SUMMARY
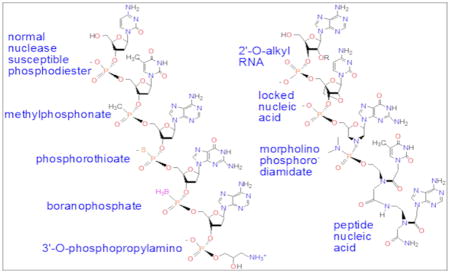
Synthetic, complementary DNA single strands and short interfering RNA double strands have been found to inhibit the expression of animal, plant, and viral genes in cells, animals, and patients, in a dose dependent and sequence specific manner. DNAs and RNAs, however, are readily digested in biological systems. Hence, chemists are obliged to design and synthesize nuclease-resistant analogs of normal DNA (Fig. 1).
Graphical abstract
I. Antisense DNA and RNA Inhibitors of Gene Expression
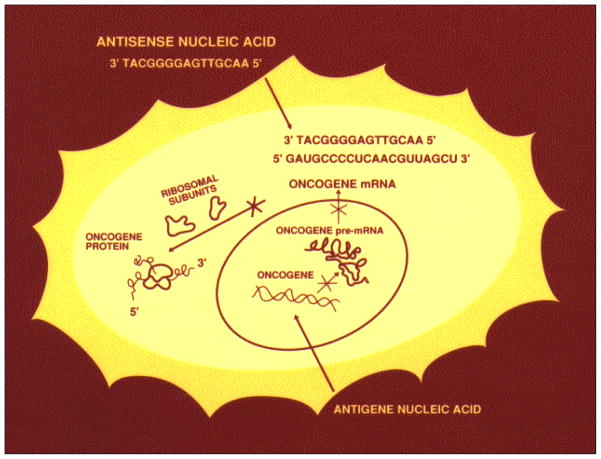
The fundamental concept of using synthetic nucleic acids as drugs against specific DNA or RNA sequences was envisioned in a study reporting synthesis and activity of an alkylating derivative of an RNA dinucleotide [1]. Antisense DNA was first successfully utilized to prevent Rous sarcoma virus mRNA translation in chick embryo fibroblast cells [2]. Antisense DNA binding to an RNA target in the nucleus forms a hybrid double strand that is attacked by nuclear ribonuclease H (RNase H), cleaving the RNA in the middle of the DNA-bound sequence (Fig. 2) [3]. Antisense DNAs have been applied since then to interdict the expression of a wide variety of viral, bacterial, and animal mRNAs, or miRNAs, in cells, animals, and patients [4–10].
Fig. 2.
Antisense DNA interdiction of mRNA translation in cells, from [4] (cover).
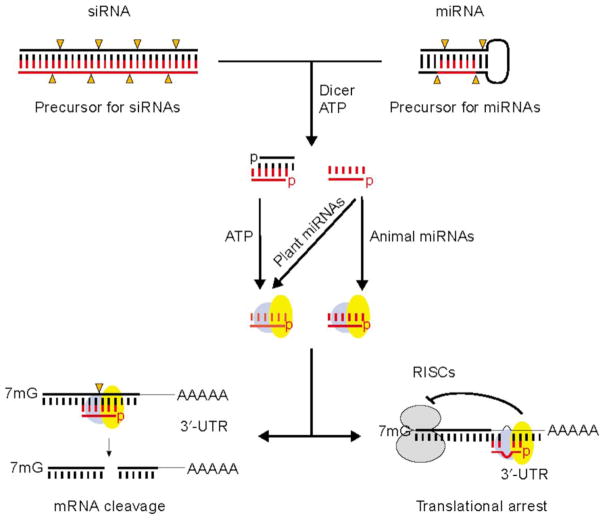
Double stranded RNA (dsRNA) in the form of small interfering RNA (siRNA) or microRNA (miRNA) displays much more potent mRNA silencing than single strand antisense DNA; this mode is called RNA interference (RNAi) [11, 12]. In cells, long dsRNA hairpin loops are cleaved out of transcripts by Drosha in the nucleus, then cleaved by Dicer in the cytoplasm to 20–22 bp duplexes [13]. However, introduction of dsRNA longer than 30 bp into mammalian cells activates a dsRNA-dependent protein kinase, activating the type-1 interferon-response, global shutdown of translation, and ultimately dramatic alteration in cellular metabolism [14]. Chemical synthesis of siRNA duplexes that imitate the Dicer products largely circumvents the non-gene-specific effects [15], yielding gene-specific-silencing in mammalian cells without activating non-specific effects. Processed siRNAs or miRNAs are then trafficked by the dsRNA-binding protein R2D2 to form RNA-induced silencing complexes (RISCs) [11, 13, 16]. RISC is a ribonucleoprotein complex that contains only one of the two strands of the siRNA or miRNA precursor, and proteins of the Argonaut (Ago) family [17]. RISCs then direct mRNA cleavage by siRNA in the middle of the bound mRNA target, or translational inhibition by miRNA (Fig. 3).
Fig. 3.
Actions of small silencing RNAs in cells. (Left) mRNA cleavage specified by a siRNA. Orange arrowhead indicates site of cleavage. (Right) Translational arrest specified by miRNAs or siRNA, from [18]. 7 mG: 7-methyl guanine; AAAAA: poly-adenosine tail; p: 5′ phosphate.
Predictions of mRNA secondary structures suggest the existence of loop and bulge sites that might be particularly susceptible to hybridization by antisense DNAs or siRNAs. Secondary structure prediction of antisense DNA correlated with activity against human MYCC oncogene mRNA [19–22], human HRAS oncogene mRNA [23], and human immunodeficiency virus [24]. With siRNA targeting, sophisticated calculations of mRNA secondary structure, and other sequence considerations, have yielded useful sets of siRNAs [25] that are commercially available. Alternatively, one can walk along the mRNA primary sequence with overlapping DNA or siRNA guide sequences, then screen for the activity of each sequence to determine a lead [9].
II. Nuclease Resistant DNA and RNA Derivatives
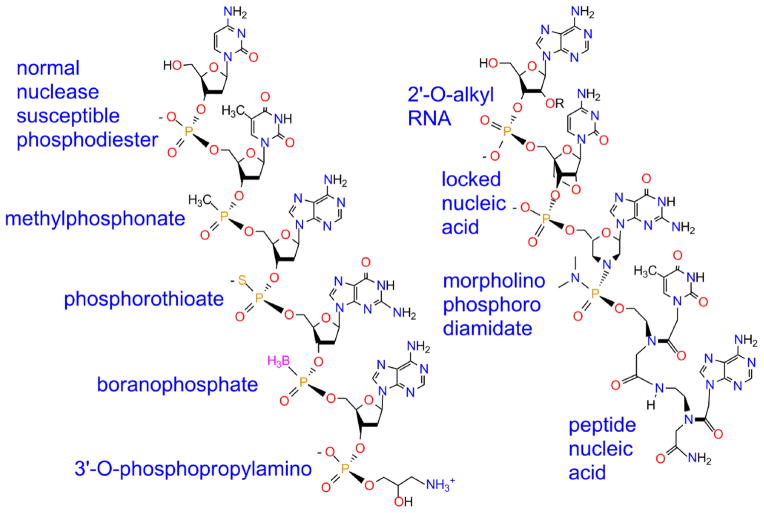
In an effort to circumvent rapid nuclease degradation of normal phosphodiester DNA in naturally occurring biological systems [26], a wide variety of backbone modifications of complementary DNA or RNA (Fig. 1) have been synthesized [27]. This spectrum of DNA or RNA analogs all improve the biological stability, solubility, cellular uptake and/or ease of synthesis [28]. The simplest DNA modification involves blocking the 3′ terminus, as with a propylamino adduct (Fig. 1) [29], to prevent attack by 3′ exonucleases, the predominant extracellular degradative mechanism for single strand DNAs [30].
Fig. 1.
Examples of DNA and RNA backbone derivatives.
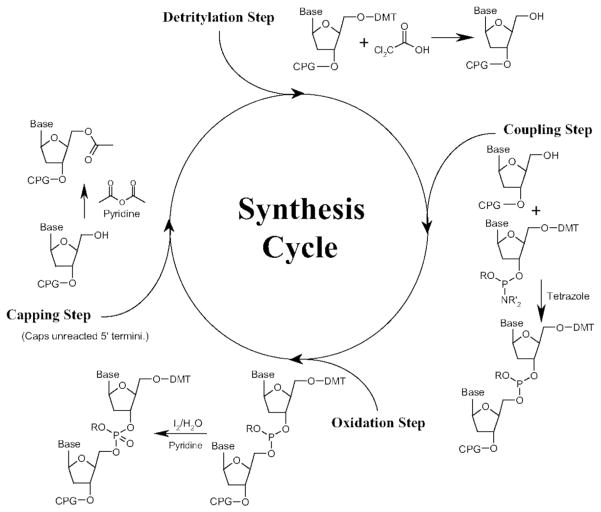
DNA oligomers were first synthesized block by block in solution [31], but are now synthesized by solid phase stepwise synthesis [32] (Fig. 4). The phosphite linkage is then oxidized with iodine in water [32]. Using P(III) phosphoramidite intermediates enables rapid coupling, compared with P(V) phosphotriester intermediates [33].
Fig. 4.
Stepwise solid phase synthesis of short DNAs by phosphoramidite method, from [34].
Phosphodiester modifications to protect the internucleoside linkage include methylphosphonates [35], phosphorothioates [36], or boranophosphates [37] (Fig. 1). Although these modifications increase the in vivo half-life of oligonucleotides, they also weaken hybridization to the RNA target sites due to the creation of chiral phosphorus diastereomers [38].
Nucleases can also be inhibited by replacing deoxyriboses with modified riboses (Fig. 1). Notable examples include 2′-O-methyl ribose [39], 2′-O-methoxyethyl ribose [40], 2′-fluoro ribose [41], 2′-fluoro arabinose [42], and 2′-4′-cyclo-methoxy ribose (locked nucleic acid, LNA) [43]. All those modifications strengthen hybridization, as well as providing nuclease resistance [44]. On the other hand, RNase H susceptibility is lost, limiting the antisense effect to steric blocking.
