Background: Lentiviral Vpx binding to primate SAMHD1 is under positive selection.
Results: Different Vpx protein variants interact with the N-terminal domain or the C-terminal tail of SAMHD1 in ubiquitin-ligase-substrate receptor complexes in a unique fashion.
Conclusion: Vpx antagonizes SAMHD1 by recruiting it via two separate regions for proteasomal degradation.
Significance: Our findings shed light on how lentivirus virulence factors intersect with host innate immunity.
Keywords: human immunodeficiency virus (HIV), nuclear magnetic resonance (NMR), SAM domain and HD domain-containing protein 1 (SAMHD1), ubiquitylation (ubiquitination), X-ray crystallography, SIV, restriction factor
Abstract
Sterile α motif (SAM) and histidine/aspartate (HD)-containing protein 1 (SAMHD1) restricts human/simian immunodeficiency virus infection in certain cell types and is counteracted by the virulence factor Vpx. Current evidence indicates that Vpx recruits SAMHD1 to the Cullin4-Ring Finger E3 ubiquitin ligase (CRL4) by facilitating an interaction between SAMHD1 and the substrate receptor DDB1- and Cullin4-associated factor 1 (DCAF1), thereby targeting SAMHD1 for proteasome-dependent down-regulation. Host-pathogen co-evolution and positive selection at the interfaces of host-pathogen complexes are associated with sequence divergence and varying functional consequences. Two alternative interaction interfaces are used by SAMHD1 and Vpx: the SAMHD1 N-terminal tail and the adjacent SAM domain or the C-terminal tail proceeding the HD domain are targeted by different Vpx variants in a unique fashion. In contrast, the C-terminal WD40 domain of DCAF1 interfaces similarly with the two above complexes. Comprehensive biochemical and structural biology approaches permitted us to delineate details of clade-specific recognition of SAMHD1 by lentiviral Vpx proteins. We show that not only the SAM domain but also the N-terminal tail engages in the DCAF1-Vpx interaction. Furthermore, we show that changing the single Ser-52 in human SAMHD1 to Phe, the residue found in SAMHD1 of Red-capped monkey and Mandrill, allows it to be recognized by Vpx proteins of simian viruses infecting those primate species, which normally does not target wild type human SAMHD1 for degradation.
Introduction
Critical components in the intricate interplay between viruses and the host machinery are restriction factors and innate immune effectors as well as virulence and permissivity factors (1–4). In SIV4/HIV infection, five restriction factors have been identified to date: Tripartite motif-containing protein 5 α (Trim5α) (5), apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G (APOBEC3G) (6, 7), bone stromal tumor protein 2 (BST2)/Tetherin (8, 9), sterile alpha motif and histidine-aspartic acid (HD)-containing protein 1 (SAMHD1) (10–12), and myxovirus resistance 2 (MX2) proteins (13–15). To counteract their anti-viral activities, HIV and SIV encode virulence factors that target many of these for proteasome-dependent degradation: viral infectivity factor (Vif) targets APOBEC3G, viral protein u (Vpu) targets BST2/Tetherin, and a subset of viral protein r (Vpr) or viral protein X (Vpx) targets SAMHD1 using the same general mechanism (16–19). In particular, the substrate specificity of E3 ubiquitin ligases is altered and exploited either by interaction with the substrate adaptors or substrate receptor components of the ligases. For example, Vpx binds DDB1- and Cullin4-associated factor 1 (DCAF1), a substrate receptor of Cullin4 RING E3 ubiquitin ligase (CRL4) and loads SAMHD1 for polyubiquitination, the first step toward degradation (10, 11, 20).
Host-pathogen co-evolution leads to positive selection, the degree of which can be identified through interspecies sequence alignments (21, 22). Positive selection at the interfaces of host-pathogen complexes is present if the ratio of non-synonymous changes to the number of synonymous changes per site is greater than 1 (23). Thus, interfaces that are used in the interaction between host and pathogen proteins accumulate more frequent amino acid changes than would occur at random. Evolutionary signatures resulting from positive selection permit differentiation of broad acting anti-viral proteins from narrowly effective virus-specific antagonists. In particular, strong positive selection signals at several sites in a single host gene suggest that the gene product is targeted by different viruses at multiple regions. This is noted for SAMHD1, in which two separate regions, the SAM domain and the C terminus, interact with Vpx from distinct HIV/SIV strains. For example, Vpx proteins from the clade that includes HIV-2 and SIVsm (isolated from the sooty mangabey monkey) target a sequence element in the C terminus of SAMHD1, whereas those from the clade that includes SIVrcm (isolated from the Red-capped monkey (RCM)) and SIVmnd (isolated from the Mandrill monkey) interact with the SAM domain, with key residues mapping to different sites of the protein (20, 24–29).
Here, we investigated the positioning and distribution of selection signals using biochemical and structural approaches to elucidate the molecular basis of Vpx/SAMHD1 co-evolution. Our data show that the large N-terminal region of MND SAMHD1, including the SAM domain, interacts with VpxSIVrcm and VpxSIVmnd in the context of DCAF1. Changing several amino acids residues in the N-terminal region of human (Hu) SAMHD1 to those found at equivalent positions in the simian proteins revealed that substitution of only a single residue, Ser-52 to Phe, is sufficient to allow recognition by SIVrcm and SIVmndVpx of and recruitment to the CRL4-DCAF1 E3 ubiquitin ligase, whereas most other changes were invariant for recruitment. Our combined structural and biochemical results provide important molecular insights into how Vpx proteins from different HIV/SIV clades engage different elements in the cellular SAMHD1 protein to overcome the cell's antiviral defense.
Experimental Procedures
Cloning and Plasmid Construction
The N-terminal regions (NTD, residues 1–115) of Hu, RCM, and MND SAMHD1 were cloned into pET41 (EMD Biosciences) with GST and His6 tags at their N termini and C termini, respectively. The NTDs were also cloned into pET21 (EMD Biosciences) with His6 tags at their C termini. The C terminus of RCM SAMHD1 (residues 595–626, CTD) was cloned into a modified pET32 vector containing a tobacco etch virus protease site at the C terminus of His6-tagged thioredoxin fusion. Full-length VpxSIVrcm (residues 1–108) or residues 1–90 (Vpx(ΔC)SIVrcm) from red-capped monkey were cloned into pET43 vectors with a C-terminal His6 tag modified to include a tobacco etch virus protease site between the N-terminal NusA fusion the Vpx proteins, as described previously (20). Full-length VpxSIVmnd (residues 1–99) and residues 1–89 (Vpx(ΔC)SIVmnd) were also cloned into pET43 vector as described above. DCAF1, human and monkey SAMHD1, VpxSIVmnd, and VpxSIVrcm were also cloned, separately, into pCDNA with a HA tag at the N terminus. The cDNAs for monkey SAMHD1, VpxSIVmnd, and VpxSIVrcm were provided by M. Emerman (Fred Hutchison Cancer Center, Seattle, WA) (25, 27). Site-specific mutants of Hu and RCM SAMHD1 were prepared using the QuikChange mutagenesis kit (Agilent Technologies). All other clones were described previously (30).
