Background: The structure of the HIV glycoprotein transmembrane anchor is unknown.
Results: NMR spectroscopy reveals two helices connected by a flexible segment. The N-terminal helix constitutes a scaffold for neutralizing antibodies.
Conclusion: The HIV transmembrane sequence combines two subdomains involved in fusion and immune response modulation during infection.
Significance: These data may guide the rational design of vaccines and inhibitors.
Keywords: AIDS, Human Immunodeficiency Virus (HIV), Nuclear Magnetic Resonance (NMR), Peptide Conformation, Transmembrane Domain, Vaccine Development, Immunogenic Domain Structure, Immunosuppressive Domain Structure, MPER Vaccine
Abstract
The membrane-proximal external region (MPER) C-terminal segment and the transmembrane domain (TMD) of gp41 are involved in HIV-1 envelope glycoprotein-mediated fusion and modulation of immune responses during viral infection. However, the atomic structure of this functional region remains unsolved. Here, based on the high resolution NMR data obtained for peptides spanning the C-terminal segment of MPER and the TMD, we report two main findings: (i) the conformational variability of the TMD helix at a membrane-buried position; and (ii) the existence of an uninterrupted α-helix spanning MPER and the N-terminal region of the TMD. Thus, our structural data provide evidence for the bipartite organization of TMD predicted by previous molecular dynamics simulations and functional studies, but they do not support the breaking of the helix at Lys-683, as was suggested by some models to mark the initiation of the TMD anchor. Antibody binding energetics examined with isothermal titration calorimetry and humoral responses elicited in rabbits by peptide-based vaccines further support the relevance of a continuous MPER-TMD helix for immune recognition. We conclude that the transmembrane anchor of HIV-1 envelope is composed of two distinct subdomains: 1) an immunogenic helix at the N terminus also involved in promoting membrane fusion; and 2) an immunosuppressive helix at the C terminus, which might also contribute to the late stages of the fusion process. The unprecedented high resolution structural data reported here may guide future vaccine and inhibitor developments.
Introduction
The HIV-1 envelope (Env)8 glycoprotein forms trimers of noncovalently associated heterodimers at the surface of the infectious virus (1–3). Furin-like proteases cleave the gp160 precursor to generate gp120 (surface) and gp41 (transmembrane) subunits, which mediate receptor binding and virus-cell fusion, respectively (1–4). Insights into the structural organization of the gp41 ectodomain have been recently obtained from cryo-electron microscopy and crystallographic studies of soluble cleaved gp140 trimers (Fig. 1A) (5–7). In contrast, structure-function relationships for the Env transmembrane domain (TMD) anchor and its connection to the ectodomain remain poorly defined (8–11).
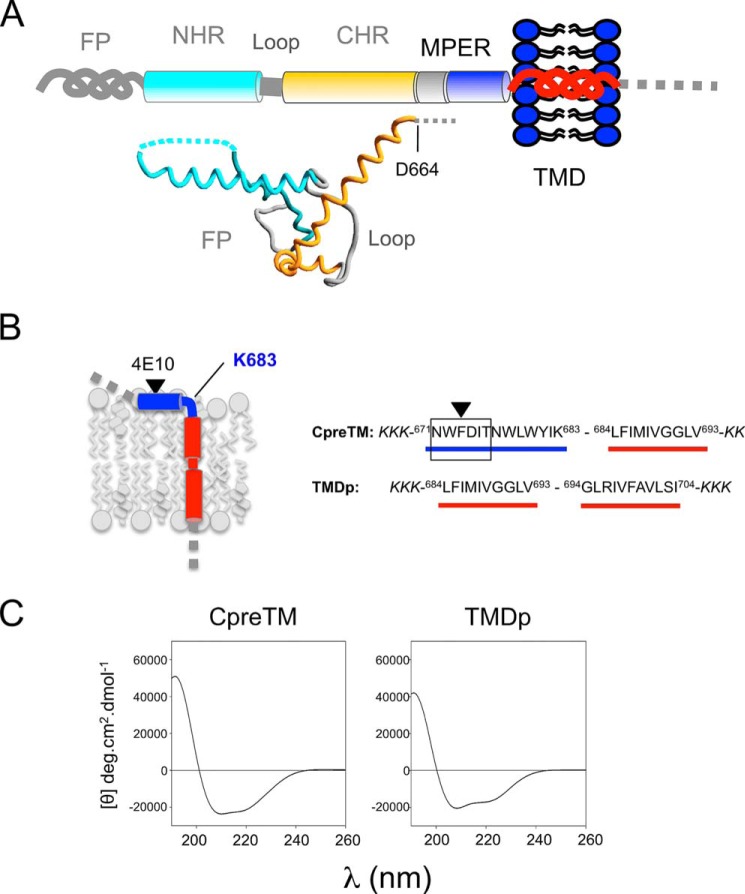
FIGURE 1.
Designation of peptides covering the HIV-1 gp41 TMD sequence and its connection to MPER. A, diagram illustrating the most important structural constituents of membrane-anchored gp41 ectodomain. FP, fusion peptide; NHR and CHR, N- and C-terminal helical regions, respectively. The same color code has been used to depict the gp41 ectodomain pre-fusion structure below (PDB accession code 4TVP). B, left panel, membrane topology predicted for the TMD and its connection to MPER. Right panel, designation of the overlapping CpreTM and TMDp peptides. C, circular dichroism spectra of CpreTM and TMDp peptides in the low polarity medium provided by 25% HFIP.
Computational techniques have provided evidence for TMD self-oligomerization in the membrane milieu (12, 13). These studies suggest that inter-chain hydrogen bonds may stabilize membrane-embedded trimers. Molecular dynamics simulation (MDS) studies on monomers revealed potential kinks and/or metastable hinges at or around Arg-696, a residue located near the center of TMD (14, 15). According to these calculations, the most stable conformation for TMD would be a complete α-helical conformation. This continuous helix can nevertheless bend and acquire metastable conformations in the membrane (14). In line with a potential conformational plasticity, several mutagenesis studies support an active role for the TMD in gp41-mediated fusion during viral entry (9, 16–18). Particularly, the specific sequence of the highly conserved N-terminal region (residues 684LFIMIVGGLVGL695), the presence of basic residues flanking or within the TMD, and the spacing between these charged residues at the C terminus of the TMD appear to be critical for the fusogenicity of Env complexes and infectivity of HIV-1 (9, 16, 18).
In addition, the TMD and the adjacent membrane-proximal external region (MPER) C-terminal segment have been involved in modulation of innate and adaptive immune responses during HIV infection. The boundary between MPER and TMD constitutes a site of vulnerability on the Env glycoprotein, which is targeted by broadly neutralizing antibodies (bNAb-s) 4E10 and 10E8 (19–21). The peptide epitopes are bound to these Fabs in a highly unusual helical conformation (22, 23). Consistent with the existence of a defined MPER-TMD structural motif, the gp41 TMD orients the epitope recognized by the 4E10 bNAb for optimal binding and overall modulates MPER epitope exposure (13). However, most of the current models do not include the MPER C-terminal 4E10/10E8 epitopes as constituents of the TMD anchor (24–29). Those models further posit that the TMD starts at Lys-683, where a sharp bend is predicted to occur in the helix backbone (Fig. 1B, left panel).
In addition, the C-terminal section of the TMD comprises a distinct immunosuppressive domain (30, 31). Studies motivated by the similarity between the sequence 694GLRIVFAV701 and a transmembrane section of the T-cell receptor α subunit demonstrated the interaction of the gp41 TMD with CD3 partners in the TCR complex, resulting in the inhibition of T-cell proliferation (30). More recently, it has been reported that the gp41 TMD may also associate with the Toll-like receptor-2 TMD in the membrane, interfering with the receptor-mediated signaling and decreasing pro-inflammatory cytokine secretion (32). Collectively, these findings suggest that the sequence of the HIV-1 TMD plays critical functions beyond that of anchoring the Env complex to membranes (8–11), although the structural requirements underlying these distinct functional roles remain to be determined.
Here, we have undertaken the structural characterization of the gp41 TMD and its connection to the upstream MPER C-terminal section by using two overlapping synthetic peptides, CpreTM and TMDp, that together cover the HIV-1 Env(671–704) sequence (HXB2c numbering) (Fig. 1B, right panel). The CpreTM peptide represents the MPER-TMD boundary and is delimited between the hinge segment at position 671–674 (24, 26, 33, 34) and the membrane-buried kink around position 693, as predicted by hydrophobicity (35) and MDS (14) analyses. Thus, NMR measurements of CpreTM in membrane mimetics were expected to prove the existence of the Lys-683 kink in the middle of its sequence. The sequence and range of the TMDp peptide corresponded to that recently defined as the full-length gp41 TMD by Cohen et al. (30). To our knowledge, NMR studies on gp41 TMD and its connection to MPER have not been reported to date.
The high resolution NMR data confirmed that CpreTM and TMDp adopt α-helical structures in membrane-mimicking environments, as well as conformational flexibility around the transmembrane sequence 690GGLV693. In contrast, no evidence was found for significant changes in the orientation of the backbone of the helix at, or close to, position Lys-683, which had been proposed to mark the initiation of the TMD at the membrane interface. Supporting the relevance of a continuously helical MPER-TMD connection for immune recognition, the CpreTM peptide based on this sequence was bound with nanomolar affinity by the anti-MPER 4E10 bNAb and raised IgG in rabbits that inhibited Env-mediated cell entry. Thus, according to the high resolution data reported here, the HIV-1 Env(671–704) sequence can be described as a two-subdomain helical motif separated by a membrane-embedded flexible hinge. The N-terminal helix, longer than expected, would be involved in membrane fusion and elicitation of neutralizing antibodies, whereas the C-terminal helix would take part in modulation of T-cell immune responses and in the last stages of the fusion process.
Experimental Procedures
Materials
The peptides used in the structural and immunological studies were synthesized in C-terminal carboxamide form by solid-phase methods using Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry, purified by reverse phase HPLC, and characterized by matrix-assisted time-of-flight (MALDI-TOF) mass spectrometry (purity >95%). Peptides were routinely dissolved in dimethyl sulfoxide (DMSO, spectroscopy grade), and their concentration was determined by the bicinchoninic acid microassay (Pierce). MAb4E10 was a gift of Dietmar Katinger (Polymun Scientific, Klosterneuburg, Austria). The Experimental Procedures described in Refs. 36, 37 were followed for the production and purification of Fabs.
Circular Dichroism
Circular dichroism (CD) measurements were obtained from a thermally controlled Jasco J-810 circular dichroism spectropolarimeter calibrated routinely with (1S)-(+)-10-camphorsulfonic acid, ammonium salt. Samples consisted of lyophilized peptides dissolved at concentrations of 0.03 mm in 2 mm HEPES (pH 7.4) buffer containing 25% (v/v) 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP). Spectra were measured in a 1-mm pathlength quartz cell initially equilibrated at 25 °C. Data were taken with a 1 nm bandwidth at 100 nm/min speed, and the results of 20 scans were averaged.
