SUMMARY
Cancer develops after the acquisition of a collection of mutations that together create the cancer phenotype. How collections of mutations work together within a cell, and whether there is selection for certain combinations of mutations, are not well understood. We investigated this problem with a mathematical model of the Ras signaling network, including a “computational random mutagenesis”. Modeling and subsequent experiments revealed that mutations of the tumor suppressor gene NF1 can amplify the effects of other Ras pathway mutations, including weakly activating, noncanonical, Ras mutants. Furthermore, analyzing recently available, large, cancer genomic data sets uncovered increased co-occurrence of NF1 mutations with mutations in other Ras network genes. Overall, these data suggest that combinations of Ras pathway mutations could serve the role of cancer “driver”. More generally, this work suggests that mutations that result in “network instability” may promote cancer in a manner analogous to genomic instability.
INTRODUCTION
Cancer genomic studies support the idea that a cell becomes cancerous through the progressive acquisition of mutations that together confer the cancer phenotype. These mutations that promote cancer are commonly referred to as “driver genes” (Stratton et al., 2009). It is not well understood how the presence of one mutation influences the selection of subsequent mutations through an evolutionary process (Yates and Campbell, 2012). A statistical lack of co-occurrence between ‘canonical’ mutations within the same pathway is well established (Thomas et al., 2007; Yates and Campbell, 2012). The lack of co-occurrence is typically attributed to the assumption that there would be no selective benefit to accumulating multiple mutations within the same molecular pathway (Yeang et al., 2008). Such arguments implicitly assume that each mutation is sufficiently strong to confer a selective advantage alone (e.g. the canonical KRAS and BRAF mutations). However, there are a number of weakly activating RAS and BRAF mutations that have been observed in cancer (Wan et al., 2004), although less commonly than the canonical mutations.
It is believed that many cancers share common phenotypes, such as constitutive activation of the Ras pathway (Hanahan and Weinberg, 2000). Within some types of cancer, there is near universal presence of a mutation that confers this phenotype to the Ras pathway. For example, essentially all sequenced pancreatic adenocarcinomas have canonical KRAS mutations (Biankin et al., 2012; Jones et al., 2008), and essentially all hairy cell leukemias have the canonical BRAF V600E mutation (Tiacci et al., 2011). More commonly, a type of cancer can utilize one of several potential gene mutations. Melanomas, for example, frequently harbor either a canonical BRAF or a canonical NRAS mutation (Hodis et al., 2012). When a canonical driver mutation is not identified in a sequenced cancer, other candidate driver mutations are often proposed based upon the identification of a mutated gene within the same pathway as a common, canonical, driver mutation (Hodis et al., 2012; Jones et al., 2008). Whether or not these less common “noncanonical” mutations, which are often less strongly activating than the canonical mutations, are sufficient to serve as a surrogate for a canonical driver mutation, or whether the ability to serve as a surrogate is conditional to some other contextual influence, is not fully understood. The role of noncanonical mutants in cancer is quite important when one considers the growing number of cancers that are being genomically characterized for both research and clinical purposes.
We investigated the potential for cooperation between less commonly mutated genes within the Ras network. We used a mathematical model to investigate whether canonical and noncanonical Ras mutants are influenced by the partial loss of tumor suppressor gene product neurofibromin (NF1). We found computational evidence for greater than additive increases in Ras activation for noncanonical Ras mutants in the neurofibromin deficient context. This prediction was also supported experimentally in cells with or without neurofibromin. Further, analyzing >3900 sequenced cancer specimens from the Cancer Cell Line Encyclopedia (CCLE) and The Cancer Genome Atlas (TCGA) uncovered an increased rate of co-occurrence between mutations the model predicted could display synergy. This work suggests that NF1 mutations promote cancer not only by the direct and immediate increase in RasGTP, but also by increasing the number of possible subsequent mutations that would further increase Ras signaling. This suggests that multiple, weakly activating mutations may together serve the role of driver gene. Generalization of the “network instability” concept as presented here to cancer treatment suggests that a wide variety of mutations could confer resistance to targeted therapies. Overall, this work demonstrates that a biochemical/mechanistic understanding of cell signaling networks can be used to uncover functional combinations of mutations.
