41]. Recently, Kidokoro et al. (2015) [8] identified 14 DREB1-type transcription factors, GmDREB1s, from the soybean genome database and found DREB1 proteins play important roles in heat stress responses. The current results suggested that the PpCBF3 in the DREB1/CBF A-1 subfamily isolated from Kentucky bluegrass was specific to cold stress responses. Other members of DREB family in Kentucky bluegrass could play roles in responses to other abiotic stresses, such as drought or salinity stress, and deserves further investigation.
Enhanced tolerance to severe freezing stress of PpCBF3-transgenic Arabidopsis associated with up-regulation of downstream cold-related genes
Overexpression of DREB1/CBF transcription factors in regulating cold tolerance has been reported in agronomic crops and the model plant Arabidopsis [13–16,42]. In Arabidopsis, the expression of three DREB1 genes (CBF1, CBF2 and CBF3) was strongly induced by cold stress [43]. These DREB1 genes predominantly acted in cold stress-inducible gene expression because of the enrichment of CRT/DRE in the promoters of cold stress-responsive genes in Arabidopsis [44]. In our study, transgenic Arabidopsis plants overexpressing PpCBF3 exposed to -20°C showed significant improvement in freezing tolerance compared to the WT as manifested by increased chlorophyll content and Fv/Fm, as well as the decreases in the percentage of chlorotic leaves and EL in transgenic plants. The growth and physiological analyses strongly suggested that over-expressing PpCBF3 played positive roles in alleviating damages from extremely low freezing temperature (-20°C), unlike previous studies which have mostly reported improved tolerance to chilling stress or mild freezing stress [13,15,18,19,25].
Cellular injuries of cold stress are related to ROS (O2 .- and H2O2) production that causes oxidative damage [45]. The improvement of freezing tolerance by PpCBF3 overexpression was found to be related to the suppression of O2 .- and H2O2 accumulation as shown by the NBT and DAB staining, respectively. In addition, it is reported that high concentrations of proline is involved in stress tolerance [46–48]. Indeed, expression level of P5CS which encodes an enzyme involved in proline synthesis was up-regulated in transgenic Arabidopsis plants, although proline content was not determined in this study. Our results strongly demonstrated that PpCBF3 could play important roles in protecting leaf damages from freezing stress by protecting photosynthetic components (chlorophyll content and photochemical reactions), cell membranes, and suppressing ROS formation, as well as up-regulation of proline synthesis, ultimately leading to improved plant growth and survival under extremely low freezing temperature.
Through the binding of DRE/CRT promoters, CBFs activate many downstream COR genes encoding hydrophilic polypeptides which enhance tolerance to low temperature in plants [19]. Consistently, the expression of COR genes including COR15a, COR6.6, COR47 and COR78 were constitutively activated by PpCBF3 in transgenic plants and up-regulated under cold stress. It is notable that the fold change of COR15a expression in transgenic plants was greater than that of other COR genes under normal and freezing stress, suggesting that COR15a could play a prominent role in freezing tolerance of transgenic plants overexpressing PpCBF3. It has been reported that in transgenic Arabidopsis plants, overexpressing COR15a improved freezing tolerance of chloroplasts frozen in situ (-4°C and -5°C) and of protoplasts frozen in vitro (between -5°C and -8°C) [49]. However, COR15a expression in transgenic Arabidopsis has no significant effect on the survival of whole frozen plants. Several COR genes (i.e. COR6.6a and COR47) are cooperatively stimulated along with COR15a when subjected to low temperature [50,51], which suggests that these COR genes might act in concert with each other to increase tolerance to freezing stress in plants. It is postulated that expression of the entire batch of COR genes activated by PpCBF3 would have greater effects on freezing tolerance than COR15a expression alone.
Phenotypic alterations due to overexpression of PpCBF3 in Arabidopsis
Previous studies have reported constitutive over-expression of DREB1/CBF genes resulted in stunted shoot growth, delayed flowering, and lower yields, all of which are considered negative characteristics for agronomic crop species [3,13–15,52]. Many attempts have been reported to minimize the negative effects of DREB1/CBF over-expression by using stress-inducible promoters [14,15,20,42,53]. Overexpression of PpCBF3 in Arabidopsis also slowed growth and delayed flowering in transgenic plants. Unlike agronomic crops, slower vertical growth and delayed flowering are desirable traits in perennial grasses used as turfgrass. Slow-growing turfgrasses require less-frequent mowing, irrigation, and fertility inputs while the presence of flowering reduces turf visual quality and uniformity [26]. Therefore, PpCBF3 could be potentially useful for genetic engineering of turfgrass improvement to freezing tolerance and developing other desirable traits, such as slowed growth and late flowering.