Recording of NMR Spectra
NMR samples were prepared by dissolving the lyophilized peptides (∼1 mg) in 0.5 ml of an H2O/D2O (9:1 ratio by volume) solution containing 2 mm HEPES buffer at pH 7.0 and either 25% (v/v) 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP-D2, 98%; Cambridge Isotopes Laboratories) or 20 mm deuterated dodecylphosphocholine (DPC-D38, 98%; Cambridge Isotopes Laboratories). Peptide concentrations were ∼0.5 mm. pH was measured with a glass micro-electrode and not corrected for isotope effects. A methanol sample was used to calibrate the temperature of the NMR probe. 1H chemical shifts were referenced to internal sodium 2,2-dimethyl-2-silapentane-5-sulfonate. 13C δ-values were indirectly referenced by using the IUPAC-IUB recommended 1H/13C chemical shift ratio (0.25144953 (38)).
The NMR spectra were acquired on a Bruker Avance-600 spectrometer operating at a proton frequency of 600.13 MHz and equipped with a cryoprobe. One-dimensional spectra were acquired using 32 K data points, which were zero-filled to 64 K data points before performing the Fourier transformation. Phase-sensitive two-dimensional correlated spectroscopy (COSY), total correlated spectroscopy (TOCSY), and nuclear Overhauser enhancement spectroscopy (NOESY) spectra were recorded by standard techniques using presaturation of the water signal and the time-proportional phase incrementation mode, as reported previously (39). NOESY mixing times were 150 ms, and TOCSY spectra were recorded using 60-ms DIPSI2 with z filter spin-lock sequence. For the samples in 25% HFIP, 1H-13C heteronuclear single quantum coherence (HSQC) spectra were acquired at 13C natural abundance using standard pulse sequences (39). Acquisition data matrices were defined by 2048 × 512 points in t2 and t1, respectively. Data were processed using the TOPSPIN program (Bruker Biospin, Karlruhe, Germany). The two-dimensional data matrix was multiplied by either a square-sine-bell or a sine-bell window function with the corresponding shift optimized for every spectrum and zero-filled to a 2048 × 1024 complex matrix prior to Fourier transformation. Baseline correction was applied in both dimensions. Two-dimensional spectra were analyzed using the SPARKY software (T. D. Goddard and D. G. Kneller, SPARKY3, University of California, San Francisco).
Structure Calculation
Structures for peptide CpreTM in DPC micelles and in 25% HFIP and for TMDp in 25% HFIP were calculated from distance and dihedral angle constraints derived from NMR parameters by following the three-step protocol previously described for MPERp (34). Distance constraints were obtained from the cross-peaks present in 150-ms two-dimensional 1H-1H NOESY spectra, which were integrated using the standard SPARKY integration subroutine (SPARKY3), and the dihedral angle restraints for φ and ψ angles were derived from 1Hα and 13Cα chemical shifts using the program TALOS (40). First, we applied the standard iterative procedure for automatic NOE assignment of the program CYANA 2.1 (41), which consists of seven cycles of combined automated NOE assignment and structure calculation of 100 conformers per cycle (42). The list of distance constraints resulting from the last automatic cycle was checked by inspection of the corresponding NOESY spectra, and ambiguous constraints were removed or relaxed to generate the final list used as input for a standard simulated annealing CYANA 2.1 calculation of 100 conformers. The 20 conformers with the lowest target function values were selected and subjected to 2,000 steps of energy minimization using the generalized Born continuum solvation model with a nonbonded cutoff of 10 Å as implemented in the program AMBER9 (D. A. Case, T. A. Darden, and T. E. Cheatham, III, University of California, San Francisco). The quality of these final structures was assessed using PROCHECK/NMR (43) as implemented at the Protein Structure Validation Suite server. All of the residues were either in the most favored or allowed regions of the Ramachandran map (Table 1). The structural ensembles calculated for CpreTM and TMDp have been deposited in the PDB data bank with accession codes 2MG1 (TMDp in HFIP), 2MG2 (CpreTM in HFIP), and 2MG3 (CpreTM in DPC). The structural statistics data for these structures are provided in Table 1. The structures were visualized and examined using the programs MOLMOL (44) and Swiss-Pdb viewer (45).
TABLE 1.
Structural statistics for the ensemble of the 20 lowest energy NMR structures of CpreTM and TMDp
Structural statistics for the ensemble of the 20 lowest energy NMR structures of CpreTM in DPC (20 mm deuterated dodecylphosphocholine, 2 mm HEPES (pH 7.0), H2O/D2O, 9:1, v/v) and HFIP (25% deuterated 1,1,1,3,3,3-hexafluoro-2-propanol in 2 mm HEPES (pH 7.0), H2O/D2O, 9:1, v/v), and TMDp in HFIP. PDB codes are indicated in parentheses. r.m.s.d., root mean square deviation.
| CpreTM in DPC (2MG3) | CpreTM in HFIP (2MG2) | TMDp in HFIP (2MG1) | ||
|---|---|---|---|---|
| No. of distance restraints | Intraresidue (i − j = 0) | 126 | 156 | 129 |
| Sequential (|i − j| = 1) | 61 | 106 | 99 | |
| Medium range (1 < |i − j| < 5) | 112 | 98 | 138 | |
| Total no. | 299 | 360 | 366 | |
| Averaged total no./residue | 10.7 | 12.9 | 13.6 | |
| No. of dihedral angle constraints | φ angles | 23 | 25 | 25 |
| ψ angles | 24 | 22 | 25 | |
| Total no. | 47 | 47 | 50 | |
| Average maximum violations per structure | Distance (Å) | 0.05 ± 0.01 | 0.01 ± 0.01 | 0.03 ± 0.00 |
| Dihedral angle (°) | 1.2 ± 0.5 | 0.03 ± 0.04 | 0.5 ± 0.00 | |
| Averaged structure energies | CYANA target function value | 0.07 ± 0.02 | 0.0006 ± 0.0006 | 0.003 ± 0.00002 |
| AMBER energy (kcal/mol) | −839 | −861 | −824 | |
| van der Waals energy (kcal/mol) | −164 | −180 | −157 | |
| Electrostatic energy (kcal/mol) | −1,850 | −2,006 | −1,434 | |
| Deviations from ideal geometry | Bond length (Å) | 0.015 | 0.015 | 0.014 |
| Bond angle (°) | 1.9 | 1.8 | 1.7 | |
| Pairwise r.m.s.d. (Å) | Backbone atoms | 1.1 ± 0.5 | 1.5 ± 0.5 | 1.0 ± 0.3 |
| (residues 2–27) | All heavy atoms | 1.9 ± 0.7 | 2.3 ± 0.5 | 1.7 ± 0.3 |
| Ramachandran plot (%) | Residues in most favored regions | 95.4 | 96.7 | 100 |
| Residues in additional allowed regions | 4.6 | 3.3 | 0.0 | |
| Residues in generously allowed regions | 0.0 | 0.0 | 0.0 | |
| Residues in disallowed regions | 0.0 | 0.0 | 0.0 |
Crystallization of Peptide in Complex with Recombinant Fab 4E10
Co-crystals of Fab 4E10 in complex with the peptide 4E10ep (NWFDITNWLWYIK-KKK) were prepared by the hanging drop method. The Fab-peptide complex in the presence of detergent dihexanoyl-phosphocholine at 3 mm was concentrated to 2.5 mg/ml and mixed with a reservoir solution composed of 200 mm sodium acetate, 30% PEG 8,000, and 100 mm sodium cacodylate (pH 6.5). Suitable crystals grew to full size within a few days at 20 °C, after which they were transferred to mother liquor supplemented with 20% glycerol, and subsequently frozen and stored in a vessel containing liquid N2.
Data Collection and Refinement
Diffraction data from a single crystal was collected on beamline BL5A of the Photon Factory (Tsukuba, Japan) under cryogenic conditions (100 K). Diffraction images were processed with the program MOSFLM and merged and scaled with the program SCALA of the CCP4 suite (46). The three-dimensional structure was determined by molecular replacement using the coordinates of 4E10 (PDB code, 2FX7) with the program PHASER (47). The initial model was further refined with the programs REFMAC5 (48) and COOT (49). Validation was carried out with PROCHECK (50). Data collection and refinement statistics are given in Table 2.
TABLE 2.
X-ray data collection and refinement statistics
Values in parentheses are for highest resolution shell.
| 4E10 + peptide | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 157.3, 44.6, 85.4 |
| α, β, γ (°) | 90.0, 113.8, 90.0 |
| Resolution (Å) | 44.7-2.1 (2.21-2.10) |
| No. of reflections (total) | 166,559 (18,803) |
| No. of reflections (unique) | 30,114 (3,540) |
| Rmerge (%)a | 10.3 (37.1) |
| I/σI | 14.0 (4.3) |
| Completeness (%) | 94.2 (76.6) |
| Redundancy | 5.5 (5.3) |
| Refinement | |
| Resolution (Å) | 44.7-2.10 |
| Rwork/Rfree (%)b | 16.2 / 20.6 |
| No. of atoms | |
| Protein | 3,308 |
| Peptide | 159 |
| Other (not solvent)c | 12 |
| Solvent | 309 |
| B-Factors | |
| Protein | 30.9 |
| Peptide | 41.8 |
| Other (not solvent)c | 34.6 |
| Solvent | 33.4 |
| Ramachandran plot | |
| No. of preferred regions, % | 344, 91.2% |
| No. of allowed regions, % | 32, 8.5% |
| No. of outliers, % | 1, 0.3% |
| Root mean square deviations | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.52 |
a Rmerge = ΣhklΣi|I(hkl)i − (I(hkl))|/Σhkl Σi I(hkl).
b Rwork = Σhkl|F(hkl)o − (F(hkl)c)|/Σhkl F(hkl)o; Rfree was calculated exactly as Rwork, where F(hkl)o values were taken from 4% of data not included in the refinement.
c Data correspond to two glycerol molecules.
Isothermal Titration Calorimetry
Calorimetric titration experiments were performed with a VP-ITC microcalorimeter (MicroCal, Northampton, MA) at 25 °C. Prior to the experiment, proteins were dialyzed overnight at 4 °C against 10 mm sodium phosphate, 150 mm NaCl, and 10% glycerol at pH 7.5, and degassed just before the measurement. The buffer was supplemented with 5 mm DPC. Fab4E10 (3 μm) was titrated with 40 μm peptide (dissolved in the dialysis buffer). The volume of each injection was 10 μl. The heat corresponding to the dilution of the peptide was subtracted. The binding isotherms were fitted to a one-site binding model using the program ORIGIN 7.0. The fitting yields the stoichiometry (n), the binding constant (KD), and the enthalpy (ΔH0) of the binding reaction (Table 3).
TABLE 3.
Thermodynamic parameters of binding of peptides to 4E10 Fab
| Peptide | KD | ΔG0 | ΔH0 | −TΔS0a | nb |
|---|---|---|---|---|---|
| nm | kcal mol−1 | kcal mol−1 | kcal mol−1 | ||
| 4E10ep | 167 ± 21 | −9.2 ± 0.1 | −15.3 ± 0.3 | 6.0 | 0.8 ± 0.1 |
| 4E10ep(686) | 48 ± 4 | −10.0 ± 0.1 | −17.9 ± 0.1 | 7.9 | 0.8 ± 0.1 |
| CpreTM | 22 ± 2 | −10.4 ± 0.1 | −12.4 ± 0.1 | 1.9 | 1.3 ± 0.1 |
a Temperature = 298 K.
b n refers to the molar ratio peptide/protein.