RESULTS
Modeling predicts synergy between weakly activating Ras pathway mutants
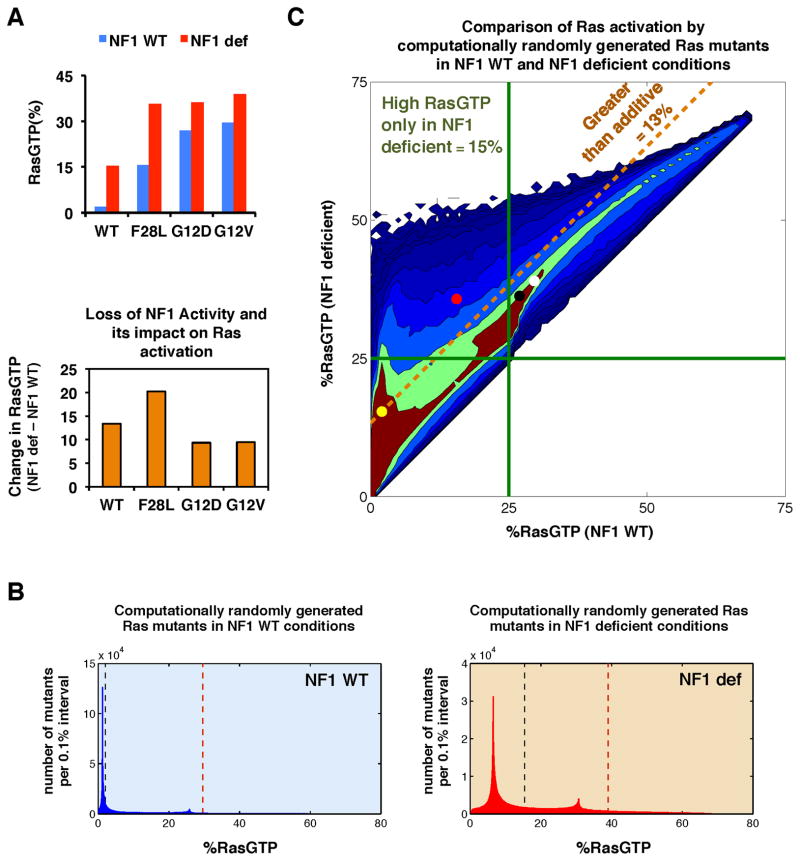
We previously developed a mathematical model based upon the biochemical reactions for the major classes of proteins that regulate Ras GTPase signaling (Stites et al., 2007). We evaluated how well our Ras model applied to neurofibromin deficient conditions, and we found that the model robustly reproduces common phenotypes of neurofibromin deficient systems (Figure S1A, Supplemental Methods). We used this model to investigate the effect of concurrent Ras and neurofibromin mutations. We initially considered the canonical, oncogenic Ras mutants RasG12D and RasG12V, and the noncanonical, weakly activating RasF28L mutant. When neurofibromin is fully present and not mutated, RasF28L was predicted to result in approximately half the RasGTP signal as RasG12D or RasG12V, which is consistent with experimental data for these mutants (Stites et al., 2007). However, when we modeled RasF28L in the neurofibromin deficient context, we observed that RasF28L generated a high level of Ras activation similar to the strong RasG12D and RasG12V mutants (Figure 1A, upper). This is due, in part, to a less-than-additive increase in Ras signals when RasG12D and RasG12V are combined with neurofibromin deficiency (Figure 1A, lower). This less-than-additive increase may be consistent with the general lack of co-occurrence between commonly observed ‘strong’ mutations in that it suggests there is a small benefit to acquiring a second strong mutation. Interestingly, RasF28L exhibited a greater-than-additive increase in Ras signals when combined with a loss of NF1 activity. We hypothesized that there may be a large number of such ‘inherently weaker’ Ras mutants beyond RasF28L that would result in high levels of Ras pathway activation in the neurofibromin deficient (NF1-deficient) conditions, but not in neurofibromin wild-type (NF1-WT) conditions.
Figure 1. Modeling predicts that weakly activating Ras mutants can appear strong within the NF1-deficient context.
(A) (upper) Simulations of the Ras network model with all wild-type Ras, a canonical oncogenic mutant (RasG12D and RasG12V), or a noncanonical, non-oncogenic mutant (RasF28L) for both NF1-WT and NF1-deficient conditions. (lower) The net change in predicted RasGTP levels going from NF1-WT to NF1-deficient conditions for the cases above.
(B) Histogram from our ‘computational mutagenesis’ displaying the number of Ras mutants with varying levels of RasGTP signal in the context of NF1-WT (blue) and NF1-deficient (red) conditions. Dashed black line shows level of RasGTP for a network with all RasWT (no mutation present); dashed brown line shows level of RasGTP for a network with a RasG12V mutation. Histogram is binned into 0.1% intervals.
(C) One million random mutants were simulated in the NF1-WT and NF1-deficient states, and the resulting levels of RasGTP are plotted for both conditions. Filled circles indicate a network containing a Ras mutant: RasG12D (black), RasG12V (white), RasF28L (red); or no Ras mutant (yellow). Green lines indicate 25% total RasGTP. Any random Ras mutant falling above the dashed gold line shows a greater net change in percent RasGTP in the NF1-deficient network compared to the NF1-WT network.
“Computational mutagenesis” predicts that some weak Ras mutants could become strong in a neurofibromin deficient context
Very few Ras mutants have been characterized as extensively as RasG12D and RasG12V. We therefore performed a “computational random mutagenesis” to more comprehensively explore the possible extent to which neurofibromin deficiency might affect potential Ras mutants (Supplemental Methods). By varying the biochemical rate constants and enzymatic parameters that characterize a Ras protein, we can model potential Ras mutants. We simulated one million such random mutants and compared the Ras signal levels (both RasGTP and effector:RasGTP complex) in the context of both the NF1-WT and NF1-deficient networks.
We analyzed the relative frequencies with which these randomly generated Ras mutants achieved different RasGTP levels (Figure 1B). In the NF1-WT context, most mutants did not cause a large increase in Ras signaling. A long, shallow tail of more strongly activating mutants was present. This suggests that there are a limited number of potential Ras mutations that result in high levels of ‘constitutive’ Ras activation and are strong enough to promote cancer by themselves. In contrast, in the NF1-deficient context, the distribution was shifted toward higher levels of RasGTP, and the relative frequencies of mutants generating higher levels of RasGTP were increased. This suggests that the number of potential Ras point mutations capable of promoting cancer may be greater in the NF1-deficient context.