In summary, PpCBF3 isolated from cold-tolerant Kentucky bluegrass was characterized as a DREB1/CBF transcriptional factor and was responsive specifically to cold stress. The physiological and gene expression analysis of transgenic Arabidopsis overexpressing PpCBF3 confirmed PpCBF3 serves roles in regulating plant tolerance to severe freezing stress and was associated with the up-regulation of downstream genes (COR15a, COR6.6, COR47, COR78 and P5CS) involved in cold tolerance. The positive physiological effects of PpCBF3 for freeing tolerance and the induction of desirable turfgrass phenotypic traits suggested that PpCBF3 could be a useful candidate gene for genetic modification of perennial grasses for high turf quality and improved freezing tolerance.
Supporting Information
Values are means ± SD of three independent experiments. The same letters atop bars indicate that there is no significant difference at P < 0.05.
(TIF)
WT plants are used as negative control. Numbers indicate transgenic line. Only 13 lines are showned here.
(TIF)
(XLSX)
Acknowledgments
We thank Guohui Yu for the subcellular localization assay, and Dr. Jingjin Yu and Dr. Jun Liu for helpful discussion of the experiment design.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no support or funding to report.
References
- 1. Kourosh V, Charles L. Abiotic Stress-Plant Responses and Applications in Agriculture In: Hasanuzzaman M, Nahar K, Fujita M, editors. Extreme Temperature Responses, Oxidative Stress and Antioxidant Defense in Plants. INTECH Open Access Publisher; 2013. pp. 169–205. [Google Scholar]
- 2. Thomashow MF. Role of cold-responsive genes in plant freezing tolerance. Plant Physiol. 1998; 118: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998; 10: 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997; 94: 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011; 62: 4731–4748. 10.1093/jxb/err210 [DOI] [PubMed] [Google Scholar]
- 6. Sanghera GS, Wani SH, Hussain W, Singh N. Engineering cold stress tolerance in crop plants. Curr Genomics 2011; 12: 30 10.2174/138920211794520178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003; 33: 751–763. [DOI] [PubMed] [Google Scholar]
- 8. Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K, Mizoi J, et al. Soybean DREB1/CBF‐type transcription factors function in heat and drought as well as cold stress‐responsive gene expression. Plant J. 2015; 81: 505–518. 10.1111/tpj.12746 [DOI] [PubMed] [Google Scholar]
- 9. Qin F, Sakuma Y, Li J, Liu Q, Li YQ, Shinozaki K, et al. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004; 45: 1042–1052. [DOI] [PubMed] [Google Scholar]
- 10. Shen Y-G, Zhang W-K, He S-J, Zhang J-S, Liu Q, Chen S-Y. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor Appl Genet. 2003; 106: 923–930. [DOI] [PubMed] [Google Scholar]
- 11. Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 2002; 14: 1675–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, et al. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. Plant J. 2004; 38: 982–993. [DOI] [PubMed] [Google Scholar]
- 13. Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006; 47: 141–153. [DOI] [PubMed] [Google Scholar]
- 14. Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 2004; 45: 346–350. [DOI] [PubMed] [Google Scholar]
- 15. Morran S, Eini O, Pyvovarenko T, Parent B, Singh R, Ismagul A, et al. Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol J. 2011; 9: 230–249. 10.1111/j.1467-7652.2010.00547.x [DOI] [PubMed] [Google Scholar]
- 16. Pellegrineschi A, Reynolds M, Pacheco M, Brito RM, Almeraya R, Yamaguchi-Shinozaki K, et al. Stress-induced expression in wheat of the Arabidopsis thaliana DREB1A gene delays water stress symptoms under greenhouse conditions. Genome 2004; 47: 493–500. [DOI] [PubMed] [Google Scholar]
- 17. Wang L, Gao J, Qin X, Shi X, Luo L, Zhang G, et al. JcCBF2 gene from Jatropha curcas improves freezing tolerance of Arabidopsis thaliana during the early stage of stress. Mol Biol Rep. 2014; 10.1007/s11033-014-3831-0 [DOI] [PubMed] [Google Scholar]
- 18. Yang S, Tang XF, Ma NN, Wang LY, Meng QW. Heterology expression of the sweet pepper CBF3 gene confers elevated tolerance to chilling stress in transgenic tobacco. J Plant Physiol. 2011; 168: 1804–1812. 10.1016/j.jplph.2011.05.017 [DOI] [PubMed] [Google Scholar]
- 19. Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998; 280: 104–106. [DOI] [PubMed] [Google Scholar]
- 20. Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999; 17: 287–291. [DOI] [PubMed] [Google Scholar]
- 21. Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, et al. Freezing‐sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing‐tolerant Arabidopsis. Plant J. 2004; 39: 905–919. [DOI] [PubMed] [Google Scholar]
- 22. Haake V, Cook D, Riechmann JL, Pineda O, Thomashow MF, Zhang JZ. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002; 130: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao H, Bughrara SS. Isolation and characterization of cold-regulated transcriptional activator LpCBF3 gene from perennial ryegrass (Lolium perenne L.). Mol Genet Genomics 2008; 279: 585–594. 10.1007/s00438-008-0335-4 [DOI] [PubMed] [Google Scholar]