Nuclease resistance also results from replacing the deoxyribose phosphodiester backbone with morpholino phosphorodiamidates [45]. Morpholino phosphorodiamidates display slightly reduced hybridization properties and reasonable base specificity [46]. Their weaker hybridization properties require long (20–25 nt) sequences for efficacy, but have shown efficacy against a broad spectrum of mRNAs, such as zebrafish embryo mRNAs [47] and Ebola virus mRNAs in mice and guinea pigs [48].
Instead of modifying the backbone, the attachment of the base may be reversed from above the deoxyribose ring to below, changing the natural β-anomer to the α-anomer, which achieves nuclease resistance without loss of base pairing effectiveness [49, 50]. The unusual α-oligodeoxynucleotides have been found capable of antisense inhibition of β-globin mRNA translation, independent of RNase H activity [51].
The most radical modifications are found in peptide nucleic acids (PNA), where both the phosphodiester linkages and sugars are replaced with a peptide-like backbone of (N-2-aminoethyl) glycine units, with the bases directly attached by methylene-carbonyl linkers (Fig. 1) [52]. Compared with other DNA or RNA derivatives, PNAs display the highest Tms for duplexes formed with single-stranded DNA or RNA [53]. Hence, sequences as short at 12 bases allow strong, specific hybridization [53]. Alternating hydroyprolyl/phosphono PNAs provide a more soluble, polyanionic version effective in zebrafish embryos [54].
Each of these structural changes affects not only nuclease susceptibility, but also cellular uptake, cellular trafficking, and RNase H activation [28]. Among the derivatives described, only phosphodiester, phosphorothioate, and boranophosphate DNAs direct RNase H degradation of hybridized RNA.
III. Methylphosphonates
Uncharged methylphosphonate DNAs (Fig. 1) are powerfully resistant to nucleases, enter animal cells, and specifically inhibited translation of rabbit globin mRNA [55], HRAS mRNA [56], and human immunodeficiency virus mRNA [57], along with several other mRNAs [35]. Methylphosphonate DNA 15mers targeted against a predicted loop at the initiation codon of murine c-myc mRNA displayed sequence-specific knockdown of c-Myc protein in the circulating lymphocytes of Eμ-myc transgenic mice [21].
However, relatively high methylphosphonate DNA concentrations are required for significant inhibition. One would have expected that the greater longevity, more efficient cellular uptake, and lack of charge on methylphosphonate DNAs would make them much more effective inhibitors of mRNA translation than normal DNAs. In cell-free extracts, nevertheless, racemic methylphosphonate DNAs are much less effective than normal DNAs [58], where nuclease sensitivity and cellular uptake are irrelevant. The key is that normal DNAs, hybridized to mRNAs, enjoy the advantage of RNase H attack on the mRNA partner in the duplex [59]. Methylphosphonate DNAs, however, are limited to steric blocking of mRNAs to ribosomal translation [60].
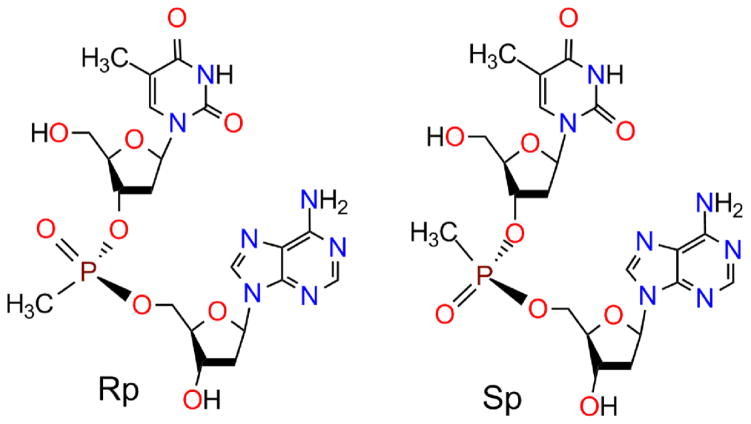
Furthermore, standard coupling of methylphosphonate DNA monomers yields racemic mixtures of Rp and Sp diastereomers at each phosphorus atom [61]. This problem occurs with every asymmetric backbone derivative.
Molecular dynamics [62], nuclear magnetic resonance spectra [63], and crystallography [64] of separated diastereomers revealed that the Sp methyl hinders base stacking by pushing against the deoxyribose ring and base (Fig. 5). The Rp methyl, on the other hand, extends away from the deoxyribose ring and base.
Fig. 5.
Stereo view of pseudoequatorial Rp (left) and pseudoaxial Sp (right) diastereomers of dTmpA.
Thus, all-Rp methylphosphonate DNAs should exhibit stronger hybridization and greater water-solubility than racemic oligomers. Stereospecific coupling by a variety of pentavalent [65–67] and trivalent [68, 69] routes have been reported. dT8 with all-Rp methylphosphonate linkages, except for a central racemic T, displayed a melting temperature of 38°C when hybridized to normal dA15, under physiological conditions where normal dT8:dA15 showed a melting temperature of 13°C, comparable to the racemic methylphosphonate dT8, and the Sp-enriched dT8 revealed a melting temperature of less than 2°C [70]. Similarly, dCCAAACA with all-Rp methylphosphonate linkages hybridized to normal dpTGTTTGGC in a physiological buffer yielded a melting temperature of 30.5°C, compared with 21°C for normal dCCAAACA, or 12.5°C for all-Sp dCCAAACA [71]. Hybridizing to normal RNAs, both all-Rp dCTCTCTCTCTCTCTA and all-Rp dAGAGAGAGAGAGAGT gave melting temperatures 10°C higher than their racemic equivalents, while the all-Sp versions displayed melting temperatures 10°C lower [72]. These results illustrate the power of stereochemistry in the hybridization of DNA derivatives with chiral linkages.
Nevertheless, stereospecific scaleup is daunting. A racemic methylphosphonate DNA linkage at the 3′ end of a 2′-O-methyl RNA-DNA-2′-O-methyl RNA chimera provided 3′-nuclease resistance to an RNase H-active sequence targeted to the 3′-side of HIV Rev response element (RRE) stem-loop IIB RNA [73]. Internal introduction of alternating 5′-O-methylphosphonate linkages in dCAGCTGCTTTTGGGATTCCGTTG hybridized to miR-191 enhanced the melting temperature while maintaining RNase H activity [74]. The development of RNase H-active DNA sequences including methylphosphonate residues that provide nuclease resistance invites translation to animal models.
IV. Phosphorothioates
Phosphorothioate DNAs [36] (Fig. 1) represent a modification with polyanionic character similar to normal phosphodiester DNAs [75], but lower nuclease sensitivity [76]. To create a 3′-5′ phosphorothioate link in DNA during solid phase synthesis, one oxidizes the P(III) phosphite intermediate with a sulfur donor such as tetraethylthiuram disulfide (TETD) [77], instead of iodine and water [32].
Phosphorothioate DNAs also lose hybridization strength due to racemic linkages, but retain RNase H activity [60]. All-Rp dAGATGTTTGAGCTCT hybridized to its RNA complement or a 475 nt RNA including the complement showed higher melting temperatures and greater RNase H cleavage of the RNA than the duplexes with racemic dAGATGTTTGAGCTCT or all-Sp dAGATGTTTGAGCTCT [78]. Due to the R/S naming convention, the S atom in an Rp phosphorothioate linkage is pseudoaxial, while the Sp S atom is pseudoequatorial, the reverse of the methylphosphonate or boranophosphonate situation.
To compensate for the lower activity of racemic phosphorothioate DNAs, sequences of 20–22 nucleotides are often chosen. Phosphorothioate DNAs were first reported to inhibit human immunodeficiency virus mRNA [79–81], influenza virus mRNA [82], protein kinase C mRNA [83], and transferrin receptor mRNA [84] in human cells, but with noticeable non-sequence-specific effects. Phosphorothioate DNA 15mers targeted against the murine c-myc mRNA initiation codon did, however, show sequence-specific knockdown of c-Myc protein and prevention of lymphoma onset in Eμ-myc transgenic mice [85].
Despite their efficacy, however, phosphorothioate DNAs exert off-target effects due to nonspecific protein binding [86], complement binding, inflammation, and inhibition of clotting [87]. The known modes of phosphorothioate toxicity invite further modifications to reduce sulfur content in therapeutic DNAs.
In clinical trials, phosphorothioate DNAs have been administered to humans to knock down mRNAs encoding Bcl-2, PKCα, c-RAF, PKA, and survivin [88], as well as apolipoprotein B-100 [89], CMV IE2 [90], HIV gag [89], ICAM1 [91], IGF1R [92], c-Myb [93], c-Myc [94], p53 [95], H-Ras [96], and VEGF [97]. The US FDA has approved two for patients: the CMV IE2 21mer fomiversen for retinitis [90], and apolipoprotein B-100 20mer mipomersen [89].
V. Boranophosphonates
Boranophosphonate DNAs [98] (Fig. 1) are isosteric with methylphosphonate DNAs, but the BH3 group exhibits a negative charge, isoelectronic with the oxygen of the phosphodiester group [37]. Aside from normal phosphodiester DNA and phosphorothioate DNA, only the boranophosphonate modification also exhibits RNase H activity [99] Further, it increases lipophilicity while maintaining binding to the targeted mRNA and exhibits a relatively low toxicity [37]. A solid phase H-phosphonate synthesis is possible [100], but enzymatic coupling has proved more facile [37]. Synthetic limitations have precluded cellular tests of boranophosphonate DNA activity.
Just as with the methylphosphonate and phosphorothioate modifications, introduction of the BH3 group creates a racemic mixture of chiral centers at the phosphorus, weakening hybridization to RNA. A successful effort to prepare an all-Sp boranophosphonate DNA, with the pseudoaxial borano group pointing towards the helix, as in the Sp methylphosphonate (Fig. 5), yielded an oligomer that retained RNase H activity [101]. Structures of DNA:RNA duplexes that include a single Rp or Sp boranophosphonate were determined by two-dimensional NMR spectroscopy, yielding helical properties midway between A-form or B-form [102]. Specific NOE evidence of base contacts placed the Sp BH3 group in the major groove. In contrast, the pseudoequatorial Rp BH3 group appeared to point away from the DNA, predicting steric clashes with critical RNase H sidechains, suggesting no RNase H activity with an all-Rp boranophosphonate DNA [102].
