Background: In chloroplasts of higher plants, a heterodimeric cpSRP43·cpSRP54 complex targets LHC proteins to the thylakoid membrane.
Results: In green algae, cpSRP43 alone forms a targeting complex with LHC proteins.
Conclusion: The coevolution of LHC proteins and cpSRP43 occurred independently of complex formation with cpSRP54.
Significance: The results provide new insights into the evolution of cpSRP-dependent protein transport.
Keywords: Chlamydomonas, chloroplast, protein evolution, protein targeting, signal recognition particle (SRP), LHCP
Abstract
In bacteria, membrane proteins are targeted cotranslationally via a signal recognition particle (SRP). During the evolution of higher plant chloroplasts from cyanobacteria, the SRP pathway underwent striking adaptations that enable the posttranslational transport of the abundant light-harvesting chlorophyll-a/b-binding proteins (LHCPs). The conserved 54-kDa SRP subunit in higher plant chloroplasts (cpSRP54) is not bound to an SRP RNA, an essential SRP component in bacteria, but forms a stable heterodimer with the chloroplast-specific cpSRP43. This heterodimeric cpSRP recognizes LHCP and delivers it to the thylakoid membrane whereby cpSRP43 plays a central role. This study shows that the cpSRP system in the green alga Chlamydomonas reinhardtii differs significantly from that of higher plants as cpSRP43 is not complexed to cpSRP54 in Chlamydomonas and cpSRP54 is not involved in LHCP recognition. This divergence is attributed to altered residues within the cpSRP54 tail and the second chromodomain of cpSRP43 that are crucial for the formation of the binding interface in Arabidopsis. These changes are highly conserved among chlorophytes, whereas all land plants contain cpSRP proteins with typical interaction motifs. These data demonstrate that the coevolution of LHCPs and cpSRP43 occurred independently of complex formation with cpSRP54 and that the interaction between cpSRP54 and cpSRP43 evolved later during the transition from chlorophytes to land plants. Furthermore, our data show that in higher plants a heterodimeric form of cpSRP is required for the formation of a low molecular weight transit complex with LHCP.
Introduction
The cytosolic signal recognition particle (SRP)2 is part of a ubiquitous protein-targeting machinery that plays a crucial role in the cotranslational targeting of proteins to the plasma membrane of prokaryotes and to the endoplasmic reticulum of eukaryotes (1, 2). The minimal functional core of all cytosolic SRPs consists of two essential conserved components, an SRP RNA and an ∼54-kDa protein (SRP54). During the evolution of higher plant chloroplasts from a cyanobacterial endosymbiont, the SRP54 component was retained (chloroplast (cp) SRP54), whereas the SRP RNA was lost. One pool of cpSRP54 cofractionates with plastid ribosomes (3) and can be cross-linked to the nascent chains of the photosystem II reaction center protein D1 (4, 5). This finding implies that it is involved in the cotranslational transport of at least some plastid-encoded proteins and functions in the absence of an SRP RNA component. Notably, a second pool of cpSRP54 forms a stable complex with cpSRP43, which is unique to chloroplasts (6–8). This complex is often referred to as cpSRP and is required for the efficient posttranslational transport of members of the nuclearly encoded light harvesting chlorophyll-a/b-binding proteins (LHCPs) to the thylakoid membrane (9). These proteins play a key role in photosynthesis and represent the most abundant transmembrane proteins in plants. After synthesis, LHCPs are translocated from the cytosol across the outer and inner envelope of chloroplasts and are subsequently routed to the cpSRP pathway by the recently identified LTD protein (10). cpSRP binds the hydrophobic LHCP to form the soluble transit complex and to deliver it to the thylakoid membrane. The integration of LHCP into the membrane requires the chloroplast SRP receptor homologue cpFtsY (11, 12), the integral translocase Alb3 (13), and GTP, which is hydrolyzed by the SRP GTPases cpSRP54 and cpFtsY. cpSRP43, which harbors four ankyrin domains and three chromodomains (CD1–CD3), plays a key role in this transport mechanism. It mediates an impressive range of protein-protein interactions as follows. (a) CD2 binds to an arginine-rich motif in the C-terminal tail of cpSRP54 (14–16), (b) the ankyrin repeats bind to the conserved L18 motif in LHCPs (17, 18), (c) an interaction between cpSRP43 and LTD is involved in delivering LHCP from the envelope to cpSRP (10), and (d) cpSRP43 binds to the translocase Alb3, a process that likely plays a central role in the docking of the cpSRP·LHCP·cpFtsY complex to Alb3 (19–22). Furthermore, cpSRP43 acts as a chaperone for LHCPs and prevents its aggregation (23, 24).
The presence of stable cpSRP54·cpSRP43 complexes in higher plant chloroplasts versus SRP54·SRP RNA complexes in prokaryotes together with the finding that the overall shape and charge distribution of cpSRP43 resemble the SRP RNA (18) supported the view that cpSRP43 replaced the ancestral SRP RNA. However, a recent study analyzing the phylogenetic distribution of SRP components in photosynthetic organisms identified conserved chloroplast SRP RNAs within a wide range of green algae and land plants, which evolved earlier than spermatophytes, showing the simultaneous presence of a cpSRP RNA and cpSRP43 in these organisms (25). These findings demonstrated that the evolution of cpSRP43 is not correlated with a loss of the SRP RNA. The moss Physcomitrella patens was studied as an example for this type of SRP system, and Physcomitrella cpSRP54 was shown to be able to form a stable complex with cpSRP43 and to bind the cpSRP RNA (25).