We then examined how the level of RasGTP for each mutant differed between NF1-WT and NF1-deficient contexts, and we considered the potential synergy between neurofibromin and Ras mutants (Figure 1C). Approximately 13% of all random mutants displayed greater-than-additive increase in RasGTP under neurofibromin deficiency. We then arbitrarily focused on mutants that resulted in 25% of total Ras bound to GTP, since ~25% of cellular RasGTP has been shown to approximate where transformation potential abruptly begins (Donovan et al., 2002). We found that 15% of all random mutants exceeded this 25% level of RasGTP in the NF1-deficient context but not in the NF1-WT context. Analysis of other threshold levels of RasGTP similarly revealed large fractions of random mutants that exceed the threshold in NF1-deficient, but not NF1-WT, conditions (Figure S1B). Similar results were found when we considered the quantity of effector:RasGTP complex as an alternative measure of Ras activation (Figure S1C). Consideration of the computational random mutants that fell within the different quadrants of Figure 1C found considerable overlap in mutant parameter values, suggesting that the behaviors of a newly discovered Ras mutant cannot be easily inferred from any single parameter measurement (Figure S1D). Overall, these simulations suggest that the strength of a mutant can be context dependent; a Ras mutant that appears “weak” in a wild-type background may appear “strong” in an NF1-deficient background. The model also predicts that the number of Ras mutants that could result in a high level of RasGTP will be increased in NF1-deficient conditions.
We highlight that these results are for modeling a loss of NF1 GAP activity, such as might occur from a deletion mutation or from a nonsense mutation (Stratton et al., 2009). Deletion mutations and truncation mutations before the GAP domain are observed in human cancers (The Cancer Genome Atlas Network, 2008). Missense point mutations are also observed for NF1, and missense mutations can result in a GAP domain with altered biochemistry (Sermon et al., 1998). We considered whether point mutants with impaired GAP activity would exhibit similar behaviors when combined with Ras mutations. Simulations of neurofibromin R1276A point mutant were performed by using measured changes to its kcat and Km. We simulated this missense mutant in combination with specific Ras mutants and with computational random Ras mutants, just as we had done for NF1 deficiency. In our simulations of the neurofibromin point mutant, we similarly observed a greater-than-additive increase in signal strength when combined with RasF28L, similarly observed a sizable proportion of greater-than-additive computational random Ras mutants, and similarly observed a large fraction of mutants that exceeded a given level of RasGTP only in the NF1-mutant conditions (Figure S1E). Together, these simulations suggest that a wide variety of NF1 mutations should create a context that amplifies the effects of noncanonical Ras mutants.
RasF28L mutant is more strongly activating in the NF1-deficient cellular context
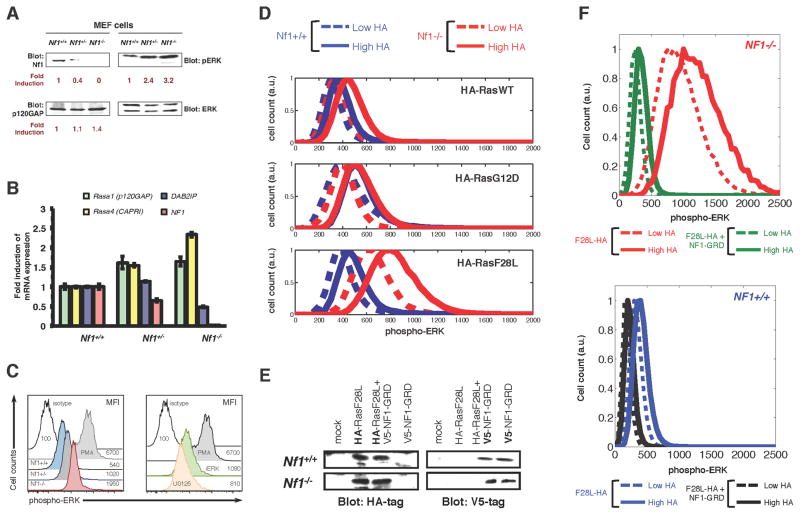
We next experimentally tested our computational prediction that some “weak” Ras mutants will more strongly activate the Ras pathway within the NF1-deficient context. We obtained mouse embryo fibroblasts (MEFs) that had been previously derived from Nf1 deficient, wild-type, and heterozygous null mice (Nf1−/−, Nf1+/+, and Nf1+/−, respectively) (Shapira et al., 2007). We confirmed that the Nf1+/− and Nf1−/− MEFs had decreased protein expression (Figure 2A) and decreased mRNA expression for NF1 (Figure 2B). Within the MEFs, we noted increased expression of RNA and protein for some Ras GAPs (Figure 2B and 2C), which is consistent with negative feedback resulting from increased Ras signaling. Negative feedback induced by Ras pathway activation has important physiologically consequences, including a role in senescence (Courtois-Cox et al., 2006), and in resistance to targeted therapies (Prahallad et al., 2012). However, levels of phosphorylated ERK (pERK) by Western blot (Figure 2A) and by flow cytometry (Figure 2C), reflected the different levels of neurofibromin deficiency. Moreover, when we tested proliferation of these MEFs in a time course using the CellTrace Violet dye (whose fluorescence dilutes by half in daughter cells), we observed enhanced level of proliferation in neurofibromin deficient MEFs compared to the control MEFs (Figure S2). These data are consistent with previous results for these MEFs (Shapira et al., 2007) and suggest that any negative feedback present in these cells is unable to overcome the anticipated phenotypic consequences of elevated Ras pathway signaling. Altogether, these observations suggest that neurofibromin plays a critical and dominant role in these MEF cells. We conclude that these MEFs could be used for our further analysis.
Figure 2. Weak Ras mutants in mammalian cells can behave as strong activators of Ras pathway signaling under the Nf1 deficient conditions.
(A) Immunoblots of Nf1+/+, Nf1+/−, and Nf1−/− mouse embryo fibroblasts (MEFs) for expression of neurofibromin, p120 Ras GAP, and phosphorylated ERK.