VI. Other Backbone Modifications
DNA oligomers with triazole internucleotide linkages show nuclease resistance, melting temperatures almost as high as unmodified DNA, and recognition by polymerases [103]. Instead of a triazole, introduction of nucleosyl-3′ amido methyl amino linkages yields a zwitterionic backbone that can hybridize with normal DNA, sensitive to mismatches [104]. For siRNA applications, RNA oligomers with neutral phosphodiester-thioester linkages displayed serum stability, albumin binding, cellular uptake, intracellular hydrolysis to normal RNA phosphodiesters, and RNAi activity in mouse livers [105].
VII. 2′-O-Alkyls
Aside from backbone modifications, the deoxyribose may also be modified to a 2′-O-alkyl ribose (Fig. 1), strengthening hybridization and resisting nuclease attack [44, 106]. The 2′-O-alkyl modifications maintain chirality of the ribose 2′ carbon. The 2′-O-alkyl RNAs differ sufficiently from DNA to preclude RNase H activity, and from RNA to preclude RISC activity. Thus, 2′-O-alkyl RNAs serve as steric inhibitors of RNA translation [44], RNA reverse transcription [107], or RNA splicing [108]. In particular, 2′-O-methyl RNAs successfully induced excision of an exon bearing a nonsense mutation from dystrophin mRNA of the mdx mouse model for Duchenne muscular dystrophy [109], demonstrating a route to therapy for muscular dystrophy in patients.
2′-O-alkyl RNA/phosphorothioate DNA/2′-O-alkyl RNA chimeras demonstrated that a combination of nuclease resistant components and parts that elicit RNase H activity, sometimes called gapmers, improve the potency of antisense DNAs [110, 111]. This chimeric approach has been applied successfully in animal trials targeting apolipoprotein B-100 mRNA in hypercholesterolemia [112], DM1 mRNA in myotonic dystrophy [113], and against huntingtin mRNA in Huntington’s disease [114], and in human trials against apolipoprotein C-III mRNA in severe hypertriglyceridemia and familial chylomicronemia [115], and against transthyretin mRNA in transthyretin-associated polyneuropathy [116].
VIII. Locked Nucleic Acids
Bridging the methyl carbon of a 2′-O-methyl ribose to the 4′ carbon yields a locked nucleic acid (LNA) (Fig. 1) [117]. The methylene bridge below the ribose ring constrains pseudorotation, favoring exclusively the C3′-endo, Northern, A-form conformer [43]. The locked-in A-form elevates 3′ stacking, thermodynamic stability, and thus melting temperatures of LNA:RNA and LNA:DNA duplexes [118].
The slightly unnatural sugar structure imparts resistance to 3′ exonucleases [119], the prominent degradative agent in blood. As with other 2′-O-alkyl RNA forms, LNA lacks RNase H or RISC activity, but LNA/DNA/RNA gapmers have displayed antisense efficacy against mRNAs [120, 121] and microRNAs [122, 123]. Exonuclease protection on the 3′ end of siRNA sense, or passenger, strands increased lifetime in blood, and thus potency [124].
On the other hand, LNA-protected siRNA also elevated off-target transcriptome effects, relative to unmodified siRNA, in mice bearing pancreatic cancer xenografts transformed to express enhanced green fluorescent protein (EGFP) mRNA as a target [125]. This is a natural result of strong LNA binding to RNA. For clinical application, one must shorten the LNA segments, and spike as many DNA residues into the guide and passenger strands as efficacy will allow.
IX. Morpholino Phosphorodiamidates
Greater resistance to nucleases and other degradative enzymes in blood, liver cells, and target cells was achieved by the design and synthesis of morpholino phosphorodiamidate oligomers (Fig. 1) [45]. Replacing the ribose with morpholine, and the phosphodiester with phosphorodiamidate, precluded nuclease recognition, or activity with RNase H or RISC, but enabled high solubility in water despite their lack of charge, due to their strong polarity [45].
Just like methylphosphonate DNA, phosphorothioate DNA, and boranophosphonate DNA, the phosphorodiamidate linkages are synthesized as a racemic mixture. Thus, hybridization with RNA is weaker than with normal DNA, resulting in the requirement for long (20–25 nt) antisense sequences for knockdown efficacy [46].
Lack of a negative charge on the neutral phosphorodiamidate linkages resulted in poor cellular uptake [46], as with methylphosphonate DNA and peptide nucleic acids. Pressure injection of antisense morpholino phosphorodiamidate oligomers targeting mRNAs of interest into the yolk or zygote of embryos of zebrafish (Danio rerio) [45], African clawed frogs (Xenopus sp.), tunicates (Ciona sp.), sea urchins (Strongylocentrotus sp.) and mice has enabled genome-wide, sequence-based, reverse genetic screens in these organisms.
Morpholino phosphorodiamidates are frequently targeted to the start codon or 5′-UTR to block translation [45], to the snRNP or splice-regulatory binding sites of pre-mRNA to redirect splicing [126], or to the guide-strand precursors of miRNA [127] to block their maturation and activity. Numerous knockdown studies have successfully phenocopied a number of mutants [128], but not all morpholino phosphorodiamidate sequences block mRNA translation efficiently, and some nonspecific mistargeting effects have been observed.
MYCC mRNA expression in tumors has been explored as a therapeutic target of morpholino phosphorodiamidate oligomers, using a 20mer version of the phosphodiester [19], methylphosphonate [21], and phosphorothioate [22, 85] 15mers used earlier. The anti-MYCC morpholino phosphorodiamidate ablated c-Myc protein in human PC-3 prostate cancer xenografts, reducing tumor burden by 75–80% [129]. The followup phase I safety trial in healthy volunteers revealed no toxicity or serious adverse events upon intravenous infusion [129]. A second phase I clinical study for pharmacokinetics and tumor bioavailability showed significant concentrations of intact anti-MYCC morpholino phosphorodiamidate oligomer in resected prostate and breast tumor tissues [130]. As before, no serious adverse events were reported [130].
Antisense morpholino phosphorodiamidate oligomers have also shown efficacy against several Ebola virus mRNAs in mice and guinea pigs [48]. Simultaneous knockdown of Ebola VP24, V35, and RNA polymerase L mRNAs protected mice and macaques from lethal infection [131]. 28 doses of the VP24 agent [48] are available. Pairs of morpholino phosphorodiamidate oligomers against Ebola and Marburg viruses were safe and well tolerated at up to 4.5 mg/kg in phase I trials with healthy male and female volunteers [132].
An antisense morpholino phosphorodiamidate oligomer, called eteplirsen, was designed to skip mutant exon 51 in DMD mRNA, which encodes dystrophin. Lack of intact dystrophin causes Duchenne muscular dystrophy, primarily in boys. A placebo-controlled phase IIb trial of eteplirsen at 50 mg/kg/week achieved 52% dystrophin-positive leg muscle fibers, and 67 meters improvement in the 6-minute walking test, in n=4 boys carrying mutant DMD after 24 weeks of treatment, and was well tolerated [133]. Participants (n=160) are currently being recruited for a phase III trial [134].
X. Peptide Nucleic Acids
The most radical modifications are found in peptide nucleic acids (PNA) (Fig. 1) where both the phosphodiester linkages and sugars are replaced with a peptide-like backbone of (N-2-aminoethyl) glycine units, with the bases directly attached by methylene-carbonyl linkers [52]. Compared with other oligonucleotide derivatives, PNAs display the highest Tms for duplexes formed with single-stranded DNA or RNA [53]. The strength and precision of hybridization by PNA 12mers enables single mismatch specificity [135–140].
PNA structure differs so much from DNA, RNA, or peptides, that proteases and nucleases fail to recognize or hydrolyze PNAs [141]. PNA-Tat peptides tested for toxicity in immunocompentent mice were non-immunogenic [142], non-mutagenic, non-clastogenic, and non-teratogenic [143]. These characteristics all favor application of PNAs in diagnosis and therapy.
While unmodified PNAs demonstrate antisense activity in vitro [144], activity in cells, however, requires microinjection of PNAs into the cytosol or nucleus. This stems from poor cellular uptake [145], which was ten times less efficient than uptake of phosphorothioate DNA in a variety of mammalian cells [146]. To alleviate this situation, cellular uptake can be improved by addition of a variety of ligands [147, 148]. Receptor-specific uptake into cells has been demonstrated for PNA-peptide chimeras [136, 138, 148, 149].
For cellular internalization without a receptor ligand, negatively charged alternating phosphonate PNA-trans-4-hydroxy-L-proline PNA analogs (HypNA-pPNA) were synthesized and characterized [150]. The negatively charged HypNA-pPNAs display excellent hybridization properties toward DNA and RNA while preserving the high single mismatch discrimination and nuclease/protease resistance of PNAs [150–153].
Strong, sequence-specific knockdown of chordin, notail, uroD, and bozozok developmental mRNAs in zebrafish embryos was achieved by microinjection of alternating phosphonate PNA monomers and trans-4-hydroxy-L-proline PNA monomers (HypNA-pPNAs) [153]. Even a single mismatch in a PNA abrogated activity, both for the latter four developmental mRNAs, and for the zebrafish orthologs of oncogenes CCND1 [154] and TP53 [155].
XI. Conjugates with targeting agents
Cellular uptake of modified DNAs can be increased by conjugation with a wide variety of ligands. For example, the activity of alkylating DNAs was enhanced by conjugation of hydrophobic, neutral cholesterol moieties at the 3′ end [156]. On the other hand, addition of a hydrophilic, positively charged poly(L-lysine) tail to the 3′ end of a single-stranded DNA elevated its activity against vesicular stomatitis virus in cell culture [157]. The same general improvement in cellular uptake of modified DNAs can be mediated by shorter positively charged cell-penetrating peptides (CPP), such as Tat 48–60 [158], penetratin [159], transportan [160], ApoE 141–150 [161], or just tetralysine [40, 162].
Receptor-mediated endocytosis of modified DNAs can be directed by conjugation of the natural ligands biotin [163] or folate [164]. Furthermore, conjugated peptides that are fragments of known protein ligands have also served as effective ligands for receptor-mediated endocytosis, such as octreotide, an octapeptide mimic of somatostatin [165, 166], JB3, a tridecapeptide mimic of insulin-like growth factor 1 [167], JB9, a tetrapeptide mimic of insulin-like growth factor 1 [135, 148, 168, 169], or DAMGO, an enkephalin analog [149].
