Abstract
Acetylation of lysine is a major posttranslational modification of proteins and is catalyzed by lysine acetyltransferases, while lysine deacetylases remove acetyl groups. Among the deacetylases, the sirtuins are NAD+-dependent enzymes, which modulate gene silencing, DNA damage repair, and several metabolic processes. As sirtuin-specific inhibitors have been proposed as drugs for inhibiting the proliferation of tumor cells, in this study, we investigated the role of these inhibitors in the growth and differentiation of Trypanosoma cruzi, the agent of Chagas disease. We found that the use of salermide during parasite infection prevented growth and initial multiplication after mammalian cell invasion by T. cruzi at concentrations that did not affect host cell viability. In addition, in vivo infection was partially controlled upon administration of salermide. There are two sirtuins in T. cruzi, TcSir2rp1 and TcSir2rp3. By using specific antibodies and cell lines overexpressing the tagged versions of these enzymes, we found that TcSir2rp1 is localized in the cytosol and TcSir2rp3 in the mitochondrion. TcSir2rp1 overexpression acts to impair parasite growth and differentiation, whereas the wild-type version of TcSir2rp3 and not an enzyme mutated in the active site improves both. The effects observed with TcSir2rp3 were fully reverted by adding salermide, which inhibited TcSir2rp3 expressed in Escherichia coli with a 50% inhibitory concentration (IC50) ± standard error of 1 ± 0.5 μM. We concluded that sirtuin inhibitors targeting TcSir2rp3 could be used in Chagas disease chemotherapy.
INTRODUCTION
Protein acetylation has recently emerged as a major posttranslational modification of proteins (1). The main targets are the reversible ε-amino group of internal lysine residues, controlling a wide range of cellular processes. This modification was initially described in the N-terminal domain of histones, which in association with methylation, phosphorylation, and other modifications regulates the chromatin structure and the association of transcriptional factors with DNA (2). Recently, proteome-wide analyses revealed a large number of acetylated proteins in the cytoplasm and mitochondria, including most of the enzymes involved in intermediate metabolism. These findings suggest a central role for acetylation in regulatory mechanisms within and outside the nucleus of the cells (3).
The regulation of lysine acetylation is mediated by the counteracting activity of two families of enzymes: the lysine acetyltransferases (KATs), which transfer the acetyl group from acetyl-coenzyme A (CoA), and the lysine deacetylases (KDACs), which remove acetyl groups of lysine residues through a deacetylation reaction. The KDACs can be divided in four classes according to their phylogenetic conservation. Classes I, II, and IV belong to the classical family or zinc-dependent enzymes, while class III enzymes are dependent on NAD+ and are called sirtuins or Sir2 (4). Sirtuins catalyze the deacetylation of lysine residues in the presence of NAD+, with the formation of nicotinamide and O-acetyl-ADP-ribose as products (5, 6). In addition, other activities have been described for these enzymes, such as lysine deglutarylation and lysine desuccinylation (7, 8). Members of the Sir2 protein family are involved in gene silencing, DNA repair, chromosomal stability, metabolic processes, and aging (9–11). In addition, sirtuin levels are increased in several cancers and have become potential targets for therapeutic approaches (12, 13). There are several compounds being tested as antitumoral agents with promising results (14).
Sirtuins are highly conserved and expressed from organisms as varied as Archaea up to higher eukaryotes (15). Different organisms have distinct Sir2 homologues. Yeast has four homologues, and humans have seven sirtuins (HSIRT1 to -7). Protozoans of the Kinetoplastida order, which are agents of parasitic diseases, also present sirtuins. Trypanosoma brucei has 3 different sirtuin homologues (TbSir2rp1 to -3) (16–19). TbSir2rp1 is localized in the nucleus and participates in DNA repair mechanisms and RNA polymerase I-mediated transcription repression of subtelomeric genes in both the insect and blood stages (18, 20). TbSir2rp2 and TbSir2rp3 are mitochondrial proteins, and their independent knockouts do not affect the proliferation of bloodstream forms (18). Among the three sirtuin genes of Leishmania, LmSir2rp1 codes for a cytoplasmic protein that presents NAD+-dependent deacetylase and ADP-ribosyltransferase activities unrelated to epigenetic silencing. It participates in the remodeling of the parasite morphology and in the parasite interaction with its mammalian host (21, 22). The other Leishmania sirtuins have not been characterized. T. cruzi has only two genes coding for sirtuins, TcSir2rp1 and TcSir2rp3, and very little is known about their function in the parasite. As new drugs are required for the treatment of these parasitic diseases, and the effect of sirtuin inhibitors has not been extensively exploited for Chagas disease treatment (23), we decided to characterize T. cruzi sirtuins and test a new compound, called salermide, which was found to be very effective in the treatment of some cancers in vitro and in vivo. We initially found that sirtuin inhibitors affected T. cruzi growth and differentiation. These would act in one or both of the two sirtuins. Therefore, we investigated their expression and cellular localization in the different stages of the parasite and the effects of the overexpression of each enzyme in the biology of the parasite. We found that these two enzymes differentially affected the proliferation and differentiation of the parasite and that salermide appears to act by inhibiting TcSir2rp3, which is quite sensitive to the inhibitor.
MATERIALS AND METHODS
Parasite cultures and metacyclogenesis.
T. cruzi strain Y epimastigotes were cultivated in liver infusion tryptose (LIT) medium containing 10% fetal bovine serum (FBS) at 28°C. Epimastigotes were seeded at a density of 2 × 106 parasites per ml and the cell numbers quantified using a Neubauer chamber. For growth inhibition assays, salermide was dissolved in dimethyl sulfoxide (DMSO) to 2 mM and nicotinamide (both from Sigma-Aldrich) in phosphate-buffered saline (PBS) to 0.5 M and added to obtain the indicated concentrations in the culture medium. Epimastigote viability was performed with alamarBlue, as described previously (24). For mammalian cell viability, we employed the PrestoBlue cell viability reagent (Life Technologies). Metacyclogenesis was induced as described in reference 25 using exponentially growing parasites in all cases. Differentiation was quantified after staining with 4-6-diamidino-2-phenylindole (DAPI) to distinguish epimastigotes from metacyclic trypomastigotes by observation under a fluorescence microscope. Intracellular and cell-derived trypomastigotes were obtained from infected LLCMK2 cells maintained in low-glucose Dulbecco's minimum essential medium (DMEM), supplemented with 10% FBS, streptomycin (100 mg/ml), and penicillin (100 U/ml) in a humidified 5% CO2 atmosphere at 37°C, as described previously (26). For invasion assays, epimastigote cultures in LIT medium were kept for 2 to 3 weeks and the metacyclics purified by DEAE-cellulose columns, as detailed in Yoshida et al. (27).
Antibodies.
Anti-hemagglutinin from influenza virus peptide (anti-HA) was obtained from Covance. The antibody against Sir2rp1 was made in-house by using the heterologous protein from Leishmania major to immunize rabbits. Anti-TcSir2rp3 was custom prepared by GenScript through immunizing rabbits with a synthetic peptide (TFRDKNGLWENHRVC). Anti-β-tubulin was prepared as described previously (28), anti-gp30 was used as ascitic fluids (29), and anti-dihydrolipoamide dehydrogenase (DHLADH) (a mitochondrion marker) was kindly provided by Kevin Tyler (United Kingdom). Anti-eIF5A and anti-methyl glutaconyl-CoA (MgCoA) antibodies were obtained using recombinant forms of the T. cruzi and T. brucei proteins, respectively, to immunize rabbits.
Generation of sirtuins overexpressing parasites.
The T. cruzi sirtuin genes, TcSir2rp1 (TcCLB.508207.150) and TcSir2rp3 (TcCLB.447255.20) (both at http://tritrypdb.org), were PCR amplified from genomic DNA of T. cruzi strain Y using the oligonucleotides TcSir2rp1Fow (5′-CCTCTAGAATGAATCAAGATAACGCC-3′), TcSir2rp1Rev (5′CCGGATCCCTAAGCGTAGTCTGGCACGTCGTAAGGGTATTTTCGGTCTGTCTGTGT-3′), TcSir2rp3Fow (5′-CCTCTAGAATGAGGCCGCGGCGTCAGGT-3′), and TcSir2rp3Rev (5′-CCGGATCCCTAAGCGTAGTCTGGCACGTCGTAAGGGTA-CACCGCGTCTTGAAGGAG-3′). The underlined residues represent cloning sites, and those in bold are the HA coding sequence. The fragments were cloned into pGEM-T Easy vector (Promega) and the expected constructs confirmed by restriction analysis and DNA sequencing. The fragments were removed by endonuclease reaction using XbaI and BamHI and inserted into p33 vector (30), which was previously digested with the same enzymes. To generate mutated TcSir2rp3 protein, we used two round of PCRs. For the first round, we employed primers TcSir2rp3-H113Y-Fov (ATCGGTGCTGCATATGTATGGGGAATTGCT) and TcSir2rp3-H113Y-Rev (ATATGCAGCACCGATACAGATCCCGCACTC) to create the point mutation. The cytosine was replaced by thymidine (in bold type). In the second round of PCR, we used the same primers described above to amplify the whole TcSir2rp3 gene. Plasmids containing TcSir2rp1-HA, TcSir2rp3-HA, and TcSir2rp3-H113Y were used to transfect T. cruzi epimastigotes using Amaxa Nucleofector (Lonza), followed by selection with 200 μg/ml Geneticin G418 (Invitrogen). The transformed parasites were confirmed by immunoblotting analysis with anti-HA antibody and the overexpression of the protein with specific antibodies raised against the respective T. cruzi sirtuins.