(B) MEFs of the Nf1+/+, Nf1+/−, and Nf1−/− genotype were analyzed by qPCR for Ras GAP genes Rasa1 (p120GAP), Rasa4 (CAPRI), DAB2IP, and Nf1. Error bars represent standard deviation from three independent experiments from three different RNA extractions/preparations.
(C) (left) Histograms present p-ERK profiles within Nf1+/+, Nf1+/−, and Nf1−/− MEF cells. Data presented are representative of at least 6 similar experiments. (right)
(D) Nf1+/+ and Nf1−/− MEFs transfected with HA-tagged H-RasWT, H-RasF28L, or H-RasG12D with HA-tag expression and pERK signal quantified by multi-color flow cytometry.
(E) Immunoblots showing expression of HA-tagged H-RasF28L and V5-tagged NF1-GRD in Nf1−/− and Nf1+/+ MEF cells following transfection, either alone or together.
(F) MEFs transfected with HA-tagged H-RasF28L or with HA-tagged H-RasF28L and NF1-GRD with HA-tag expression and pERK signal quantified by multi-color flow cytometry.. Nf1(−/−) + F28L, red; Nf1(−/−) + F28L + NF1-GRD, green; Nf1(+/+) + F28L, blue; Nf1(+/+) + F28L + NF1-GRD, black. Higher HA, solid; lower HA, dashed.
We next examined how variation in the level of RasF28L expressed could affect ERK phosphorylation in Nf1+/+ and Nf1−/− conditions. We transiently transfected HA-tagged H-RasF28L, H-RasG12D, or H-RasWT into Nf1+/+ and Nf1−/− MEFs. Flow-cytometry was used to obtain quantitative measurements of both the amount of transfected protein expression at a single cell level, and the amount of resulting Ras pathway activation within each cell. These measurements showed that RasF28L expression caused a higher level of phosphorylated ERK signal in Nf1−/− MEFs than in Nf1+/+ MEFs (Figure 2D). Furthermore, gating the RasF28L cells based on relatively lower and relatively higher expression demonstrated that the change in ERK phosphorylation for increased RasF28L expression was concomitantly enhanced in the Nf1−/− MEFs relative to the Nf1+/+ MEFs, consistent with the model predictions. We quantified the ratio of change in mean pERK signal between the high and low HA gates and found that the increase in pERK was 1.9 times higher for RasF28L in Nf1−/− MEFs compared to Nf1+/+ MEFs. In contrast, RasG12D cells experienced a smaller increase going from low to high expression between Nf1+/+ and Nf1−/− MEFs (with the ratio of change quantified at 1.1). The ratio for RasWT was in between that of RasF28L and RasG12D (1.4×).
If the greater changes in phosphorylated ERK within Nf1−/− conditions were due to the loss of GAP activity by neurofibromin, then reintroduction of neurofibromin would be expected to reverse the increased change. We expressed the GAP-related domain of neurofibromin (NF1-GRD), as this domain alone is sufficient to catalyze the hydrolysis of GTP on Ras. We co-transfected Nf1−/− and Nf1+/+ MEFs with HA-tagged RasF28L and with V5-tagged NF1-GRD (Figure 2E). The flow cytometry-based ERK activation assay revealed that co-expression of NF1-GRD reversed the large magnitude changes in pERK due to RasF28L expression in Nf1−/− MEFs (Figure 2F, upper). Furthermore, co-expression of NF1-GRD with RasF28L in Nf1+/+ MEFs had a similar but smaller effect on changing the phosphorylated ERK signal as a function of RasF28L (Figure 2F, lower). Overall, these experimental observations support the important and non-obvious prediction from the mathematical model that neurofibromin deficiency can cause some normally “weak” Ras mutants to have a stronger effect on Ras pathway activation.
Instability of the Ras signaling network in the context of NF1 deficiency
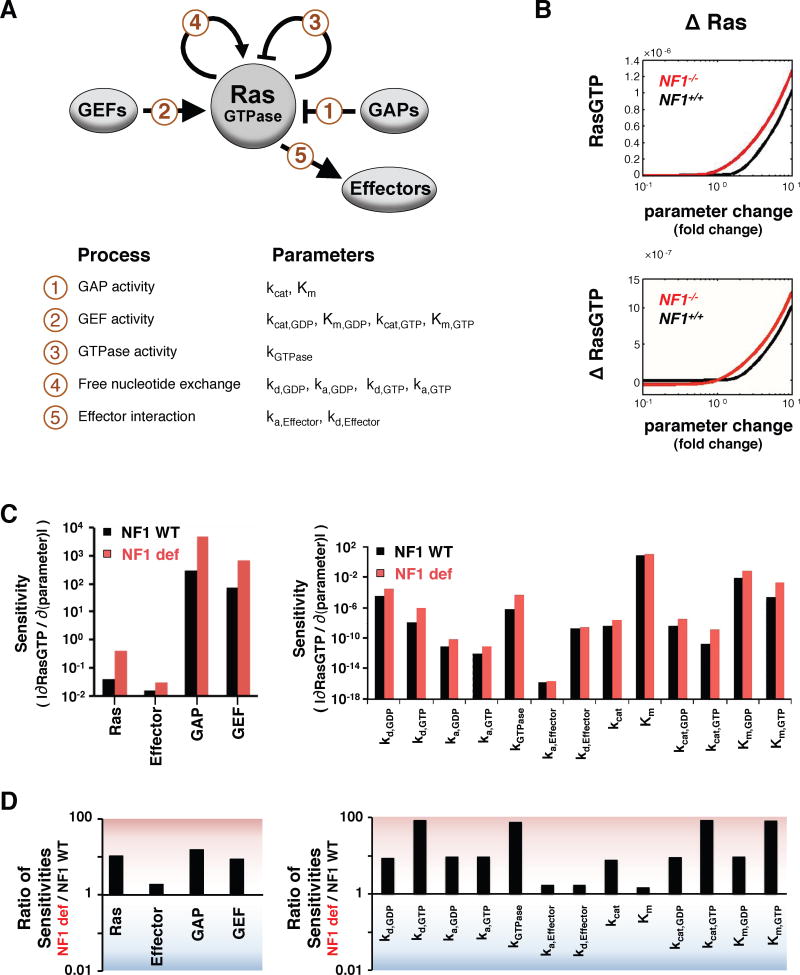
We next investigated why GAP deficient conditions might augment the effects of some Ras mutations. We hypothesized that the sensitivity of the NF1-WT and NF1-deficient Ras networks might be differentially affected by changes in Ras biochemistry. Our Ras model includes the five basic processes that regulate the nucleotide binding state of Ras proteins: GAP activity on Ras, GEF activity on Ras, intrinsic GTPase activity, spontaneous nucleotide dissociation and association, and GTP-bound Ras interactions with effector proteins (Figure 3A). Each of these processes is modeled with mass-action kinetics or enzymatic kinetics and described with the parameters listed in Figure 3A. We considered the magnitude of change in total RasGTP that would result for a wide range of changes in each parameter when it occurs in NF1-WT and in NF1-deficient conditions (Figures 3B and S3A,B). Such a sensitivity analysis approach has been valuable in a wide variety of modeling studies (Benedict et al., 2011; Chen et al., 2014; Gaudet et al., 2012; Schoeberl et al., 2002; Stites et al., 2007).