Membrane Binding Assays
Vesicle flotation experiments in sucrose gradients were performed following the method described by Yethon et al. (51). In brief, 100 μl of a sample containing rhodamine-labeled liposomes (1.5 mm lipid concentration) was adjusted to a sucrose concentration of 1.4 m in a final volume of 300 μl, and subsequently overlaid with 400- and 300-μl layers of 0.8 and 0.5 m sucrose, respectively. The gradient was centrifuged at 436,000 × g for 3 h in a TLA 120.2 rotor (Beckman Coulter, Brea CA). After centrifugation, four 250-μl fractions were collected. Material adhered to tubes was collected into a 5th fraction by washing with 250 μl of hot (100 °C) 1% (w/v) SDS.
Rabbit Immunization and Response Analyses
New Zealand White rabbits were immunized at the antibody production service from the CID-CSIC (Barcelona, Spain). For immunization in Freund's Complete (FC) adjuvant, the peptide was dissolved in 0.5 ml of PBS and mixed with an equal volume of adjuvant (Sigma). Liposome-based formulations were prepared following the methods described by Dreesman et al. (52) and Maeso et al. (53) and included MPL as adjuvant (54). Peptide dissolved in DMSO was added at a final peptide-to-lipid ratio of 1:50 (mol/mol) to a swirling solution of freeze-thaw 1-palmitoyl-2-oleoylphosphatidylcholine/cholesterol/phosphatidic acid/MPL (2.0:1.5:0.2:0.01 molar ratio) vesicles dispersed in PBS. After incubation for 1 h, the samples were lyophilized. Rabbits were inoculated intradermally at multiple sites on day 0 with 1 ml of sample reconstituted in pure water, which contained 0.5 mg of peptide. For subsequent boosting injections, 1 ml of the reconstituted liposome formulation containing 0.3 mg of peptide was used on day 21, and 0.2 mg of liposomal peptide were injected on days 42, 63, and 84. Specific antibodies against the TMD N-terminal helix were recovered from sera through affinity purification. To that end the KKK-NWFDITNWLWYIKLFI-KKK-C peptide was immobilized onto a beaded agarose support using the Sulfolink immobilization kit for peptides (Thermo Scientific, Rockford, IL). The remaining nonspecific binding sites in columns were blocked adding l-cysteine·HCl at 50 mm. Every analyzed serum was loaded on the columns after diluting and filtering it to remove the particulate material. Filtered samples were let flow through the columns five times to allow the binding of the serum antibodies that recognized specifically the immobilized peptide. After washing the columns with at least 10 bed volumes of 500 mm NaCl containing buffer to remove nonspecifically bound antibodies and most serum proteins, the fraction enriched in the specific antibodies was eluted using 100 mm glycine buffer at pH 2.5. The fraction that was not recovered using acidic pH was eluted using freshly prepared 100 mm triethylamine buffer at pH 11.5. IgG recovery was routinely determined by SDS-PAGE and Western blot analyses of the column-bound fractions.
For the cell infection inhibition assays (34, 37), HIV-1 pseudoviruses were produced by transfection of human kidney HEK293T cells with the full-length env clone JRCSF (kindly provided by Jamie K. Scott and Naveed Gulzar, Simon Fraser University, British Columbia, Canada) using calcium phosphate. Cells were co-transfected with vectors pWXLP-GFP and pCMV8.91, encoding a green fluorescent protein and an env-deficient HIV-1 genome, respectively (provided by Patricia Villace, CSIC, Madrid, Spain). After 24 h, the medium was replaced with Opti-MEM GlutaMAX II (Invitrogen) without serum. Three days after transfection, the pseudovirus particles were harvested, passed through 0.45-μm pore sterile filters (Millex® HV, Millipore NV, Brussels, Belgium), and finally concentrated by ultracentrifugation in a sucrose gradient. Neutralization was determined using TZM-bl target cells (AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health, and contributed by J. Kappes). Samples were set up in duplicate in 96-well plates and incubated for 1.5 h at 37 °C with a 10–15% tissue culture infectious dose of pseudovirus. After antibody-pseudovirus co-incubation, 11,000 target cells were added in the presence of 30 μg/ml DEAE-dextran (Sigma). Neutralization levels after 72 h were inferred from the reduction in the number of GFP-positive cells as determined by flow cytometry using a BD FACSCalibur Flow Cytometer (BD Biosciences).
Results
Models for TMD and Its Connection to MPER in Membranes
The high hydrophobicity-at-interface of the MPER C-terminal region is consistent with its insertion into the external leaflet of the viral membrane (27, 55, 56). Current models (Fig. 1B, left panel) depict the axis of the MPER C-terminal helix inserted in parallel to the membrane plane and the 4E10 epitope residues facing the membrane interface (26, 27). It is postulated that this interfacial helix is followed by a kink of ∼90° at Lys-683 to allow insertion of the TMD in a perpendicular orientation. In addition, MDS studies predict conformational flexibility for a membrane-embedded TMD stretch close to the polar Arg-696 (14, 15). However, high resolution structural data supporting these models have not been reported yet.
To obtain structural information at the atomic level on the membrane-inserting sequences of gp41, we used CD and NMR to characterize the CpreTM and TMDp synthetic peptides, encompassing the MPER C-terminal region plus the N-terminal TMD section and the full TMD, respectively (Fig. 1B, right panel). In a membrane-mimicking low polarity environment, these two peptides displayed circular dichroism minima at 208 and 222 nm (Fig. 1C), and [θ]222/[θ]208 <1 ellipticity ratios, consistent with monomeric, mainly helical structures (57). Deconvolution of the spectra using the CDPro software package (58) confirmed that the helical content in these samples amounts to ∼70%, whereas contribution of β-structures is negligible (<5%). These observations underpinned subsequent structural characterization by solution NMR.
NMR Structures of CpreTM and TMDp Peptides
Figs. 2–3 and Table 1 summarize the NMR results obtained for the partially overlapping CpreTM and TMDp sequences in DPC micelles and in 25% HFIP at pH 7.0 and 25 °C. Based on analyses of two-dimensional COSY, TOCSY, and NOESY spectra by standard sequential methods (59, 60), we completely assigned the 1H chemical shifts for peptide CpreTM in the two solvent conditions and for peptide TMDp in 25% HFIP. Most 13C chemical shifts were also assigned for the two peptides in 25% HFIP by analyzing the 1H,13C-HSQC spectra acquired at natural 13C abundance. However, because of signal broadening, we could not assign TMDp in micellar media. NMR signals of CpreTM were also slightly broader in DPC than in HFIP, but in this case only the quality of the less sensitive natural abundance 1H,13C-HSQC was greatly affected. Therefore, 13C chemical shifts for CpreTM in DPC were not assigned. The assigned chemical shifts have been deposited at BioMagResBank with accession codes 19583 (CpreTM in DPC), 19582 (CpreTM in HFIP), and 19581 (TMDp in HFIP).
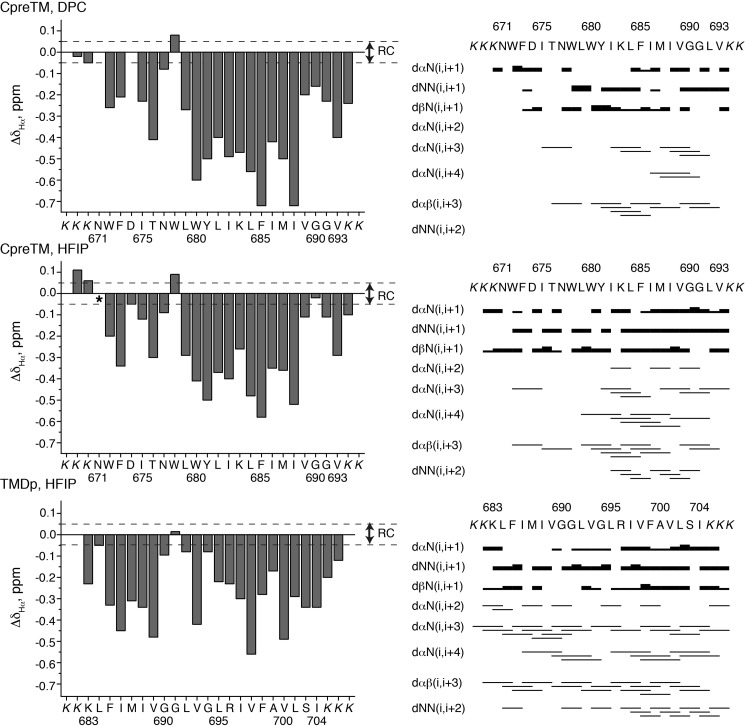
FIGURE 2.
NMR data supporting mainly helical structures for CpreTM and TMDp peptides. Peptides CpreTM and TMDp were measured in 20 mm DPC or 25% HFIP, as indicated in the panels, at pH 7.0 and 25 °C. Left panels, bar plots show the conformational shifts for the Hα protons values (ΔδHα = δHαobserved − δHαRC, ppm) as a function of peptide sequence. Random coil (RC) values for Hα protons are taken from Ref. 78. The N- and C-terminal residues are not included because of charge end effects. The asterisk in the CpreTM-HFIP panel indicates that the chemical shift for the Hα of Asn-671 was not determined. Dashed lines indicate the random coil range for ΔδHα values. Right panels, NOE summaries. In the case of the sequential NOEs, the intensities classified as strong, medium, and weak are indicated by the thickness of the lines. The Lys residues added to the wild type sequence to improve peptide solubility are shown in italics.
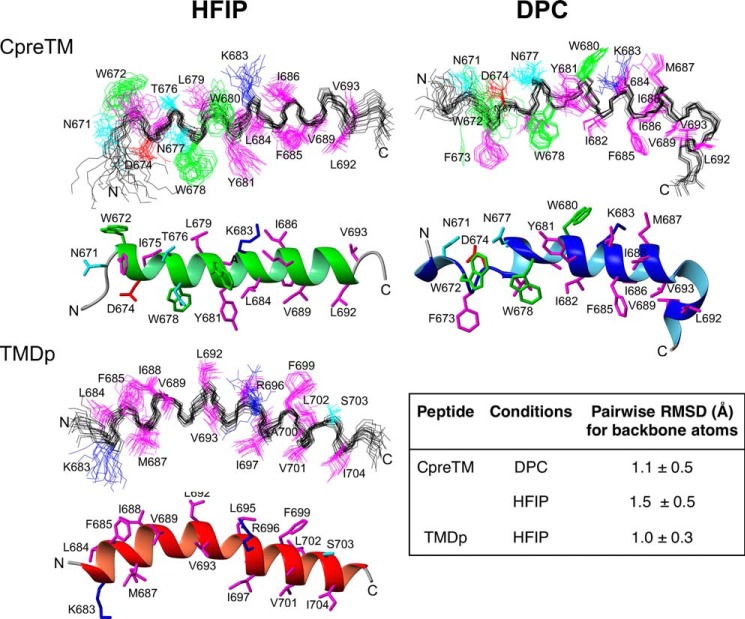
FIGURE 3.