Bifunctional conjugation of receptor ligands and imaging moieties to modified DNAs, particularly PNAs, has enabled imaging of particular mRNAs in specific cells that overexpress the target receptor. Coupling a chelator to the N-terminus of PNA 12mers against particular oncogene mRNAs, and a JB9 mimic of insulin-like growth factor 1, allowed 99mTc SPECT imaging of CCND1 and MYCC mRNAs in breast cancer xenografts [136, 137], and KRAS2 mutant mRNA in pancreatic xenografts [170]. The same strategy provided 64Cu PET images of CCND1 mRNA in breast cancer xenografts [139] and KRAS2 mutant mRNA in pancreatic xenografts [138]. Following a similar plan, chelator-PNA-octreotate yielded 111In SPECT images of BCL2 mRNA in lymphoma xenografts [165]. Applying the same bifunctional principle to fluorescence imaging, a DAMGO-PNA-fluorophore construct yielded fluorescent images of MAOA mRNA in neuronal cells that express μ-opoid receptor [149].
XII. Conclusions
Antisense DNA derivatives show great promise for gene-specific knockdown therapy. In the beginning, great skepticism existed about the possibility of achieving antisense DNA inhibition of mRNA translation in cell-free extracts. When this barrier was overcome, the question of cellular uptake arose. It should have been impossible for charged oligomers to enter cells, yet it was found that uptake mechanisms operate in all cells. Degradation of single-stranded DNAs was expected to be extremely rapid within cells, but this did not turn out to be true. Furthermore, single-stranded DNAs taken up by cells penetrated not only the cytoplasm, but also the nucleus, opening up the possibility of interdicting both mRNA processing and even transcription.
On the other hand, the observed rapid degradation of DNA by serum nucleases, and the inefficiency of cellular uptake, underscored the need for some modes of derivatization to permit application of DNA oligomers as practical therapeutics. A wide variety of bulky modifications of the 5′ and 3′ termini have been found to inhibit exonucleases, and to accelerate cellular uptake. Similarly, alteration of even a few of the internucleotide phosphodiester linkages to less susceptible derivatives, particularly at the 3′ end, markedly increases the lifetime of single-stranded DNAs in the presence of serum.
Some examples of backbone and end group modifications have been found effective in animal models, and have proceeded to clinical trials. Two sequences have been approved for clinical use by FDA; several more are in late phase III trials.
Acknowledgments
It is a pleasure to thank my numerous students, postdoctoral fellows, and collaborators who have studied DNA and RNA derivatives in my laboratory over the past 40 years. Our investigations have been supported by grants from the American Cancer Society, American Foundation for AIDS Research, Florida High Technology and Industry Council, Genta, Inc., Heritable Disease Foundation, Leukemia Society, Milligen/Biosearch, Inc., US Army Medical Research and Development Command, US Department of Energy, US National Institutes of Health, and the US National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
XIV. References
- 1.Belikova AM, Zarytova VF, Grineva NI. Synthesis of ribonucleosides and diribonucleoside phosphates containing 2-chloroethylamine and nitrogen mustard residues. Tetrahedron Lett. 1967;37:3557–3562. doi: 10.1016/s0040-4039(01)89794-x. [DOI] [PubMed] [Google Scholar]
- 2.Zamecnik PC, Stephenson ML. Inhibition of Rous sarcoma virus replication and cell transformation by a specific oligodeoxynucleotide. Proc Natl Acad Sci U S A. 1978;75:280–284. doi: 10.1073/pnas.75.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walder RY, Walder JA. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc Natl Acad Sci U S A. 1988;85:5011–5015. doi: 10.1073/pnas.85.14.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wickstrom E. Prospects for Antisense Nucleic Acid Therapy of Cancer and AIDS. Wiley-Liss; New York: 1991. p. 283. [Google Scholar]
- 5.Agrawal S. Antisense Therapeutics. Humana Press; Totowa NJ: 1996. [Google Scholar]
- 6.Wickstrom E. Clinical Trials of Genetic Therapy with Antisense DNA and DNA Vectors. Marcel Dekker; New York: 1998. [Google Scholar]
- 7.Smith JB, Wickstrom E. Preclinical antisense DNA therapy of cancer in mice. Methods in Enzymology. 2000;314:537–580. doi: 10.1016/s0076-6879(99)14128-4. [DOI] [PubMed] [Google Scholar]
- 8.Phillips MI. Antisense therapeutics: a promise waiting to be fulfilled. Methods Mol Med. 2005;106:3–10. [PubMed] [Google Scholar]
- 9.Bennett CF, Swayze EE. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annual review of pharmacology and toxicology. 2010;50:259–293. doi: 10.1146/annurev.pharmtox.010909.105654. [DOI] [PubMed] [Google Scholar]
- 10.Kole R, Krainer AR, Altman S. RNA therapeutics: beyond RNA interference and antisense oligonucleotides. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 12.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 13.Zamore PD. Ancient pathways programmed by small RNAs. Science. 2002;296:1265–1269. doi: 10.1126/science.1072457. [DOI] [PubMed] [Google Scholar]
- 14.Kaufman RJ. Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc Natl Acad Sci U S A. 1999;96:11693–11695. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 16.Tuschl T. Expanding small RNA interference. Nat Biotechnol. 2002;20:446–448. doi: 10.1038/nbt0502-446. [DOI] [PubMed] [Google Scholar]
- 17.Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- 18.Huttenhofer A, Brosius J, Bachellerie JP. RNomics: identification and function of small, non-messenger RNAs. Current opinion in chemical biology. 2002;6:835–843. doi: 10.1016/s1367-5931(02)00397-6. [DOI] [PubMed] [Google Scholar]
- 19.Wickstrom EL, Bacon TA, Gonzalez A, Freeman DL, Lyman GH, Wickstrom E. Human promyelocytic leukemia HL-60 cell proliferation and c-Myc protein expression are inhibited by an antisense pentadecadeoxynucleotide targeted against c-MYC mRNA. Proceedings of the National Academy of Sciences USA. 1988;85:1028–1032. doi: 10.1073/pnas.85.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bacon TA, Wickstrom E. Walking along human c-MYC mRNA with antisense oligodeoxynucleotides: maximum efficacy at the 5′ cap region. Oncogene Research. 1991;6:13–19. [PubMed] [Google Scholar]
- 21.Wickstrom E, Bacon TA, Wickstrom EL. Down-regulation of c-Myc antigen expression in lymphocytes of Em-c-myc transgenic mice treated with anti-c-myc DNA methylphosphonates. Cancer Research. 1992;52:6741–6745. [PubMed] [Google Scholar]
- 22.Smith JB, Wickstrom E. Antisense c-myc and immunostimulatory oligonucleotide inhibition of tumorigenesis in a murine B-cell lymphoma transplant model. Journal of the National Cancer Institute. 1998;90:1146–1154. doi: 10.1093/jnci/90.15.1146. [DOI] [PubMed] [Google Scholar]
- 23.Daaka Y, Wickstrom E. Target dependence of antisense oligodeoxynucleotide inhibition of c-Ha-Ras p21 expression and focus formation in T24-transformed NIH3T3 cells. Oncogene Research. 1990;5:267–275. [PubMed] [Google Scholar]
- 24.Looney DJ, Ojwang JO, Harper ME, Wickstrom E, Wong-Staal F. Inhibition of HIV-1 by deoxyribonucleotides directed against regulatory gene messages and response elements. Journal of Cellular Biochemistry. 1991;15:36. [Google Scholar]
- 25.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 26.Wickstrom E. Oligodeoxynucleotide stability in subcellular extracts and culture media. Journal of Biochemical & Biophysical Methods. 1986;13:97–102. doi: 10.1016/0165-022x(86)90021-7. [DOI] [PubMed] [Google Scholar]
- 27.Knorre DG, Vlassov VV, Zarytova VF, Lebedev AV, Fedorova OS. Design and Targeted Reactions of Oligonucleotide Derivatives. CRC Press; Boca Raton, Florida: 1994. [Google Scholar]
- 28.Wickstrom E. Strategies for administering targeted therapeutic oligodeoxynucleotides. Trends In Biotechnology. 1992;10:281–287. doi: 10.1016/0167-7799(92)90245-q. [DOI] [PubMed] [Google Scholar]
- 29.Fu ZF, Wickstrom E, Jiang M, Corisdeo S, Yang J, Dietzschold B, Koprowski H. Inhibition of rabies virus infection by an oligodeoxynucleotide complementary to rabies virus genomic RNA. Antisense and Nucleic Acid Drug Development. 1996;6:87–93. doi: 10.1089/oli.1.1996.6.87. [DOI] [PubMed] [Google Scholar]
- 30.Zendegui JG, Vasquez KM, Tinsley JH, Kessler DJ, Hogan ME. In vivo stability and kinetics of absorption and disposition of 3′ phosphopropyl amine oligonucleotides. Nucleic Acids Res. 1992;20:307–314. doi: 10.1093/nar/20.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ralph RK, Smith RA, Khorana HG. Studies on polynucleotides. XV. Enzymic degradation. The mode of action of pancreatic deoxyribonuclease on thymidine, deoxycytidine, and deoxyadenosine polynucleotides. Biochemistry. 1962;1:131–137. doi: 10.1021/bi00907a020. [DOI] [PubMed] [Google Scholar]
- 32.Beaucage SL, Caruthers MH. Deoxynucleoside phosphoramidites-A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Letters. 1981;22:1859–1862. [Google Scholar]