Recombinant TcSir2rp3 and activity assay.
TcSir2rp3 gene was amplified from the genomic DNA of T. cruzi Y strain using the oligonucleotides TcSir2rp3-pET(F) (5′-CTGGATCCATGAGGCCGCGGCGTCAGG) and TcSir2rp3-pET(R) (5′-CTCGAGCTACACCGCGTCTTGAAGGAGCT) and cloned into pGEM-T-easy vector (Promega). The expected BamHI-XhoI fragment was then inserted in the respective sites of pET28a (Novagen) and used to transform ArcticExpress DE3 bacteria (Agilent Technologies). The protein was obtained by diluting 50-fold a stationary-culture growth at 37°C in Luria broth medium containing 20 μg/ml gentamicin and 50 μg/ml kanamycin, following 3 h of growth at 37°C in the same medium without antibiotics and induction with 0.1 mM isopropyl β-d-1-thiogalactopyranoside at 12°C. Bacterial cells were collected by centrifugation (5,000 × g for 10 min) and the pellet resuspended in 20 mM Tris-HCl (pH 8.0) and centrifuged again. The washed cells were resuspended in one-tenth of the culture volume in cold lysis buffer (200 mM NaCl, 5% glycerol, 5 mM 2-mercaptoethanol, and 25 mM HEPES-NaOH [pH 7.5]) and submitted to three passages on a French press apparatus (Thermo Electron). The lysates were cleared by 30 min of centrifugation at 16,000 × g and incubated for 2 h at 4°C with one-tenth the lysate volume of nickel nitrilotriacetic acid (Ni-NTA) Superflow beads (Qiagen). The resin was washed with 50 volumes of the lysis buffer containing 20 mM imidazole, and the enzyme was eluted with one volume of the same buffer containing 0.25 M imidazole and stored at −20°C. Deacetylase assays were performed at 37°C in 50 μl of 25 mM Tris-HCl (pH 8.0), 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2 containing 0.6 mM NAD+ (Sigma-Aldrich), and 20 μM synthetic peptide Abz-Gly-Pro-acetyl-Lys-Ser-Gln-EDDnp, where Abz is ortho-aminobenzoic acid and EDDnp is N-[2,4-dinitrophenyl]ethylenediamine. The peptide was synthesized as described previously (31), further purified by chromatography on a C18 column, dried, and resuspended in DMSO. A recombinant protein corresponding to the T. brucei RNA triphosphatase cloned in pET28a (32) was prepared similarly and used as a negative control, and a total HeLa cell extract was used as a positive control in the activity assays. In the experiments containing salermide, the addition of substrate started the reaction. At the end of incubation, the reactions were stopped with 50 μl of a solution containing 12 mM nicotinamide in 100 mM NaCl and 50 mM Tris-HCl (pH 8.0), in the absence or presence of 0.6 mg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Worthington). After further incubation for 30 min, the fluorescence at 420 nm was read (excitation, 320 nm) in a SpectraMax M3 plate reader (Molecular Devices).
Protein sample preparation and immunoblotting.
Parasites were collected and lysed in PBS buffer with 1% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride (PMSF), and 1× Complete protease (EDTA-free) inhibitor cocktail (Roche). For digitonin extraction, parasites were collected in a similar manner and lysed with the indicated concentrations of digitonin for 5 min at 37°C, as described previously (33). Each fraction corresponding to supernatants of 10,000 g (5 min) was mixed with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer, applied to 10% polyacrylamide gels, and transferred to nitrocellulose membranes. The membranes were treated with 5% nonfat milk in PBS containing 0.01% Tween 20 (PBS-T) for 60 min and incubated with specific primary antibodies for 1 h in the same buffer. The membranes were washed with PBS-T and incubated for 1 h with goat anti-mouse or rabbit IgG-horseradish peroxidase (HRP) (Life Technologies) at a 1:10,000 dilution. Bound antibodies were detected using ECL reagent (Millipore) and chemiluminescence detected using Odyssey Li-Cor equipment.
Immunofluorescence analysis.
The collected parasites were washed in PBS and attached to glass slides pretreated with 0.01% poly-l-lysine (Sigma-Aldrich). The slides were then incubated with 2% paraformaldehyde in PBS at room temperature for 15 min and treated for 5 min in PBS containing 0.1% Triton X-100. Fixed and permeabilized cells were washed with PBS, and the slides were incubated for 1 h at room temperature with primary antibodies diluted in PBS with 2% bovine serum albumin (BSA) containing primary antibodies as follows: anti-HA was diluted 1:1,000, anti-Sir2rp1 was diluted 1:1,000, anti-Sir2rp3 was diluted 1:2,000, and anti-DHLADH was diluted 1:10,000. The slides were washed with PBS, incubated for 1 h with the secondary antibody (anti-mouse IgG-Alexa 488 and/or anti-rabbit IgG-Alexa 594; Invitrogen), washed, and mounted with ProLong gold antifade reagent (Invitrogen) in the presence of 10 μg of DAPI per ml. Z-stacks were acquired with an Olympus (BX-61) microscope equipped with a 100× plan apo-oil objective (numerical aperture [NA], 1.4), followed by blind deconvolution employing the AutoQuant X2.2 software (Media Cybernetics).
Measurement of intracellular ATP levels and parasite movement.
Cellular ATP concentration was determined by a bioluminescence method using the ATP bioluminescence assay kit (Sigma-Aldrich). The assay is based on the requirement of ATP for producing light by adding exogenous luciferase (emission maximum, 560 nm [pH 7.8]). Briefly, 1 × 107 parasites from exponentially growing cells were collected and lysed, according to the manufacturer's instructions. An aliquot of the samples was added to the assay mix, and the luminescence intensity was immediately measured in a Molecular Devices M3 plate reader. The amount of ATP was calculated based on a standard curve and the values expressed as milligrams of protein.
For parasite movement quantification, exponentially growing epimastigotes were added to nontreated slides for 10 min, covered with a glass coverslip, and immediately sealed for microscopy analyses, using the differential interference contrast optics of an Olympus (BX-61) microscope equipped with a 60× plan objective (NA, 1.4). Thirty sequential images were obtained at 4.75 s each. The images were further used for the movement quantification using MTrackJ from the ImageJ software package.
Infection assays.
Cell invasion assays were carried out by seeding the parasites onto 13-mm diameter glass coverslips coated with nonconfluent LLCMK2 cells plated 24 h earlier in 24-well plates (27). After 1 h of incubation with the parasites, the cells were washed once with PBS and fixed in Bouin solution, stained with Giemsa, and sequentially dehydrated in acetone, a graded series of acetone to xylol (9:1, 7:3, 3:7), and xylol. In some assays, after 1 h of incubation with parasites, LLCMK2 cells were washed and further incubated for the indicated periods in DMEM containing 5% FBS before fixation and staining, as described above. For salermide inhibition assays, trypomastigotes were pretreated with the indicated amounts of salermide dissolved in DMSO for 4 h in DMEM containing 10% FBS at 37°C in 5% CO2 before addition to the cells. Otherwise, to follow the intracellular development, the wells were washed, and fresh DMEM containing the drug was added. The drug was added again to the medium after 24 h. Alternatively, trypomastigotes were incubated with LLCMK2 cells for 2 h, washed, and incubated for 24 h. Next, salermide was added to the cell cultures to concentrations of 0.2, 0.5, 1, and 2 µM, as indicated (see Fig. 7). The controls contained the same amount of DMSO. Infection was quantified by microscopic examination of duplicate coverslips. Intracellular parasites were distinguished morphologically in the host cell cytoplasm (34), and the numbers indicated (see Fig. 6C and 7) are means from triplicate experiments.
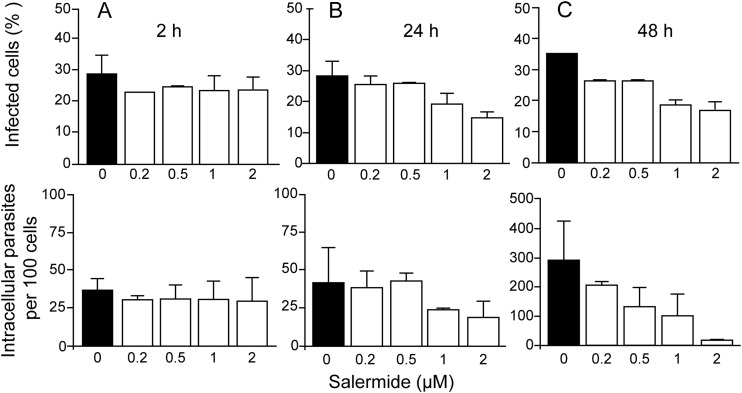
FIG 7.
Salermide inhibits T. cruzi multiplication in mammalian cells. (A) Wild-type trypomastigotes derived from infected mammalian cells were pretreated for 4 h with the indicated concentrations of salermide and then incubated for 2 h with LLCMK2 cells. The upper graphs show the percentage of infected cells, and the bottom graphs show the number of intracellular parasites after 2 h of incubation. (B and C) Number of infected cells and parasites at 24 and 48 h after infection. The values are means ± SE (n = 3).