NMR structures calculated for CpreTM and TMDp peptides in DPC 20 mm or in 25% HFIP. Superposition of the 20 lowest energy conformers and ribbon representation of the lowest energy conformer are displayed in each panel (top and bottom structures, respectively). Backbone atoms are shown in black, and side chains are colored according to residue type, i.e. the positively charged residues (R and K) are in blue; the negatively charged residues (D and E) in red; the polar residues (S, T, N, and Q) in cyan; the aromatic Trp residues in green; and all the other in magenta. Ribbons are colored as follows: green for CpreTM in HFIP, blue for CpreTM in DPC, and red for TMDp in HFIP. The inset lists the pairwise root mean square deviation values for the backbone atoms of the three structural ensembles.
Having assigned the NMR spectra, we performed a qualitative analysis of the NMR parameters. First, we examined the conformational shift ΔδHα values, which are the differences between the δ values observed for the Hα protons and those typical of random coil (RC) peptides (ΔδHα = δHαobserved − δHαRC, ppm). As seen in Fig. 2 (left panels), most ΔδHα values in peptide CpreTM in the two examined solvent conditions and in peptide TMDp in HFIP lie outside the range typical of random coil peptides (|ΔδHα| >0.05 ppm), which is an indication that the two peptides adopt ordered structures. It is empirically well established that the ΔδHα values are related to the φ and ψ dihedral angles (61), with positive values being characteristic of β-strands and negative values of α-helices. Hence, the negative ΔδHα values shown by most residues in CpreTM and TMDp (Fig. 2, left panels) are indicative of helical structure, which is highly populated according to the values' large magnitudes. The populations of helix estimated from the averaged ΔδHα value (62) (60% for CpreTM and 70% for TMDp in HFIP) are in accordance with the helix contents deduced from CD data (see above). The positive and large ΔδCα displayed by the two peptides in HFIP are also in agreement with the formation of helical structures (data not shown). It is interesting to note the similitude of the profiles observed for CpreTM in DPC and in HFIP. In both conditions, Trp-678 exhibits a positive ΔδHα value. This positive conformational shift could indicate some irregularity in the helical structure or could be explained by the anisotropy effects of the aromatic rings. Analogously, the almost null ΔδHα values observed in HFIP for Gly-691 in both peptides, CpreTM and TMDp, is compatible with the distortion of the helix around this residue (see below).
The strongest evidence on the formation of helical structures comes from the set of observed NOEs, because only pairs of spatially closed protons (approximately at a distance shorter than 5 Å) give rise to NOE cross-peaks. Thus, CpreTM in both DPC and HFIP and TMDp in HFIP showed nonsequential NOEs characteristic of helical structures, i.e. dαN(i,i + 2), dαN(i,i + 3), dαN(i,i + 4), dαβ(i,i + 3), and dNN(i,i + 2) (Fig. 2, right panel, and supplemental spectra), and medium range NOEs (i,i + 3 and i,i + 4) involving side-chain protons, for example between the aromatic protons of Phe-685 and the methyl groups of Val-689, which are observed in the two peptides, and between the Trp-678 aromatic protons and the δ methyl group of Ile-682 (supplemental spectra).
To visualize and get detail into the features of the helices, we performed structural calculations (see under “Experimental Procedures”). As reported previously, for the MPERp peptide (34), the superposition of CpreTM and TMDp structures show that the conformations are well defined (Fig. 3, top and bottom panels, respectively). The pairwise root mean square deviations of the 20 lowest energy conformers calculated for each peptide and condition are also indicative of well defined structures (Fig. 3, inset). Not only the backbone, but also most side chains can be considered ordered according to the usual criterion of the χ1 angle variation being less than ±30°.
The CpreTM structure solved in DPC displayed a distinct pattern. At the N terminus, the hinge segment 671NWFD674 (33) is partially extended, consistent with the conformational flexibility put forward by previous NMR studies (24, 26, 29, 33, 34). This element is followed by a continuous α-helix extending to Gly-690. This residue exhibits a positive dihedral φ angle (48 ± 1°), typical of residues at position C′ in C-terminal helix-capping motifs (63). At this position, a kink induces a sharp change in the direction of the helix. The following Gly-691 displays again a negative φ angle (−72 ± 2°) and helical conformation.
The main axes of 675–689 and 691–693 helices form an angle of 133 ± 2°, based on calculations using the 20 models and the CalcHelix routine of the MOLMOL program (44). This helix-kink-helix motif appears to be stabilized by nonpolar interactions between side chains of Phe-685, Ile-686, Val-689, and Val-693 that cluster at the inner side of the elbow and are mostly buried from the solvent (mean accessible solvent surface areas are 31.5, 27.4, 7.9, and 25.7%, respectively).
A less abrupt, but significant change in the helix direction is also observed around the Gly-690/Gly-691 dipeptide in the CpreTM and TMDp structures solved in HFIP. In this medium, the helical axes form angles of 52 ± 3 and 34 ± 6° in CpreTM and TMDp, respectively. In line with the observed bending, the helix H-bond networks are also disrupted around Gly-690/Gly-691. Thus, instead of bonding to Met-687, the Gly-691 amide is H-bonded to Ile-688 CO in CpreTM, whereas Gly-691 and Leu-692 amides are not H-bonded in TMDp. Nonetheless, in both HFIP structures, the φ angles of Gly-690/Gly-691 residues remained negative (−56 ± 1°/−107 ± 3° in CpreTM-HFIP, and −61 ± 3°/−66 ± 4° in TMDp), indicating that bending occurs without local unfolding of the helical conformation. As compared with the residue burial observed for the kinked structure in CpreTM-DPC, the more exposed side chains of residues Ile-686, Val-689, and Val-693 further supported preservation of the overall secondary structure in the bent HFIP structures.
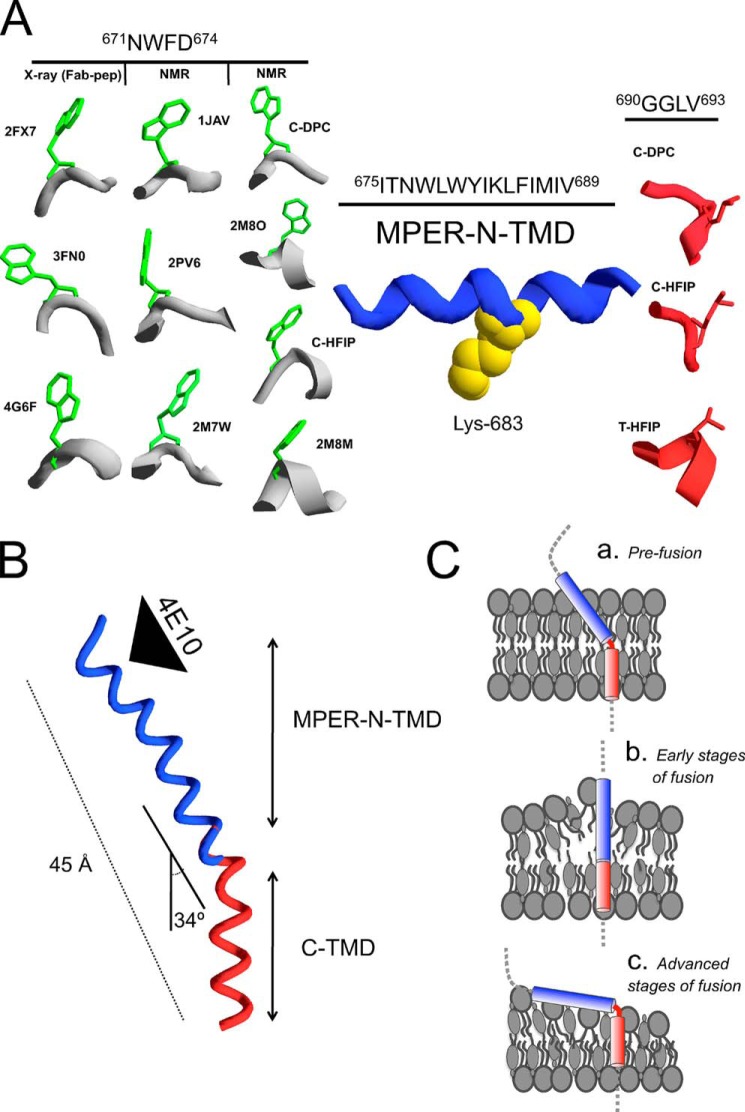
Designation of MPER-N-TMD and C-TMD Subdomains
The high order level of the calculated structures together with the coincident 1Hα chemical shift deviations at the overlapping sites warranted the superposition of the different structures calculated for MPERp (34), CpreTM, and TMDp peptides (Fig. 4). The overarching structure resulting from this reconstruction process reveals novel information on the organization of the gp41 TMD anchor and its connection to the upstream MPER region. Overall, the data confirm the adoption of chiefly α-helical conformations for these sequences in a membrane milieu. However, the first-time high resolution data on the stretches covered by CpreTM and TMDp reveal the presence of a kink or the bending of the helix in a membrane-buried position. Unexpectedly, no change in the direction of the helix axis is observed at position Lys-683, which was previously proposed to initiate the TMD domain (26, 27, 64).
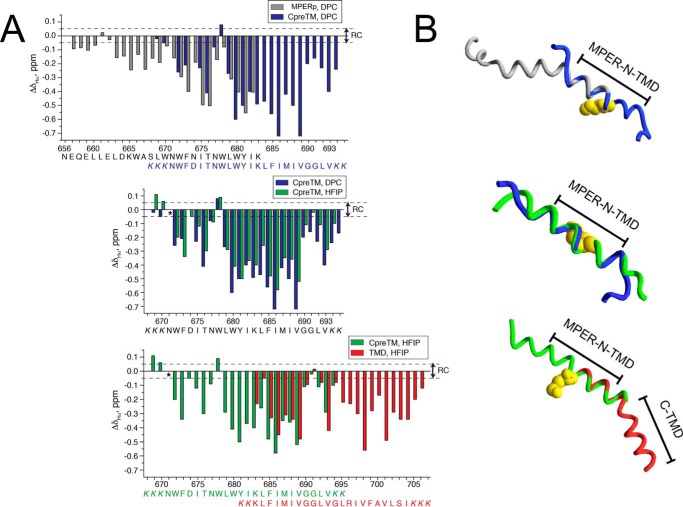
FIGURE 4.