In this study, we analyzed the molecular characteristics of the chloroplast SRP system of the unicellular green alga Chlamydomonas reinhardtii, which represents a chlorophyte that contains cpSRP pathway homologs but has lost the cpSRP RNA (25). Surprisingly, we showed that cpSRP43 does not form a complex with cpSRP54 in Chlamydomonas. Structural modeling combined with mutational analysis enabled the identification of residues in the Chlamydomonas cpSRP54 and cpSRP43 proteins that interfere with complex formation. These alterations are conserved among chlorophytes, indicating that complex formation between cpSRP43 and cpSRP54 developed later during evolution. Furthermore, our data show that cpSRP54 is not involved in LHCP recognition but very likely required for LHCP insertion in Chlamydomonas. For higher plants, we confirmed the importance of a heterodimeric cpSRP for efficient transit complex formation with LHCP.
Experimental Procedures
Culture Conditions
Chlamydomonas reinhardtii strain CC-406 cw15 mt− was grown under orbital shaking in Tris acetate-phosphate medium (26) at 25 °C and 30 microeinsteins/m2s.
Gel Filtration of a Total Protein Extract of Chlamydomonas
Concentrated Chlamydomonas cells from a 400-ml culture (∼2 × 106 cells/ml) were sonicated for 60 s on ice in 4 ml of lysis buffer (20 mm Hepes-KOH, pH 7.5, 155 mm NaCl supplemented with Complete Mini protease inhibitor mixture (Roche Diagnostics)). Insoluble material was removed by ultracentrifugation for 30 min at 120,000 × g through a sucrose cushion (0.6 m sucrose in 20 mm Hepes-KOH, pH 7.5). Soluble proteins were filtered through a 0.22-μm filter, and 500 μl of the total soluble protein extract was loaded onto a Superose 6 10/300 GL gel filtration column (GE Healthcare). The column was run with a flow rate of 0.5 ml/min in 20 mm Hepes-KOH, pH 8.0, 180 mm NaCl, 5 mm MgCl2.
Plasmid Construction
The cDNA clone coding for Cr-cpSRP54 was obtained from the Kazusa DNA Research Institute (clone number AV640228). The cDNA sequence for Cr-cpSRP43 was determined by RT-PCR as described previously (25) (GenBankTM accession number KC331036.1) and synthesized in an optimized form for Escherichia coli codon usage by GenScript. The coding sequence of mature and precursor Cr-LhcbM3 (Cr-LHCP) was amplified by RT-PCR using total cDNA.
For the yeast two-hybrid analyses, the coding sequences for the mature form of Cr-cpSRP54 starting with IRSAMFDS and the M-domain of Cr-cpSRP54 starting with MGDVLTLY were cloned into the NcoI/SalI restriction site of pGBKT7 (Clontech). Full-length Cr-cpSRP43 was cloned into pGBKT7 and pACT2 (Clontech) using the NcoI/SalI and NcoI/BamHI restriction sites, respectively. The fusion construct Cr-cpSRP54M/At-cpSRP54C-term (Cr-54M/At-54C-term) was synthesized by overlap PCR and encodes Cr-cpSRP54M, the 28 C-terminal residues (starting with KKVAPG) of which were replaced with the C terminus of At-cpSRP54 (residues 527–564). The fusion construct was then cloned into pGBKT7 as described above. All other yeast two-hybrid constructs were published previously (14).
For the overexpression of Cr-cpSRP54-His, full-length Cr-His-cpSRP43 and Cr-GST-cpSRP43, the corresponding coding sequences were cloned into the NcoI/SalI site of pET29b (Novagen), the BamHI/SalI site of pCOLATMDuet-1 (Novagen), and the BamHI/XhoI site of pGEX4T3 (GE Healthcare), respectively. To obtain Cr-His-cpSRP54M and the fusion construct Cr-His-54M/At-54C-term, the coding sequences were cloned into the BamHI/SalI site of pETTMDuet-1 (Novagen). At-cpSRP54M (amino acids 371–564) and At-cpSRP54MΔC-term (amino acids 371–529) were cloned into the BamHI/HindIII site of pETDuet-1. The constructs encoding At-cpSRP54M-His, At-His-cpSRP43, and At-GST-cpSRP43 were described previously (8, 14, 19). The coding sequence of Cr-LHCP was cloned into the pDS12 vector (kindly provided by H. Paulsen, University of Mainz, Germany) using SphI and PstI restriction sites.
For in vitro transcription and translation the coding sequence of mature and precursor Cr-LHCP was cloned into pIVEX1.3 (5Prime) using NcoI/SalI restriction sites. Site-directed mutagenesis constructs were generated using the QuikChange XL site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer's protocol.
Yeast Two-hybrid Analysis
The yeast two-hybrid assays were performed as described previously (14). The growth of the yeast cells on medium lacking leucine, tryptophan, and histidine (−LTH) was classified as ++, +, and − whereby ++ indicates that most colonies have a diameter of >1.5 mm, + indicates the growth of a few colonies with a diameter of up to 1.0 mm, and − indicates normal background growth (whitish colonies <0.6 mm). The filter lifts to measure β-galactosidase activity were incubated for 1.5 h to develop a blue color (+) or no blue color (−).
Protein Expression and Purification
His fusion constructs were expressed in the E. coli strain BL21(DE3) or Rosetta2(DE3) (Novagen) and purified under native conditions using nickel-nitrilotriacetic acid resin (Qiagen) as suggested by the manufacturer. After purification, all of the proteins were desalted using PD-10 columns (GE Healthcare) and eluted into buffer A (20 mm Hepes, pH 8.0, 300 mm NaCl). Recombinant GST fusion constructs were expressed in the E. coli strain BL21(DE3). The overexpressed proteins were purified using glutathione-Sepharose (GE Healthcare) as suggested by the manufacturer. Proteins were eluted from the glutathione-Sepharose with 50 mm Tris-HCl, pH 8.0, 10 mm reduced glutathione.