Figure 3. Mathematical model of the Ras network predicts the NF1-deficient Ras network is generally more sensitive to perturbations.
(A) The components of the Ras network considered are Ras, Ras GEFs, Ras GAPs, and Ras effectors, along with the modeled reactions and their biochemical parameters.
(B) Sample steady-state RasGTP and net change in steady-state RasGTP levels for a range of fold-changes in model parameters. Ras expression level is shown here; all parameters are presented in Figure S3. ΔRasGTP levels are normalized to the total amount of Ras.
(C) The magnitude of the immediate rate of change in RasGTP for a change in parameter is a measure of the sensitivity of the Ras network to that parameter.
(D) The ratio of the sensitivities determined in C.
When we considered the amount of RasGTP that results from a change in a single reaction parameter, our sensitivity analysis found qualitatively similar results for NF1-WT and NF1-deficient networks. It was notable, however, that the magnitude of change in RasGTP for a small change in a parameter was always greater for the NF1-deficient network (Figures 3B and SA,B). This was true for all 17 of the model’s biochemical properties (P<1.6×10−5 by the two-tail exact binomial test).
To further investigate the relevance of this observation, we analytically solved for the change in total RasGTP for a change in parameter for both NF1-WT and NF1-deficient networks (i.e. we found the slope of the curves relating steady-state RasGTP to parameter values taken through the point corresponding to the baseline Ras network parameters). As inferred from the graphical relationship, the analytical approach revealed that the net change in RasGTP induced by a small parameter change is higher in the NF1-deficient network (Figure 3C). Indeed, the NF1-deficient network was often 10–100 times more sensitive to the same change in a network reaction rate constant and/or protein concentration (Figure 3D). Thus, any small perturbation to the Ras network should cause a larger magnitude change in Ras signal if the network is also NF1-deficient. The increased sensitivity is robust to the level of GAP deficiency considered (Figure S3C) and to the concentrations of Ras network proteins modeled (Figure S3D). This suggests that the predicted increased sensitivity of neurofibromin deficient cells could occur in a wide variety of cell types, regardless of specific levels of protein expression and of specific levels of neurofibromin deficiency.
Instability of the Ras signaling network in the context of GAP and GEF deregulation
Since GAPs and GEFs have opposite effects on Ras, we considered the possibility that partial activation of Ras GEFs will be analogous to partial loss of Ras GAPs. Our computational random mutagenesis found that increased basal GEF activity (such as what might follow from an upstream mutation, or from a Ras GEF mutation) could also potentiate the effects of some (but not all) Ras mutants (Figure S3E), similar to what we found for neurofibromin in Figure 1. We also performed a sensitivity analysis for increased basal Ras GEF activity like we did for neurofibromin deficiency in Figure 3C. We found that increased basal GEF activity also makes the Ras network more sensitive to perturbations (Figure S3F). That is, increased sensitivity to perturbations should be a more general feature of cells harboring Ras GAP and Ras GEF mutations.