Protein Expression and Purification
The various SAMHD1 proteins, including the GST fusions and thioredoxin fusion, and NusA-Vpx fusion proteins were expressed in Escherichia coli Rosetta 2 (DE3) grown in Luria-Bertani medium with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside at 18 °C for 16 h. Alternatively, the same Rosetta 2 (DE3) cells were grown, and proteins were expressed in autoinduction medium at 18 °C for 24 h. All proteins were purified as described previously (20, 30). Protein complexes, including DDB1-DCAF1CA (DCAF1 residues 1045–1396), DDB1-DCAF1CA-VpxSIVrcm, and DDB1-DCAF1CA-Vpx(ΔC)SIVrcm were prepared as described previously (20, 27, 30). Mixtures of DCAF1CA, NusA-Vpx(ΔC)SIVmnd, and MND SAMHD1-NTD at molar ratio of 1:2.5:5 were incubated and digested with tobacco etch virus protease. The complex was purified over an 8-ml Mono Q column (GE Healthcare) at pH 7.5 using a 0–1 m NaCl gradient with a buffer containing 25 mm Tris-HCl, 1 mm Tris(2-carboxyethyl)phosphine, 5% glycerol, and 0.02% sodium azide. For isotopic labeling of SAMHD1 NTDs, proteins were expressed in modified minimal media using 15NH4Cl and [U-13C6]glucose or [U-13C6,2H7]glucose as the sole nitrogen and/or carbon sources, respectively. Protein concentrations were determined using theoretical molecular extinction coefficients based on amino acid sequences using the ExPASy Proteomics Server.
Isothermal Titration Calorimetry
Typically, protein complexes, DDB1-DCAF1CA-VpxSIVrcm, were placed in the sample cell of the calorimeter (MicroCal Inc., Northampton, MA), and aliquots of SAMHD1 NTD or CTD were added at 25 °C. Titrations were carried out in 10 mm HEPES buffer, pH 8.0, 1 mm Tris(2-carboxyethyl)phosphine, 100 mm NaCl, and 0.02% sodium azide. All samples were dialyzed against this buffer and concentrated using Amicon concentrators (Millipore) before titrations. Titration of SAMHD1 NTD or CTD into the buffer was used for base-line correction. Data analyses were performed using Origin 7 software (OriginLab Corp).
NMR Spectroscopy
NMR data were collected on Bruker 600-, 700-, 800-, and 900-MHz AVANCE spectrometers. All spectrometers were equipped with a z axis gradient, triple resonance cryoprobes. Experiments were performed at 298 K. Heteronuclear two-dimensional 1H,15N HSQC and TROSY-HSQC and three-dimensional HNCACB, TROSY-HNCACB, TROSY-HN(CO)CACB, and HNCA experiments were performed on a 13C,2H,15N-labeled MND SAMHD1NTD sample to obtain the backbone assignments. For mapping the DCAF1CA-VpxSIVmnd and DCAF1CA-VpxSIVrcm binding sites on SAMHD1 NTD, two-dimensional 1H,15N TROSY-HSQC spectra were recorded for 13C,2H,15N-labeled MND SAMHD1 NTD in the presence of unlabeled DCAF1CA-VpxSIVmnd or DCAF1CA-VpxSIVrcm at 1:0.5, 1:1, and 1:1.5 molar ratios. All data were processed with NMRPipe and analyzed with Collaborative Computing Project for NMR.
X-ray Crystallography
Crystallization of the ternary complex consisting of DCAF1CA, Vpx(ΔC)SIVmnd, and MND SAMHD1 NTD was carried out at room temperature by the sitting drop vapor diffusion method using drops comprising 2 μl of complex (∼2 mg/ml) and 2 μl of reservoir solution (1.6 m NaH2PO4, 0.4 m K2HPO4, 0.1 m sodium phosphate/citrate buffer, pH 4.2). A well diffracting crystal was obtained after ∼2 weeks. X-ray diffraction data were collected on the flash-cooled (−180 °C) crystal at the SER-CAT facility sector 22-BM beamline at the Advance Photon Source at Argonne National Laboratory, Chicago, IL, up to 2.6 Å resolution. All diffraction data were processed, integrated, and scaled using XDS (31). The crystallographic phase was determined by molecular replacement using the coordinates of DCAF1 and VpxSIVsm in the ternary complex of DCAF1-Vpx-SAMHD1 CTD (PDB code 4CC9) (29), and the coordinates of the SWISS-MODEL-generated SAM domain model (32) based on the solution structure of the N terminus (residues 23–118) of SAMHD1 (PDB code 2E8O) (33) as structural probes in PHASER (34). After generation of the initial model, the chain was rebuilt using the program Coot (35). Iterative refinement was carried out by alternating between manual re-building in Coot (35), refinement in BUSTER (36), and refinement in PHENIX (37). The final model, refined in PHENIX, exhibited clear electron density for residues Arg-1073–Gln-1314 and Ser-1328–Gly1390 of DCAF1, Glu-10–Cys-88 of VpxSIVmnd, and Gln-2–Arg-20 and Gly-35–Ser-107 (or Gln-2–Thr-21 and Gly-35–Gln-109) of MND SAMHD1 NTD, with an R-factor of 22.9% and a free R of 28.0%. Note that the electron density for backbone atoms of MND SAMHD1 NTD residues residing in helices α3 and α4 were traceable but that of side chains was weakly observed. Of all the residues, a total of 97% were located in both favored and allowed regions of the Ramachandran plot, respectively, as evaluated by MOLPROBITY (38). A summary of pertinent data collection and refinement is provided in Table 1.
TABLE 1.
Data collection, refinement, and Ramachandran statistics for DCAF1-VPXSIVmnd-MND SAMHD1 NTD
| Complexa | |
|---|---|
| Data collection | |
| Space group | P31 |
| Cell dimensions | |
| a, b, c (Å) | 74.49/74.49/178.18 |
| α, β, γ (°) | 90/90/120 |
| Asymmetric unit content | 2 |
| Wavelength (Å) | 1.00 |
| Resolution (Å) | 36.72-2.60 (2.72-2.60)b |
| Rmerge | 0.201 (1.818)b |
| Rmeas | 0.214 (1.989)b |
| 〈I/σI 〉 | 12.7 (1.4)b |
| Completeness (%) | 98.6 (89.3)b |
| Redundancy | 9.1 (6.2)b |
| Refinement | |
| Resolution (Å) | 36.72-2.60 (2.74-2.60)b |
| No. unique reflections | 33,447 |
| No. test reflectionsc | 1,005 |
| Rwork/Rfree | 0.199/0.228 (0.316/0.395)b |
| No. atoms | |
| Protein | 7,712 |
| Ligand/ion | 2 |
| Water | 114 |
| Average B-factors (Å2) | |
| Protein | 49.19 |
| Ligand/ion | 62.57 |
| Water | 38.76 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 1.428 |
| MolProbity statisticsd | |
| All atom clashscore | 23.54 |
| Rotamer outliers (%) | 2.96 |
| Cβ deviation | 0 |
| Ramachandrand | |
| Favored and allowed region (%) | 97.4 |
| Outliers (%) | 2.6 |
| PDB accession code | 4Z8L |
a Data were obtained from the best diffracting crystal according to crystallization condition (see “Experimental Procedures” for details) and collected at −180 °C. The crystallographic phase problem was solved by molecular replacement using PHASER (see “Experimental Procedures” for details). Refinement was carried out using PHENIX.
b Values in parentheses are for highest resolution shell.
c Random selection.
d Values are obtained from MOLPROBITY.