Expression and Isolation of Cr-LHCP Inclusion Bodies
Cr-LHCP was expressed in E. coli JM101, and cells were harvested by centrifugation. The cell pellet was resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 5 mm EDTA) for subsequent sonification (5 min with 50% duty cycle). Inclusion bodies were harvested by centrifugation (20 min, 14,000 rpm, 4 °C) and washed three times with washing buffer 1 (lysis buffer + 0.5% (w/v) Triton X-100, 0.1 mm PMSF, 1 mm DTT) and three times with washing buffer 2 (lysis buffer + 1% (w/v) Triton X-100, 1% (w/v) sodium deoxycholate, 0.1 mm PMSF, 1 mm DTT). In each washing step, the resuspended inclusion bodies were sonified again. Finally the inclusion bodies were resuspended and stored at −20 °C in lysis buffer.
In Vitro Transcription and Translation of Cr-LHCP
In vitro transcription and translation of 35S-labeled Cr-LHCP were done using the TranscriptAid T7 High Yield Transcription kit (Thermo Scientific) and wheat germ extract (Promega) according to the manufacturer's instructions.
Protein Pulldown Analysis
20 μg of the indicated His and GST fusion constructs were incubated in 120 μl of PBS buffer (300 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 20 mm imidazole, 2 mm DTT, pH 7.3) for 30 min at room temperature. For further experiments, 25 μl of a Chlamydomonas stromal extract (corresponding to 2.5 mg of chlorophyll/ml) and 25 μl of non-labeled in vitro translation product of Cr-LHCP were added to the His and GST fusion constructs and treated as described above. Subsequently, the samples were incubated with glutathione-Sepharose for 1 h at 4 °C and washed two times with PBS buffer. Bound proteins were eluted with 10 mm glutathione in 50 mm Tris-HCl, pH 8.0. The eluted fractions were analyzed by SDS-PAGE and detected by Coomassie staining or Western blotting.
Formation and Analysis of Protein Complexes
Equimolar amounts of At-His-cpSRP43 and At-His-cpSRP54M constructs were mixed and incubated for 5 min at room temperature. cpSRP complex formation was analyzed by size exclusion chromatography using a SuperdexTM 200 10/300 GL column (GE Healthcare) in buffer A with a flow rate of 0.4 ml/min.
To analyze transit complex formation, we first solubilized and heated Pisum sativum Lhcb1 (LHCP) (kindly provided by H. Paulsen, University of Mainz, Germany) or Cr-LHCP inclusion bodies in buffer A that contained 5% (w/v) SDS for 2 min to 98 °C. After cooling to room temperature, At-His-cpSRP43, Cr-His-cpSRP43, or a combination of the indicated concentrations of At-His-cpSRP43 and At-His-cpSRP54M (or At-His-cpSRP54MΔ530–564) or Cr-His-cpSRP43 and Cr-His-cpSRP54M were added. In two subsequent steps, the SDS concentration was diluted 5-fold in each step using buffer A, and the sample was incubated for 30 and 60 min, respectively. For SDS precipitation, 250 mm KCl was added, and the sample was incubated on ice for 10 min. Precipitated SDS was removed by centrifugation (10 min, 14,000 rpm, 4 °C). Alternatively, Cr-His-cpSRP54M was added to the Cr-His-cpSRP43·Cr-LHCP complex after SDS removal. Protein complexes in the supernatant were analyzed by size exclusion chromatography as described above. The method for de- and renaturation of LHCP was adapted from Paulsen et al. (27).
Isolation of Chloroplasts and Insertion-competent Thylakoids from Chlamydomonas reinhardtii
Chloroplasts were isolated from Chlamydomonas cells as described previously (28) with the modification that cells were grown in Tris acetate-phosphate medium. Isolated chloroplasts were resuspended in SH buffer (50 mm Hepes, pH 8.0, 10 mm MgCl2, 300 mm sorbitol) and stored on ice until use. Thylakoids were isolated from the chloroplasts by osmotic lysis in HM buffer (10 mm Hepes, pH 8.0, 10 mm MgCl2) for 30 min and repeated grinding using a micropestle at a final concentration of 1.5–2 mg of chlorophyll/ml. Thylakoid membranes were harvested by centrifugation (5 min, 3000 rpm, 4 °C), and the supernatant was collected as the corresponding stromal fraction. Thylakoids were resuspended in SH buffer at a final concentration of 1.5 mg of chlorophyll/ml.
In Vitro Insertion Assays
Chlamydomonas thylakoids equal to 20 μg of chlorophyll were incubated with 5 μl of 35S-labeled mature or precursor Cr-LHCP in vitro translation product in SH buffer supplemented with 10 mm methionine in a total volume of 100 μl in the presence or absence of 5 mm GTP or ATP. Stromal extract equal to 90 μg of chlorophyll of the corresponding thylakoids was added as indicated. The insertion reaction was incubated for 30 min at 25 °C. The samples were split into two aliquots to subject one of these aliquots to thermolysin digestion on ice for 1 h. 10 mm EDTA and 0.13 n NaOH were added to all samples, and thylakoids were recovered by centrifugation (1 min, 8000 rpm). After washing the thylakoids in SH buffer supplemented with 10 mm EDTA, thylakoids were resuspended in SDS-PAGE sample buffer and subjected to SDS-PAGE. The presence of Cr-LHCP in the thylakoid fractions was analyzed by autoradiography.
Structural Figures
A homology model of Cr-cpSRP43 was generated using the PHYRE2 server (29) on the basis of the complex structure between At-cpSRP43ΔCD3 and the At-cpSRP54 tail region (Protein Data Bank code 3UI2 (16)). Figures were drawn using PyMOL, version 1.5.0.4 (Schrödinger, LLC).