Comparison of the overlapping regions of MPERp, CpreTM, and TMDp peptides. A, bar graphs of ΔδHα values (ΔδHα = δHαobserved − δHαRC, ppm) as a function of peptide sequence for MPERp and CpreTM in 20 mm DPC (top panel), for CpreTM in 20 mm DPC and in 25% HFIP (center panel), and for CpreTM and TMDp in 25% HFIP (bottom panel). pH 7.0 and 25 °C in all the cases. The bars are colored as follows: gray for peptide MPERp; green for CpreTM in HFIP; blue for CpreTM in DPC; and red for TMDp in HFIP. Random coil (RC) values for Hα protons were taken from Ref. 78, and values for MPERp in DPC come from Ref. 34. The values for the N- and C-terminal residues are not included because of charge end effects. Peptide sequences are indicated below, and colored as the corresponding bars in the plots comparing MPERp versus CpreTM and CpreTM versus TMDp. The Lys residues added to the wild type sequence to improve peptide solubility are shown in italics. Dashed lines indicate the random coil range for ΔδHα values. B, superposition of the actual structures in ribbon representation displayed following the same color code that in A. Side chain of Lys-683 residue is depicted in space-filling representation (yellow) and marks the predicted starting point for the TMD region. The segments indicate the approximate range of MPER-N-TMD and C-TMD subdomains.
Thus, the structurally variable segments at positions 671NWFD674 and 690GGLV693 delimit an unforeseen continuous helix at the N-terminal section of the TMD, spanning residues 675ITNWLWYIKLFIMIV689 (Fig. 4). The presence of a membrane-buried flexible element further underpins the existence of the C-terminal TMD section (Fig. 4). We propose that the TMD segments uncovered by the superposition of the NMR structures represent structurally defined functional subdomains, and we have designated them as MPER-N-TMD and C-TMD (Fig. 4).
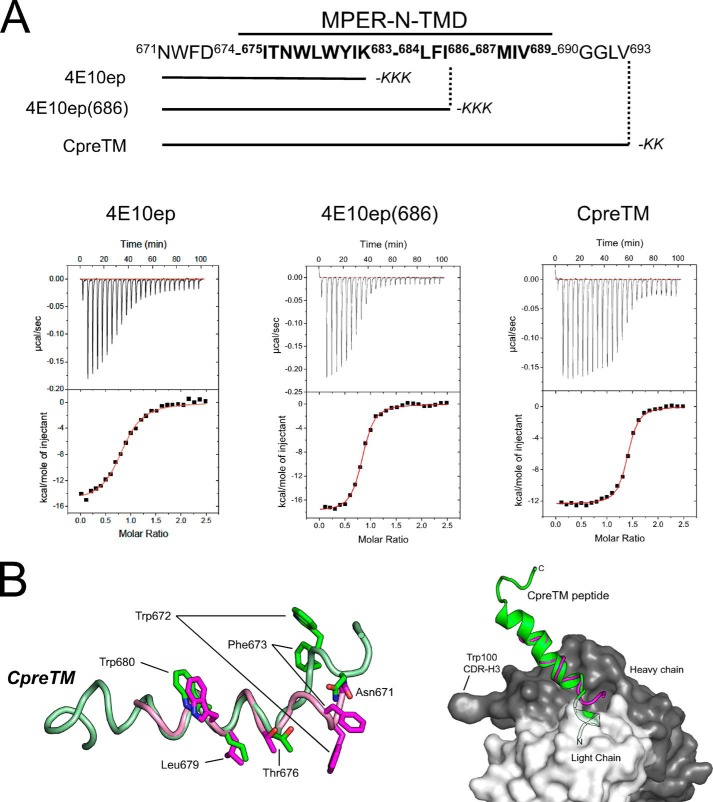
Effects of MPER-N-TMD Residues on the Energetics of 4E10 Binding
The NMR structures determined above reveal the presence of an uninterrupted N-terminal α-helix that encompasses neutralizing epitopes located at MPER C-terminal region. As noted above, in membrane-inserted constructs, gp41 TMD residues specifically enhance the binding of the bNAb 4E10 to the epitope (13). Together, both observations suggest that the continuous MPER-N-TMD helix might embody the natural scaffold structure for the helical 4E10 epitope in the membrane environment. To support this hypothesis, we determined the energetic basis of antibody-peptide binding using peptide epitopes progressively elongated at the C terminus in the presence of the membrane mimetic detergent DPC (Fig. 5). To examine the binding thermodynamics, we employed isothermal titration calorimetry (ITC) using a monovalent 4E10 Fab expressed in Escherichia coli. To validate this recombinant Fab, we crystallized it in complex with the peptide NWFDITNWLWYIK-KKK (4E10ep) (Table 2). The crystal structure of the recombinant 4E10 Fab-peptide complex determined at 2.10 Å was essentially indistinguishable from that of the Fab obtained by papain digestion of the recombinant IgG (root mean square deviation = 0.20 Å) (22).
FIGURE 5.
Binding of peptides to Fab4E10. A, top, overview of the sequences of the peptide epitopes displaying increasingly elongated C termini. The three peptides shared C-terminal solubilization Lys tags but otherwise differed on the number of MPER-N-TMD residues that they incorporated at the C terminus. Bottom, binding isotherms of the peptide epitopes to Fab4E10 examined by ITC. The upper panels indicate the heat released upon successive injections (10 μl) of peptide solution (40 μm) into the calorimeter cell containing Fab4E10 (3 μm). Solutions were supplemented with 5 mm DPC. Fab was titrated with 4E10ep (left panel), 4E10(686) (middle panel), and CpreTM (right panel). Lower panels display the integrated heat (symbols) and the nonlinear least squares fit (red line) of the data to a one-site binding model with the program ORIGIN. B, modeled orientation of the MPER-N-TMD subdomain in the binding region of the recombinant Fab. Peptide 4E10ep is depicted as a magenta ribbon. The modeled CpreTM peptide is depicted in green. In the antibody, the light and dark surface corresponds to the heavy and light chains, respectively. The position of the peptide was modeled by superposing the crystallographic coordinates of main-chain atoms of residues 674–683 of 4E10ep with the coordinates of the equivalent atoms of CpreTM obtained by NMR. Given the comparable side-chain orientations (Fig. 3), only the first coordinate set of the NMR ensemble obtained in DPC was used.
The titration of Fab with peptides gave rise to well defined exothermic peaks that, after integration, defined classical sigmoidal binding isotherms (Fig. 5A). The fitting of the binding isotherms to a one-site model yielded three key thermodynamic parameters in a single experiment (dissociation constant (KD), change of enthalpy (ΔH0), and stoichiometry (n), see Table 3). The titration experiments demonstrate that in the presence of DPC all peptides engage with the Fab with high affinity (nanomolar range) driven by favorable noncovalent interactions (ΔH0 <0). However, significant differences are observed in the relative contribution of the enthalpy (ΔH0) and entropy (−TΔS0) terms to the binding free energy (ΔG0) of each peptide to the Fab (Table 3). Specifically, the entropy loss of the peptide CpreTM upon binding to the antibody is greatly reduced in comparison with that of the peptides with shorter C-terminal tails. This observation suggests that preservation of the full MPER-N-TMD helix results in the optimization of hydrophobic surfaces in contact with Fab and/or in lower levels of conformational and roto-translational restrictions upon binding. The thermodynamic signature of CpreTM (high affinity and pre-organization of the helical structure in a membrane-mimetic environment) could be a desirable property for the design of better vaccines, as previously discussed for the case of alternative 4E10 epitope scaffolds (65) or stapled peptides (66).
To obtain a molecular model of the MPER-N-TMD subdomain bound to the Fab, the constant helical region (residues 674–683) was employed to superpose the coordinates of the CpreTM peptide in DPC (determined by NMR above) with those of the shorter 4E10ep peptide-epitope obtained from the crystal structure with Fab (Fig. 5B, left panel). The fact that, according to the χ1 criterion (see above), the side chains of the considered residues are ordered in the NMR structure supports the validity of this structural comparison. As shown above, the coordinates of the CpreTM in DPC show a disorganized hinge segment at its N-terminal region (Fig. 3). Therefore, the superposition of CpreTM structure with 4E10ep peptide in the Fab-bound structure suggests that the segment 671NWF673 of CpreTM must reorient to bind adequately to the antibody. Conversely, the other residues (674–683) superpose with those of the Fab-bound peptide with remarkable exactitude. It is also observed that the additional residues at the C terminus of CpreTM and the long CDR-H3 loop protrude from the same side of the Fab (Fig. 5B, right panel). This interaction mode might enable the peptide to engage in additional noncovalent interactions with the antibody. Thus, our data suggest that a preformed MPER-N-TMD helix might primarily promote antibody binding by reducing the entropic cost, while the flexible 671NWF673 segment would adjust to engage the paratope through stereospecific interactions.
Immunogenicity of the MPER-N-TMD in Rabbits
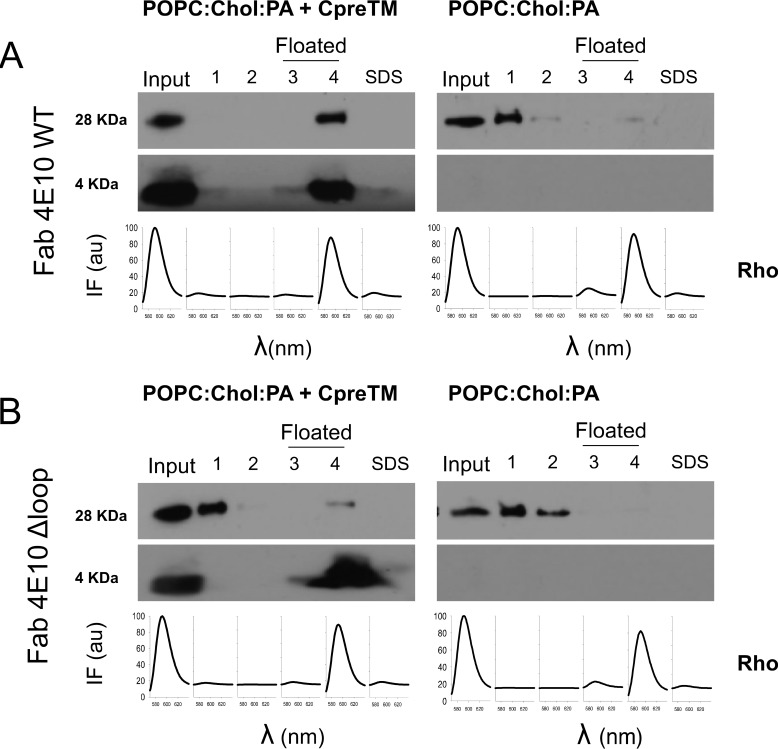
To further corroborate the implication of the MPER-N-TMD subdomain in gp41 immunogenicity, we carried out rabbit immunizations using CpreTM peptide as a vaccine. CpreTM was formulated in the nonpolar media provided by the standard FC adjuvant or liposomes. Fig. 6 displays experiments conducted to establish the effective insertion of CpreTM into the liposomes and its antigenicity at the membrane surface. Following a strategy similar to that described in our previous study (34), those experiments included a Fab4E10 mutant with the CDR-H3 loop tip ablated as a negative control. The Fab4E10-Δloop mutant binds peptide epitopes in solution with comparable affinity, but it is not neutralizing (36), and therefore allows discriminating functional versus nonfunctional binding.
FIGURE 6.