- 33.Letsinger RL, Ogilvie KK, Miller PS. Developments in syntheses of oligodeoxyribonucleotides and their organic derivatives. Journal of the American Chemical Society. 1969;91:3360–3365. [Google Scholar]
- 34.Hogrefe RI. A Short History of Oligonucleotide Synthesis. Trilink Biotechnologies; San Diego, CA: 2014. p. 6. [Google Scholar]
- 35.Miller PS, Agris CH, Aurelian L, Blake KR, Murakami A, Reddy MP, Spitz SA, Ts’o PO. Control of ribonucleic acid function by oligonucleoside methylphosphonates. Biochimie. 1985;67:769–776. doi: 10.1016/s0300-9084(85)80166-8. [DOI] [PubMed] [Google Scholar]
- 36.Stec WJ, Zon G. Stereochemical studies of the formation of chiral interneucleotide linkages by phosphoramidite coupling in the synthesis of oligodeoxyribonucleotides. Tetrahedron Letters. 1984;25:5279–5282. [Google Scholar]
- 37.Shaw BR, Sergueev D, He K, Porter K, Summers J, Sergueeva Z, Rait V. Boranophosphate backbone: a mimic of phosphodiesters, phosphorothioates, and methyl phosphonates. Methods Enzymol. 2000;313:226–257. doi: 10.1016/s0076-6879(00)13015-0. [DOI] [PubMed] [Google Scholar]
- 38.Lebedev AV, Wickstrom E. The chirality problem in P-substituted oligonucleotides. In: Trainor G, editor. Perspectives in Drug Discovery and Design. ESCOM Science Publishers; Leiden: 1996. pp. 17–40. [Google Scholar]
- 39.Inoue H, Hayase Y, Imura A, Iwai S, Miura K, Ohtsuka E. Synthesis and Hybridization Studies on 2 Complementary Nona(2′-O-Methyl)Ribonucleotides. Nucleic Acids Research. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sazani P, Astriab-Fischer A, Kole R. Effects of base modifications on antisense properties of 2′-O-methoxyethyl and PNA oligonucleotides. Antisense Nucleic Acid Drug Dev. 2003;13:119–128. doi: 10.1089/108729003768247583. [DOI] [PubMed] [Google Scholar]
- 41.Kawasaki AM, Casper MD, Freier SM, Lesnik EA, Zounes MC, Cummins LL, Gonzalez C, Cook PD. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease-resistant antisense compounds with high affinity and specificity for RNA targets. J Med Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 42.Watts JK, Katolik A, Viladoms J, Damha MJ. Studies on the hydrolytic stability of 2′-fluoroarabinonucleic acid (2′F-ANA) Org Biomol Chem. 2009;7:1904–1910. doi: 10.1039/b900443b. [DOI] [PubMed] [Google Scholar]
- 43.Vester B, Wengel J. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 44.Iribarren AM, Sproat BS, Neuner P, Sulston I, Ryder U, Lamond AI. 2′-O-alkyl oligoribonucleotides as antisense probes. Proc Natl Acad Sci U S A. 1990;87:7747–7751. doi: 10.1073/pnas.87.19.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev. 1997;7:187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- 46.Summerton J, Stein D, Huang SB, Matthews P, Weller D, Partridge M. Morpholino and phosphorothioate antisense oligomers compared in cell-free and in-cell systems. Antisense Nucleic Acid Drug Dev. 1997;7:63–70. doi: 10.1089/oli.1.1997.7.63. [DOI] [PubMed] [Google Scholar]
- 47.Heasman J. Morpholino oligos: making sense of antisense? Dev Biol. 2002;243:209–214. doi: 10.1006/dbio.2001.0565. [DOI] [PubMed] [Google Scholar]
- 48.Iversen PL, Warren TK, Wells JB, Garza NL, Mourich DV, Welch LS, Panchal RG, Bavari S. Discovery and early development of AVI-7537 and AVI-7288 for the treatment of Ebola virus and Marburg virus infections. Viruses. 2012;4:2806–2830. doi: 10.3390/v4112806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morvan F, Rayner B, Imbach JL, Thenet S, Bertrand JR, Paoletti J, Malvy C, Paoletti C. alpha-DNA II. Synthesis of unnatural alpha-anomeric oligodeoxyribonucleotides containing the four usual bases and study of their substrate activities for nucleases. Nucleic Acids Res. 1987;15:3421–3437. doi: 10.1093/nar/15.8.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bacon TA, Morvan F, Rayner B, Imbach JL, Wickstrom E. alpha-Oligodeoxynucleotide stability in serum, subcellular extracts and culture media. Journal of Biochemical & Biophysical Methods. 1988;16:311–318. doi: 10.1016/0165-022x(88)90065-6. [DOI] [PubMed] [Google Scholar]
- 51.Boiziau C, Kurfurst R, Cazenave C, Roig V, Thuong NT, Toulme JJ. Inhibition of translation initiation by antisense oligonucleotides via an RNase-H independent mechanism. Nucleic Acids Res. 1991;19:1113–1119. doi: 10.1093/nar/19.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen PE, Egholm M, Berg RH, Buchardt O. Peptide nucleic acids (PNAs): potential antisense and anti-gene agents. Anticancer Drug Des. 1993;8:53–63. [PubMed] [Google Scholar]
- 53.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 54.Wickstrom E, Choob M, Urtishak KA, Tian X, Sternheim N, Talbot S, Archdeacon J, Efimov VA, Farber SA. Sequence specificity of alternating hydroyprolyl/phosphono peptide nucleic acids against zebrafish embryo mRNAs. J Drug Target. 2004;12:363–372. doi: 10.1080/10611860412331285242. [DOI] [PubMed] [Google Scholar]
- 55.Blake KR, Murakami A, Spitz SA, Glave SA, Reddy MP, Ts’o PO, Miller PS. Hybridization arrest of globin synthesis in rabbit reticulocyte lysates and cells by oligodeoxyribonucleoside methylphosphonates. Biochemistry. 1985;24:6139–6145. doi: 10.1021/bi00343a016. [DOI] [PubMed] [Google Scholar]
- 56.Chang EH, Miller PS, Cushman C, Devadas K, Pirollo KF, Ts’o PO, Yu ZP. Antisense inhibition of ras p21 expression that is sensitive to a point mutation. Biochemistry. 1991;30:8283–8286. doi: 10.1021/bi00098a001. [DOI] [PubMed] [Google Scholar]
- 57.Sarin PS, Agrawal S, Civeira MP, Goodchild J, Ikeuchi T, Zamecnik PC. Inhibition of acquired immunodeficiency syndrome virus by oligodeoxynucleoside methylphosphonates. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:7448–7451. doi: 10.1073/pnas.85.20.7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maher LJ, 3rd, Dolnick BJ. Comparative hybrid arrest by tandem antisense oligodeoxyribonucleotides or oligodeoxyribonucleoside methylphosphonates in a cell-free system. Nucleic Acids Res. 1988;16:3341–3358. doi: 10.1093/nar/16.8.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dagle JM, Weeks DL, Walder JA. Pathways of degradation and mechanism of action of antisense oligonucleotides in Xenopus laevis embryos. Antisense Res Dev. 1991;1:11–20. doi: 10.1089/ard.1991.1.11. [DOI] [PubMed] [Google Scholar]
- 60.Furdon PJ, Dominski Z, Kole R. RNase H cleavage of RNA hybridized to oligonucleotides containing methylphosphonate, phosphorothioate and phosphodiester bonds. Nucleic Acids Res. 1989;17:9193–9204. doi: 10.1093/nar/17.22.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kan LS, Cheng DM, Miller PS, Yano J, Ts’o PO. Proton nuclear magnetic resonance studies on dideoxyribonucleoside methylphosphonates. Biochemistry. 1980;19:2122–2132. doi: 10.1021/bi00551a020. [DOI] [PubMed] [Google Scholar]
- 62.Ferguson DM, Kollman PA. Application of free-energy decomposition to determine the relative stability of R and S oligodeoxyribonucleotide methylphosphonates. Antisense Res Dev. 1991;1:243–254. [PubMed] [Google Scholar]
- 63.Löschner T, Engels JW. Diastereomeric dinucleoside-methylphosphonates: Determination of configuration with the 2-D NMR ROESY technique. Nucleic Acids Research. 1990;18:5083–5088. doi: 10.1093/nar/18.17.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chacko KK, Lindner K, Saenger W, Miller PS. Molecular structure of deoxyadenylyl-3′-methylphosphonate-5′-thymidine dihydrate, (d-ApT x 2H2O), a dinucleoside monophosphate with neutral phosphodiester backbone. An X-ray crystal study. Nucleic Acids Res. 1983;11:2801–2814. doi: 10.1093/nar/11.9.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller PS, Reddy MP, Murakami A, Blake KR, Lin SB, Agris CH. Solid-Phase Syntheses of Oligodeoxyribonucleoside Methylphosphonates. Biochemistry. 1986;25:5092–5097. doi: 10.1021/bi00366a017. [DOI] [PubMed] [Google Scholar]
- 66.Lebedev AV, Rife JP, Seligsohn HW, Wenzinger GR, Wickstrom E. Stereospecific coupling reaction for internucleotide methylphosphono-5′-thioate linkage. Tetrahedron Letters. 1990;31:855–858. [Google Scholar]
- 67.Le Bec C, Wickstrom E. Stereospecific Grignard-Activated Solid Phase Synthesis of DNA Methylphosphonate Dimers. J Org Chem. 1996;61:510–513. doi: 10.1021/jo9517499. [DOI] [PubMed] [Google Scholar]
- 68.Löschner T, Engels J. One pot RP-diastereoselective synthesis of dinucleoside methylphosphonates using methyldichlorophosphine. Tetrahedron Letters. 1989;30:5587–5590. [Google Scholar]
- 69.Lebedev AV, Wenzinger GR, Wickstrom E. A new DMAP catalyzed phosphonamidite coupling reaction for synthesis of oligodeoxynucleoside methylphosphonate derivatives. Tetrahedron Letters. 1990;31:851–854. [Google Scholar]