Results
cpSRP54 Does Not Form a Complex with cpSRP43 in C. reinhardtii
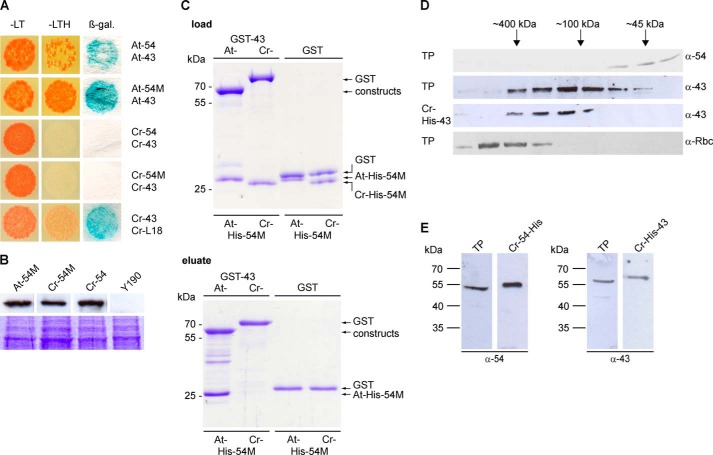
The cpSRP54 and cpSRP43 proteins of C. reinhardtii (Cr-cpSRP54 and Cr-cpSRP43) exhibited the same domain arrangement as the Arabidopsis thaliana homologs and a sequence similarity of 79 and 56%, respectively (Fig. 1). To analyze the binding between Cr-cpSRP54 and Cr-cpSRP43, yeast two-hybrid studies were conducted (Fig. 2A). Surprisingly, in contrast to the Arabidopsis proteins, Cr-cpSRP43 interacted neither with full-length Cr-cpSRP54 nor with the M-domain of Cr-cpSRP54. However, we observed a clear interaction between Cr-cpSRP43 and the L18 motif of Cr-LHCP, which demonstrates the functional expression of Cr-cpSRP43 in yeast cells (Fig. 2A). The expression of the Cr-cpSRP54 constructs was verified by Western blot analysis of yeast cell extracts (Fig. 2B).
FIGURE 1.
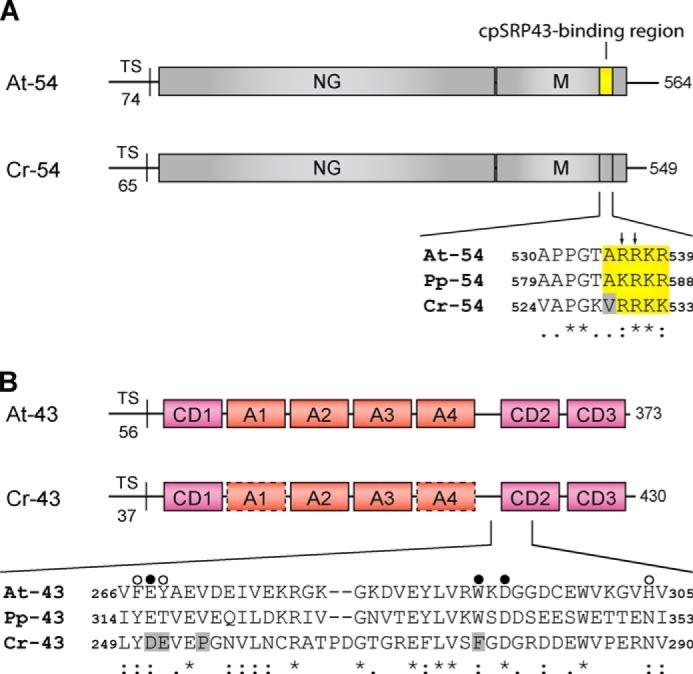
Domain organization of the cpSRP54 and cpSRP43 proteins and sequence alignment of the binding regions. A, cpSRP54 consists of an N-terminal GTPase-containing domain (NG) and a C-terminal methionine-rich domain (M). The sequence alignment shows the cpSRP43-binding motif of A. thaliana cpSRP54 (At-54) and the corresponding regions in cpSRP54 of P. patens (Pp-54) and C. reinhardtii (Cr-54). The conserved residues that are important for binding are depicted in yellow. Arrows indicate the twin arginine motif (Arg-536/Arg-537) that is crucial for cpSRP43 binding in Arabidopsis. The gray box indicates Val-529 in Cr-cpSRP54 that interferes with cpSRP43 binding. TS, transit sequence. B, cpSRP43 consists of three chromodomains (CD1–CD3) and an ankyrin repeat domain (A1–A4). The sequence alignment shows the region of CD2 composed of the twinned cages that recognize Arg-536 and Arg-537 in At-cpSRP54 (16). The cage 1- and cage 2-forming residues of At-cpSRP43 and the corresponding positions in Pp-cpSRP43 (Pp-43) and Cr-cpSRP43 (Cr-43) are marked with filled and open circles, respectively. The gray boxes indicate residues in Cr-cpSRP43 that differ in these positions from At-cpSRP43 (At-43) and Pp-cpSRP43. In addition, Pro-255 in Cr-cpSRP43 that interferes with cpSRP54 binding is marked by a gray box. Symbols display the degree of conservation: identical residues (asterisk), conserved substitution (colon), semiconserved substitution (dot).
FIGURE 2.