GST Pulldown Assays
Typically, 100 pmol of GST-tagged SAMHD1 NTD was incubated with 100 pmol of DDB1-DCAF1CA-Vpx in 250 μl of buffer containing 25 mm sodium phosphate, pH 7.5, 150 mm NaCl, 2 mm DTT, 10% glycerol, 0.02% sodium azide, 0.3% Nonidet P-40, and 1% Tween (binding buffer) for 30 min with rocking at 4 °C. Glutathione-Sepharose 4B beads (50 μl of a 50% slurry) were added to the protein mixtures and further agitated for 1 h. The beads were washed 4 times with 1 ml of binding buffer, and bound proteins were eluted with 50 μl of Laemmli sample buffer, separated by 4–20% SDS-PAGE, transferred to PVDF membrane, and detected by immunoblotting with anti-His antibody (Millipore).
Surface Plasmon Resonance (SPR)
SPR experiments were carried out using a BIAcore 2000 instrument (GE Healthcare) as described previously (30).
In Vitro Ubiquitination Assays
Ubiquitination of Hu SAMHD1 wild type (WT) and the S52F mutant proteins were carried out using CRL4-DCAF1CA-Vpx(ΔC)SIVrcm as the E3 ubiquitin ligase in the presence of UBA1, UbcH5, and ubiquitin as reported previously (20). The degree of ubiquitination was assessed by immunoblotting and detection with anti-T7 antibody (EMD Biosciences).
Mammalian Cell Lines, Transfection, and Immunoblotting
HEK293T cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics. Cells were plated on 6-cm plates (4.5 × 106 cells per plate) and transfected with 7 μg of a mixture of pCDNA plasmids encoding SAMHD1, DCAF1CB (residues 1040–1400), and Vpx using LipofectAMINES 2000 (Life Technologies). After 48 h cells were lysed with a buffer containing 25 mm sodium phosphate, 300 mm NaCl, 1 mm EDTA, 0.3% Nonidet P-40, 1% Tween, 5% glycerol, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor mixture (Roche Applied Science). For immunoprecipitation, agarose beads conjugated with anti-FLAG antibodies (Sigma) were added to the cell lysates. Proteins were analyzed as described for GST pulldown assays, with appropriate antibodies, including anti-HA (Covance) and anti-actin (Sigma) antibodies.
Results
VpxSIVrcm Facilitates the Interaction between SAMHD1 and DDB1-DCAF1 via the N-terminal Domain of SAMHD1
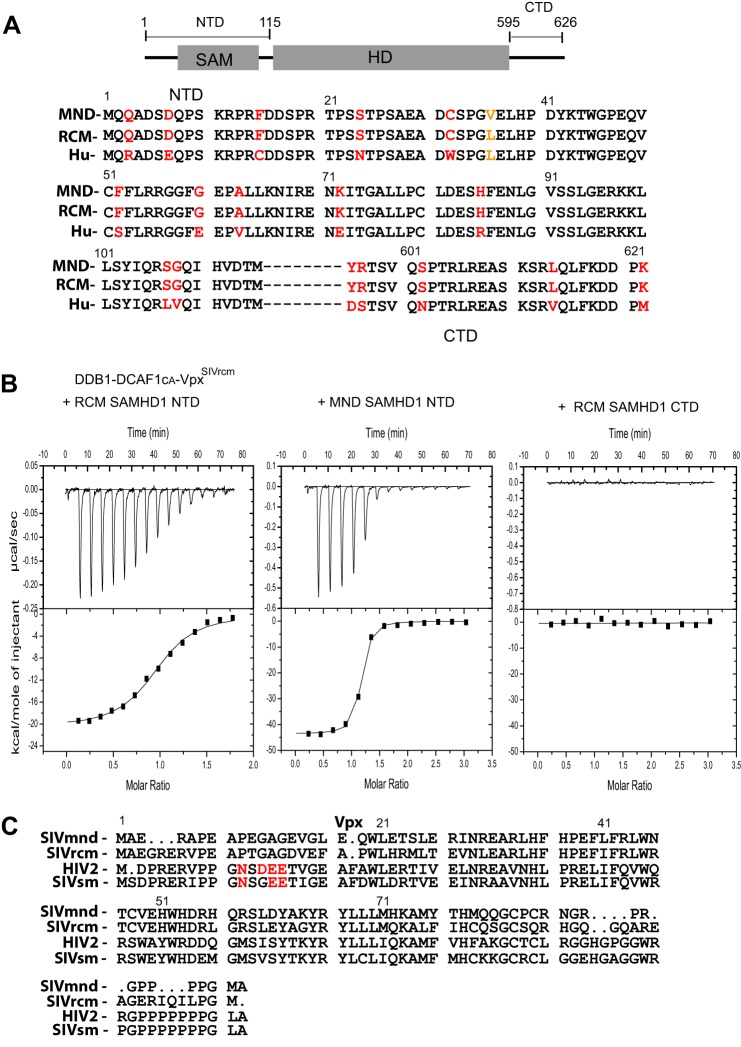
We and others previously reported that VpxSIVsm and VpxHIV-2 preferentially interact with the CTD of SAMHD1, whereas VpxSIVrcm and VpxSIVmnd interact with the NTD of SAMHD1 (Fig. 1A) (20, 25–28). The interactions between these particular Vpx and the respective SAMHD1 proteins require the substrate adaptor and receptor component (DDB1-DCAF1) of CRL4, with the E3 ligase mediating proteasome-dependent down-regulation of SAMHD1. Because RCM and MND SAMHD1, but not Hu SAMHD1, were effectively down-regulated by VpxSIVrcm and VpxSIVmnd (25), we speculated that the NTD of RCM and MND SAMHD1 share a sequence motif that is targeted by VpxSIVrcm and VpxSIVmnd. To identify this motif, we aligned the amino acid sequences of the NTD of Hu, RCM, and MND SAMHD1 proteins (Fig. 1A). Despite being highly homologous, several residues, including amino acids at positions 3, 7, 15, 24, 32, 52, 60, 63, 72, 85, 107, and 108 are different between the monkey and human sequences. The two monkey sequences, on the other hand, contain only one conservative change at position 36 (Leu to Val). Thus, the NTD of RCM SAMHD1 and MND SAMHD1 are essentially identical. We, therefore, hypothesized that DDB1-DCAF1CA-VpxSIVrcm could bind to the NTD of both RCM and MND SAMHD1. To quantitatively determine binding affinities between SAMHD1 and DDB1-DCAF1-VpxSIVrcm, we performed isothermal titration calorimetry (Fig. 1B). As expected, both the NTDs of RCM and MND SAMHD1 bound tightly to DDB1-DCAF1-VpxSIVrcm, with Kd values of 170 ± 22 nm and 22 ± 2 nm, respectively. In contrast, the CTD of RCM SAMHD1, which is identical to that of MND SAMHD1, did not show any measurable interaction with DDB1-DCAF1-VpxSIVrcm. These isothermal titration calorimetry binding data quantitatively confirm our previous results obtained by analytical gel filtration analysis (27). Furthermore, the data suggest that VpxSIVmnd and VpxSIVrcm contain a common sequence motif(s) for recruitment of the NTD of primate SAMHD1 proteins. Indeed, the amino acid sequences of VpxSIVmnd and VpxSIVrcm are highly homologous, and the sequence motif, essential for recruitment of the C terminus of SAMHD1 to DCAF1, is not present in either of the two Vpx proteins (Fig. 1C).