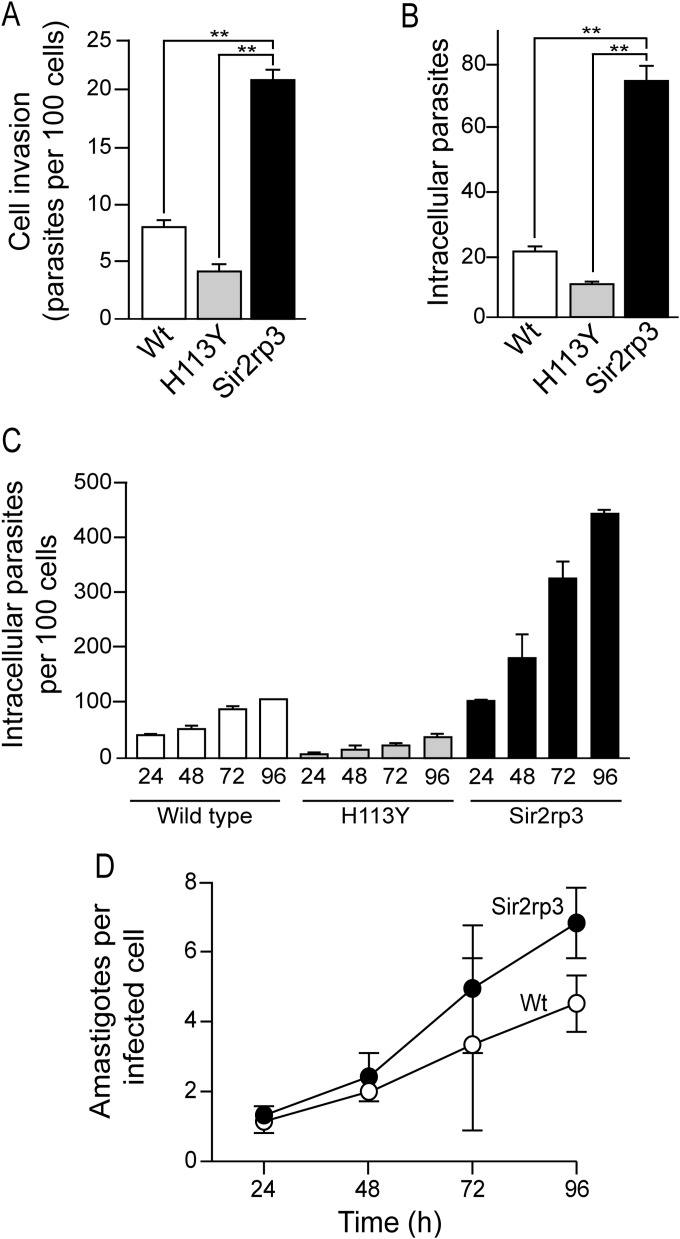
FIG 6.
TcSir2rp3 overexpression increases parasite invasion and multiplication inside mammalian cells. One half a milliliter containing 1 × 106 wild-type (white bars), TcSir2rp3H113Y (gray bars), or TcSir2rp3-ox (black bars) metacyclic trypomastigotes was incubated for 1 h with LLCMK2 epithelial cells previously seeded on glass coverslips in 24-well plates. The number of internalized parasites per 100 cells (A) and total intracellular parasites (B) was then quantified. (C) Mean number of intracellular parasites after further incubation at the indicated times. (D) Normalized growth of intracellular amastigotes. All numbers are means ± standard deviations (SD) (n = 3). The asterisks indicate statistically significant differences based on the Student t test (**, P < 0.01).
In vivo infection assays were carried out with trypomastigote forms of Y strain of T. cruzi obtained from infected LLCMK2 cells that were washed and resuspended in DMEM without FBS. One hundred microliters was injected intraperitoneally into 4-week-old female BALB/c mice. Parasitemia was measured in blood collected by tail vein puncture in a microscope. For this procedure, 5 μl of blood was deposited in each slide and covered with a 22 by 22-mm coverslip. The number of parasites were counted in ≥20 microscope fields and corrected by a factor predetermined using a calibrated coverslip. Treatments with salermide started after 1 h or 24 h postinfection and were maintained daily.
Statistical analysis.
All quantitative assays were biological triplicates, and the values reported in this work are means ± standard errors (SE) of three measurements from each triplicate experiment. Statistical significance was obtained with Prism 6 (GraphPad) using the Student t test.
Ethical statements.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Brazilian National Council of Animal Experimentation (http://www.cobea.org.br/). The protocol was approved by the Committee on the Ethics of Animal Experiments of the Institutional Animal Care and Use at the Federal University of São Paulo (ID no. CEP 1982/11).
RESULTS
Sirtuin inhibitors affect epimastigote growth and viability.
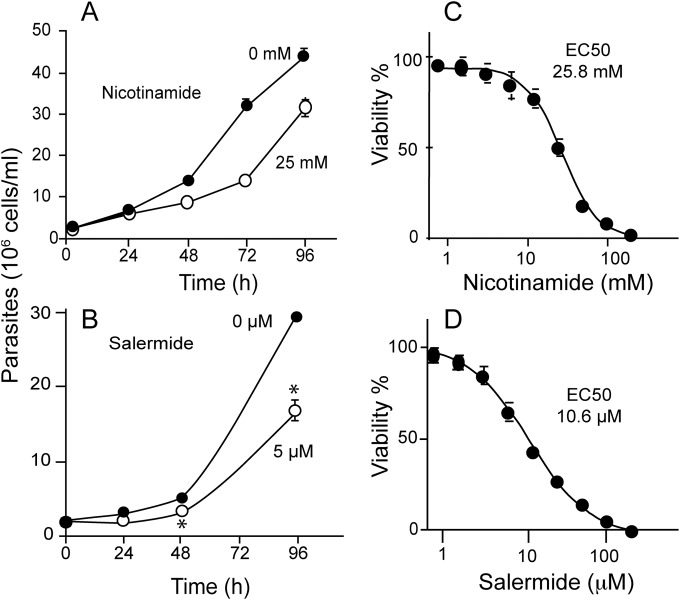
Several compounds have been shown to inhibit the KDAC activity and have been exploited as drugs to treat a variety of human diseases (35, 36). Nicotinamide, an amide of nicotinic acid, has been described as the classical sirtuin inhibitor (37, 38). Salermide, a derivative of sirtinol, is a novel sirtuin inhibitor that acts through binding in the C-pocket of the enzyme (39, 40). Nicotinamide and salermide affected the proliferation of the parasite at concentrations of >25 mM and 5 μM, respectively (Fig. 1A and B), causing viability loss, as detected by alamarBlue assays, with 50% effective concentrations (EC50s) of 25.8 mM and 10.6 μM, respectively (Fig. 1C and D).
FIG 1.
Effect of sirtuin inhibitors in the parasite growth and viability. Epimastigotes were cultivated in the presence of the indicated concentrations of nicotinamide (A) and salermide (B). (C and D) Viability of epimastigotes treated with nicotinamide and salermide for 72 h, determined by the alamarBlue viability assay in a 96-well plate. The values are means ± standard errors (SE) (n = 3). The EC50 values are indicated in each panel.
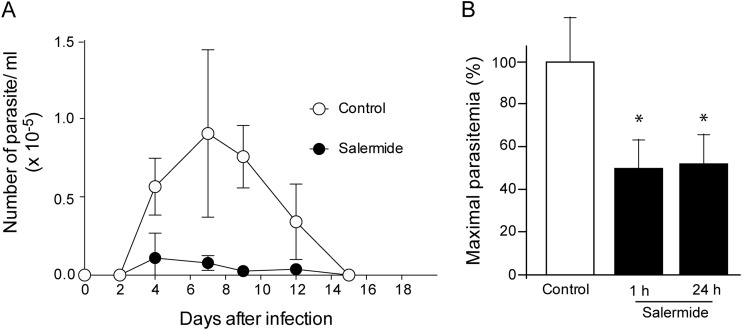
Salermide treatment leads to the control of parasite infection in vivo.
To investigate if the effect caused by salermide could be extended to an in vivo infection, BALB/c mice were intraperitoneally injected with 0.5 × 105 to 2 × 105 cell-derived trypomastigotes from Y strain. One hour after infection and every day following infection up to day 12, mice were treated with 100 μM salermide in 100 μl, a concentration that had been demonstrated to not cause any side effect (39). The parasitemia was followed every 2 days, comparing with infected mice that received DMSO only, which was used as a vehicle. A reduction in parasitemia was observed after salermide treatment (Fig. 2A). A reduction in parasitemia was also observed in a second experiment when the treatment started at 1 or 24 h after infection (Fig. 2B). In this case, the maximal parasitemia reached 2.5 × 105 parasites per ml, higher than the parasitemia observed in the experiment shown in Fig. 2A.
FIG 2.
In vivo T. cruzi infection is reduced by salermide. Parasitemia of BALB/c mice infected with cell-derived trypomastigotes submitted to salermide treatment (●) or to DMSO alone (○), both injected 1 h after infection. The values are means ± SE (n = 10). (B) Maximal percentage of parasitemia (mean values, n = 5) of mice treated with DMSO (white bar) or with salermide. Treatment started 1 or 24 h after infection (black bars). One hundred percent in the controls corresponds to 2.5 × 105 parasites/ml. The asterisks indicate significant differences (P < 0.05) compared to control animals, as determined by the Student t test.