Unlike higher plants, Chlamydomonas cpSRP54 and cpSRP43 do not form a stable complex in vitro and in vivo. A, for yeast two-hybrid assays, the yeast strain Y190 was cotransformed with pGBKT7 constructs encoding the full-length cpSRP54 (54) or its M-domain (54M) of Arabidopsis (At) and Chlamydomonas (Cr) and pACT2 constructs encoding cpSRP43 (43) of these organisms. Cotransformed cells were dotted onto minimal media lacking Leu and Trp (−LT) to check for cotransformation or −LTH to assess the extents of interaction. β-Galactosidase (β-gal.) activity was visualized using filter assays. B, expression of the pGBKT7 encoded fusion proteins in the yeast cells was verified by Western blot analysis of total yeast protein extracts (equivalent to 1 ml of a culture with an A600 of 1) using antibodies against the c-Myc epitope (BD Biosciences). Untransformed yeast cells were used as negative controls. Equal loading was controlled by a duplicate gel stained with Coomassie Blue. C, in vitro pulldown assays were performed with recombinant GST-cpSRP43 (At- and Cr-GST-43) proteins and the His-tagged M-domain of cpSRP54 proteins (At- and Cr-His-54M) as indicated using glutathione-Sepharose. Control reactions were performed with recombinant GST. One-tenth of the loaded and one-third of the eluted proteins (upper and lower panels) were separated by SDS-PAGE and detected by Coomassie staining. D, either total soluble protein extracts (TP) of Chlamydomonas or recombinant Cr-His-cpSRP43 (Cr-43) was fractionated by size exclusion chromatography, and the levels of Cr-cpSRP54, Cr-cpSRP43, and the large subunit of Rubisco (Rbc) were evaluated by Western blot analyses. E, Western blot analysis of 30 μg of a total soluble protein extract (TP) from Chlamydomonas and recombinant Cr-His-cpSRP43 (Cr-43) or Cr-cpSRP54-His (Cr-54) using antibodies directed against Cr-cpSRP54M (generated against recombinant Cr-cpSRP54M; Seqlab) or At-cpSRP43 (7).
To confirm the inability of the cpSRP proteins of Chlamydomonas to form a complex, in vitro pulldown experiments were performed using recombinant Cr-GST-cpSRP43 and the His-tagged M-domain of cpSRP54 (Cr-His-cpSRP54M). As a positive control, the corresponding proteins from Arabidopsis were used. Negative control reactions were conducted with recombinant GST. Whereas At-GST-cpSRP43 clearly coprecipitated At-His-cpSRP54M, we did not observe binding of the corresponding Chlamydomonas proteins (Fig. 2C).
Previously, in higher plants, stromal cpSRP43 was found to coelute with cpSRP54 as a ∼200-kDa species by size exclusion chromatography (8), whereas stromal cpSRP43 of the Arabidopsis cpSRP54 knock-out mutant ffc1-2 eluted at ∼70 kDa, which is identical to the elution profile of purified recombinant cpSRP43 (12). To determine the native molecular weight of Cr-cpSRP43 and Cr-cpSRP54, a total protein extract of Chlamydomonas cells was fractionated by size exclusion chromatography, and Cr-cpSRP54 and Cr-cpSRP43 were detected using antibodies directed against Cr-cpSRP54M or At-cpSRP43 (Fig. 2D). These antibodies recognized recombinant Cr-cpSRP54-His or Cr-His-cpSRP43 and cross-reacted with a single protein of the expected apparent molecular weight in a total Chlamydomonas cell extract (Fig. 2E). Whereas stromal Cr-cpSRP54 eluted as a monomer at ∼45 kDa, stromal Cr-cpSRP43 eluted at ∼100 kDa, which corresponded to the elution profile of recombinant Cr-His-cpSRP43 (Fig. 2D). As Cr-cpSRP43 has a predicted molecular mass of 48 kDa, the migration as an ∼100-kDa species on the gel filtration column reflects the elution behavior of At-cpSRP43, which elutes at ∼70 kDa as mentioned above and has a predicted molecular mass of 35 kDa. To prove the integrity of the Chlamydomonas cell extract, the elution of Rubisco as a high molecular weight complex is shown (Fig. 2D). These data clearly demonstrate that, contrary to Arabidopsis proteins, stromal Cr-cpSRP54 and Cr-cpSRP43 do not form a stable complex.
The ARR Motif in the cpSRP54 Tail Region Is Crucial for Binding cpSRP43 in Arabidopsis
In the At-cpSRP54 protein, the At-cpSRP43-binding motif comprises a positively charged 10-amino acid region (positions 530–539) that is located in close proximity to the C terminus (14). Within this tail region, two neighboring arginines, Arg-536 and Arg-537, play crucial roles in the formation of the binding interface, and mutations of either residue abolish cpSRP complex formation (14, 16). The At-cpSRP54 tail region appears to be conserved in the Physcomitrella and Chlamydomonas cpSRP54 proteins (Fig. 1A), and the ability of Physcomitrella cpSRP54 (Pp-cpSRP54) to bind to Physcomitrella cpSRP43 (Pp-cpSRP43) or At-cpSRP43 has been recently demonstrated (25). Here, however, we show that Cr-cpSRP54 is unable to interact with At-cpSRP43 in yeast two-hybrid experiments and in vitro pulldown assays, whereas a fusion construct in which the C terminus of Cr-cpSRP54 was replaced with the portion of the At-cpSRP54 tail region that is crucial for At-cpSRP43 binding (Cr-54M/At-54C-term) clearly bound to At-cpSRP43 (Table 1 and Fig. 3A). These data suggested that the twin arginine residues within the cpSRP54 tail region are necessary but not sufficient to facilitate cpSRP complex formation. To identify additional crucial residues, point mutations were introduced in At-cpSRP54M at those positions within the tail that differed in the Arabidopsis and Physcomitrella proteins from those of the Chlamydomonas protein. These positions in the Arabidopsis protein are Ala-530, Thr-534, and Ala-535 (Fig. 1A). The ability of the At-cpSRP54M mutants to bind to At-cpSRP43 was tested in yeast two-hybrid experiments (Table 1). No change in binding intensity to At-cpSRP43 was observed when using the At-cpSRP54M constructs containing mutations in positions 530 (A530M and A530V) and 534 (T534M). However, the mutation of alanine at position 535 (A535M and A535V) resulted in a complete loss of interaction with At-cpSRP43, indicating that the ARR motif in At-cpSRP54 is crucial for binding to At-cpSRP43. Consistently, the mutation V529A in Cr-cpSRP54M that restores the ARR motif resulted in an interaction between Cr-cpSRP54M (V529A) and At-cpSRP43 as demonstrated by yeast two-hybrid experiments and in vitro pulldown assays (Table 1 and Fig. 3A). Furthermore, in the Arabidopsis cpSRP complex structure (At-cpSRP43 with At-cpSRP54 tail) (16), the Ala-535 residue of the At-cpSRP54 protein is located within a tight hydrophobic pocket that is formed by the central β-sheet of CD2 of At-cpSRP43 (Fig. 4A). This pocket is unable to accommodate the bulkier side chains of valine or methionine, which would sterically prevent the interaction.