FIGURE 1.
Vpx from SIVrcm (VpxSIVrcm) binds to the N terminus of RCM and MND SAMHD1 and recruits SAMHD1 to DDB1-DCAF1. A, schematic representation of the SAMHD1 structure and amino acid sequence alignment of the NTD (residues 1–115) and the CTD (residues 595–626) of MND, RCM, and Hu SAMHD1. Amino acid residues that vary between monkey and human SAMHD1 are colored in red, and the single residue that differs between MND and RCM SAMHD1 is colored in orange. B, isothermal titration calorimetry traces for RCM SAMHD1 NTD, MND SAMHD1 NTD, and RCM SAMHD1 CTD binding to DDB1-DCAF1CA-VpxSIVrcm. Aliquots of RCM SAMHD1 NTD (30 μm) were added to the DDB1-DCAF1CA-VpxSIVrcm complex (4.6 μm) in a buffer containing 20 mm HEPES, pH 7.4, 150 mm NaCl, and 0.02% sodium azide at 12 °C (left panel). Aliquots of MND SAMHD1 NTD (30 μm) and RCM SAMHD1 CTD (residues 595–626 at 30 μm) were added to the DDB1-DCAF1CA-VpxSIVrcm complex (2 μm) in the same buffer, respectively (middle panel and right panel). The heat of dilution at each concentration and buffer condition was determined and subtracted. The data in the left panel and middle panel were best fitted to a single-site binding isotherm yielding a Kd of 170 ± 22 nm and 22 ± 2 nm, respectively. C, amino sequence alignment of VpxSIVmnd, VpxSIVrcm, VpxHIV-2, and VpxSIVsm. The sequence motif essential for SAMHD1 C-terminal binding is colored in red (29, 30).
Structural Mapping of DCAF1-VpxSIVmnd and DCAF1-VpxSIVrcm Binding to the SAM Domain of SAMDH1
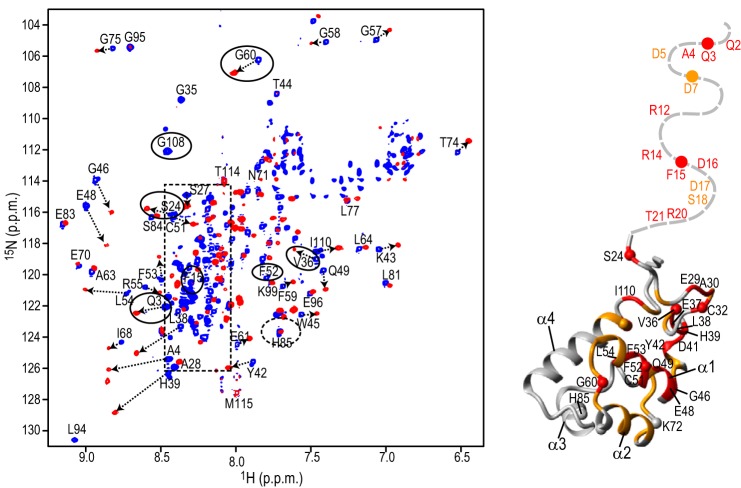
To identify the region of the SAM domain involved in the binding by DCAF1-VpxSIVmnd or DCAF1-VpxSIVrcm, we utilized NMR spectroscopy. Fig. 2 displays a superposition of the 1H,15N TROSY-HSQC spectra of the MND SAMHD1 NTD in the absence (blue spectrum) and presence (red spectrum) of DCAF1-VpxSIVmnd at a 1:1 molar ratio. More than half of the 1H,15N resonances from the SAM domain (55 out of 105 residues) exhibited noticeable chemical shift perturbations (> 0.070 ppm), indicating that binding affects a large portion of the SAM domain. Furthermore, when DCAF1-VpxSIVmnd was added at a submolar amount (∼1:0.5) to the NTD, the 1H,15N resonances from the free and bound states are in slow exchange on the NMR chemical time scale (data not shown). Thus, NMR (Fig. 2) and isothermal titration calorimetry (Fig. 1B) analyses yielded consistent results. Residues whose resonances experience chemical shift changes upon binding (e.g. Gln-3, Ser-24, Val-36, Phe-52, Gly-60, and Gly-108 resonances, marked with solid ovals in Fig. 2) are indicated on the SAM domain structure (amino acids 23–118) of Hu SAMHD1 (PDB code 2E8O), with changes colored in red (>0.15 ppm) and orange (between 0.07 and 0.15 ppm). As can be easily appreciated, binding involves one face of the structure, forming a large extended binding site for DCAF1-VpxSIVmnd (Fig. 2). Several of the affected residues are those that are different between monkey and human SAM (Fig. 1A), some of which reside on α helices α1 and α2. However, not all divergent residues are affected by the DCAF1-VpxSIVmnd binding, in particular those residing on α helices α3 and α4. For example, His-85 in the monkey sequences is replaced by Arg-85 in human (Fig. 1A), but its amide resonance exhibited negligible (<0.035 ppm) chemical shift changes upon complex formation (dashed oval in Fig. 2), demonstrating that His-85 and the surrounding region are not part of the binding interface.
FIGURE 2.
Structural mapping of the DCAF1-VpxSIVmnd interaction onto the SAM domain. A Superposition of the 900-MHz 1H,15N TROSY-HSQC spectra of the MND SAMHD1 NTD (52 μm) in the absence (blue) and presence (red) of 52 μm DCAF1-VpxSIVmnd at 298 K (left panel). Selected assignments are provided for several representative, well separated resonances, labeled on free MND SAMHD1 NTD spectrum. Resonances in DCAF1-VpxSIVmnd-bound state are tentatively assigned and connected to the free resonances by arrows. Resonances of the residues that are different in monkey and human are enclosed by ovals. The random coil region of the spectrum is shown by a dashed rectangle. Structural mapping of residues that exhibit large 1H,15N chemical shift perturbations upon DCAF1-VpxSIVmnd binding is provided (right panel). Residues that experience perturbations >0.15 ppm and between 0.07 and 0.15 ppm are colored in red and orange, respectively, onto the backbone structure of the human NTD region (residues 23–118; PDB code 2E8O). The N-terminal flexible tail (residues Met-1–Pro22) for which no coordinates are available is represented by a gray dashed line.
In our NMR analysis, we also obtained information on the 30-residue N-terminal tail of the SAM domain that is flexible in the solution structure. The associated amide resonances appear as sharp and intense cross-peaks in the central, poorly dispersed, random coil region of the 1H,15N TROSY-HSQC spectrum of the free protein (blue resonances, enclosed by the dashed rectangle in Fig. 2). Interestingly, many of these N-terminal tail resonances exhibited large chemical shift perturbations upon binding DCAF1-VpxSIVmnd, indicating that the N-terminal region is involved in complex formation and changes conformation in the complex.