- 70.Lesnikowski ZJ, Jaworska M, Stec WJ. Octa(thymidine methanephosphonates) of partially defined stereochemistry: synthesis and effect of chirality at phosphorus on binding to pentadecadeoxyriboadenylic acid. Nucleic Acids Res. 1990;18:2109–2115. doi: 10.1093/nar/18.8.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vyazovkina EV, Savchenko EV, Lokhov SG, Engels JW, Wickstrom E, Lebedev AV. Synthesis of specific diastereomers of a DNA methylphosphonate heptamer, d(CpCpApApApCpA), and stability of base pairing with the normal DNA octamer d(TPGPTPTPTPGPGPC) Nucleic Acids Research. 1994;22:2404–2409. doi: 10.1093/nar/22.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reynolds MA, Hogrefe RI, Jaeger JA, Schwartz DA, Riley TA, Marvin WB, Daily WJ, Vaghefi MM, Beck TA, Knowles SK, Klem RE, Arnold LJ., Jr Synthesis and thermodynamics of oligonucleotides containing chirally pure R(P) methylphosphonate linkages. Nucleic Acids Res. 1996;24:4584–4591. doi: 10.1093/nar/24.22.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Prater CE, Saleh AD, Wear MP, Miller PS. Chimeric RNase H-competent oligonucleotides directed to the HIV-1 Rev response element. Bioorg Med Chem. 2007;15:5386–5395. doi: 10.1016/j.bmc.2007.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Šípová H, Špringer T, Rejman D, Šimák O, Petrová M, Novák P, Rosenbergová Š, Páv O, Liboska R, Barvík I, Štěpánek J, Rosenberg I, Homola J. 5′-O-Methylphosphonate nucleic acids—new modified DNAs that increase the Escherichia coli RNase H cleavage rate of hybrid duplexes. Nucleic Acids Research. 2014;42:5378–5389. doi: 10.1093/nar/gku125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eckstein F. Nucleoside phosphorothioates. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 76.Campbell JM, Bacon TA, Wickstrom E. Oligodeoxynucleoside phosphorothioate stability in subcellular extracts, culture media, sera and cerebrospinal fluid. Journal of Biochemical & Biophysical Methods. 1990;20:259–267. doi: 10.1016/0165-022x(90)90084-p. [DOI] [PubMed] [Google Scholar]
- 77.Vu H, Hirschbein BL. Internucleotide phosphite sulfurization with tetraethylthiuram disulfide. Phosphorothioate oligonucleotide synthesis via phosphoramidite chemistry. Tetrahedron Letters. 1991;32:3005–3008. [Google Scholar]
- 78.Koziolkiewicz M, Krakowiak A, Kwinkowski M, Boczkowska M, Stec WJ. Stereodifferentiation--the effect of P chirality of oligo(nucleoside phosphorothioates) on the activity of bacterial RNase H. Nucleic Acids Res. 1995;23:5000–5005. doi: 10.1093/nar/23.24.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Matsukura M, Shinozuka K, Zon G, Mitsuya H, Reitz M, Cohen JS, Broder S. Phosphorothioate analogs of oligodeoxynucleotides: inhibitors of replication and cytopathic effects of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1987;84:7706–7710. doi: 10.1073/pnas.84.21.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Agrawal S, Goodchild J, Civeira MP, Thornton AH, Sarin PS, Zamecnik PC. Oligodeoxynucleoside phosphoramidates and phosphorothioates as inhibitors of human immunodeficiency virus [published erratum appears in Proc Natl Acad Sci U S A 1989 Mar;86(5):1504] Proceedings of the National Academy of Sciences of the United States of America. 1988;85:7079–7083. doi: 10.1073/pnas.85.19.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Agrawal S, Ikeuchi T, Sun D, Sarin PS, Konopka A, Maizel J, Zamecnik PC. Inhibition of human immunodeficiency virus in early infected and chronically infected cells by antisense oligodeoxynucleotides and their phosphorothioate analogues. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:7790–7794. doi: 10.1073/pnas.86.20.7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Leiter JM, Agrawal S, Palese P, Zamecnik PC. Inhibition of influenza virus replication by phosphorothioate oligodeoxynucleotides. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:3430–3434. doi: 10.1073/pnas.87.9.3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Farese RV, Standaert ML, Ishizuka T, Yu B, Hernandez H, Waldron C, Watson J, Farese JP, Cooper DR, Wickstrom E. Antisense DNA downregulates protein kinase C isozymes (beta and alpha) and insulin-stimulated 2-deoxyglucose uptake in rat adipocytes. Antisense Research & Development. 1991;1:35–42. doi: 10.1089/ard.1991.1.35. [DOI] [PubMed] [Google Scholar]
- 84.Ho PT, Ishiguro K, Wickstrom E, Sartorelli AC. Non-sequence-specific inhibition of transferrin receptor expression in HL-60 leukemia cells by phosphorothioate oligodeoxynucleotides. Antisense Research & Development. 1991;1:329–342. doi: 10.1089/ard.1991.1.329. [DOI] [PubMed] [Google Scholar]
- 85.Huang Y, Snyder R, Kligshteyn M, Wickstrom E. Prevention of tumor formation in a mouse model of Burkitt’s lymphoma by 6 weeks of treatment with anti-c-myc DNA phosphorothioate. Molecular Medicine. 1995;1:647–658. [PMC free article] [PubMed] [Google Scholar]
- 86.Agrawal S. Antisense oligonucleotides: towards clinical trials. Trends Biotechnol. 1996;14:376–387. doi: 10.1016/0167-7799(96)10053-6. [DOI] [PubMed] [Google Scholar]
- 87.Iversen PL, Copple BL, Tewary HK. Pharmacology and toxicology of phosphorothioate oligonucleotides in the mouse, rat, monkey and man. Toxicology Letters. 1995;82–83:425–430. doi: 10.1016/0378-4274(95)03572-9. [DOI] [PubMed] [Google Scholar]
- 88.Gleave ME, Monia BP. Antisense therapy for cancer. Nature reviews Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 89.Food and Drug Administration. FDA approves new orphan drug Kynamro to treat inherited cholesterol disorder. In: Liscinsky M, editor. FDA NEWS RELEASE. Food and Drug Administration; Silver Spring, MD: 2013. [Google Scholar]
- 90.Food and Drug Administration. Vitravene (Fomivirsen Sodium Intravitreal Injectable) Injection, Drug Approval Package. Food and Drug Administration; Silver Spring, MD: 1998. [Google Scholar]
- 91.van Deventer SJ, Tami JA, Wedel MK. A randomised, controlled, double blind, escalating dose study of alicaforsen enema in active ulcerative colitis. Gut. 2004;53:1646–1651. doi: 10.1136/gut.2003.036160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andrews DW, Resnicoff M, Flanders AE, Kenyon L, Curtis M, Merli G, Baserga R, Iliakis G, Aiken RD. Results of a pilot study involving the use of an antisense oligodeoxynucleotide directed against the insulin-like growth factor type I receptor in malignant astrocytomas. J Clin Oncol. 2001;19:2189–2200. doi: 10.1200/JCO.2001.19.8.2189. [DOI] [PubMed] [Google Scholar]
- 93.Luger SM, O’Brien SG, Ratajczak J, Ratajczak MZ, Mick R, Stadtmauer EA, Nowell PC, Goldman JM, Gewirtz AM. Oligodeoxynucleotide-mediated inhibition of c-myb gene expression in autografted bone marrow: a pilot study. 2002 doi: 10.1182/blood.v99.4.1150. [DOI] [PubMed] [Google Scholar]
- 94.Gelmon KA, Batist G, Chi K, Sandor K, Webb M, D’Aloisio S, Burge C, Saltman D, Goldie J, Miller W. A dose escalation phase I study of c-MYC antisense in combination with cisplatin in the treatment of solid tumours and lymphomas. AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics; Miami Beach FL. 2001. [Google Scholar]
- 95.Bishop MR, Iversen PL, Bayever E, Sharp JG, Greiner TC, Copple BL, Ruddon R, Zon G, Spinolo J, Arneson M, Armitage JO, Kessinger A. Phase I trial of an antisense oligonucleotide OL(1)p53 in hematologic malignancies [see comments] Journal of Clinical Oncology. 1996;14:1320–1326. doi: 10.1200/JCO.1996.14.4.1320. [DOI] [PubMed] [Google Scholar]
- 96.Adjei AA, Rowinsky EK. Novel anticancer agents in clinical development. Cancer Biol Ther. 2003;2:S5–15. [PubMed] [Google Scholar]
- 97.Levine AM, Tulpule A, Quinn DI, Gorospe G, 3rd, Smith DL, Hornor L, Boswell WD, Espina BM, Groshen SG, Masood R, Gill PS. Phase I study of antisense oligonucleotide against vascular endothelial growth factor: decrease in plasma vascular endothelial growth factor with potential clinical efficacy. J Clin Oncol. 2006;24:1712–1719. doi: 10.1200/JCO.2005.03.4801. [DOI] [PubMed] [Google Scholar]
- 98.Sood A, Shaw BR, Spielvogel BF. Boron-containing nucleic acids. 2. Synthesis of oligodeoxynucleoside boranophosphates. Journal of the American Chemical Society. 1990;112:9000–9001. [Google Scholar]
- 99.Rait VK, Shaw BR. Boranophosphates support the RNase H cleavage of polyribonucleotides. Antisense Nucleic Acid Drug Dev. 1999;9:53–60. doi: 10.1089/oli.1.1999.9.53. [DOI] [PubMed] [Google Scholar]
- 100.Iwamoto N, Oka N, Wada T. Stereocontrolled synthesis of oligodeoxyribonucleoside boranophosphates via stereodefined H-phosphonate intermediates. Nucleic Acids Symposium Series. 2009;53:9–10. doi: 10.1093/nass/nrp005. [DOI] [PubMed] [Google Scholar]
- 101.Li P, Sergueeva ZA, Dobrikov M, Shaw BR. Nucleoside and Oligonucleoside Boranophosphates: Chemistry and Properties. Chemical Reviews. 2007;107:4746–4796. doi: 10.1021/cr050009p. [DOI] [PubMed] [Google Scholar]
- 102.Johnson CN, Spring AM, Sergueev D, Shaw BR, Germann MW. Structural Basis of the RNase H1 Activity on Stereo Regular Borano Phosphonate DNA/RNA Hybrids. Biochemistry. 2011;50:3903–3912. doi: 10.1021/bi200083d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Varizhuk AM, Kaluzhny DN, Novikov RA, Chizhov AO, Smirnov IP, Chuvilin AN, Tatarinova ON, Fisunov GY, Pozmogova GE, Florentiev VL. Synthesis of triazole-linked oligonucleotides with high affinity to DNA complements and an analysis of their compatibility with biosystems. J Org Chem. 2013;78:5964–5969. doi: 10.1021/jo400651k. [DOI] [PubMed] [Google Scholar]