T. cruzi sirtuins.
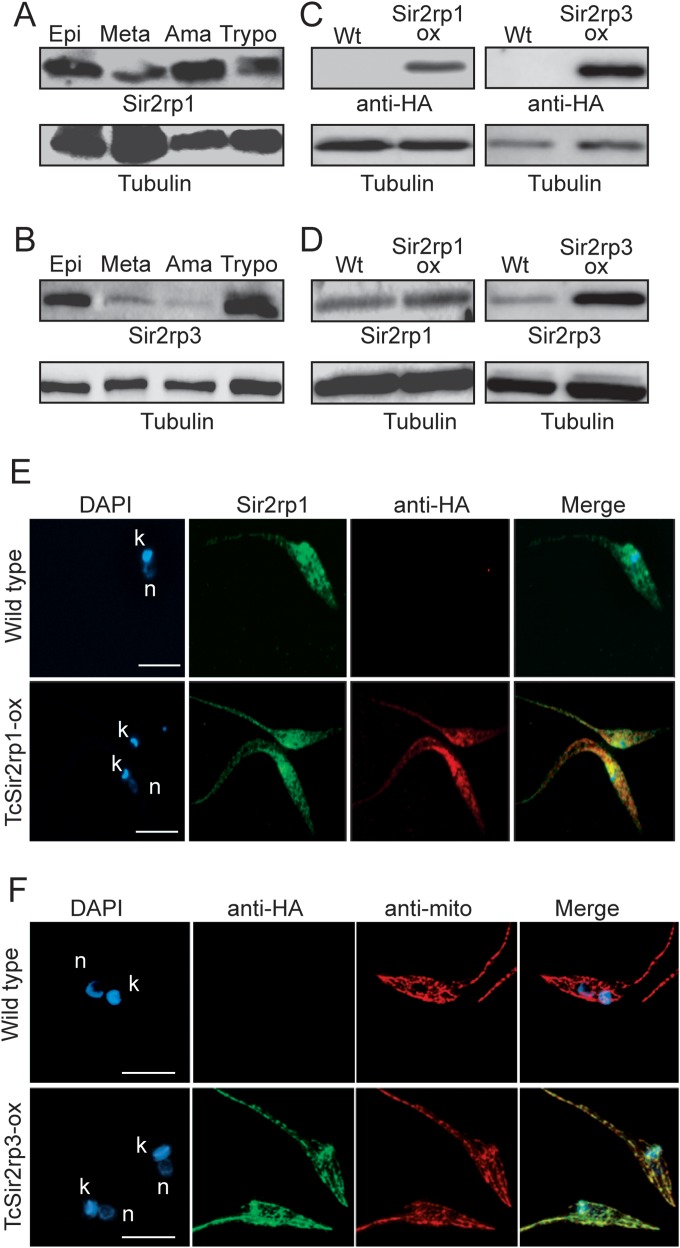
The two genes coding for sirtuins in T. cruzi, TcSir2rp1 (TcCLB.508207.150, predicted size of about 40 kDa), and TcSir2rp3 (TcCLB.447255.20, predicted size of about 27 kDa), contain the putative sirtuin domain (see Fig. S1A in the supplemental material) and share 42% and 18% identity, respectively, with the sirtuin domain of Saccharomyces cerevisiae Sir2. Both proteins contain critical histidine residues for catalytic activity, a NAD+ binding domain, and a noncanonical CX2CX20CX2C Zn2+ binding motif (see Fig. S1B in the supplemental material). Phylogenetic analysis based on the neighbor-joining (NJ) method grouped TcSir2rp1 with TbSir2rp1 and LmSir2rp1, indicating a possible cytosolic localization for TcSir2rp1, as we did not find a nuclear localization signal in its amino acid sequence (see Fig. S1C in the supplemental material). TcSir2rp3 clustered with the mitochondrial T. brucei protein (TbSir2rp3). The antibody against the recombinant Sir2rp1 of Leishmania major recognized a similar 40-kDa band in T. cruzi Y and several other strains (see Fig. S1D in the supplemental material). Antibodies prepared against a synthetic peptide of TcSir2rp3 reacted in Western blots with two bands: one band with the expected size for TcSir2rp3 (27 kDa) that increased in size in samples from parasites overexpressing TcSir2rp3, and another 55-kDa band (the reaction with this band was probably nonspecific) (see Fig. S1E in the supplemental material).
When we analyzed the expression of TcSir2rp1 and TcSir2rp3 in different parasite stages, using these antibodies, we found that TcSir2rp1 was highly expressed in replicative parasite forms (epimastigotes and amastigotes) and less in metacyclic trypomastigotes and trypomastigotes derived from infected mammalian cells (Fig. 3A). In contrast, TcSir2rp3 was found largely expressed in epimastigotes and cell-derived trypomastigotes and much less in intracellular amastigotes and metacyclic trypomastigotes (Fig. 3B).
FIG 3.
Expression and cellular localization of T. cruzi sirtuins. (A and B) Immunoblotting of total proteins of epimastigotes (Epi), purified metacyclic trypomastigotes (Meta), intracellular amastigotes (Ama), and trypomastigotes (Trypo) derived from infected mammalian cells probed with anti-Sir2rp1 (A) or anti-TcSir2rp3 antibodies (B). (C and D) Immunoblotting of wild-type (Wt) epimastigotes or epimastigotes expressing HA-tagged TcSir2rp1 (TcSir2pr1-ox) or TcSir2rp3 (TcSir2pr3-ox) and probed with anti-HA (C) or with antibodies specific for each one of the sirtuins (D). Anti-β-tubulin was used as the loading control. (E) Indirect immunofluorescences of wild-type or TcSir2rp1-ox epimastigotes using anti-HA (red) and anti-Sir2rp1 (green) antibodies. (F) Indirect immunofluorescences of wild-type and TcSir2rp3-ox epimastigotes using anti-HA (green) and anti-DHLADH, a mitochondrial marker (anti-mito) (red). The figures also show the DAPI labeling and the merged fluorescence. n, nucleus; k, kinetoplast. Scale bars = 5 μm.
To better characterize the proteins of TcSir2rp in the parasite, we investigated their cellular localization by generating cell lines constitutively expressing C-terminal HA-tagged versions of these proteins (TcSir2rp1-ox and TcSir2rp3-ox). Protein expression was confirmed by immunoblotting with anti-HA (Fig. 3C and D). We found that TcSir2rp1 was expressed twice as much of the endogenous enzyme, while TcSir2rp3 was overexpressed 20-fold more, as judged by densitometry of Western blotting using specific antibodies for both proteins (Fig. 3D). Both endogenous and tagged TcSir2rp1 were localized in the cytosol of the parasite, as assessed by immunofluorescence using anti-HA and anti-Sir2rp1 antibodies (Fig. 3E), and the protein localization was maintained during the parasite life cycle, except on trypomastigotes that presented a more clustered localization (see Fig. S2A in the supplemental material). The cytosolic localization was confirmed by digitonin fractionation that revealed that most of the protein was present in fractions corresponding to cytoplasmic markers (see Fig. S2B in the supplemental material). In contrast, labeling with anti-HA and anti-DHLADH revealed that TcSir2rp3 tagged with HA had a mitochondrial localization (Fig. 3F). In addition, digitonin fractionation assays confirmed the mitochondrial localization of TcSir2rp3, as it was coextracted with MgCoA (see Fig. S2C in the supplemental material), a mitochondrial matrix protein in trypanosomes (M. L. Lima and S. Schenkman, unpublished data).
TcSir2p has antagonistic effects in epimastigote growth and metacyclogenesis.
The increased expression of TcSir2rp1 led to a delay in parasite growth, while the overexpression of TcSir2rp3 improved it compared with wild-type cells (Fig. 4A). This effect appeared to be related to the metabolic state of these parasites, as the TcSir2rp3-ox cell line presented a 20% reduction in intracellular levels of ATP and an increased motility compared with that of TcSir2rp1-ox and wild-type parasites (see Fig. S3A and B in the supplemental material).
FIG 4.