TABLE 1.
Interaction of various Chlamydomonas and Arabidopsis cpSRP54 M constructs with Arabidopsis cpSRP43
The yeast strain Y190 was cotransformed with the indicated combinations of bait and prey constructs. In the Cr-54M/At-54C-term fusion construct, the C terminus of Chlamydomonas cpSRP54 M was replaced with that of Arabidopsis containing the cpSRP43-binding motif. The growth of the transformants on selective medium (−LTH) and β-galactosidase activity was analyzed.
| Constructs (bait) | At-43 (prey) |
|
|---|---|---|
| −LTH | β-Gal | |
| Cr-54M | − | − |
| Cr-54M/At-54C-term | ++ | + |
| At-54M(A530M) | ++ | + |
| At-54M(A530V) | ++ | + |
| At-54M(T534M) | ++ | + |
| At-54M(A535M) | − | − |
| At-54M(A535V) | + | − |
| Cr-54M(V529A) | ++ | + |
FIGURE 3.
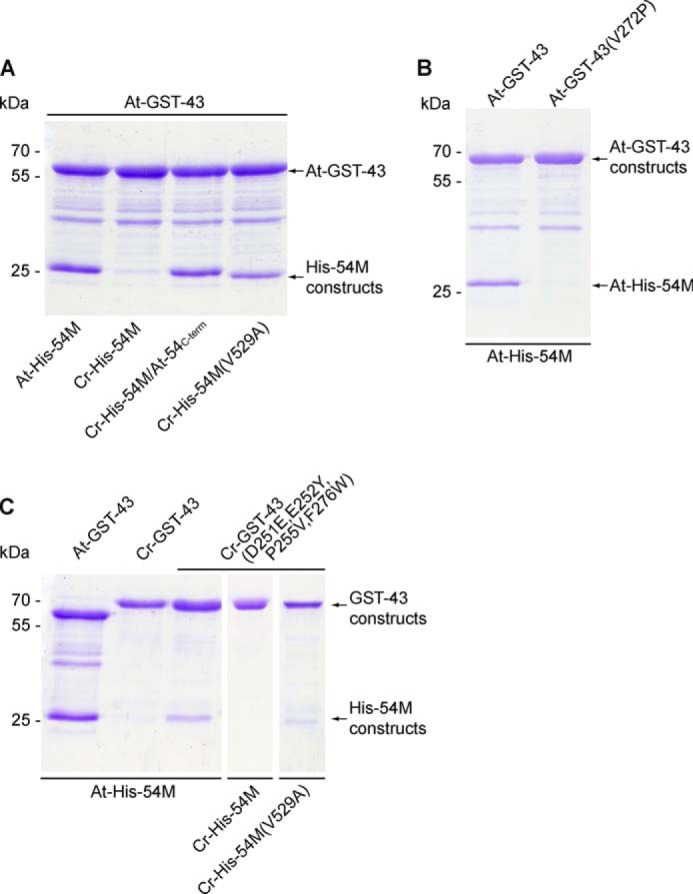
Interaction analyses between various cpSRP43 and cpSRP54M constructs of Arabidopsis and Chlamydomonas. In vitro pulldown assays were performed with recombinant GST-cpSRP43 (GST-43) and His-tagged cpSRP54M (His-54M) proteins as indicated using glutathione-Sepharose.
FIGURE 4.
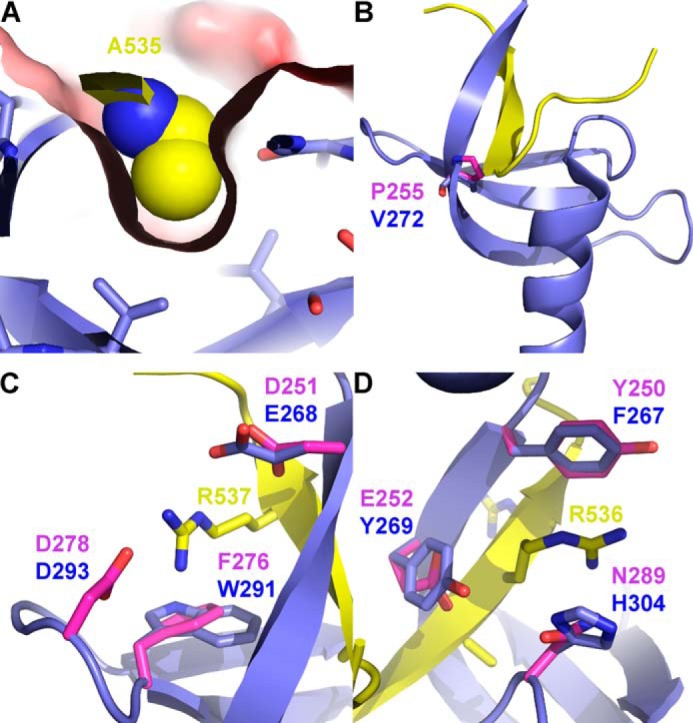
Binding of Arabidopsis cpSRP54 by cpSRP43-CD2. A, the contact between Ala-535 of At-cpSRP54 (yellow) and At-cpSRP43-CD2 (blue) as observed in the complex structure (Protein Data Bank code 3UI2 (16)). Ala-535 is shown as space-filling spheres, whereas CD2 is shown as a stick model with the solvent-accessible surface colored by surface potential. B, the CD2 from At-cpSRP43 (blue) with the bound tail region of At-cpSRP54 (yellow) in a schematic representation. Val-272 of At-cpSRP43 is shown as sticks. In addition, Pro-255 from a homology model of Cr-cpSRP43 is shown as pink sticks. C and D, the twinned aromatic cages of cpSRP43. Superposition of the Arabidopsis complex structure of At-cpSRP43-ΔCD3 (blue) and the At-cpSRP54 tail region (yellow) (Protein Data Bank code 3UI2 (16)) with the homology model of Cr-cpSRP43 (pink) is shown. Residues forming the aromatic cages are shown as sticks and labeled accordingly (C, cage 1; D, cage 2). The two arginine residues (Arg-536 and Arg-537) from the At-cpSRP54 ARR motif are also shown as sticks.