A similar NMR study, mapping the binding of DCAF1-VpxSIVrcm to MND SAMHD1 NTD, shows that essentially the same regions of the SAM domain are involved in both complexes (data not shown). Note that the MND SAMHD1 and RCM SAMHD1 exhibit identical amino acid sequences for the SAM domain, except for the L36V change.
Crystal Structure of the DCAF1-VpxSIVmnd-SAMHD1 NTD Complex
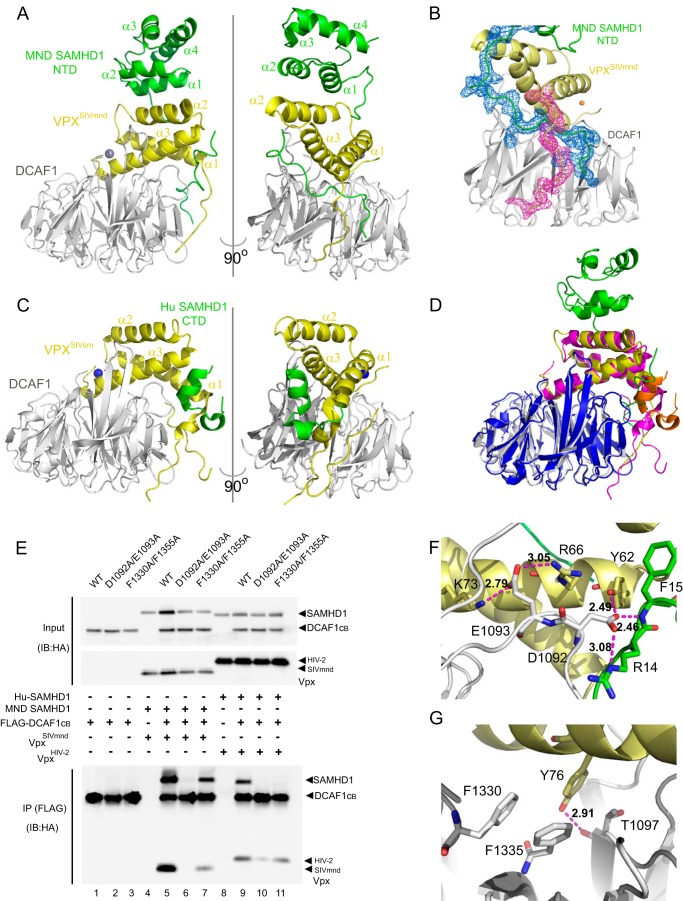
To further delineate the details of SAMHD1 recruitment via its NTD for degradation by VpxSIVmnd, we determined the crystal structure of the DCAF1-VpxSIVmnd-SAMHD1 NTD complex at 2.6 Å resolution. Two ternary complexes are found in the asymmetric unit (data not shown), and given their near identity, only one is described below. Several important features are noted: two critical anchoring points are used by the SAMHD1 NTD to interact with DCAF1 and Vpx, involving the N-terminal tail (residues Gln-2–Arg-20) and the core SAM domain, in particular residues residing in helices α1 and α2. The N-terminal tail up to residue Arg-20 contacts both DCAF1 and Vpx residues and is intertwined between these two proteins (Fig. 3, A and B), whereas residues Gly-35–His-39 and the first two helices (α1, residues Gly-46–Gly-57, and α2, residues Gly-60–Glu-70) of the SAMHD1 NTD interact solely with Vpx (Fig. 3A). This mode of interaction is different from what was observed in the structure of the DCAF1-VpxSIVsm-SAMHD1 CTD (Fig. 3C) (29). On the other hand, despite sequence differences between VpxSIVmnd and VpxSIVsm (Fig. 1C), both Vpx proteins interact with DCAF1 in a very similar way (Fig. 3D) for recruitment of SAMHD1 to DCAF1 via the SAMHD1 NTD and CTD, respectively.
FIGURE 3.
Complex formation between DCAF1-VpxSIVmnd and MND SAMHD1. A, crystal structures of the DCAF1-VpxSIVmnd-MND SAMHD1 NTD complex. All proteins are displayed in ribbon representation. DCAF1, VpxSIVmnd, and MND SAMHD1 NTD are colored in light gray, yellow, and green, respectively. B, electron density for residues Lys-11–Glu-30 of MND SAMHD1 NTD and residues Glu-7–Leu-17 of VpxSIVmnd, contoured at 1σ. C, crystal structures of the DCAF1-VpxSIVsm-Hu SAMHD1 CTD complexes (29). DCAF1, VpxSIVsm, and Hu SAMHD1 CTD are colored in light gray, yellow, and green, respectively. D, superposition of the structures of DCAF1-VpxSIVmnd-MND SAMHD1 NTD and DCAF1-VpxSIVsm-Hu SAMHD1 CTD. In the DCAF1-VpxSIVmnd-MND SAMHD1 NTD complex, the individual components DCAF1, VpxSIVmnd, and MND SAMHD1 NTD are colored in light gray, yellow, and green, respectively. In the DCAF1-VpxSIVsm-Hu SAMHD1 CTD complex, the individual components DCAF1, VpxSIVsm, and Hu SAMHD1 CTD are colored in blue, magenta, and orange, respectively. E, immunoprecipitation (IP) of complexes. FLAG-tagged WT or mutant DCAF1 proteins (residues 1040–1400) were transiently expressed in the presence of MND SAMHD1 and VpxSIVmnd or Hu SAMHD1 and VpxHIV-2 as indicated. Cells were treated with MG132 for 6 h before cell lysates were subjected to immunoprecipitation with anti-FLAG antibody and analyzed by immunoblotting (IB). F and G, hydrogen bonds formed by DCAF1 residues Asp-1092/Glu-1093 (F) and hydrophobic interactions involving DCAF1 residues Phe-1330/Phe-1355 (G) in the DCAF1-VpxSIVmnd-MND SAMHD1 NTD complex. DCAF1, VpxSIVsm, and Hu SAMHD1 NTD are colored in light gray, yellow, and green, respectively.
The above structural observations were validated in co-immunoprecipitation experiments. In particular, VpxSIVmnd co-immunoprecipitated with MND SAMHD1 and WT DCAF1 as well as the F1330A/F1355A mutant DCAF1, whereas VpxHIV-2 failed to immunoprecipitate Hu SAMHD1 with the F1330A/F1355A DCAF1 mutant (Fig. 3E). On the other hand, the D1092A/E1093A DCAF1 mutation abolished both VpxSIVmnd-SAMHD1 and VpxHIV-2-SAMHD1 interactions (Fig. 3E). These co-immunoprecipitation results can be explained based on the DCAF1-VpxSIVmnd interface in the crystal structure. In the structure, the side chain of DCAF1 Asp-1092 is hydrogen-bonded to the backbone amide of Phe-15 and the Nϵ of the Arg-14 side chain of MND SAMHD1 NTD as well as to the hydroxyl group of the Tyr-62 side chain of VpxSIVmnd, whereas the side chain of DCAF1 Glu-1093 hydrogen bonds with the NH2 group of Arg-66 and the Nζ group of Lys-73 of MND SAMHD1 NTD (Fig. 3F). Similar hydrogen-bonding patterns are present in the DCAF1-VpxSIVsm structure (29). Therefore, changing Asp-1092 and Glu-1093 to Ala eliminates these H-bonds (Fig. 3F). Similarly, residues Phe-1330 and Phe-1335 of DCAF1 engage in hydrophobic contacts with Tyr-76 in VpxSIVmnd (Fig. 3F). The same hydrophobic triad can be formed with Phe-80 in VpxHIV-2 (29), and removal of the two aromatic side chains on DCAF1 will abrogate the interaction with VpxHIV-2. This loss may possibly be compensated for in VpxSIVmnd, where the hydroxyl group of the equivalent tyrosine is close to the Oγ hydroxyl group of Thr-1097 in DCAF1 (Fig. 3G). All together, the structural data clearly visualize how the distinct regions of SAMHD1 are targeted by clade-specific Vpx proteins.