- 104.Schmidtgall B, Spork AP, Wachowius F, Hobartner C, Ducho C. Synthesis and properties of DNA oligonucleotides with a zwitterionic backbone structure. Chemical Communications. 2014;50:13742–13745. doi: 10.1039/c4cc06371f. [DOI] [PubMed] [Google Scholar]
- 105.Meade BR, Gogoi K, Hamil AS, Palm-Apergi C, van den Berg A, Hagopian JC, Springer AD, Eguchi A, Kacsinta AD, Dowdy CF, Presente A, Lonn P, Kaulich M, Yoshioka N, Gros E, Cui XS, Dowdy SF. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol. 2014;32:1256–1261. doi: 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Freier SM, Altmann KH. The ups and downs of nucleic acid duplex stability: structure-stability studies on chemically-modified DNA:RNA duplexes. Nucleic Acids Res. 1997;25:4429–4443. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boiziau C, Larrouy B, Sproat BS, Toulme JJ. Antisense 2′-O-alkyl oligoribonucleotides are efficient inhibitors of reverse transcription. Nucleic Acids Res. 1995;23:64–71. doi: 10.1093/nar/23.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barabino SM, Blencowe BJ, Ryder U, Sproat BS, Lamond AI. Targeted snRNP depletion reveals an additional role for mammalian U1 snRNP in spliceosome assembly. Cell. 1990;63:293–302. doi: 10.1016/0092-8674(90)90162-8. [DOI] [PubMed] [Google Scholar]
- 109.Mann CJ, Honeyman K, Cheng AJ, Ly T, Lloyd F, Fletcher S, Morgan JE, Partridge TA, Wilton SD. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci U S A. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Monia BP, Lesnik EA, Gonzalez C, Lima WF, McGee D, Guinosso CJ, Kawasaki AM, Cook PD, Freier SM. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J Biol Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- 111.Agrawal S, Jiang Z, Zhao Q, Shaw D, Cai Q, Roskey A, Channavajjala L, Saxinger C, Zhang R. Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: in vitro and in vivo studies. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:2620–2625. doi: 10.1073/pnas.94.6.2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu RZ, Kim TW, Hong A, Watanabe TA, Gaus HJ, Geary RS. Cross-Species Pharmacokinetic Comparison from Mouse to Man of a Second-Generation Antisense Oligonucleotide, ISIS 301012, Targeting Human Apolipoprotein B-100. Drug Metabolism and Disposition. 2007;35:460–468. doi: 10.1124/dmd.106.012401. [DOI] [PubMed] [Google Scholar]
- 113.Wheeler TM, Leger AJ, Pandey SK, MacLeod AR, Nakamori M, Cheng SH, Wentworth BM, Bennett CF, Thornton CA. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kordasiewicz Holly B, Stanek Lisa M, Wancewicz Edward V, Mazur C, McAlonis Melissa M, Pytel Kimberly A, Artates Jonathan W, Weiss A, Cheng Seng H, Shihabuddin Lamya S, Hung G, Bennett CÂF, Cleveland Don W. Sustained Therapeutic Reversal of Huntington’s Disease by Transient Repression of Huntingtin Synthesis. Neuron. 2012;74:1031–1044. doi: 10.1016/j.neuron.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Graham MJ, Lee RG, Bell TA, 3rd, Fu W, Mullick AE, Alexander VJ, Singleton W, Viney N, Geary R, Su J, Baker BF, Burkey J, Crooke ST, Crooke RM. Antisense oligonucleotide inhibition of apolipoprotein C-III reduces plasma triglycerides in rodents, nonhuman primates, and humans. Circ Res. 2013;112:1479–1490. doi: 10.1161/CIRCRESAHA.111.300367. [DOI] [PubMed] [Google Scholar]
- 116.Ackermann EJ, Guo S, Booten S, Alvarado L, Benson M, Hughes S, Monia BP. Clinical development of an antisense therapy for the treatment of transthyretin-associated polyneuropathy. Amyloid : the international journal of experimental and clinical investigation : the official journal of the International Society of Amyloidosis. 2012;19(Suppl 1):43–44. doi: 10.3109/13506129.2012.673140. [DOI] [PubMed] [Google Scholar]
- 117.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. LNA (Locked Nucleic Acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 118.Kierzek E, Pasternak A, Pasternak K, Gdaniec Z, Yildirim I, Turner DH, Kierzek R. Contributions of stacking, preorganization, and hydrogen bonding to the thermodynamic stability of duplexes between RNA and 2′-O-methyl RNA with locked nucleic acids. Biochemistry. 2009;48:4377–4387. doi: 10.1021/bi9002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Frieden M, Hansen HF, Koch T. Nuclease stability of LNA oligonucleotides and LNA-DNA chimeras. Nucleosides Nucleotides Nucleic Acids. 2003;22:1041–1043. doi: 10.1081/NCN-120022731. [DOI] [PubMed] [Google Scholar]
- 120.Kurreck J, Wyszko E, Gillen C, Erdmann VA. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 2002;30:1911–1918. doi: 10.1093/nar/30.9.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Frieden M, Orum H. Locked nucleic acid holds promise in the treatment of cancer. Curr Pharm Des. 2008;14:1138–1142. doi: 10.2174/138161208784246234. [DOI] [PubMed] [Google Scholar]
- 122.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 123.Fabani MM, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, Koch T, Wahlestedt C. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mook O, Vreijling J, Wengel SL, Wengel J, Zhou C, Chattopadhyaya J, Baas F, Fluiter K. In vivo efficacy and off-target effects of locked nucleic acid (LNA) and unlocked nucleic acid (UNA) modified siRNA and small internally segmented interfering RNA (sisiRNA) in mice bearing human tumor xenografts. Artif DNA PNA XNA. 2010;1:36–44. doi: 10.4161/adna.1.1.12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: a quantifiable method for gene knockdown. Genesis. 2001;30:154–156. doi: 10.1002/gene.1053. [DOI] [PubMed] [Google Scholar]
- 127.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 129.Iversen PL, Arora V, Acker AJ, Mason DH, Devi GR. Efficacy of antisense morpholino oligomer targeted to c-myc in prostate cancer xenograft murine model and a Phase I safety study in humans. Clin Cancer Res. 2003;9:2510–2519. [PubMed] [Google Scholar]
- 130.Devi GR, Beer TM, Corless CL, Arora V, Weller DL, Iversen PL. In vivo bioavailability and pharmacokinetics of a c-MYC antisense phosphorodiamidate morpholino oligomer, AVI-4126, in solid tumors. Clin Cancer Res. 2005;11:3930–3938. doi: 10.1158/1078-0432.CCR-04-2091. [DOI] [PubMed] [Google Scholar]
- 131.Warfield KL, Swenson DL, Olinger GG, Nichols DK, Pratt WD, Blouch R, Stein DA, Aman MJ, Iversen PL, Bavari S. Gene-specific countermeasures against Ebola virus based on antisense phosphorodiamidate morpholino oligomers. PLoS pathogens. 2006;2:e1. doi: 10.1371/journal.ppat.0020001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heald AE, Iversen PL, Saoud JB, Sazani P, Charleston JS, Axtelle T, Wong M, Smith WB, Vutikullird A, Kaye E. Safety and pharmacokinetic profiles of phosphorodiamidate morpholino oligomers with activity against ebola virus and marburg virus: results of two single-ascending-dose studies. Antimicrob Agents Chemother. 2014;58:6639–6647. doi: 10.1128/AAC.03442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mendell JR, Rodino-Klapac LR, Sahenk Z, Roush K, Bird L, Lowes LP, Alfano L, Gomez AM, Lewis S, Kota J, Malik V, Shontz K, Walker CM, Flanigan KM, Corridore M, Kean JR, Allen HD, Shilling C, Melia KR, Sazani P, Saoud JB, Kaye EM G Eteplirsen Study. Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol. 2013;74:637–647. doi: 10.1002/ana.23982. [DOI] [PubMed] [Google Scholar]
- 134.Anonymous. Confirmatory Study of Eteplirsen in DMD Patients (PROMOVI) National Institutes of Health; 2015. clinicaltrials.gov. [Google Scholar]
- 135.Tian X, Wickstrom E. Continuous solid-phase synthesis and disulfide cyclization of peptide-PNA-peptide chimeras. Organic Letters. 2002;4:4013–4016. doi: 10.1021/ol026676b. [DOI] [PubMed] [Google Scholar]
- 136.Tian X, Aruva MR, Qin W, Zhu W, Duffy KT, Sauter ER, Thakur ML, Wickstrom E. External imaging of CCND1 cancer gene activity in experimental human breast cancer xenografts with 99mTc-peptide-peptide nucleic acid-peptide chimeras. Journal of Nuclear Medicine. 2004;45:2070–2082. [PubMed] [Google Scholar]
- 137.Tian X, Aruva MR, Qin W, Zhu W, Sauter ER, Thakur ML, Wickstrom E. Noninvasive molecular imaging of MYC mRNA expression in human breast cancer xenografts with a [99mTc]peptide-peptide nucleic acid-peptide chimera. Bioconjugate Chemistry. 2005;16:70–79. doi: 10.1021/bc0497923. [DOI] [PubMed] [Google Scholar]
- 138.Chakrabarti A, Zhang K, Aruva MR, Cardi CA, Opitz AW, Wagner NJ, Thakur ML, Wickstrom E. Radiohybridization PET imaging of KRAS G12D mRNA expression in human pancreas cancer xenografts with [64Cu]DO3A-peptide nucleic acid-peptide nanoparticles. Cancer Biology & Therapy. 2007;6:948–956. doi: 10.4161/cbt.6.6.4191. [DOI] [PubMed] [Google Scholar]
- 139.Tian X, Aruva MR, Zhang K, Cardi CA, Thakur ML, Wickstrom E. PET imaging of CCND1 mRNA in human MCF7 estrogen receptor-positive breast cancer xenografts with an oncogene-specific [64Cu]DO3A-PNA-peptide radiohybridization probe. Journal of Nuclear Medicine. 2007;48:1699–1707. doi: 10.2967/jnumed.107.042499. [DOI] [PubMed] [Google Scholar]