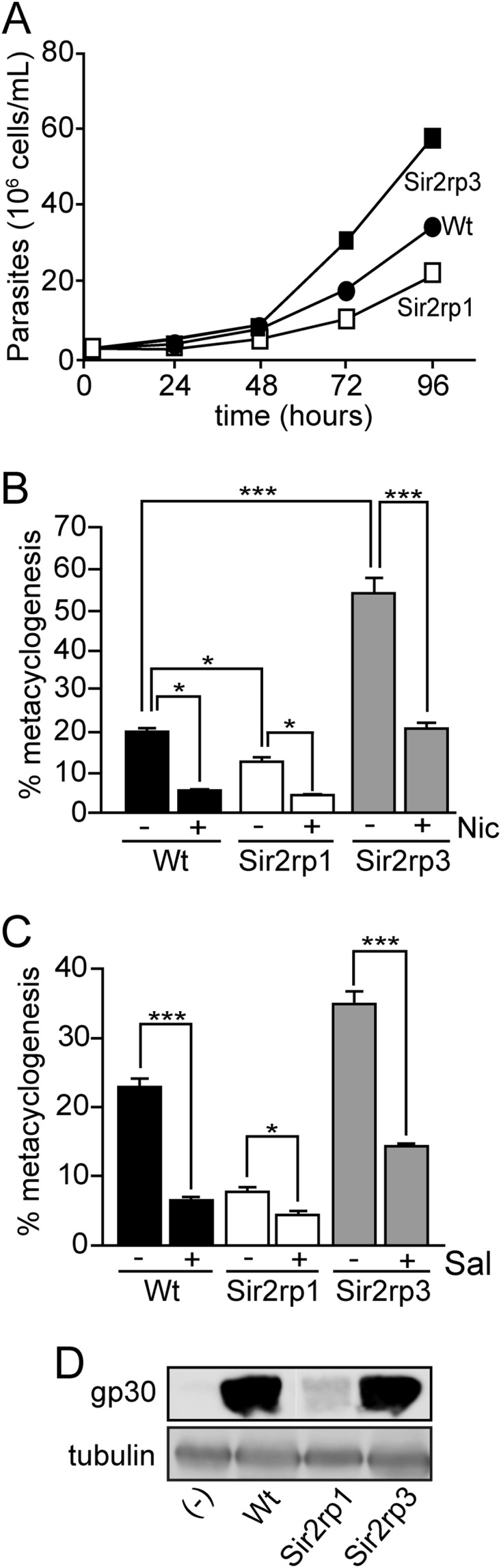
T. cruzi sirtuins have opposite effects on parasite growth and differentiation. (A) Growth curves of wild-type (●), TcSir2rp1-ox (□), and TcSir2rp3-ox (■) epimastigotes. (B and C) Percentage of differentiation of epimastigotes into metacyclics by the wild-type (black bars), TcSir2rp3-ox (gray bars), and TcSir2rp1-ox (white bars) parasites in the absence (−) or presence (+) of 15 mM nicotinamide (Nic) (B) and 2 μM salermide (Sal) (C). All values are means ± SE (n = 3). The asterisks indicate statistically significant differences based on the Student t test (*, P < 0.01; ***, P < 0.003). (D) Immunoblotting of parasites after differentiation induction probed with anti-gp30. An epimastigote extract was used as the negative control (−), and anti-β-tubulin was used as the loading control.
Next, we examined the differentiation of these epimastigotes in the exponentially growing phase into metacyclic trypomastigotes induced by starvation. TcSir2rp1-ox differentiated less and TcSir2rp3-ox cell lines differentiated more than wild-type parasites (Fig. 4B and C). The reduced differentiation of the TcSir2rp1-ox line was further confirmed by the weak expression of gp30, a specific metacyclic protein (Fig. 4D). To know if these effects were due the activity of overexpressed sirtuins, metacyclogenesis was conducted in the presence of two sirtuin inhibitors (nicotinamide and salermide). As shown in Fig. 4B and C, both compounds decreased the differentiation in all cell lines. It affected relatively more the differentiation of wild-type parasites and largely reverted the increased differentiation of TcSir2rp3-ox. Importantly, it does not revert the negative effect of TcSir2rp1-ox parasites, suggesting that salermide is acting preferentially on Sir2rp3. In contrast, class I and II deacetylase inhibitors (trichostatin A and butyric acid) caused no changes in metacyclogenesis (see Fig. S3C and D in the supplemental material).
Overexpression of a mutated TcSir2rp3 version does not affect parasite growth and differentiation.
To verify if the activity of the TcSir2rp3 protein was causing the increase in parasite growth and differentiation, we generated a parasite cell line overexpressing a version of the protein substituting the histidine (H) residue at position 113 by a tyrosine (Y) (TcSir2rp3-H113Y) (Fig. 5A). This modification created an inactive version of the enzyme, as described for other sirtuins (41). The overexpression of the protein was confirmed by immunoblotting with anti-HA and anti-Sir2rp3 antibodies using total protein lysates (Fig. 5B). Similarly, mitochondrial localization was observed for the mutated version of TcSir2rp3, as accessed by immunofluorescence assays using anti-HA and anti-Sir2rp3 (see Fig. S4 in the supplemental material). Indeed, TcSir2rp3-H113Y parasites behaved as nontransfected cells and did not grow as well as TcSir2rp3-ox (Fig. 5C). The mutation also impaired the increased differentiation promoted by the wild-type gene (Fig. 5D). These results indicate that the effects observed for the overexpression of TcSir2rp3 were due to its activity.
FIG 5.
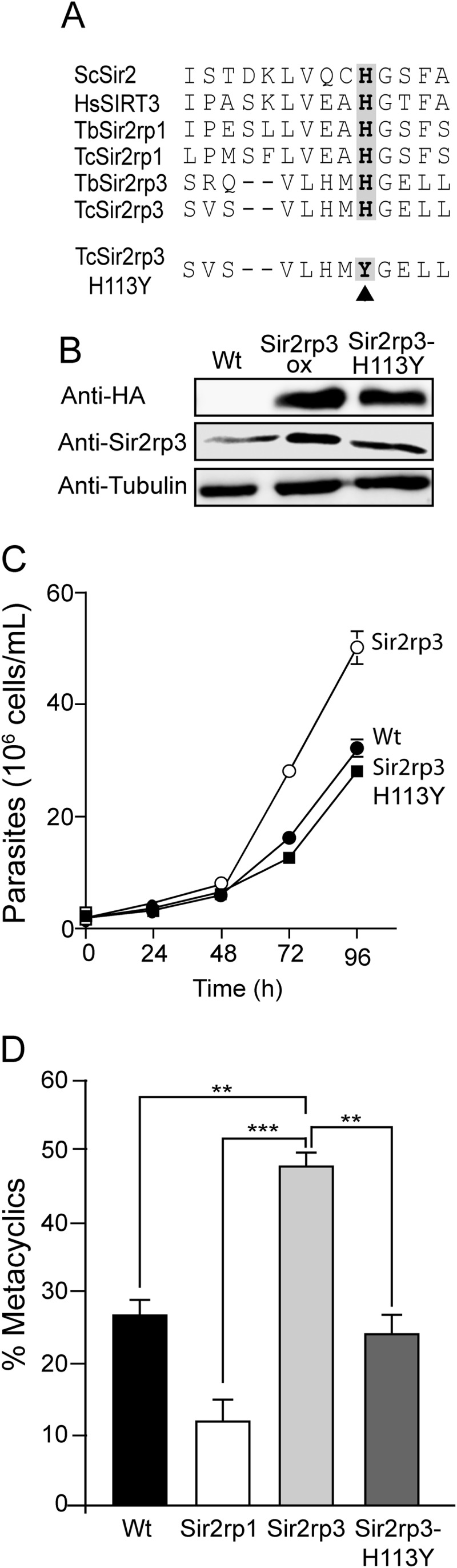
Mutated version of TcSir2rp3 does not affect parasite growth and differentiation. (A) Amino acid sequences in the active site of sirtuins of S. cerevisiae (ScSir2), Homo sapiens (HsSIRT3), T. brucei (TbSir2rp1, TbSir2rp3), and T. cruzi (TcSir2rp1 and TcSir2rp3) and the position of the substitution of the His 113 by tyrosine in TcSir2rp3 (highlighted in gray). The accession numbers are shown in Fig. S1 in the supplemental material. (B) Western blotting of whole lysates of wild-type (Wt), TcSir2rp3-ox, and TcSir2rp3-His113Y parasite lines probed with anti-HA and anti-TcSir2rp3 antibodies. (C) Growth curves of wild-type (●), TcSir2rp3-ox (○), and TcSir2rp3-His113Y (■). (D) Percentage of differentiation in metacyclic trypomastigotes by the wild-type (black bar), TcSir2rp3-ox (light gray bar), TcSir2rp1-ox (white bar), and TcSir2rp3-His113Y (dark gray bar). All values are means ± SE (n = 3), and the asterisks indicate statistically significant differences based on the Student t test (**, P < 0.05; ***, P < 0.001).
TcSir2rp3 overexpression increases parasite invasion and multiplication.
Subsequently, we asked if overexpression of those proteins could affect parasite infectivity and multiplication inside the host cell. These experiments were performed using TcSir2rp3-ox parasites because we were not able to generate enough metacyclic trypomastigotes from the TcSir2rp1-ox cell line. Metacyclic trypomastigote forms of wild-type, TcSir2rp3-ox, and TcSir2rp3 H113Y parasites were incubated with LLCMK2 epithelial cells for 1 h, and the numbers of infected cells and internalized parasites were determined. As observed in the Fig. 6A and B, overexpression of TcSir2rp3 increased invasion about 2.5-fold and resulted in 4-fold more internalized parasites than nontransfected parasites. In contrast, metacyclic trypomastigotes with H113Y did not invade and proliferate more than wild-type parasites (Fig. 6A and B). These mutants showed a significant decrease in cell invasion, suggesting that the mutated protein probably was acting as a dominant negative mutant in metacyclic trypomastigotes.