Co-occurrence between NF1 mutations and noncanonical Ras mutations in cancer genomic data sets
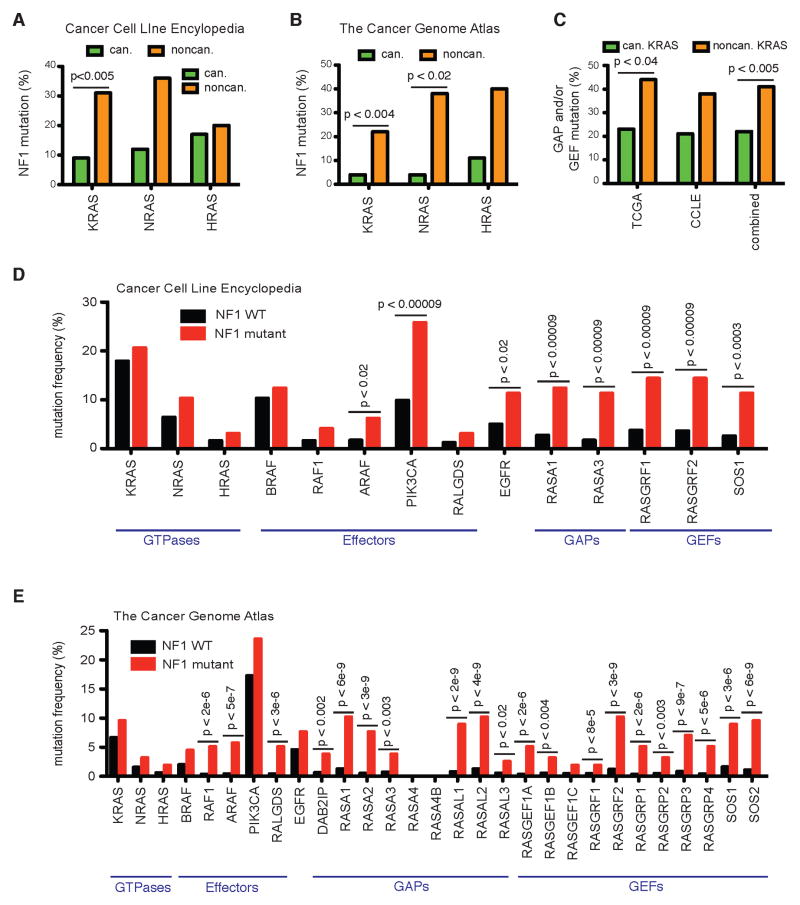
Although co-occurrence of strongly activated, canonical Ras pathway mutations (like KRAS and BRAF mutations) are known to co-occur much less than would be expected from their individual frequencies, much less is known about the co-occurrence patterns of the much less frequently mutated genes in the Ras network. As noncanonical RAS mutations are much less commonly observed than canonical RAS mutations, an analysis of increased co-occurrence requires a large dataset. We first considered the Cancer Cell Line Encyclopedia (CCLE), which includes massively parallel sequencing data for greater than nine hundred cancer cell lines (Barretina et al., 2012). We considered the mutations to KRAS, NRAS, HRAS, and NF1 within this dataset (Table S1). We investigated the frequency with which NF1 mutations co-occurred with noncanonical RAS mutations (i.e. KRAS, NRAS, and HRAS mutations that are not at codon 12, 13, or 61) and with canonical RAS mutations (codon 12, 13, or 61) (Figure 4A). Within the population of cancer cells containing a canonical KRAS mutation, NF1 mutations occurred at a rate similar to their 9% overall rate of occurrence in this data set. In contrast, 31% of cell lines containing noncanonical KRAS mutations also had an NF1 mutation (p<0.005). Within this dataset, we note that BRAF V600E and canonical KRAS mutations tend not to co-occur (p<0.0001).
Figure 4. Co-occurrence of noncanonical Ras mutants with NF1, Ras GAP, and Ras GEF mutants.
(A) Percentage of canonical and noncanonical KRAS, NRAS, and HRAS mutants that co-occur with an NF1 mutation within the CCLE dataset (Barretina et al., 2012) or
(B) within the TCGA dataset (Kandoth et al., 2013).
(C) Percentage of canonical and noncanonical KRAS mutant samples from the CCLE and TCGA that also harbor at least one GAP or GEF mutant.
(D) Mutation frequency for Ras network genes within NF1 mutant and NF1 WT subsets of the CCLE dataset (Barretina et al., 2012).
(E) Mutation frequency for Ras network genes within NF1 mutant and NF1 WT subsets of the TCGA dataset (Kandoth et al., 2013).
N.S., not significant. The p-value for all panels is by Fisher’s exact test.
We next analyzed the recently published dataset from The Cancer Genome Atlas (TCGA), which included sequencing for more than three thousand different human cancer samples (Kandoth et al., 2013). Noncanonical KRAS, NRAS, and HRAS mutations were found to co-occur much more frequently with an NF1 mutation (Figure 4B, Table S2). The increase was statistically significant for KRAS (p<0.004) and NRAS (p<0.02). The rate of NF1 mutation in cancers with a noncanonical KRAS mutation (22%) was higher than the overall 5% rate of mutation for NF1 in the entire data set and higher than the 4% rate of NF1 mutation in cancers with a canonical KRAS mutation. Of note, within this TCGA data set there was no apparent trend for or against co-occurrence of canonical KRAS mutations with the BRAF V600E mutation.
Within both data sets, both canonical and noncanonical KRAS mutations co-occurred with TP53 mutations at similar rates (Figure S4A). Rates of TP53 mutations were not significantly different between NF1 mutant and NF1 WT samples for the CCLE and for the TCGA data (Figure S4B). Many of the NF1 mutations observed in cancer samples have also been observed in patients with neurofibromatosis (Table S3), which demonstrates a clear functional consequence for these NF1 mutations. The overall number of mutations per sample for cancers containing noncanonical KRAS mutations or canonical KRAS mutations were not significantly different (Figure S4C). Co-occurrences with an NF1 mutation were more likely to be observed in samples with a higher number of mutations reported for both canonical (p = 0.025) and noncanonical (p = 0.0047) KRAS mutations (Figure S4D, p-values by the Mann-Whitney test). Overall, the increased co-occurrence of NF1 mutations with noncanonical RAS mutations is consistent with our prediction that a large fraction of less potent RAS mutations cause strong Ras pathway activation if they co-occur with an NF1 mutation.