cpSRP43 of Chlamydomonas Is Unable to Bind to the Canonical Tail Region of cpSRP54
In At-cpSRP43, a twinned aromatic cage in CD2 that is formed by six residues (three residues per cage) mediates the binding of cpSRP54 (16). As these residues appear to be conserved throughout the green lineage (16), we expected that Cr-cpSRP43 can bind to cpSRP54 proteins that contain the canonical cpSRP43-binding motif. Remarkably, however, no interaction was detected between Cr-cpSRP43 and At-cpSRP54M (Table 2). To unravel the structural differences between Cr-cpSRP43 and At-cpSRP43, a sequence alignment of CD2 of Cr-, At-, and Pp-cpSRP43 was performed. As shown in Fig. 1B, two residues of cage 1 in Cr-cpSRP43 (Asp-251 and Phe-276) and one residue of cage 2 in Cr-cpSRP43 (Glu-252) differed from the corresponding residues in At- and Pp-cpSRP43. To test whether Cr-cpSRP43 is still compatible with the formation of the twinned aromatic cage, we generated a homology model of Cr-cpSRP43 that was based on the complex structure between At-cpSRP43 and the At-cpSRP54 tail (16). Indeed, in the structural model, the six aligned cage-forming residues are positioned in nearly identical positions around the two arginine residues from the At-cpSRP54 ARR motif (Fig. 4, C and D).
TABLE 2.
Interaction of various Chlamydomonas cpSRP43 constructs with Arabidopsis cpSRP54M
The yeast strain Y190 was cotransformed with the indicated combinations of bait and prey constructs. The growth of the transformants on selective medium (−LTH) and β-galactosidase activity was analyzed.
| Constructs (prey) | At-54M (bait) |
|
|---|---|---|
| −LTH | β-Gal | |
| Cr-43 | − | − |
| Cr-43(D251E,E252Y,F276W) | − | − |
| Cr-43(P255V) | − | − |
| Cr-43(D251E,E252Y,P255V,F276W) | + | + |
To analyze whether the above mentioned residue differences in the cages cause the inability of Cr-cpSRP43 to interact with the canonical cpSRP54 tail, we generated a mutant construct, Cr-cpSRP43(D251E,E252Y,F276W), containing the same cage residues as that of At-cpSRP43. However, these mutations were not sufficient to restore binding to At-cpSRP54M (Table 2). For At-cpSRP43, a contiguous mixed β-sheet involving the first β-strand of CD2 has previously been identified as a stabilizing factor in complex formation with At-cpSRP54 (16). Strikingly, in Cr-cpSRP43, a proline (Pro-255) is present in this region, which would destabilize this β-sheet, thereby inhibiting the β-completion required for efficient interaction (Fig. 4B). The detrimental effect of proline in this position was confirmed by the observation that the mutation V272P in At-cpSRP43 resulted in a complete loss of At-cpSRP54M binding (Fig. 3B). Indeed, although the single P255V mutation did not affect the binding behavior of Cr-cpSRP43, the quadruple mutation (D251E,E252Y,P255V,F276W) restoring both aromatic cages and removing the crucial proline residue led to a clear interaction with At-cpSRP54M, although this interaction was significantly weaker than the interaction between At-cpSRP43 and At-cpSRP54M (Table 2 and Fig. 3C). Contrary, no interaction was observed between the mutated Cr-cpSRP43 and Cr-cpSRP54, confirming that Cr-cpSRP54 does not contain the canonical cpSRP43-binding motif as described above (Fig. 3C). A fairly weak interaction was detected between the mutated Cr-cpSRP43 and Cr-cpSRP54M (V529A) (Fig. 3C). Our previous observation that the binding between mutated Cr-cpSRP43 and At-cpSRP54M is considerably weaker than the binding between At-cpSRP43 and At-cpSRP54M and that the binding between the mutated Cr-cpSRP54M and At-cpSRP43 is also less efficient than between the Arabidopsis proteins indicated that the mutations in the Chlamydomonas proteins do not fully restore the canonical binding interface. Therefore it is conceivable that the combination of the two Chlamydomonas proteins resulted in a rather low binding affinity.
The cpSRP Complex Formation Evolved in Streptophytes
The identification of residues within the tail region of cpSRP54 and CD2 of cpSRP43 crucial for the formation of a binding interface enabled a thorough prediction of the phylogenetic distribution of cpSRP complex formation within chloroplasts of the green lineage. The cpSRP43-binding motif consisting of an alanine residue followed by the positively charged (R/K)R motif is highly conserved within the cpSRP54 tail region of streptophytes including land plants and charophytes (Fig. 5A). In contrast, all cpSRP54 proteins of chlorophytes displayed valine instead of alanine upstream of the (R/K)R motif (Fig. 5A).
FIGURE 5.
Residues that are important for the formation of the cpSRP54-cpSRP43 binding interface are conserved in land plants but not in chlorophytes. Sequence alignment of the cpSRP43-binding motif in cpSRP54 (A) and the twinned cage-forming region of cpSRP43-CD2 (B) from various organisms of the green lineage (further details are given in the legend to Fig. 1).