- 24. Tang M, Lu S, Jing Y, Zhou X, Sun J, Shen S. Isolation and identification of a cold-inducible gene encoding a putative DRE-binding transcription factor from Festuca arundinacea. Plant Physiol Biochem. 2005; 43: 233–239. [DOI] [PubMed] [Google Scholar]
- 25. Xiong Y, Fei SZ. Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 2006; 224: 878–888. [DOI] [PubMed] [Google Scholar]
- 26. Fry J, Huang B. Applied turfgrass science and physiology J. Wiley; 2004. [Google Scholar]
- 27. Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. Calif Agric Exp Sta Circ. 1950; 347:1–32. [Google Scholar]
- 28. Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol plantarum 1962; 15: 473–497. [Google Scholar]
- 29. Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. Tape-Arabidopsis Sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 2009; 5: 16 10.1186/1746-4811-5-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006; 45: 616–629. [DOI] [PubMed] [Google Scholar]
- 31. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998; 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 32. Li XL, Yang X, Hu YX, Yu XD, Li QL. A novel NAC transcription factor from Suaeda liaotungensis K. enhanced transgenic Arabidopsis drought, salt, and cold stress tolerance. Plant Cell Rep. 2014; 33: 767–778. 10.1007/s00299-014-1602-y [DOI] [PubMed] [Google Scholar]
- 33. Nilsen ET, Orcutt DM. Physiology of plants under stress Abiotic factors. J. Wiley; 2004. [Google Scholar]
- 34. Arnon DI. Copper Enzymes in Isolated Chloroplasts. Polyphenoloxidase in Beta Vulgaris. Plant Physiol. 1949; 24: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Marcum KB. Cell membrane thermostability and whole-plant heat tolerance of Kentucky bluegrass. Crop Sci. 1998; 38: 1214–1218. [Google Scholar]
- 36. Lee BH, Lee H, Xiong L, Zhu JK. A mitochondrial complex I defect impairs cold-regulated nuclear gene expression. Plant Cell 2002; 14: 1235–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002; 290: 998–1009. [DOI] [PubMed] [Google Scholar]
- 38. Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998; 16: 433–442. [DOI] [PubMed] [Google Scholar]
- 39. Agarwal PK, Agarwal P, Reddy M, Sopory SK. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 2006; 25: 1263–1274. [DOI] [PubMed] [Google Scholar]
- 40. Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. BBA-Gene Regul Mech. 2012; 1819: 86–96. [DOI] [PubMed] [Google Scholar]
- 41. Magome H, Yamaguchi S, Hanada A, Kamiya Y, Oda K. dwarf and delayed-flowering 1, a novel Arabidopsis mutant deficient in gibberellin biosynthesis because of overexpression of a putative AP2 transcription factor. Plant J. 2004; 37: 720–729. [DOI] [PubMed] [Google Scholar]
- 42. James VA, Neibaur I, Altpeter F. Stress inducible expression of the DREB1A transcription factor from xeric, Hordeum spontaneum L. in turf and forage grass (Paspalum notatum Flugge) enhances abiotic stress tolerance. Transgenic Res. 2008; 17: 93–104. [DOI] [PubMed] [Google Scholar]
- 43. Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, et al. AnArabidopsisGene Family Encoding DRE/CRT Binding Proteins Involved in Low-Temperature-Responsive Gene Expression. Biochem Bioph Res Co. 1998; 250: 161–170. [DOI] [PubMed] [Google Scholar]
- 44. Maruyama K, Todaka D, Mizoi J, Yoshida T, Kidokoro S, Matsukura S, et al. Identification of cis-acting promoter elements in cold-and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 2012; 19: 37–49. 10.1093/dnares/dsr040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Asada K. Ascorbate peroxidase–a hydrogen peroxide‐scavenging enzyme in plants. Physiol Plantarum 1992; 85: 235–241. [Google Scholar]
- 46. Igarashi Y, Yoshiba Y, Sanada Y, Yamaguchi-Shinozaki K, Wada K, Shinozaki K. Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant Mol Biol. 1997; 33: 857–865. [DOI] [PubMed] [Google Scholar]
- 47. Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, et al. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J. 2002; 29: 417–426. [DOI] [PubMed] [Google Scholar]
- 48. Wanner LA, Junttila O. Cold-induced freezing tolerance in Arabidopsis. Plant Physiol. 1999; 120: 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF. Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA. 1996; 93: 13404–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hajela RK, Horvath DP, Gilmour SJ, Thomashow MF. Molecular cloning and expression of cor (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol. 1990; 93: 1246–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF. Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1998; 95: 14570–14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, et al. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002; 129: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xue GP, Way HM, Richardson T, Drenth J, Joyce PA, McIntyre CL. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol Plant 2011; 4: 697–712. 10.1093/mp/ssr013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Values are means ± SD of three independent experiments. The same letters atop bars indicate that there is no significant difference at P < 0.05.
(TIF)
WT plants are used as negative control. Numbers indicate transgenic line. Only 13 lines are showned here.
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.