Noncanonical Ras mutations commonly co-occur with Ras GAP and Ras GEF mutations
We considered noncanonical KRAS mutations for co-occurrence with non-NF1 Ras GAP mutations and with Ras GEF mutations. Within both the CCLE and TCGA data we find increased rates of co-occurrence between Ras GAPs and noncanonical KRAS mutations (Figure S4E). We also find increased rates of co-occurrence between Ras GEFs and noncanonical KRAS mutations in both the CCLE and TCGA data (Figure S4F). Overall, noncanonical KRAS mutations co-occurred with at least one Ras GAP or Ras GEF mutation approximately twice as often as canonical KRAS mutations in both the CCLE and TCGA data (Figure 4C). The trend for an increased rate of GAP or GEF mutations with noncanonical KRAS mutations in the combined data set was highly significant (p<0.005 by Fisher’s Exact test). Overall, the increased co-occurrence between noncanonical RAS mutations and Ras GAP or Ras GEF mutations is consistent with our computational model-based prediction that Ras GAP and Ras GEF mutations can synergize with some weakly activating Ras mutations.
General increase in co-occurrence of Ras pathway mutations in cancer genomes
We hypothesized that the increased sensitivity to perturbation should also result in an increased co-occurrence of NF1 mutations with mutations to the genes of Ras network proteins (GEFs, GAPs, and effectors). Within the CCLE data the frequency of Ras network mutations appeared higher for NF1 mutant cells than for NF1 wild-type cells (Figure 4D). The increased co-occurrence was statistically significant for Ras effectors PIK3CA and ARAF, as well as for upstream EGFR mutations, and for several Ras GAPs and Ras GEFs. Within the larger TCGA data set we also found a trend for increased co-occurrence of NF1 mutations with less commonly mutated Ras pathway genes (Figure 4E). Of note, we found increased rates of co-occurrence between NF1 mutations and noncanonical Ras effector mutations, RAF1, ARAF, and RALGDS, as well as multiple other Ras GAPs and Ras GEFs were quite enriched.
DISCUSSION
It has been argued that better methods are needed for analyzing the relationship between mutations and the perturbed cell signaling networks that drive cancer (Yaffe, 2013). We have presented here the use of mass-action modeling to uncover previously unappreciated relationships between pairs of mutations. Our model was based on the traditional understanding of Ras network biochemistry and the available, quantitative measurements that characterize Ras network biochemistry. Model predictions have been prospectively validated experimentally in mammalian cells. Importantly, insights gained from these computational and experimental studies generated specific hypotheses that we could test in existing, large, cancer genomic data sets. Such an approach should be more generally applicable to other signaling networks.
The finding that the Ras network with a weakly activating mutation is generally more sensitive to subsequent perturbations has important implications in cancer development. The cancer phenotype results from the acquisition of multiple somatic mutations that ultimately result in altered levels of protein expression (including complete loss of expression) and/or the expression of a protein with altered biochemistry (Stratton et al., 2009). The computational model-based predictions and experimental work presented here suggest that the number of biochemical perturbations (and causal genetic aberrations) with a large effect on Ras pathway signal increases in the context of neurofibromin deficiency. This would expand the number of potential “driver genes” that promote cancer in the neurofibromin-deficient context. The net rate of acquiring a cancer-promoting mutation is proportional to both the rate of mutation and the proportion of mutations that offer a selective advantage to the cell. Genetic instability is a common feature of cancer that results in an increased rate of mutations that could promote tumorigenesis (Beckman and Loeb, 2005). Our work suggests an alternative, yet complementary mechanism of “network instability” that results in an increased net rate of acquiring cancer-promoting mutations through the acquisition of a state where a greater proportion of mutations would offer a selective advantage.
The concept of network instability as demonstrated here could also have important implications for the treatment of diseases. Targeted therapies against a cancer oncogene function by inhibiting the signal produced by the oncogene. If the patterns of instability observed for Ras (e.g. Figure S3C) are more general for other signaling proteins, it would suggest that partial reduction of signal may result in a network that remains unstable in the sense that the effects of other mutations are amplified. This instability would enable a wide variety of potential mutations to restore activation of the targeted pathway. This consequence of network instability is consistent with patterns of resistance to targeted therapies. For example, it has been shown that BRAFV600E cells being treated with vemurafenib can become resistant through increased COT expression (Johannessen et al., 2010). COT is MAPKKK, like BRAF, but it has rarely been observed mutated in cancers. The increased importance of a gene mutation that is not commonly associated with the cancer is consistent with the concept of network instability. Of note, one of the commonly found mechanisms for resistance to vemurafenib is loss of neurofibromin function (Maertens et al., 2013; Shalem et al., 2014; Whittaker et al., 2013). Those findings further highlight the role of NF1 mutations as promoters of network instability that amplify the consequences of other Ras pathway mutations.
EXPERIMENTAL PROCEDURES
Mathematical model
The mathematical model of the Ras signaling network has been previously described extensively (Stites and Ravichandran, 2012; Stites et al., 2007) and is elaborated upon in the Supplementary Methods. The model was developed, simulated, and analyzed in MATLAB v7.11.0.584 (R2010b) (MathWorks). Algebraic manipulations and numerical evaluations of the algebraic equations were performed in Mathematica 8 (Wolfram). For NF1-WT conditions, we used the full concentration of Ras GAP identified in our original Ras network model, and we used 50% of this value to model NF1-deficient conditions. Simulations and/or analytical calculations were used to determine levels of RasGTP.