Since TcSir2rp3-ox parasites were able to infect a higher number of cells, we investigated whether sirtuin expression also affected intracellular multiplication. The number of intracellular parasites was monitored every 24 h up to 96 h. As shown in Fig. 6C, the overexpression of TcSir2rp3, but not the H13Y mutant, also caused an increased rate of proliferation compared with that of nontransfected parasites. This phenomenon was more evident when we assessed the late time points (72 and 96 h). This was due to an increased multiplication of the TcSir2rp3 parasites inside host cells, about 1.5 times more than wild-type parasites (Fig. 6D). We also compared rates of parasite invasion by cell-derived trypomastigote forms. The parasites had similar invasion rates, in agreement with the fact that TcSir2rp3 is highly expressed in trypomastigotes. In contrast, the proliferation of amastigotes inside the cells was higher in the case of TcSir2rp3-ox parasites, seen especially in the late stages of infection (72 and 96 h) (see Fig. S5A, to C in the supplemental material), which normally presented low levels of Sir2rp3 expression (see Fig. 3). In fact, we confirmed that TcSir2rp3 was highly overexpressed in amastigotes and much less in trypomastigotes (see Fig. S5E and F in the supplemental material). These results agree with the fact that TcSir2rp3 expression is low in wild-type amastigotes and that an increase in expression might cause an increased rate of proliferation. More importantly, during the progress of the infection, a high number of extracellular amastigotes was observed in the supernatant of cells infected with TcSir2rp3-ox parasites compared with wild-type parasites (see Fig. S5D in the supplemental material), indicating an imbalance in parasite multiplication and further differentiation to trypomastigotes. These data imply that TcSir2rp3 might affect parasite functions in different ways, depending on the parasite form.
Salermide affects trypomastigote invasion and multiplication. All phenotypes observed with TcSir2rp3-ox parasites suggested that this enzyme would be involved in the proliferation of intracellular parasites. Therefore, we evaluated the effect of salermide in parasite cell invasion. When cell-derived wild-type trypomastigotes were pretreated for 4 h with different concentrations of the drug and incubated for 2 h with LLCMK2 cells, we did not observe any significant effect in the number of infected cells or intracellular parasites for all tested concentrations (Fig. 7A). However, we found a decrease in intracellular parasite numbers at 24 h (Fig. 7B) and 48 h (Fig. 7C) after invasion, suggesting that the drug interfered with the development of amastigotes. The drug did not affect cell viability up to 2.5 μM for the mammalian cell, as determined by a PrestoBlue assay upon 48 h of incubation (see Fig. S6 in the supplemental material). The effect of 2 μM salermide was also observed on transgenic parasites, promoting 30% inhibition of invasion (the number of metacyclic trypomastigotes that invade the cells after 2 h of incubation with LLCMK2 cells) and 50% of amastigote proliferation (the number of intracellular amastigotes 24 and 48 h after infection of LLCMK2 cells), indicating that the overexpressors might also be targeted by salermide.
Salermide inhibits TcSir2rp3.
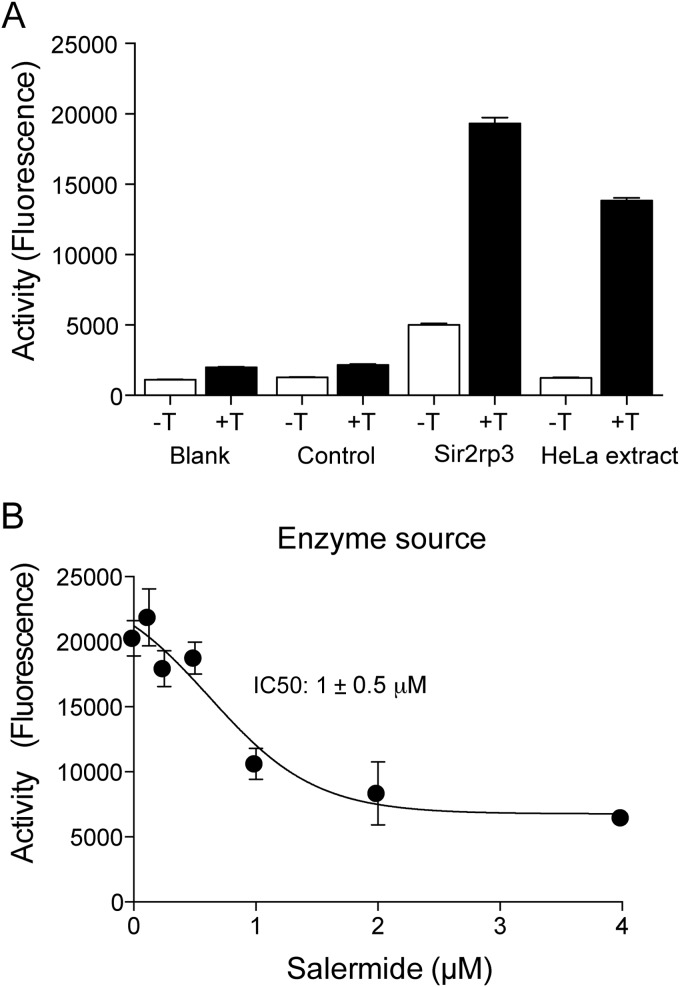
The gene corresponding to the Sir2rp3 was cloned and expressed in bacteria as a recombinant protein containing a 6×His tag in the N terminus. The protein was then purified by Ni-agarose affinity chromatography and tested for its deacetylase activity toward the Abz-Gly-Pro-acetylLys-Ser-Gln-EDDnp synthetic peptide. This Förster resonance energy transfer substrate was then incubated with or without trypsin. When the acetyl group was removed, the trypsin hydrolyzed the Pro-Lys bond to generate fluorescence. Indeed, the recombinant enzyme was highly active compared to a control recombinant and a HeLa cell extract (Fig. 8A). More importantly, salermide was able to inhibit the activity of the recombinant TcSir2rp3, with an IC50 in the range of 1 μM (Fig. 8B), supporting the notion that effects of salermide in the parasite were accomplished through this enzyme.
FIG 8.
Recombinant TcSir2rp3 is inhibited by salermide. (A) Mean fluorescence ± SE (n = 3) of buffer (Blank), an unrelated recombinant (Control), the recombinant TcSir2rp3, and a HeLa cell extract incubated with the Abz-Gly-Pro-acetyl-Lys-Ser-Gln-EDDnp, as described in Materials and Methods, for 3 h at 37°C. The reactions were stopped with the addition of nicotinamide, and reaction mixtures were incubated for 30 min without (white bars) or with (black bars) trypsin (T). (B) Recombinant TcSir2rp3 was preincubated with the indicated amounts of salermide in the reaction buffer without the Abz-Gly-Pro-acetylLys-Ser-Gln-EDDnp. After 15 min, the deacetylase reactions were initiated by adding the peptide substrate. Similar results were obtained with two different enzyme preparations using at least two different batches of salermide.
DISCUSSION
It is becoming clear that protein acetylation is a key posttranslational modification that regulates several biological processes in higher eukaryotes (42–44) and protozoan parasites (45, 46). Here, we present evidence that sirtuins can be targets for Chagas disease therapy, as these enzymes are important factors controlling T. cruzi growth and differentiation during infection. The idea that sirtuins play a key role derives from our results showing that specific inhibitors largely affect growth and mainly differentiation of T. cruzi.
T. cruzi contains and expresses two sirtuins, one in the cytosol and the other in the mitochondrion, based on immunofluorescence analysis and epitope tagging. We were not able to detect labeling for both enzymes in the nucleus in the different parasite stages or under different growing conditions for either the endogenous or the tagged proteins. The cytosolic localization of the rp1 homologue was also found in Leishmania (22) and differs from what was observed in T. brucei, which contains three enzymes, one reported to be located in the nucleus affecting DNA repair and telomeric silencing and two present in the mitochondrion (18).
We found that exogenous expression of these T. cruzi sirtuins had opposing effects. TcSir2rp1 affected parasite growth and differentiation to infective forms in a negative way, as observed for T. brucei (18). TcSir2rp1 did not dramatically affect growth, as observed in T. brucei, which relocalized from the nucleus to the cytosol upon overexpression (18), because it was minimally overexpressed. In contrast, TcSir2rp3 stimulated both growth and differentiation of epimastigotes. This could be explained by different targets being deacetylated by each one of the enzymes in either the cytosol or the mitochondrion. For example, TcSir2rp1 deacetylated in the cytosol cytoskeleton proteins, such as tubulin, which has been described as a main target for trypanosomatid cytoplasmic sirtuins (22). Alpha- and beta-tubulin monomers form microtubules, which are involved in several eukaryotic cellular functions, such as cell division, intracellular transport, and migration (47). As tubulin acetylation stabilizes neighboring tubulin protofilament interactions (48), the overexpression of TcSir2rp1 might destabilize microtubules, causing reduced cellular division and metacyclogenesis, processes that depend on changes in parasite morphology (49). In contrast, the increase in parasite proliferation, movement, and differentiation observed for TcSir2rp3-ox cell lines might be explained in part by changes in the activity of key enzymes of the metabolic control and oxidative stress response, known to be regulated by acetylation in the mitochondria of different organisms (50–52). For example, acetylation controls the activity of superoxide dismutase (53, 54), ATP synthase (F1F0 ATPase) (55), and acetyl-CoA synthetase (56, 57). The fact that overexpression of a mutated version of TcSir2rp3 without enzymatic activity does not affect growth and differentiation supports the notion that deacetylation of mitochondrial targets is responsible for the observed phenotypes.