Correlation between 4E10 antibody function and binding to liposomal vaccines. In the vesicle flotation experiments, rhodamine emission (spectra depicted at the bottom of each panel) denoted the presence of liposomes in the 3rd and 4th fractions (i.e. floated fractions). The presence of Fabs and CpreTM peptide in the different fractions was probed in Western blot (28- and 4-kDa bands, respectively). A, left panel, Fab4E10-WT was incubated with CpreTM-containing liposomes. Consistent with Fab binding to membrane-inserted CpreTM, both peptide and Fab4E10-WT co-floated with liposomes in these samples. Right panel, Fab4E10-WT was incubated with liposomes devoid of peptide and predominantly recovered from the pellets (1st and 2nd fractions). B, samples containing the Fab4E10-ΔLoop were incubated as in the previous panels. Under both conditions Fab4E10-ΔLoop was predominantly recovered from the pellet fractions.
In these experiments, CpreTM (4 kDa) and liposomes (Rho emission) were recovered together from the floating fractions, demonstrating quantitative and irreversible CpreTM binding to membranes (Fig. 6, A and B, left panels). Consistent with efficient binding to the peptide epitope inserted into the membrane, the Fab4E10-WT (28 kDa) co-floated with the liposomes that contained the CpreTM peptide (Fig. 6A, left panel). In contrast, the Fab4E10-Δloop was mainly recovered from pellets under these conditions (Fig. 6B, left panel), indicating that this nonfunctional mutant Fab has lost the capacity to bind to the membrane-inserted epitope. Finally, none of the Fabs seemed to co-float appreciably with control liposomes lacking peptide under otherwise similar experimental conditions (Fig. 6, A and B, right panels). Thus, the existence of an antibody-binding function correlation in the case of the liposome-CpreTM vaccine suggests that this formulation could be relevant to target 4E10-like B-cell receptors.
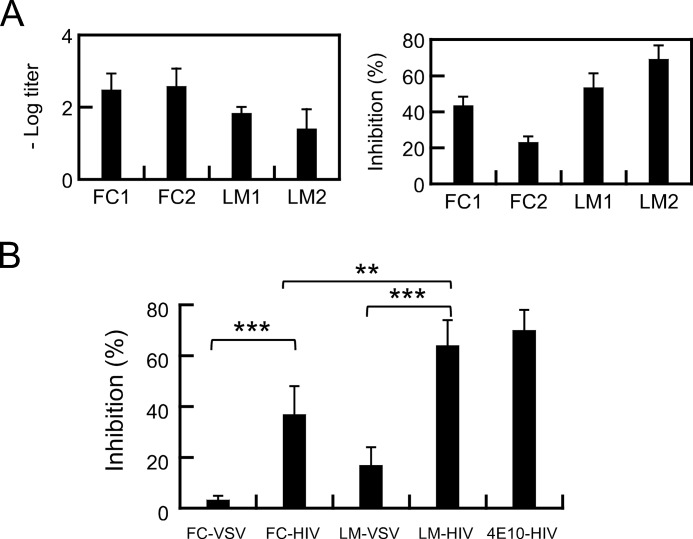
In Fig. 7 the immunogenicity of CpreTM-FC and CpreTM-liposome-MPL formulations was compared (FC1/FC2 and LM1/LM2, respectively). Antigen-specific IgGs could be recovered upon immunization of rabbits with both formulations. The midpoint titers in the FC sera were approximately an order of magnitude higher than in the Liposome-MPL sera (Fig. 7A, left panel). However, measurements of cell entry inhibition by CpreTM-specific antibodies isolated from the sera of the four rabbits resulted in a different pattern (Fig. 7A, right panel). The most effective antibodies in these experiments were those isolated from LM sera. Thus, when grouped according to the adjuvant (Fig. 7B), the inhibitory capacity of LM antibodies was significantly higher than that displayed by the FC antibodies. Moreover, the inhibition levels of HIV-Env-mediated cell entry were significant as compared with those of the negative controls (VSV-GP pseudotypes). However, the observation that a fraction of the pseudovirus population used as negative control is indeed inhibited might indicate certain level of nonspecific reactivity of the raised antibodies with lipids or unrelated proteins.
FIGURE 7.
Immune response after rabbit vaccination with peptide-based formulations. A, left panel, midpoint IgG titers (negative log EC50 dilution values). Sera from rabbits immunized with FC or LM adjuvants were titrated in ELISA using 1.4 μm CpreTM. Right panel, inhibition of cell entry by MPER-N-TMD-specific antibodies in the sera of the different rabbits. HIV JRCSF (tier-2) pseudoviruses were preincubated with antibodies purified from sera of rabbits immunized with the CpreTM-based formulations. Infection of TZM-bl target cells was subsequently monitored by flow cytometry as described previously (34, 37). Purified polyclonal IgGs were used at 30 μg/ml in these assays. B, levels of inhibition of cell entry mediated by HIV-1-Env or VSV-GP controls. Rabbits were grouped according to the employed adjuvant. Significant inhibition of Env-HIV-1 versus VSV-GP was observed for both groups, although LM IgG-mediated inhibition was significantly higher than that mediated by FC IgG (***, p < 0.0005; **, p < 0.005). MAb4E10 (2 μg/ml) was included as positive control. Means ± S.D. of six determinations are shown.
Discussion
Recent crystallographic studies have shed light on the structural basis sustaining gp41 ectodomain metastability in the pre-fusion state (5, 7, 67). Remarkably, the gp41 native-like structure is preserved in the absence of the membrane anchors, which appear to contribute only modestly to the overall stability of the trimeric Env complex (68, 69). Thus, the gp41 membrane regions may fold independently from the ectodomain to exert their anchoring function. However, the Env(671–704) sequence covering the entire gp41 TMD and a fraction of the adjacent MPER shows a high degree of conservation (18, 26, 36), consistent with the additional functional roles ascribed to this region during the infectious cycle, i.e. membrane fusion promotion (17, 18, 36) and immune response modulation (30–32). Despite these important functional roles, high resolution structural data on the gp41 TMD and its junction to MPER were lacking (Table 4).
TABLE 4.
Atomic structures of peptides that include MPER-N-TMD and C-TMD residues
Residues outside MPER-N-TMD and C-TMD subdomains are in italics.
| Method | Peptide sequencea | PDB accession codesb |
|---|---|---|
| X-rayc | 671NWFDITNWLWYIK683-KKK | 2FX7/4WY7 |
| 670W-NWFDITN677-KKKK | 3FN0 | |
| RRR-656NEQELLELDKWASLW-NWFDITNWLWYIR683-RRR | 4G6F | |
| Solution NMR | 665KWASLW-NWFNITNWLWYIK683 | 1JAV |
| 662ELDKWASLW-NWFNITNWLWYIK683 | 2PV6 | |
| 656NEQELLELDKWASLW-NWFNITNWLWYIK683 | 2LP7/2M7W/2M8O/2M8M | |
| KKK-671NWFDITNWLWYIKLFIMIVGGLV693-KK | 2MG2/2MG3 | |
| KK-683KLFIMIVGGLVGLRIVFAVLSI704-KKK | 2MG1 |
a Numbering and sequences are based on B subtype sequences.
b Structures described in this work are underlined.
c Structures were solved in complex with Fabs.
The NMR structures calculated herein for the overlapping CpreTM and TMDp peptides exhibited remarkably well defined main-chain conformations and little variability among residue side-chain orientations (Fig. 3). The fact that overlapping helical regions display comparable conformational shift values strongly suggests that the two peptides adopt similar structures and hence warranted the superposition of the calculated structures (Fig. 4). This reconstruction process, which included the previously solved MPERp peptide structure (34), provides insights into the HIV-1 gp41 TMD structure and its connection to the immunogenic MPER. Most notably, the structures calculated for the CpreTM peptide in membrane-mimetic environments do not show evidence of the predicted helix kink at position Lys-683 (Figs. 3 and 4). On the contrary, CpreTM structures display a continuous helix that extends into the TMD to Gly-690. Furthermore, the structures calculated for the TMDp peptide corroborated the existence of a membrane-buried stretch that presents conformational variability. Thus, the structures solved here support the existence of two functional TMD subdomains as follows: MPER-N-TMD, which encompasses the N-terminal TMD helix, is longer than expected; and the shorter helical C-TMD, which is immersed in the inner leaflet of the membrane (Fig. 8).
FIGURE 8.
Models for gp41 TMD organization in membranes as inferred from the calculated NMR structures. A, disparate structures adopted by the 671NWFD674 and 690GGLV693 hinges, which flank the MPER-N-TMD helix at the N and C terminus, respectively. The side chains of Trp-672 (green) and Leu-692 (red) are displayed to facilitate comparison. Side chain of Lys-683 residue is depicted in space-filling representation (yellow). B, bipartite TMD organization as inferred from the combination of CpreTM and TMDp structures measured in HFIP. C, models for TMD insertion into membranes (see text for details).
Although the molecular mechanism remains unsolved, MPER-N-TMD promotes membrane fusion as evidenced by mutagenesis studies and peptide-based assays (18, 36, 64, 70). Consistent with a pivotal role in the fusion process, bNAbs raised against this element in response to infection exhibit in vitro and in vivo efficacy and exceptional neutralization breadth (21, 71, 72). The existence of a continuous MPER-N-TMD helix further implies that peptides ending at Lys-683 (Table 4), such as MPERp (see also Fig. 4), contain a truncated structural motif at their C termini. This truncation might explain the variety of conformations observed for the MPER C terminus (24, 26, 29, 34), as well as the failure of the MPERp-like peptides to direct humoral responses to the MPER C-terminal epitopes (34, 53, 54, 73–75).
Consistent with the existence of a longer than expected natural scaffold structure, ITC assays revealed that pre-formation of the MPER-N-TMD helix in a membrane mimetic medium may be required for optimal 4E10 antibody binding to its helical epitope (Fig. 5). In addition, the peptide encompassing this element was immunogenic in rabbits when formulated in the nonpolar environments provided by FC (reverse micelles) or LM (lipid bilayers) adjuvants, and it elicited antibodies with the capacity of blocking cell infection (Fig. 7). These results support the importance of the MPER-N-TMD helix for functional antibody and B-cell receptor binding, and therefore, preservation of this element should be considered in the future design of anti-MPER immunogens.
In the CpreTM structure solved in DPC, two flexible elements flanked the MPER-N-TMD helix (Fig. 8A). The variety of conformations reported for the N-terminal 671NWFD674 segment supports its structural metastability (Fig. 8A, left panel, and Table 4) (33). Our findings further suggest that the C-terminal 690GGLV693 residues might function as a second articulated joint embedded in the membrane core (Fig. 8A, right panel). The CpreTM structure in DPC displayed a helix kinked at position Gly-690 (Fig. 8A, C-DPC). This finding is consistent with Gly being the most common residue at position C′ in capping motifs found at the helix C termini (63). In addition, it gives structural support to previous MDS of monomers immersed into the lipid bilayer milieu, which suggested that the midspan polar Arg and the Gly residues might provoke an inherent metastability and be capable of adopting alternating kinked forms (14).
The CpreTM and TMDp structures solved in HFIP also exhibited a change in the orientation of the helix axis at position 690GGLV693 (Fig. 8A, C-HFIP and T-HFIP, respectively). In this case, although the calculated angles indicated a significant degree of helix bending, the overall helical conformation is preserved in both peptides.