A Single Residue Change in the Human SAMHD1 NTD Mediates Proteasome-dependent Down-regulation by VpxSIVrcm
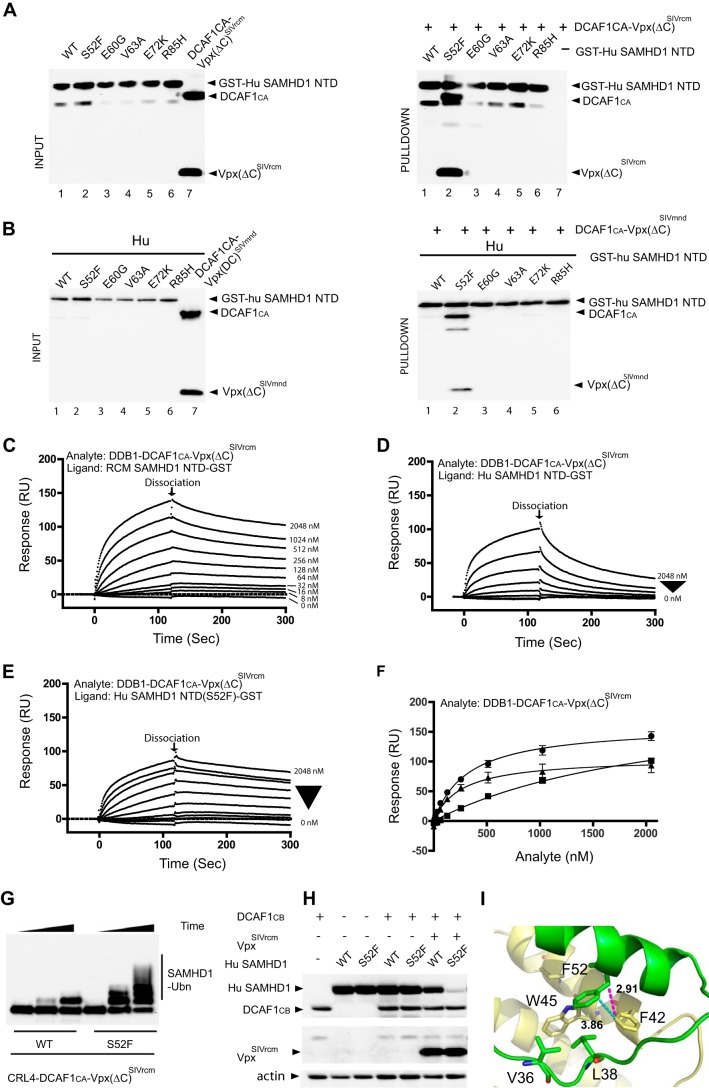
To identify the human SAMHD1 residue(s) that is responsible for the lack of recognition by VpxSIVrcm and VpxSIVmnd, N-terminal SAMHD1 residues that varied between human and monkey were exchanged, and GST pulldown assays with mixtures of GST-Hu SAMHD1 NTD and DDB1-DCAF1-Vpx(ΔC)SIVrcm or DDB1-DCAF1-Vpx(ΔC)SIVmnd were performed (Fig. 4, A and B). Specifically, Hu SAMHD1 NTD amino acids were individually changed to those at the corresponding position in RCM and MND SAMHD1, i.e. S52F, E60G, V63A, E72K, and R85H (Fig. 1A). The S52F change substantially enhanced the binding of GST-Hu SAMHD1 NTD with DDB1-DCAF1-Vpx(ΔC)SIVrcm as well as with DDB1-DCAF1-Vpx(ΔC)SIVmnd (Fig. 4, A and B), whereas none of the other changes produced a similar effect. We verified that the mutation at residue 52 does not affect the structural integrity of the SAM domain by NMR and ascertained that no significant changes in the 1H,15N TROSY-HSQC spectrum of the S52F mutant protein, compared with that of WT human SAMHD1 NTD, occurred (data not shown). Similarly, the 1H,15N TROSY-HSQC spectra of WT and F52S MND were also essentially identical (data not shown).
FIGURE 4.
A single residue in the SAM domain of human SAMHD1 is critical for binding to DDB1-DCAF1-VpxSIVrcm and DDB1-DCAF1-VpxSIVmnd. A and B, GST-pulldown assay of GST-Hu SAMHD1 NTD interacting with DDB1-DCAF1CA-Vpx(ΔC)SIVrcm (A) and DDB1-DCAF1CA-Vpx(ΔC)SIVmnd (B). Hu SAM domain residues that differ from the simian protein were individually changed to those present in MND SAMHD1 and RCM SAMHD1 (see Fig. 1A). Pulldown assays of DDB1-DCAF1CA-Vpx(ΔC)SIVrcm or DDB1-DCAF1CA-Vpx(ΔC)SIVmnd protein complexes with GST-Hu SAMHD1 NTD were immunoblotted with the appropriate antibodies. C–E, real-time kinetic analysis DDB1-DCAF1-VpxSIVrcm binding to NTD. SPR sensorgrams of DDB1-DCAF1CA-Vpx(ΔC)SIVrcm binding to RCM SAMHD1 NTD-GST (C), Hu SAMHD1 NTD-GST (D), and Hu SAMHD1 NTD(S52F)-GST (E). The concentrations of DDB1-DCAF1CA-Vpx(ΔC)SIVrcm were 8, 16, 32, 64, 128, 256, 512, 1024, and 2048 nm. F, binding isotherms of DDB1-DCAF1CA-Vpx(ΔC)SIVrcm binding to RCM SAMHD1 NTD-GST (solid circles, Kd = 322 ± 25 nm), to Hu SAMHD1 NTD-GST (solid squares, Kd = 2260 ± 370 nm), and to Hu SAMHD1 NTD(S52F)-GST (solid triangles, Kd = 256 ± 42 nm) were generated by plotting the response levels with different analyte concentrations at 120 s (marked as dissociation in C, D, and E). RU, response units. Dissociation constants were determined from two independent series of experiments. G, in vitro ubiquitination (Ubn) of full-length Hu SAMHD1 WT or S52F with CRL4-DCAF1CA-Vpx(ΔC)SIVrcm E3 ubiquitin ligase. T7-epitope-tagged SAMHD1 proteins were incubated with E1, E2, ubiquitin, and the appropriate E3 ligases in ubiquitination buffer, as described under “Experimental Procedures.” H, HEK293 cells were transiently co-transfected with DCAF1CB (residues 1040–1400), Hu WT or S52F SAMHD1, and VpxSIVrcm as indicated. The levels of expressed proteins were determined by immunoblotting with appropriate antibodies. I, residue Phe-52 of MND SAMHD1 NTD (green, displayed in stick representation) is involved in hydrophobic contacts with Phe-42 and Trp-45 of VpxSIVmnd (yellow, displayed in stick representation).