- 140.Amirkhanov NV, Zhang K, Aruva MR, Thakur ML, Wickstrom E. Imaging human pancreatic cancer xenografts by targeting mutant KRAS2 mRNA with [(111)In]DOTA(n)-poly(diamidopropanoyl)(m)-KRAS2 PNA-D(Cys-Ser-Lys-Cys) nanoparticles. Bioconjug Chem. 2010;21:731–740. doi: 10.1021/bc900523c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Buchardt O, Egholm M, Berg RH, Nielsen PE. Peptide nucleic acids and their potential applications in biotechnology. Trends Biotechnol. 1993;11:384–386. doi: 10.1016/0167-7799(93)90097-S. [DOI] [PubMed] [Google Scholar]
- 142.Cutrona G, Boffa LC, Mariani MR, Matis S, Damonte G, Millo E, Roncella S, Ferrarini M. The peptide nucleic acid targeted to a regulatory sequence of the translocated c-myc oncogene in Burkitt’s lymphoma lacks immunogenicity: follow-up characterization of PNAEmu-NLS. Oligonucleotides. 2007;17:146–150. doi: 10.1089/oli.2007.9999. [DOI] [PubMed] [Google Scholar]
- 143.Boffa LC, Menichini P, Bolognesi C, Cutrona G, Roncella S, Damonte GL, Millo E, Mariani MR, Matis S, Russo D, Ciliutti P, Ferrarini M. Lack of mutagenicity and clastogenicity of PNAEmu-NLS targeted to a regulatory sequence of the translocated c-myc oncogene in Burkitt’s lymphoma. Mutat Res. 2007;628:129–137. doi: 10.1016/j.mrgentox.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 144.Hanvey JC, Peffer NJ, Bisi JE, Thomson SA, Cadilla R, Josey JA, Ricca DJ, Hassman CF, Bonham MA, Au KG, et al. Antisense and antigene properties of peptide nucleic acids. Science. 1992;258:1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- 145.Bonham MA, Brown S, Boyd AL, Brown PH, Bruckenstein DA, Hanvey JC, Thomson SA, Pipe A, Hassman F, Bisi JE, et al. An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res. 1995;23:1197–1203. doi: 10.1093/nar/23.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gray GD, Basu S, Wickstrom E. Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′-alkylamino oligodeoxynucleotides, 2′-O-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochemical Pharmacology. 1997;53:1465–1476. doi: 10.1016/s0006-2952(97)82440-9. [DOI] [PubMed] [Google Scholar]
- 147.Good L, Nielsen PE. Inhibition of translation and bacterial growth by peptide nucleic acid targeted to ribosomal RNA. Proc Natl Acad Sci U S A. 1998;95:2073–2076. doi: 10.1073/pnas.95.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Basu S, Wickstrom E. Synthesis and characterization of a peptide nucleic acid conjugated to a D-peptide analog of insulin-like growth factor 1 for increased cellular uptake. Bioconjugate Chemistry. 1997;8:481–488. doi: 10.1021/bc9700650. [DOI] [PubMed] [Google Scholar]
- 149.Sethi D, Chen CP, Jing RY, Thakur ML, Wickstrom E. Fluorescent peptide-PNA chimeras for imaging monoamine oxidase A mRNA in neuronal cells. Bioconjugate Chemistry. 2012;23:158–163. doi: 10.1021/bc2004507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Efimov VA, Buryakova AA, Choob MV, Chakhmakhcheva OG. Peptide nucleic acids and their phosphonate analogues: II. Synthesis and physicochemical properties of hybrids containing serine and 4-hydroxyproline residues. Bioorganicheskaya Khimia. 1999;25:611–622. [Google Scholar]
- 151.Efimov VA, Buryakova AA, Chakhmakhcheva OG. Synthesis of polyacrylamides N-substituted with PNA-like oligonucleotide mimics for molecular diagnostic applications. Nucleic Acids Res. 1999;27:4416–4426. doi: 10.1093/nar/27.22.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Phelan D, Hondorp K, Choob M, Efimov V, Fernandez J. Messenger RNA isolation using novel PNA analogues. Nucleosides Nucleotides Nucleic Acids. 2001;20:1107–1111. doi: 10.1081/NCN-100002499. [DOI] [PubMed] [Google Scholar]
- 153.Urtishak KA, Choob M, Tian X, Sternheim N, Talbot WS, Wickstrom E, Farber SA. Targeted gene knockdown in zebrafish using negatively charged peptide nucleic acid mimics. Developmental Dynamics. 2003;228:405–413. doi: 10.1002/dvdy.10394. [DOI] [PubMed] [Google Scholar]
- 154.Duffy KT, McAleer MF, Davidson WR, Kari L, Kari C, Liu CG, Farber SA, Cheng KC, Mest JR, Wickstrom E, Dicker AP, Rodeck U. Coordinate control of cell cycle regulatory genes in zebrafish development tested by cyclin D1 knockdown with morpholino phosphorodiamidates and hydroxyprolyl-phosphono peptide nucleic acids. Nucleic Acids Research. 2005;33:4914–4921. doi: 10.1093/nar/gki799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Duffy KT, Wickstrom E. Zebrafish tp53 knockdown extends the survival of irradiated zebrafish embryos more effectively than the p53 inhibitor pifithrin-a. Cancer Biology & Therapy. 2007;6:675–678. doi: 10.4161/cbt.6.5.3956. [DOI] [PubMed] [Google Scholar]
- 156.Boutorin AS, Gus’kova LV, Ivanova EM, Kobetz ND, Zarytova VF, Ryte AS, Yurchenko LV, Vlassov VV. Synthesis of alkylating oligonucleotide derivatives containing cholesterol or phenazinium residues at their 3′-terminus and their interaction with DNA within mammalian cells. FEBS Lett. 1989;254:129–132. doi: 10.1016/0014-5793(89)81023-3. [DOI] [PubMed] [Google Scholar]
- 157.Lemaitre M, Bayard B, Lebleu B. Specific antiviral activity of a poly(L-lysine)-conjugated oligodeoxyribonucleotide sequence complementary to vesicular stomatitis virus N protein mRNA initiation site. Proc Natl Acad Sci U S A. 1987;84:648–652. doi: 10.1073/pnas.84.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Koppelhus U, Awasthi SK, Zachar V, Holst HU, Ebbesen P, Nielsen PE. Cell-dependent differential cellular uptake of PNA, peptides, and PNA-peptide conjugates. Antisense Nucleic Acid Drug Dev. 2002;12:51–63. doi: 10.1089/108729002760070795. [DOI] [PubMed] [Google Scholar]
- 159.Turner JJ, Ivanova GD, Verbeure B, Williams D, Arzumanov AA, Abes S, Lebleu B, Gait MJ. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Research. 2005;33:6837–6849. doi: 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kilk K, Elmquist A, Saar K, Pooga M, Land T, Bartfai T, Soomets U, Langel U. Targeting of antisense PNA oligomers to human galanin receptor type 1 mRNA. Neuropeptides. 2004;38:316–324. doi: 10.1016/j.npep.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 161.O’Donovan L, Okamoto I, Arzumanov AA, Williams DL, Deuss P, Gait MJ. Parallel Synthesis of Cell-Penetrating Peptide Conjugates of PMO Toward Exon Skipping Enhancement in Duchenne Muscular Dystrophy. Nucleic acid therapeutics. 2014 doi: 10.1089/nat.2014.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sun X, Fang H, Li X, Rossin R, Welch MJ, Taylor JS. MicroPET imaging of MCF-7 tumors in mice via unr mRNA-targeted peptide nucleic acids. Bioconjug Chem. 2005;16:294–305. doi: 10.1021/bc049783u. [DOI] [PubMed] [Google Scholar]
- 163.Pardridge WM, Boado RJ, Kang YS. Vector-mediated delivery of a polyamide (“peptide”) nucleic acid analogue through the blood-brain barrier in vivo. Proc Natl Acad Sci U S A. 1995;92:5592–5596. doi: 10.1073/pnas.92.12.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Matulic-Adamic J, Serebryany V, Haeberli P, Mokler VR, Beigelman L. Synthesis of N-acetyl-D-galactosamine and folic acid conjugated ribozymes. Bioconjug Chem. 2002;13:1071–1078. doi: 10.1021/bc025525q. [DOI] [PubMed] [Google Scholar]
- 165.Jia F, Figueroa SD, Gallazzi F, Balaji BS, Hannink M, Lever SZ, Hoffman TJ, Lewis MR. Molecular imaging of bcl-2 expression in small lymphocytic lymphoma using 111In-labeled PNA-peptide conjugates. J Nucl Med. 2008;49:430–438. doi: 10.2967/jnumed.107.045138. [DOI] [PubMed] [Google Scholar]
- 166.Balkin ER, Jia F, Miller WH, Lewis MR. In vitro evaluation of targeted antisense 177Lu radiotherapy. Anticancer Res. 2011;31:3143–3149. [PubMed] [Google Scholar]
- 167.Basu S, Wickstrom E. Solid phase synthesis of a D-peptide-phosphorothioate oligodeoxynucleotide conjugate from two arms of a polyethylene glycol-polystyrene support. Tetrahedron Letters. 1995;36:4943–4946. [Google Scholar]
- 168.Tian X, Chakrabarti A, Amirkhanov NV, Aruva MR, Zhang K, Mathew B, Cardi C, Qin W, Sauter ER, Thakur ML, Wickstrom E. External imaging of CCND1, MYC, and KRAS oncogene mRNAs with tumor-targeted radionuclide-PNA-peptide chimeras. Annals of the New York Academy of Sciences. 2005;1059:106–144. doi: 10.1196/annals.1339.038. [DOI] [PubMed] [Google Scholar]
- 169.Cesarone G, Edugupanti OP, Chen CP, Wickstrom E. Insulin receptor substrate 1 knockdown in human MCF7 estrogen receptor-positive breast cancer cells by nuclease-resistant IRS1 siRNA conjugated to a disulfide-bridged D-peptide analog of insulin-like growth factor 1. Bioconjugate Chemistry. 2007;18:1831–1840. doi: 10.1021/bc070135v. [DOI] [PubMed] [Google Scholar]
- 170.Chakrabarti A, Aruva MR, Sajankila SP, Thakur ML, Wickstrom E. Synthesis of novel peptide nucleic acid-peptide chimera for non-invasive imaging of cancer. Nucleosides Nucleotides & Nucleic Acids. 2005;24:409–414. doi: 10.1081/ncn-200061865. [DOI] [PubMed] [Google Scholar]