Our determination of the high resolution structure of TMDp confirms the predominantly helical conformation of the TMD sequence in a low polarity environment (Fig. 8B), in agreement with previous MDS and molecular modeling reports (13, 14). Helix bending around position 690GGLV693 also supports the existence of the C-TMD subdomain. The C-TMD includes the immunosuppressive 694GLRIVFAVL702 motif, which resembles part of the TCRα TMD (30). The gp41 TMD co-localizes with CD3 within the TCR complex and functions by inhibiting CD3 antibody-stimulated in vitro T-cell proliferation (30, 31). Our data here emphasize the uniqueness of this motif within TMD and provide the first detailed view of its helical structure at atomic resolution.
The segmentation into MPER-N-TMD and C-TMD subdomains and the existence of a membrane-embedded flexible site suggest different models for MPER-TMD organization in membranes (Fig. 8C). In principle, a flexible joint may allow alternating angles for MPER-N-TMD helix insertion (Fig. 8C, blue cylinders). Tilted insertion in the “a” model would adjust the length of the TMD anchor to the membrane thickness and satisfy most of the aromatic interactions with the membrane interface at its N terminus. The helix is proposed to twist within the membrane at position 691GL692 and then continue in a direction parallel to the bilayer normal. In the pre-fusion state, the Asp-664 residues at the C terminus of the C-terminal helical region (CHR in Fig. 1A) are splayed apart by ∼30 Å at the base of the trimeric gp41 ectodomain (5, 7). Thus, a tilted MPER-N-TMD might be required to link the TMD with the native ectodomain. A continuous helical segment comprising MPER and TMD segments was indeed suggested by a cryo-EM study (76). In addition, this arrangement might habilitate the C-TMD (red, Fig. 8B) for functional interaction with the TCR complex (30). Finally, a wedge between MPER-N-TMD and C-TMD helices might enable the formation of water defects in the bilayer as suggested by recent MDS studies (15).
The “b” model postulates MPER-N-TMD helix insertion occurring perpendicular to the membrane plane. Following the molecular modeling reported by Montero et al. (13), this arrangement would be stabilized through more extensive oligomeric contacts. We have recently proposed that, in this configuration, the MPER-N-TMD would be membrane-active and capable of extracting phospholipid from the viral envelope (36). Thus, we speculate that this arrangement might be related to fusion intermediates, emerging upon activation of the process, and proposed to promote the initial lipid exchange between viral and cell membranes (2, 77).
Finally, the “c” model incorporates the sharp kink observed in the CpreTM-DPC structure. The possibility for occurrence of kinked metastable membrane structures was previously suggested by MDS analyses (14). We infer that such metastable arrangements could be disruptive for the lipid bilayer organization, and thus, we hypothesize that its occurrence might be also confined to some of the structural intermediates emerging along the fusion pathway (2, 3). Mutagenic analysis revealed that the Env(696–707) stretch encompassing the C-TMD subdomain is important for fusion (16). Thus, we speculate that within a kinked structure, the C-TMD residing in the inner leaflet of the viral membrane might gain access to the hemifusion diaphragm (2, 3) and cause its destabilization. In this way, the flexible joint between MPER-N-TMD and C-TMD may facilitate the formation of a fusion pore within a hemifusion diaphragm.
Concluding Remarks
The NMR structures determined in this and the previous study (34) suggest that gp41 MPER-TMD region is organized in contact with the viral membrane as a continuous, but distinctly jointed, helical structure (Fig. 4). Most importantly, the NMR data reveal the functional segmentation of the gp41 membrane anchor into two subdomains as follows: MPER-N-TMD, encompassing a helix, longer than expected and flanked by two hinges, which is involved in fusion and elicitation of humoral responses; and C-TMD, which includes a C-terminal helix involved in down-regulation of the TCR-mediated signaling and possibly in the promotion of the last steps of the fusion process. Thus, the membrane interaction model emerging from these studies diverges from the current view that considers MPER as an interfacially adsorbed helix, bent at position Lys-683 and followed by the TMD inserting perpendicularly to the viral membrane plane. Moreover, the experimental results indicate that the MPER-N-TMD may function as a helical scaffold to increase affinity of anti-MPER antibody in membrane mimetics and suggest that its preservation may be a prerequisite for the immunogenic efficacy of peptide vaccines targeting the MPER C-terminal region.
Supplementary Material
Acknowledgments
Access to beamline BL5A was granted by the Photon Factory Advisory Committee (Proposal Number 2013G738). Technical assistance by Miguel García is greatly appreciated. Critical reading by Dr. Douglas V. Laurents is also acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI097051 (to J. L. N.). This work was also supported by Canadian Institutes of Health Research Grant MOP-114941 (to J. L. N.), Spanish MINECO Grant CTQ2011-22514 (to M. A. J.), Basque Government Grant IT838-13 (to J. L. N.), and Japan Society for the Promotion of Science Project 25249115 (to K. T. and J. M. M. C.).

This article contains supplemental spectra.
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 2MG1, 2MG2, 2MG3, and 4WY7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Chemical shifts have been deposited in the BioMagResBank Databank with accession numbers 19583, 19582, and 19581.
- Env
- envelope
- bNAb
- broadly neutralizing antibody
- DPC
- dodecylphosphocholine
- HFIP
- 1,1,1,3,3,3-hexafluoro-2-propanol
- MPER
- membrane-proximal external region
- TMD
- transmembrane domain
- PDB
- Protein Data Bank
- MDS
- molecular dynamics simulation
- HSQC
- heteronuclear single quantum coherence
- FC
- Freund's Complete
- ITC
- isothermal titration calorimetry
- TCR
- T-cell receptor.
References
- 1. Wyatt R., Sodroski J. (1998) The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 [DOI] [PubMed] [Google Scholar]
- 2. Melikyan G. B. (2011) Membrane fusion mediated by human immunodeficiency virus envelope glycoprotein. Curr. Top. Membr. 68, 81–106 [DOI] [PubMed] [Google Scholar]
- 3. Blumenthal R., Durell S., Viard M. (2012) HIV entry and envelope glycoprotein-mediated fusion. J. Biol. Chem. 287, 40841–40849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eckert D. M., Kim P. S. (2001) Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 [DOI] [PubMed] [Google Scholar]
- 5. Julien J. P., Cupo A., Sok D., Stanfield R. L., Lyumkis D., Deller M. C., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Ward A. B., Wilson I. A. (2013) Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lyumkis D., Julien J. P., de Val N., Cupo A., Potter C. S., Klasse P. J., Burton D. R., Sanders R. W., Moore J. P., Carragher B., Wilson I. A., Ward A. B. (2013) Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pancera M., Zhou T., Druz A., Georgiev I. S., Soto C., Gorman J., Huang J., Acharya P., Chuang G. Y., Ofek G., Stewart-Jones G. B., Stuckey J., Bailer R. T., Joyce M. G., Louder M. K., et al. (2014) Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haffar O. K., Dowbenko D. J., Berman P. W. (1988) Topogenic analysis of the human immunodeficiency virus type 1 envelope glycoprotein, gp160, in microsomal membranes. J. Cell Biol. 107, 1677–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Helseth E., Olshevsky U., Gabuzda D., Ardman B., Haseltine W., Sodroski J. (1990) Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 64, 6314–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yue L., Shang L., Hunter E. (2009) Truncation of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein defines elements required for fusion, incorporation, and infectivity. J. Virol. 83, 11588–11598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu S., Kondo N., Long Y., Xiao D., Iwamoto A., Matsuda Z. (2010) Membrane topology analysis of HIV-1 envelope glycoprotein gp41. Retrovirology 7, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim J. H., Hartley T. L., Curran A. R., Engelman D. M. (2009) Molecular dynamics studies of the transmembrane domain of gp41 from HIV-1. Biochim. Biophys. Acta 1788, 1804–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Montero M., Gulzar N., Klaric K. A., Donald J. E., Lepik C., Wu S., Tsai S., Julien J. P., Hessell A. J., Wang S., Lu S., Burton D. R., Pai E. F., Degrado W. F., Scott J. K. (2012) Neutralizing epitopes in the membrane-proximal external region of HIV-1 gp41 are influenced by the transmembrane domain and the plasma membrane. J. Virol. 86, 2930–2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gangupomu V. K., Abrams C. F. (2010) All-atom models of the membrane-spanning domain of HIV-1 gp41 from metadynamics. Biophys. J. 99, 3438–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baker M. K., Gangupomu V. K., Abrams C. F. (2014) Characterization of the water defect at the HIV-1 gp41 membrane spanning domain in bilayers with and without cholesterol using molecular simulations. Biochim. Biophys. Acta 1838, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Owens R. J., Burke C., Rose J. K. (1994) Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J. Virol. 68, 570–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyauchi K., Komano J., Yokomaku Y., Sugiura W., Yamamoto N., Matsuda Z. (2005) Role of the specific amino acid sequence of the membrane-spanning domain of human immunodeficiency virus type 1 in membrane fusion. J. Virol. 79, 4720–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shang L., Yue L., Hunter E. (2008) Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection. J. Virol. 82, 5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zwick M. B. (2005) The membrane-proximal external region of HIV-1 gp41: a vaccine target worth exploring. AIDS 19, 1725–1737 [DOI] [PubMed] [Google Scholar]
- 20. Montero M., van Houten N. E., Wang X., Scott J. K. (2008) The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol. Mol. Biol. Rev. 72, 54–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kwong P. D., Mascola J. R. (2012) Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity 37, 412–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cardoso R. M., Brunel F. M., Ferguson S., Zwick M., Burton D. R., Dawson P. E., Wilson I. A. (2007) Structural basis of enhanced binding of extended and helically constrained peptide epitopes of the broadly neutralizing HIV-1 antibody 4E10. J. Mol. Biol. 365, 1533–1544 [DOI] [PubMed] [Google Scholar]
- 23. Huang J., Ofek G., Laub L., Louder M. K., Doria-Rose N. A., Longo N. S., Imamichi H., Bailer R. T., Chakrabarti B., Sharma S. K., Alam S. M., Wang T., Yang Y., Zhang B., Migueles S. A., et al. (2012) Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schibli D. J., Montelaro R. C., Vogel H. J. (2001) The membrane-proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well defined helix in dodecylphosphocholine micelles. Biochemistry 40, 9570–9578 [DOI] [PubMed] [Google Scholar]
- 25. Sáez-Cirión A., Nir S., Lorizate M., Agirre A., Cruz A., Pérez-Gil J., Nieva J. L. (2002) Sphingomyelin and cholesterol promote HIV-1 gp41 pretransmembrane sequence surface aggregation and membrane restructuring. J. Biol. Chem. 277, 21776–21785 [DOI] [PubMed] [Google Scholar]