Once recruited to DDB1-DCAF1, a substrate adaptor-receptor component of CRL4 E3 ubiquitin ligase, by Vpr or Vpx, SAMHD1 is polyubiquitinated and degraded via the ubiquitin-proteasome system (UPS). HIV-2 Vpx significantly decreases the dissociation rate of SAMHD1 from DDB1-DCAF1, as seen by SPR (30). The increased residence time in the complex may contribute to a more efficient polyubiquitination by the CRL4 E3 ubiquitin ligase. We hypothesized that the interaction between DDB1-DCAF1-Vpx and RCM SAMHD1 may also influence the dissociation rate, and we, therefore, performed similar SPR analyses using the NTD of RCM SAMHD1 and its cognate DDB1-DCAF1-VpxSIVrcm complex (Fig. 4C). VpxSIVrcm substantially decreased the dissociation of the complex from the RCM SAMHD1 NTD, whereas the same virus-host protein complex dissociated from the human SAMHD1 NTD much faster (Fig. 4D). After introduction of the S52F amino acid change into human SAMHD1 NTD, a reduced dissociation rate from the DDB1-DCAF1-Vpx(ΔC)SIVrcm complex was observed (Fig. 4E). Analysis of the overall dissociation curves also suggests that the S52F amino acid change in human SAMHD1 increased the binding affinity to the virus-host protein complexes (Fig. 4F). Because the S52F mutant of human SAMHD1 was able to bind to the DDB1-DCAF1-VpxSIVrcm complex, we also tested whether in vitro and in vivo polyubiquitination by the CRL4-DCAF1-Vpx E3 ubiquitin ligase occurs. As illustrated in Fig. 4, G and H, this is clearly the case. The importance of a Phe residue at position 52 is supported by the structure of the DCAF1-VpxSIVmnd-SAMHD1 NTD complex. As illustrated in Fig. 4I, Phe-52 of MND SAMDH1-NTD engages in hydrophobic contacts with Phe-42 (as well as Trp-45) of VpxSIVmnd (distances <4 Å), and identical contacts should be made with VpxSIVrcm, as no differences in sequence in this region of VpxSIVmnd and VpxSIVrcm exist (Fig. 1B). Therefore, replacement by a Ser, the amino acid present at this position in human SAMHD1, will interfere with this interaction.
Discussion
One of the hallmarks revealing the presence of the evolutionary interplay between virulence factors and host restriction factors is the presence of positive selection at protein-protein interaction interfaces. This is seen by an adaptive mutation(s) in the viral proteins and localized sequence variation of the restriction factors in different host species (for reviews, see Refs. 23, 39, and 40). In particular, the co-evolution of primate SAMHD1 and Vpx of various SIVs resulted in the use of two distinctive protein-protein interfaces: NTD/Vpx and the CTD/Vpx (27). Specifically, Vpx from SIVrcm and SIVmnd preferentially interact with the SAMHD1 NTD, whereas Vpx from SIVsm and HIV-2 requires the SAMHD1 CTD to establish antagonism, and a stable interaction only occurs when a third component, DCAF1, is present (30). The crystal structure of the DCAF1-VpxSIVsm-SAMHD1 CTD elegantly illustrated how Vpx binding to DCAF1 creates a new molecular interface for the SAMHD1 CTD and corroborated previous biochemical findings (29). In our current work, NMR studies reveal that a large portion of the SAMHD1 NTD is affected by binding to the DCAF1-Vpx complex (Fig. 2). The crystal structure of DCAF1-VpxSIVmnd-SAMHD1 NTD complex confirmed the NMR data. The distinct differences in the interaction of Vpx from SIVmnd and SIVsm with SAMHD1 are in stark contrast to the very similar mode observed for binding of these Vpx proteins to DCAF1 (Fig. 3, A, C, and D). Therefore, the distinct modes of SAMHD1 recruitment most likely are the result of sequence variations in Vpx, which have led to creation of unique interfaces for different regions of SAMHD1 upon DCAF1 binding. In addition to structural causes for the distinct engagement of SAMHD1 by the Vpx variants from the two species, dynamics may also play a differentiating role; real-time kinetic analyses by SPR suggest that Vpx substantially decreases the dissociation rate of SAMHD1 from the CRL4-DCAF1 E3 ubiquitin ligase, depending on whether SAMHD1 is recruited via its SAM domain (Fig. 4) or the C terminus (30).
In terms of co-evolution, it appears that the NTDs of primate SAMHD1 proteins exhibit more dramatic signatures of positive selection than the CTDs. Specifically, Gly-46 and Arg-69 are not conserved between African green monkey and MND SAMHD1s and were shown to be essential for interaction with Vpx proteins of their cognate lentiviral strains (25). Interestingly, Phe-52, which is located not far from Gly-46 and Arg-69 in the structure of the SAM domain, is affected by DCAF1-Vpx binding (Fig. 2), and the importance of a Phe residue at position 52 is clearly supported by the structure of the complex (Fig. 4I), which positions it close to the aromatic side chains of Phe-42 and Trp-45 in VpxSIVmnd. Because the human SAMHD1 sequence possesses a serine at the corresponding position, this interaction is not possible but can be restored by the replacement of Ser-52 with Phe-52 in human SAMHD1, which renders it susceptible to down-regulation by VpxSIVrcm (Fig. 4). These critical interface residues appear to be conserved in primate SAMHD1s, including MND and RCM. Consistent with our data, phylogenetic studies of primate SAMHD1 suggested that residue 52, along with residue 15, was under positive selective pressure (28).
In conclusion, our studies presented here, in conjunction with several previous reports (20, 25–28), provide mechanistic and structural insight at the molecular level into how Vpx proteins from different SIV lineages have adapted to sequence variations in their cellular target.
While this manuscript was in preparation a similar structure of DCAF1-VpxSIVmnd-SAMHD1 NTD complex was reported (41).
Author Contributions
J. A. and A. M. G. conceived and coordinated the study. Y. W. designed, performed, and analyzed the experiments shown in Figs. 1 and 4. C. H. B. and I. L. B. designed, performed, and analyzed the experiments shown in Fig. 2., L. M. I. K., J. M., M. D., and G. C. designed research and performed and analyzed the experiments shown in Fig. 3. Y. W., L. M. I. K., I. L. B., J. A., and A. M. G. wrote the paper, and all authors approved its final version.
Acknowledgments
We thank Mike Delk and Doowon Lee for NMR and x-ray technical support and the staff of the SER-CAT facility sector 22 beam line of the Advanced Photon Source at Argonne National Laboratory (Chicago, IL) for help during x-ray data collection. Dr. Teresa Brosenitsch is gratefully acknowledged for editorial help. We also thank Jacek Skowronski for critical reading of the manuscript and communication.
Note Added in Proof
Many of the values in Table 1 in the version of the article that was published as a Paper in Press on June 4, 2015 have been changed because PDB entry 4Z8L was re-refined with better data processing and refinement statistics. The updated Table 1 is now available. These changes do not affect the interpretation of the results or the conclusions.
This work was supported, in whole or in part, by National Institutes of Health Grant P50GM082251 (to A. M. G.). The authors declare that they have no conflicts of interest with the contents of this article.