Flow cytometry
Immortalized Nf1+/+, Nf1+/−, and Nf1−/− MEF cells were transiently transfected with plasmid constructs using Lipofectamine (Invitrogen). Cells were starved for 12 hours before cytometry analysis. MEF cell staining for phospho-Erk1/2 (referred to as phoshpo-ERK or pERK) and HA-tagged H-Ras was performed as described previously (Stites et al., 2007). Data were acquired on a FACS CantoII (Becton Dickinson). Cells were gated based on the intensity of the HA signal to define “high” and “low” HA expressing populations. The same intensity gates were used for comparisons between Nf1+/+ and Nf1−/− MEFs, as well as between cells transfected with different Ras constructs. Mean pERK intensity was quantified for high and low HA expression, and the ratio of differences between high and low HA expression,
was used as a measure of the change in sensitivity between Nf1−/− and Nf1+/+ conditions.
Genomic analysis
Mutations for the genes of Ras network proteins were obtained from the CCLE portal (http://www.broadinstitute.org/ccle) and/or from published cancer genome publications (Barretina et al., 2012; Kandoth et al., 2013). Exonic missense and nonsense mutation, and exonic insertions and deletions were considered. The mutations considered from the CCLE and TCGA are listed in Tables S1 and S2. Fisher’s exact test was used to determine p-values for co-occurring mutations. Calculations were performed in R version 2.13.0.
Supplementary Material
Acknowledgments
This work was supported by DOD Award W81XWH-09-1-0087 and by NIGMS award GM55761, both to K.S.R. E.C.S. was partially funded by the TGen Foundation as the Randy Pausch Scholar and also by the following NIH T32 training grants: Cell and Molecular Biology; MSTP; and Molecular Biology of Cancer. K.S.R. is Bill Benter Senior Fellow of the American Asthma Foundation and thanks the support via the Harrison Professorship.
Footnotes
Supplemental information includes Supplemental Methods, four figures, and three tables.
AUTHOR CONTRIBUTIONS
ECS, PTK, and KSR designed the study; ECS performed the computational analyses; PTK performed the flow cytometry experiments; PTK and LBH performed the cell culture work; PTK, LBH, and SFW performed the molecular biology; ECS, PTK, and KSR wrote the manuscript with contributions from LBH and SFW; KSR supervised the research.
References
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckman RA, Loeb LA. Genetic instability in cancer: theory and experiment. Semin Cancer Biol. 2005;15:423–435. doi: 10.1016/j.semcancer.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Benedict KF, Mac Gabhann F, Amanfu RK, Chavali AK, Gianchandani EP, Glaw LS, Oberhardt MA, Thorne BC, Yang JH, Papin JA, et al. Systems analysis of small signaling modules relevant to eight human diseases. Ann Biomed Eng. 2011;39:621–635. doi: 10.1007/s10439-010-0208-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, Miller DK, Wilson PJ, Patch AM, Wu J, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yue H, Ouyang Q. Correlation between oncogenic mutations and parameter sensitivity of the apoptosis pathway model. PLoS Comput Biol. 2014;10:e1003451. doi: 10.1371/journal.pcbi.1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois-Cox S, Genther Williams SM, Reczek EE, Johnson BW, McGillicuddy LT, Johannessen CM, Hollstein PE, MacCollin M, Cichowski K. A negative feedback signaling network underlies oncogene-induced senescence. Cancer Cell. 2006;10:459–472. doi: 10.1016/j.ccr.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the contextual sensitivity of factors that determine cell-to-cell variability in receptor-mediated apoptosis. PLoS Comput Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, Johnson B, Hollstein P, Frederick DT, Cooper ZA, Messiaen L, Bronson RT, McMahon M, Granter S, Flaherty K, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A, Bernards R. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Schoeberl B, Eichler-Jonsson C, Gilles ED, Muller G. Computational modeling of the dynamics of the MAP kinase cascade activated by surface and internalized EGF receptors. Nature biotechnology. 2002;20:370–375. doi: 10.1038/nbt0402-370. [DOI] [PubMed] [Google Scholar]
- Sermon BA, Lowe PN, Strom M, Eccleston JF. The importance of two conserved arginine residues for catalysis by the ras GTPase-activating protein, neurofibromin. J Biol Chem. 1998;273:9480–9485. doi: 10.1074/jbc.273.16.9480. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S, Barkan B, Friedman E, Kloog Y, Stein R. The tumor suppressor neurofibromin confers sensitivity to apoptosis by Ras-dependent and Ras-independent pathways. Cell Death Differ. 2007;14:895–906. doi: 10.1038/sj.cdd.4402057. [DOI] [PubMed] [Google Scholar]
- Stites EC, Ravichandran KS. Mathematical investigation of how oncogenic ras mutants promote ras signaling. Methods Mol Biol. 2012;880:69–85. doi: 10.1007/978-1-61779-833-7_5. [DOI] [PubMed] [Google Scholar]
- Stites EC, Trampont PC, Ma Z, Ravichandran KS. Network analysis of oncogenic Ras activation in cancer. Science. 2007;318:463–467. doi: 10.1126/science.1144642. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas RK, Baker AC, Debiasi RM, Winckler W, Laframboise T, Lin WM, Wang M, Feng W, Zander T, MacConaill L, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- Tiacci E, Trifonov V, Schiavoni G, Holmes A, Kern W, Martelli MP, Pucciarini A, Bigerna B, Pacini R, Wells VA, et al. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, Schadendorf D, Root DE, Garraway LA. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer Discov. 2013;3:350–362. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB. The scientific drunk and the lamppost: massive sequencing efforts in cancer discovery and treatment. Sci Signal. 2013;6:pe13. doi: 10.1126/scisignal.2003684. [DOI] [PubMed] [Google Scholar]
- Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. 2012;13:795–806. doi: 10.1038/nrg3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeang CH, McCormick F, Levine A. Combinatorial patterns of somatic gene mutations in cancer. Faseb J. 2008;22:2605–2622. doi: 10.1096/fj.08-108985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.