- 26. Sun Z. Y., Oh K. J., Kim M., Yu J., Brusic V., Song L., Qiao Z., Wang J. H., Wagner G., Reinherz E. L. (2008) HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity 28, 52–63 [DOI] [PubMed] [Google Scholar]
- 27. Huarte N., Lorizate M., Maeso R., Kunert R., Arranz R., Valpuesta J. M., Nieva J. L. (2008) The broadly neutralizing anti-human immunodeficiency virus type 1 4E10 monoclonal antibody is better adapted to membrane-bound epitope recognition and blocking than 2F5. J. Virol. 82, 8986–8996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dennison S. M., Stewart S. M., Stempel K. C., Liao H. X., Haynes B. F., Alam S. M. (2009) Stable docking of neutralizing human immunodeficiency virus type 1 gp41 membrane-proximal external region monoclonal antibodies 2F5 and 4E10 is dependent on the membrane immersion depth of their epitope regions. J. Virol. 83, 10211–10223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reardon P. N., Sage H., Dennison S. M., Martin J. W., Donald B. R., Alam S. M., Haynes B. F., Spicer L. D. (2014) Structure of an HIV-1-neutralizing antibody target, the lipid-bound gp41 envelope membrane proximal region trimer. Proc. Natl. Acad. Sci. U.S.A. 111, 1391–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cohen T., Cohen S. J., Antonovsky N., Cohen I. R., Shai Y. (2010) HIV-1 gp41 and TCRα trans-membrane domains share a motif exploited by the HIV virus to modulate T-cell proliferation. PLoS Pathog. 6, e1001085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ashkenazi A., Faingold O., Shai Y. (2013) HIV-1 fusion protein exerts complex immunosuppressive effects. Trends Biochem. Sci. 38, 345–349 [DOI] [PubMed] [Google Scholar]
- 32. Reuven E. M., Ali M., Rotem E., Schwarzter R., Gramatica A., Futerman A. H., Shai Y. (2014) The HIV-1 envelope transmembrane domain binds TLR2 through a distinct dimerization motif and inhibits TLR2-mediated responses. PLoS Pathog. 10, e1004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun Z. Y., Cheng Y., Kim M., Song L., Choi J., Kudahl U. J., Brusic V., Chowdhury B., Yu L., Seaman M. S., Bellot G., Shih W. M., Wagner G., Reinherz E. L. (2014) Disruption of helix-capping residues 671 and 674 reveals a role in HIV-1 entry for a specialized hinge segment of the membrane proximal external region of gp41. J. Mol. Biol. 426, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Serrano S., Araujo A., Apellániz B., Bryson S., Carravilla P., de la Arada I., Huarte N., Rujas E., Pai E. F., Arrondo J. L., Domene C., Jiménez M. A., Nieva J. L. (2014) Structure and immunogenicity of a peptide vaccine, including the complete HIV-1 gp41 2F5 epitope: implications for antibody recognition mechanism and immunogen design. J. Biol. Chem. 289, 6565–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Apellániz B., Nir S., Nieva J. L. (2009) Distinct mechanisms of lipid bilayer perturbation induced by peptides derived from the membrane-proximal external region of HIV-1 gp41. Biochemistry 48, 5320–5331 [DOI] [PubMed] [Google Scholar]
- 36. Apellániz B., Rujas E., Carravilla P., Requejo-Isidro J., Huarte N., Domene C., Nieva J. L. (2014) Cholesterol-dependent membrane fusion induced by the gp41 membrane-proximal external region-transmembrane domain connection suggests a mechanism for broad HIV-1 neutralization. J. Virol. 88, 13367–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Julien J. P., Huarte N., Maeso R., Taneva S. G., Cunningham A., Nieva J. L., Pai E. F. (2010) Ablation of the complementarity-determining region H3 apex of the anti-HIV-1 broadly neutralizing antibody 2F5 abrogates neutralizing capacity without affecting core epitope binding. J. Virol. 84, 4136–4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Markley J. L., Bax A., Arata Y., Hilbers C. W., Kaptein R., Sykes B. D., Wright P. E., Wüthrich K. (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 280, 933–952 [DOI] [PubMed] [Google Scholar]
- 39. Mirassou Y., Santiveri C. M., Perez de Vega M. J., Gonzalez-Muniz R., Jimenez M. A. (2009) Disulfide bonds versus TrpTrp pairs in irregular β-hairpins: NMR structure of vammin loop 3-derived peptides as a case study. Chembiochem 10, 902–910 [DOI] [PubMed] [Google Scholar]
- 40. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 41. Güntert P., Mumenthaler C., Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 [DOI] [PubMed] [Google Scholar]
- 42. Güntert P. (2004) Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 [DOI] [PubMed] [Google Scholar]
- 43. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 44. Koradi R., Billeter M., Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 45. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 46. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 47. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 49. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK–a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 51. Yethon J. A., Epand R. F., Leber B., Epand R. M., Andrews D. W. (2003) Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J. Biol. Chem. 278, 48935–48941 [DOI] [PubMed] [Google Scholar]
- 52. Dreesman G. R., Sanchez Y., Ionescu-Matiu I., Sparrow J. T., Six H. R., Peterson D. L., Hollinger F. B., Melnick J. L. (1982) Antibody to hepatitis B surface antigen after a single inoculation of uncoupled synthetic HBsAg peptides. Nature 295, 158–160 [DOI] [PubMed] [Google Scholar]
- 53. Maeso R., Huarte N., Julien J. P., Kunert R., Pai E. F., Nieva J. L. (2011) Interaction of anti-HIV type 1 antibody 2F5 with phospholipid bilayers and its relevance for the mechanism of virus neutralization. AIDS Res. Hum. Retroviruses 27, 863–876 [DOI] [PubMed] [Google Scholar]
- 54. Alving C. R., Rao M., Steers N. J., Matyas G. R., Mayorov A. V. (2012) Liposomes containing lipid A: an effective, safe, generic adjuvant system for synthetic vaccines. Expert Rev. Vaccines 11, 733–744 [DOI] [PubMed] [Google Scholar]
- 55. Suárez T., Gallaher W. R., Agirre A., Goñi F. M., Nieva J. L. (2000) Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74, 8038–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sáez-Cirión A., Arrondo J. L., Gómara M. J., Lorizate M., Iloro I., Melikyan G., Nieva J. L. (2003) Structural and functional roles of HIV-1 gp41 pretransmembrane sequence segmentation. Biophys. J. 85, 3769–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Litowski J. R., Hodges R. S. (2002) Designing heterodimeric two-stranded α-helical coiled-coils. Effects of hydrophobicity and α-helical propensity on protein folding, stability, and specificity. J. Biol. Chem. 277, 37272–37279 [DOI] [PubMed] [Google Scholar]
- 58. Sreerama N., Woody R. W. (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 287, 252–260 [DOI] [PubMed] [Google Scholar]
- 59. Wüthrich K., Billeter M., Braun W. (1984) Polypeptide secondary structure determination by nuclear magnetic resonance observation of short proton-proton distances. J. Mol. Biol. 180, 715–740 [DOI] [PubMed] [Google Scholar]
- 60. Wuthrich K. (1986) NMR of Proteins and Nucleic Acids, John Wiley & Sons, Inc., New York [Google Scholar]
- 61. Wishart D. S., Sykes B. D., Richards F. M. (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222, 311–333 [DOI] [PubMed] [Google Scholar]
- 62. Vila R., Ponte I., Jiménez M. A., Rico M., Suau P. (2000) A helix-turn motif in the C-terminal domain of histone H1. Protein Sci. 9, 627–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aurora R., Rose G. D. (1998) Helix capping. Protein Sci. 7, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Salzwedel K., West J. T., Hunter E. (1999) A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73, 2469–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Correia B. E., Ban Y. E., Holmes M. A., Xu H., Ellingson K., Kraft Z., Carrico C., Boni E., Sather D. N., Zenobia C., Burke K. Y., Bradley-Hewitt T., Bruhn-Johannsen J. F., Kalyuzhniy O., Baker D., et al. (2010) Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18, 1116–1126 [DOI] [PubMed] [Google Scholar]
- 66. Bird G. H., Irimia A., Ofek G., Kwong P. D., Wilson I. A., Walensky L. D. (2014) Stapled HIV-1 peptides recapitulate antigenic structures and engage broadly neutralizing antibodies. Nat. Struct. Mol. Biol. 21, 1058–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sanders R. W., Moore J. P. (2014) HIV: A stamp on the envelope. Nature 514, 437–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Khayat R., Lee J. H., Julien J. P., Cupo A., Klasse P. J., Sanders R. W., Moore J. P., Wilson I. A., Ward A. B. (2013) Structural characterization of cleaved, soluble HIV-1 envelope glycoprotein trimers. J. Virol. 87, 9865–9872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Klasse P. J., Depetris R. S., Pejchal R., Julien J. P., Khayat R., Lee J. H., Marozsan A. J., Cupo A., Cocco N., Korzun J., Yasmeen A., Ward A. B., Wilson I. A., Sanders R. W., Moore J. P. (2013) Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J. Virol. 87, 9873–9885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Reuven E. M., Dadon Y., Viard M., Manukovsky N., Blumenthal R., Shai Y. (2012) HIV-1 gp41 transmembrane domain interacts with the fusion peptide: implication in lipid mixing and inhibition of virus–cell fusion. Biochemistry 51, 2867–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. van Gils M. J., Sanders R. W. (2014) In vivo protection by broadly neutralizing HIV antibodies. Trends Microbiol. 22, 550–551 [DOI] [PubMed] [Google Scholar]
- 72. Trkola A., Kuster H., Rusert P., von Wyl V., Leemann C., Weber R., Stiegler G., Katinger H., Joos B., Günthard H. F. (2008) In vivo efficacy of human immunodeficiency virus neutralizing antibodies: estimates for protective titers. J. Virol. 82, 1591–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dennison S. M., Sutherland L. L., Jaeger F. H., Anasti K. M., Parks R., Stewart S., Bowman C., Xia S. M., Zhang R., Shen X., Scearce R. M., Ofek G., Yang Y., Kwong P. D., Santra S., et al. (2011) Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS One 6, e27824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kim M., Song L., Moon J., Sun Z. Y., Bershteyn A., Hanson M., Cain D., Goka S., Kelsoe G., Wagner G., Irvine D., Reinherz E. L. (2013) Immunogenicity of membrane-bound HIV-1 gp41 membrane-proximal external region (MPER) segments is dominated by residue accessibility and modulated by stereochemistry. J. Biol. Chem. 288, 31888–31901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Venditto V. J., Watson D. S., Motion M., Montefiori D., Szoka F. C., Jr. (2013) Rational design of membrane proximal external region lipopeptides containing chemical modifications for HIV-1 vaccination. Clin. Vaccine Immunol. 20, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mao Y., Wang L., Gu C., Herschhorn A., Désormeaux A., Finzi A., Xiang S.-H., Sodroski J. G. (2013) Molecular architecture of the uncleaved HIV-1 envelope glycoprotein trimer. Proc. Natl. Acad. Sci. U.S.A. 110, 12438–12443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. de la Vega M., Marin M., Kondo N., Miyauchi K., Kim Y., Epand R. F., Epand R. M., Melikyan G. B. (2011) Inhibition of HIV-1 endocytosis allows lipid mixing at the plasma membrane, but not complete fusion. Retrovirology 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wishart D. S., Bigam C. G., Holm A., Hodges R. S., Sykes B. D. (1995) 1H, 13C and 15N random coil NMR chemical shifts of the common amino acids. I. Investigations of nearest-neighbor effects. J. Biomol. NMR 5, 67–81 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.