The overexpression of TcSir2rp3 increased the invasion capacity of metacyclic trypomastigotes but not of cell-derived trypomastigotes. In fact, metacyclic forms express much less TcSir2rp3 than cell-derived trypomastigotes, indicating that their invasion capacity might be limited by energy supply, which is shown to be required for adhesion and invasion (58). On the other hand, after invasion, both forms differentiate into amastigotes, which also have small amounts of TcSir2rp3. Overexpression of the enzyme increased the proliferation rate, probably by accelerating parasite metabolism in comparison with the wild type. Moreover, by growing faster, the overexpressors burst the cells before the differentiation of amastigotes back to trypomastigotes. Perhaps the low levels of TcSir2rp3 in amastigotes guarantee full differentiation due to a more controlled replication rate. Accordingly, these effects were not observed when we performed the assays with metacyclic trypomastigotes derived from TcSir2rp3-H113Y, indicating that the deacetylase activity was involved in these processes. In addition, the overexpression of the mutated form decreased the invasion and intracellular multiplication relative to those of the controls. This pronounced effect might be due to the low expression levels of endogenous protein in metacyclic trypomastigote and amastigote forms of the parasite in which the overexpression of a mutated version of the protein acted as a dominant negative in these forms, perhaps by competing with protein substrates and cofactors, such as NAD+.
The key role of sirtuins in the regulation of vital cellular processes in T. cruzi supports their use as targets for drug development, as proposed recently (23, 59–61). Here, we used salermide, a specific sirtuin inhibitor that had been tested for treatment of some kinds of cancers in vitro and in vivo, with high efficacy (39). Salermide impaired epimastigote proliferation and differentiation to infective forms. This effect was observed in vitro and in vivo. We were able to detect a reduction in amastigotes after 48 h posttreatment of cell infection in vitro by using concentrations that have little effect on host cells. This in vitro effect was more pronounced when treating the cells just after cell invasion, when the trypomastigotes still express large amounts of TcSir2rp3, and before the differentiation into amastigotes. We also obtained a reduction in parasitemia with treatments at doses lower than the ones shown to be effective for eliminating cancer cells in vivo, without side effects in the mice. However, we could not prevent mortality with the employed doses, arguing that further developments are required to obtain more-potent sirtuin inhibitors for T. cruzi. Importantly, the concentration of salermide required to inhibit differentiation is much lower than the ones required for growth. This might be due to deacetylation, which might be triggered by stress responses through the activation of the above-mentioned enzymes, also suggesting that sirtuin inhibitors could be used in combination with other drugs.
Salermide might act also on TcSir2rp1 and TcSir2rp3, with opposite effects. Therefore, it would be important to develop more specific and safe inhibitors. In this sense, we have initially expressed a recombinant and active form of TcSir2rp3 and showed that it might be inhibited by salermide at doses much lower than those used to inhibit the human enzyme. This finding might indicate that TcSir2rp3 is the target of salermide in T. cruzi. Several sirtuin inhibitors have recently been developed, aiming to obtain highly specific inhibitors for each one of the sirtuins (62, 63). Therefore, it would be possible to develop more potent inhibitors to T. cruzi sirtuins.
In conclusion, in this work, we demonstrated for the first time the involvement of sirtuins in the control of several cellular processes in all parasite forms and extend the opportunity to explore sirtuins with specificity toward each one of the T. cruzi enzymes as promising drug targets for Chagas disease treatment.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by FundaçaÞo de Amparo à Pesquisa do Estado de SaÞo Paulo (FAPESP) (grant 2011/51973-3 to S.S., 2012/09403-8 to N.S.M., 2009/54364-8 to L.D.S.A, and 2013/16211-0 to R.P.P.A) and by the Conselho Nacional de Desenvolvimento Científico e Tecnologico-CNPq (grant 477143/2011-3 to S.S, Instituto Nacional de Ciência e Tecnologia de Vacinas) from Brazil.
We thank Claudio Rogério Oliveira and Claudeci Medeiros for their technical help.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.04694-14.
REFERENCES
- 1.Norris KL, Lee JY, Yao TP. 2009. Acetylation goes global: the emergence of acetylation biology. Sci Signal 2:pe76. doi: 10.1126/scisignal.297pe76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. 2000. The language of covalent histone modifications. Nature 403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.Norvell A, McMahon SB. 2010. Cell biology. Rise of the rival. Science 327:964–965. doi: 10.1126/science.1187159. [DOI] [PubMed] [Google Scholar]
- 4.Gregoretti IV, Lee YM, Goodson HV. 2004. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol 338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 5.Landry J, Slama JT, Sternglanz R. 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun 278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 6.Tanny JC, Moazed D. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci U S A 98:415–420. doi: 10.1073/pnas.98.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan M, Peng C, Anderson KA, Chhoy P, Xie Z, Dai L, Park J, Chen Y, Huang H, Zhang Y, Ro J, Wagner GR, Green MF, Madsen AS, Schmiesing J, Peterson BS, Xu G, Ilkayeva OR, Muehlbauer MJ, Braulke T, Muhlhausen C, Backos DS, Olsen CA, McGuire PJ, Pletcher SD, Lombard DB, Hirschey MD, Zhao Y. 2014. Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab 19:605–617. doi: 10.1016/j.cmet.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, Lombard DB, Zhao Y. 2013. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell 50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb S, Esposito RE. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 10.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. 1990. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell 63:751–762. [DOI] [PubMed] [Google Scholar]
- 11.Lin SJ, Defossez PA, Guarente L. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 12.Liu T, Liu PY, Marshall GM. 2009. The critical role of the class III histone deacetylase SIRT1 in cancer. Cancer Res 69:1702–1705. doi: 10.1158/0008-5472.CAN-08-3365. [DOI] [PubMed] [Google Scholar]
- 13.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. 2007. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res 67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 14.Yuan H, Su L, Chen WY. 2013. The emerging and diverse roles of sirtuins in cancer: a clinical perspective. Onco Targets Ther 6:1399–1416. doi: 10.2147/OTT.S37750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev 9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 16.Ivy JM, Klar AJ, Hicks JB. 1986. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol 6:688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rine J, Herskowitz I. 1987. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics 116:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alsford S, Kawahara T, Isamah C, Horn D. 2007. A sirtuin in the African trypanosome is involved in both DNA repair and telomeric gene silencing but is not required for antigenic variation. Mol Microbiol 63:724–736. doi: 10.1111/j.1365-2958.2006.05553.x. [DOI] [PubMed] [Google Scholar]
- 19.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. 2005. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-Salcedo JA, Gijón P, Nolan DP, Tebabi P, Pays E. 2003. A chromosomal SIR2 homologue with both histone NAD-dependent ADP-ribosyltransferase and deacetylase activities is involved in DNA repair in Trypanosoma brucei. EMBO J 22:5851–5862. doi: 10.1093/embojcdg553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zemzoumi K, Sereno D, François C, Guilvard E, Lemesre JL, Ouaissi A. 1998. Leishmania major: cell type dependent distribution of a 43 kDa antigen related to silent information regulatory-2 protein family. Biol Cell 90:239–245. [DOI] [PubMed] [Google Scholar]
- 22.Tavares J, Ouaissi A, Santarem N, Sereno D, Vergnes B, Sampaio P, Cordeiro-da-Silva A. 2008. The Leishmania infantum cytosolic SIR2-related protein 1 (LiSIR2RP1) is an NAD+-dependent deacetylase and ADP-ribosyltransferase. Biochem J 415:377–386. doi: 10.1042/BJ20080666. [DOI] [PubMed] [Google Scholar]
- 23.Veiga-Santos P, Reignault LC, Huber K, Bracher F, De Souza W, De Carvalho TM. 2014. Inhibition of NAD+-dependent histone deacetylases (sirtuins) causes growth arrest and activates both apoptosis and autophagy in the pathogenic protozoan Trypanosoma cruzi. Parasitology 141:814–825. doi: 10.1017/S0031182013001704. [DOI] [PubMed] [Google Scholar]
- 24.Demir O, Labaied M, Merritt C, Stuart K, Amaro RE. 2014. Computer-aided discovery of Trypanosoma brucei RNA-editing terminal uridylyl transferase 2 inhibitors. Chem Biol Drug Des 84:131–139. doi: 10.1111/cbdd.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Camargo EP. 1964. Growth and differentiation in Trypanosoma cruzi. I. Origin of metacyclic trypomastigotes in liquid media. Rev Inst Med Tropical S Paulo 6:93–100. [PubMed] [Google Scholar]