The atomic coordinates and structure factors (code 4Z8L) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SIV
- simian immunodeficiency virus
- SIVsm
- SIV from the sooty mangabey monkey
- RCM
- red-capped monkey
- SIVrcm
- SIV from Red-capped monkey
- MND
- mandril monkey
- SIVmnd
- SIV from the Mandrill monkey
- SAMHD1
- sterile α motif (SAM) and histidine-aspartate (HD) domain-containing protein 1
- Hu
- human
- Vpx
- viral protein X
- DDB1
- DNA damage-binding protein 1
- DCAF1
- DDB1- and Cullin4-associated factor 1
- SAMHD1 NTD
- residues 1–115
- SAMHD1 CTD
- residues 595–626
- APOBEC3G
- apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G
- BST2
- bone stromal tumor protein 2
- Vpr
- viral protein r
- CRL4
- Cullin4 RING E3 ubiquitin ligase
- HSQC
- heteronuclear single quantum correlation
- TROSY
- transverse relaxation optimized spectroscopy
- SPR
- surface plasmon resonance.
References
- 1. Harris R. S., Hultquist J. F., Evans D. T. (2012) The restriction factors of human immunodeficiency virus. J. Biol. Chem. 287, 40875–40883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Luban J. (2012) Innate immune sensing of HIV-1 by dendritic cells. Cell Host Microbe 12, 408–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mashiba M., Collins K. L. (2013) Molecular mechanisms of HIV immune evasion of the innate immune response in myeloid cells. Viruses 5, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng Y. H., Jeang K. T., Tokunaga K. (2012) Host restriction factors in retroviral infection: promises in virus-host interaction. Retrovirology 9, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., Sodroski J. (2004) The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 [DOI] [PubMed] [Google Scholar]
- 6. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418, 646–650 [DOI] [PubMed] [Google Scholar]
- 7. Harris R. S., Bishop K. N., Sheehy A. M., Craig H. M., Petersen-Mahrt S. K., Watt I. N., Neuberger M. S., Malim M. H. (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113, 803–809 [DOI] [PubMed] [Google Scholar]
- 8. Van Damme N., Goff D., Katsura C., Jorgenson R. L., Mitchell R., Johnson M. C., Stephens E. B., Guatelli J. (2008) The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3, 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neil S. J., Zang T., Bieniasz P. D. (2008) Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451, 425–430 [DOI] [PubMed] [Google Scholar]
- 10. Laguette N., Sobhian B., Casartelli N., Ringeard M., Chable-Bessia C., Ségéral E., Yatim A., Emiliani S., Schwartz O., Benkirane M. (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474, 654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hrecka K., Hao C., Gierszewska M., Swanson S. K., Kesik-Brodacka M., Srivastava S., Florens L., Washburn M. P., Skowronski J. (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474, 658–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Z., Pan Q., Ding S., Qian J., Xu F., Zhou J., Cen S., Guo F., Liang C. (2013) The Interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14, 398–410 [DOI] [PubMed] [Google Scholar]
- 14. Kane M., Yadav S. S., Bitzegeio J., Kutluay S. B., Zang T., Wilson S. J., Schoggins J. W., Rice C. M., Yamashita M., Hatziioannou T., Bieniasz P. D. (2013) MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502, 563–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goujon C., Moncorgé O., Bauby H., Doyle T., Ward C. C., Schaller T., Hué S., Barclay W. S., Schulz R., Malim M. H. (2013) Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502, 559–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Strebel K. (2013) HIV accessory proteins versus host restriction factors. Curr. Opin. Virol. 3, 692–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Romani B., Cohen E. A. (2012) Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase. Curr. Opin. Virol. 2, 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng Y., Baig T. T., Love R. P., Chelico L. (2014) Suppression of APOBEC3-mediated restriction of HIV-1 by Vif. Front. Microbiol. 5, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blanchet F. P., Mitchell J. P., Piguet V. (2012) β-TrCP dependency of HIV-1 Vpu-induced downregulation of CD4 and BST-2/tetherin. Curr. HIV Res. 10, 307–314 [DOI] [PubMed] [Google Scholar]
- 20. Ahn J., Hao C., Yan J., DeLucia M., Mehrens J., Wang C., Gronenborn A. M., Skowronski J. (2012) HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J. Biol. Chem. 287, 12550–12558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sawyer S. L., Wu L. I., Emerman M., Malik H. S. (2005) Positive selection of primate TRIM5α identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U.S.A. 102, 2832–2837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sawyer S. L., Emerman M., Malik H. S. (2004) Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2, E275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emerman M., Malik H. S. (2010) Paleovirology: modern consequences of ancient viruses. PLoS Biol. 8, e1000301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang C., de Silva S., Wang J. H., Wu L. (2012) Co-evolution of primate SAMHD1 and lentivirus Vpx leads to the loss of the vpx gene in HIV-1 ancestor. PLoS ONE 7, e37477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lim E. S., Fregoso O. I., McCoy C. O., Matsen F. A., Malik H. S., Emerman M. (2012) The ability of primate lentiviruses to degrade the monocyte restriction factor SAMHD1 preceded the birth of the viral accessory protein Vpx. Cell Host Microbe 11, 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Laguette N., Rahm N., Sobhian B., Chable-Bessia C., Münch J., Snoeck J., Sauter D., Switzer W. M., Heneine W., Kirchhoff F., Delsuc F., Telenti A., Benkirane M. (2012) Evolutionary and functional analyses of the interaction between the myeloid restriction factor SAMHD1 and the lentiviral Vpx protein. Cell Host Microbe 11, 205–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fregoso O. I., Ahn J., Wang C., Mehrens J., Skowronski J., Emerman M. (2013) Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition and degradation of the restriction factor SAMHD1. PLoS Pathog. 9, e1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wei W., Guo H., Gao Q., Markham R., Yu X. F. (2014) Variation of two primate lineage-specific residues in human SAMHD1 confers resistance to N terminus-targeted SIV Vpx proteins. J. Virol. 88, 583–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schwefel D., Groom H. C., Boucherit V. C., Christodoulou E., Walker P. A., Stoye J. P., Bishop K. N., Taylor I. A. (2014) Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505, 234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. DeLucia M., Mehrens J., Wu Y., Ahn J. (2013) HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 catalysis prior to proteasome-dependent degradation. J. Biol. Chem. 288, 19116–19126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 33. Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000) The Protein Data Bank. Nucleic Acids Res. 28, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 36. Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Sharff A., Smart O. S., Vonrhein C., Womack T. O. (2011) autoBUSTER. Global Phasing Ltd., Cambridge, UK [Google Scholar]
- 37. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J., Chen K., Wang J. H., Zhang C. (2010) Molecular evolution of the primate antiviral restriction factor tetherin. PLoS ONE 5, e11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kirchhoff F. (2010) Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8, 55–67 [DOI] [PubMed] [Google Scholar]
- 41. Schwefel D., Boucherit V. C., Christodoulou E., Walker P. A., Stoye J. P., Bishop K. N., Taylor I. A. (2015) Molecular determinants for recognition of divergent SAMHD1 proteins by the lentiviral accessory protein Vpx. Cell Host Microbe 17, 489–499 [DOI] [PMC free article] [PubMed] [Google Scholar]