- 26.Abuin G, Freitas-Junior LHG, Colli W, Alves MJ, Schenkman S. 1999. Expression of trans-sialidase and 85 kDa glycoprotein genes in Trypanosoma cruzi is differentially regulated at the post-transcriptional level by labile protein factors. J Biol Chem 274:13041–13047. doi: 10.1074/jbc.274.19.13041. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida N, Mortara RA, Araguth MF, Gonzalez JC, Russo M. 1989. Metacyclic neutralizing effect of monoclonal antibody 10D8 directed to the 35- and 50-kilodalton surface glycoconjugates of Trypanosoma cruzi. Infect Immun 57:1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cornejo A, Oliveira CR, Wurtele M, Chung J, Hilpert K, Schenkman S. 2012. A novel monoclonal antibody against the C-terminus of beta-tubulin recognizes endocytic organelles in Trypanosoma cruzi. Protein Pept Lett 19:636–643. doi: 10.2174/092986612800494075. [DOI] [PubMed] [Google Scholar]
- 29.Araya JE, Cano MI, Yoshida N, da Silveira JF. 1994. Cloning and characterization of a gene for the stage-specific 82-kDa surface antigen of metacyclic trypomastigotes of Trypanosoma cruzi. Mol Biochem Parasitol 65:161–169. doi: 10.1016/0166-6851(94)90124-4. [DOI] [PubMed] [Google Scholar]
- 30.Ramirez MI, Yamauchi LM, de Freitas LH Jr, Uemura H, Schenkman S. 2000. The use of the green fluorescent protein to monitor and improve transfection in Trypanosoma cruzi. Mol Biochem Parasitol 111:235–240. doi: 10.1016/S0166-6851(00)00309-1. [DOI] [PubMed] [Google Scholar]
- 31.Angelo PF, Lima AR, Alves FM, Blaber SI, Scarisbrick IA, Blaber M, Juliano L, Juliano MA. 2006. Substrate specificity of human kallikrein 6: salt and glycosaminoglycan activation effects. J Biol Chem 281:3116–3126. doi: 10.1074/jbc.M510096200. [DOI] [PubMed] [Google Scholar]
- 32.Massayuki Kikuti C, Tersariol IL, Schenkman S. 2006. Divalent metal requirements for catalysis and stability of the RNA triphosphatase from Trypanosoma cruzi. Mol Biochem Parasitol 150:83–95. doi: 10.1016/j.molbiopara.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 33.Chow C, Cloutier S, Dumas C, Chou MN, Papadopoulou B. 2011. Promastigote to amastigote differentiation of Leishmania is markedly delayed in the absence of PERK eIF2alpha kinase-dependent eIF2alpha phosphorylation. Cell Microbiol 13:1059–1077. doi: 10.1111/j.1462-5822.2011.01602.x. [DOI] [PubMed] [Google Scholar]
- 34.Martins RM, Alves RM, Macedo S, Yoshida N. 2011. Starvation and rapamycin differentially regulate host cell lysosome exocytosis and invasion by Trypanosoma cruzi metacyclic forms. Cell Microbiol 13:943–954. doi: 10.1111/j.1462-5822.2011.01590.x. [DOI] [PubMed] [Google Scholar]
- 35.Minucci S, Pelicci PG. 2006. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 36.Milne JC, Denu JM. 2008. The sirtuin family: therapeutic targets to treat diseases of aging. Curr Opin Chem Biol 12:11–17. doi: 10.1016/j.cbpa.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 37.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem 277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 38.Jackson MD, Schmidt MT, Oppenheimer NJ, Denu JM. 2003. Mechanism of nicotinamide inhibition and transglycosidation by Sir2 histone/protein deacetylases. J Biol Chem 278:50985–50998. doi: 10.1074/jbc.M306552200. [DOI] [PubMed] [Google Scholar]
- 39.Lara E, Mai A, Calvanese V, Altucci L, Lopez-Nieva P, Martinez-Chantar ML, Varela-Rey M, Rotili D, Nebbioso A, Ropero S, Montoya G, Oyarzabal J, Velasco S, Serrano M, Witt M, Villar-Garea A, Imhof A, Mato JM, Esteller M, Fraga MF. 2009. Salermide, a sirtuin inhibitor with a strong cancer-specific proapoptotic effect. Oncogene 28:781–791. doi: 10.1038/onc.2008.436. [DOI] [PubMed] [Google Scholar]
- 40.Rotili D, Tarantino D, Nebbioso A, Paolini C, Huidobro C, Lara E, Mellini P, Lenoci A, Pezzi R, Botta G, Lahtela-Kakkonen M, Poso A, Steinkuhler C, Gallinari P, De Maria R, Fraga M, Esteller M, Altucci L, Mai A. 2012. Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J Med Chem 55:10937–10947. doi: 10.1021/jm3011614. [DOI] [PubMed] [Google Scholar]
- 41.Schwer B, North BJ, Frye RA, Ott M, Verdin E. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158:647–657. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 43.Kaluarachchi Duffy S, Friesen H, Baryshnikova A, Lambert JP, Chong YT, Figeys D, Andrews B. 2012. Exploring the yeast acetylome using functional genomics. Cell 149:936–948. doi: 10.1016/j.cell.2012.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. 2006. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell 23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 45.Jeffers V, Sullivan WJ Jr. 2012. Lysine acetylation is widespread on proteins of diverse function and localization in the protozoan parasite Toxoplasma gondii. Eukaryot Cell 11:735–742. doi: 10.1128/EC.00088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miao J, Lawrence M, Jeffers V, Zhao F, Parker D, Ge Y, Sullivan WJ Jr, Cui L. 2013. Extensive lysine acetylation occurs in evolutionarily conserved metabolic pathways and parasite-specific functions during Plasmodium falciparum intraerythrocytic development. Mol Microbiol 89:660–675. doi: 10.1111/mmi.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nogales E. 2000. Structural insights into microtubule function. Annu Rev Biochem 69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 48.Cueva JG, Hsin J, Huang KC, Goodman MB. 2012. Posttranslational acetylation of α-tubulin constrains protofilament number in native microtubules. Curr Biol 22:1066–1074. doi: 10.1016/j.cub.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rondinelli E, de Moura-Neto RS, Silva R, de Almeida Soares CM, Carvalho JF, Torres de Castro F. 1986. Control of tubulin gene expression during metacyclogenesis of Trypanosoma cruzi. FEBS Lett 208:379–385. doi: 10.1016/0014-5793(86)81053-5. [DOI] [PubMed] [Google Scholar]
- 50.Amado FM, Barros A, Azevedo AL, Vitorino R, Ferreira R. 2014. An integrated perspective and functional impact of the mitochondrial acetylome. Expert Rev Proteomics 11:383–394. doi: 10.1586/14789450.2014.899470. [DOI] [PubMed] [Google Scholar]
- 51.Hebert AS, Dittenhafer-Reed KE, Yu W, Bailey DJ, Selen ES, Boersma MD, Carson JJ, Tonelli M, Balloon AJ, Higbee AJ, Westphall MS, Pagliarini DJ, Prolla TA, Assadi-Porter F, Roy S, Denu JM, Coon JJ. 2013. Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol Cell 49:186–199. doi: 10.1016/j.molcel.2012.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newman JC, He W, Verdin E. 2012. Mitochondrial protein acylation and intermediary metabolism: regulation by sirtuins and implications for metabolic disease. J Biol Chem 287:42436–42443. doi: 10.1074/jbc.R112.404863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tao R, Coleman MC, Pennington JD, Ozden O, Park SH, Jiang H, Kim HS, Flynn CR, Hill S, Hayes McDonald W, Olivier AK, Spitz DR, Gius D. 2010. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol Cell 40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Zhang J, Lin Y, Lei Q, Guan KL, Zhao S, Xiong Y. 2011. Tumour suppressor SIRT3 deacetylates and activates manganese superoxide dismutase to scavenge ROS. EMBO Rep 12:534–541. doi: 10.1038/embor.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vassilopoulos A, Pennington JD, Andresson T, Rees DM, Bosley AD, Fearnley IM, Ham A, Flynn CR, Hill S, Rose KL, Kim HS, Deng CX, Walker JE, Gius D. 2014. SIRT3 deacetylates ATP synthase F complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal 21:551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. 2006. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A 103:10224–10229. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hallows WC, Lee S, Denu JM. 2006. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A 103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schenkman S, Robbins ES, Nussenzweig V. 1991. Attachment of Trypanosoma cruzi to mammalian cells requires parasite energy, and invasion can be independent of the target cell cytoskeleton. Infect Immun 59:645–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soares MB, Silva CV, Bastos TM, Guimarães ET, Figueira CP, Smirlis D, Azevedo WF Jr. 2012. Anti-Trypanosoma cruzi activity of nicotinamide. Acta Trop 122:224–229. doi: 10.1016/j.actatropica.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 60.Sacconnay L, Smirlis D, Queiroz EF, Wolfender JL, Soares MB, Carrupt PA, Nurisso A. 2013. Structural insights of SIR2rp3 proteins as promising biotargets to fight against Chagas disease and leishmaniasis. Mol Biosyst 9:2223–2230. doi: 10.1039/c3mb70180h. [DOI] [PubMed] [Google Scholar]
- 61.Sacconnay L, Angleviel M, Randazzo GM, Queiroz MM, Queiroz EF, Wolfender JL, Carrupt PA, Nurisso A. 2014. Computational studies on sirtuins from Trypanosoma cruzi: structures, conformations and interactions with phytochemicals. PLoS Negl Trop Dis 8:e2689. doi: 10.1371/journal.pntd.0002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seifert T, Malo M, Kokkola T, Engen K, Fridén-Saxin M, Wallén EAA, Lahtela-Kakkonen M, Jarho EM, Luthman K. 2014. Chroman-4-one- and chromone-based sirtuin 2 inhibitors with antiproliferative properties in cancer cells. J Med Chem 57:9870–9888. doi: 10.1021/jm500930h. [DOI] [PubMed] [Google Scholar]
- 63.Cui H, Kamal Z, Ai T, Xu Y, More SS, Wilson DJ, Chen L. 2014. Discovery of potent and selective sirtuin 2 (SIRT2) inhibitors using a fragment-based approach. J Med Chem 57:8340–8357. doi: 10.1021/jm500777s. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.