Within the cpSRP43 proteins, all land plants contain the canonical cpSRP54-binding motif in CD2. However, all chlorophyte cpSRP43 proteins display the proline in the first β-strand of CD2 that is detrimental to cpSRP54 binding (Fig. 5B). Furthermore, cpSRP43 proteins of chlorophytes exhibit a negatively charged residue (Glu-252 in Chlamydomonas) instead of an aromatic tyrosine (Tyr-269 in Arabidopsis) in cage 2 of CD2 that probably also interferes with cpSRP complex formation (see above). Similar to chlorophytes, this residue is also negatively charged in charophytes (Fig. 5B). These data strongly suggest that the ability of cpSRP complex formation is not present in chlorophytes and evolved in streptophytes either with the appearance of charophytes or land plants.
In Chlamydomonas, cpSRP54 Is Not Involved in Transit Complex Formation
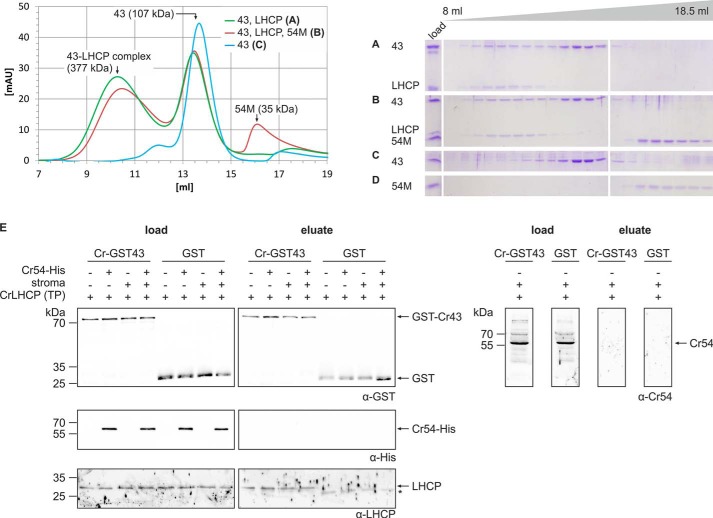
We next aimed to analyze the role of cpSRP43 and cpSRP54 in the formation of soluble LHCP complexes in chlorophytes and to answer the question whether non-cpSRP43-bound cpSRP54 might contribute to transit complex formation by contacting LHCP directly. Therefore, Cr-LHCP was expressed as inclusion bodies, denatured by SDS, and incubated with equimolar amounts of recombinant Cr-cpSRP43 or a combination of Cr-cpSRP43 and Cr-cpSRP54M. After removal of the detergent, the resulting complexes were separated by size exclusion chromatography. In a second, alternative experimental setup, Cr-cpSRP54M was added to the Cr-cpSRP43/Cr-LHCP combination after removal of the detergent to avoid a possible detrimental effect of the de- and renaturing procedure on Cr-cpSRP54M. As both types of experiments generated the same results, only the second set of data are presented (Fig. 6, A–D). Cr-cpSRP43 coeluted with Cr-LHCP from the column in a soluble complex with an apparent molecular mass of ∼377 ± 97 kDa (Fig. 6, A and B), whereas free Cr-cpSRP43 eluted at about ∼100 kDa (Fig. 6, A, B, and C). Cr-cpSRP54M was not present in the Cr-cpSRP43·LHCP complex fractions and eluted separately in the same fractions as recombinant Cr-cpSRP54M alone (Fig. 6, B and D). It should be noted that Cr-LHCP alone could not be separated on a gel filtration column as it formed insoluble aggregates in the absence of Cr-cpSRP43 after removal of the detergent.
FIGURE 6.
Cr-cpSRP54 is not involved in transit complex formation in Chlamydomonas. A–D, protein complex formation using the following equimolar combinations of recombinant proteins was analyzed by size exclusion chromatography: Cr-His-cpSRP43 and Cr-LHCP (A) and Cr-His-cpSRP43 (43), Cr-LHCP (LHCP), and Cr-His-cpSRP54M (54M) (B). As controls, the single proteins Cr-His-cpSRP43 (C) and Cr-His-cpSRP54M (D) were analyzed. Elution fractions ranging from 8 to 18.5 ml were analyzed by SDS-PAGE and Coomassie staining (A–D). Cr-His-cpSRP54M was added to the combination of Cr-His-cpSRP43 and Cr-LHCP after dilution and removal of SDS (B). The same elution profiles were obtained when Cr-His-cpSRP54M was added to this combination before SDS removal.3 Single proteins were always treated in the same way as in assays analyzing complex formation. E, pulldown assays were conducted by incubation of recombinant Cr-GST-cpSRP43 and the Cr-LHCP in vitro translation product (TP) with the indicated combinations of recombinant Cr-cpSRP54-His and a stromal extract from Chlamydomonas. Control reactions were performed with recombinant GST. Proteins were enriched using glutathione-Sepharose. Samples of the load and the eluted fractions were analyzed by Western blotting using antibodies directed against the GST tag, His tag, or LHCP. The asterisk (*) marks signals caused by an unspecific cross-reaction of the α-Cr-LHCP antibody with the eluted GST protein. To detect endogenous stromal Cr-cpSRP54 (Cr-54) in the load and eluted fractions, the samples from pulldown assays conducted in the presence of Cr-GST-cpSRP43, Cr-LHCP, and stromal extract were blotted and analyzed using an antibody directed against Cr-cpSRP54M. mAU, milliabsorbance units.
To further analyze whether cpSRP54 might contribute to transit complex formation, in vitro translated Cr-LHCP was incubated with recombinant Cr-GST-cpSRP43 in the presence and absence of Cr-cpSRP54-His. We also analyzed these two combinations in the presence of stroma to test whether a stromal component is required for recruiting Cr-cpSRP54-His to the transit complex or whether native instead of recombinant cpSRP54 might be able to contribute to transit complex formation. Proteins were purified using glutathione-Sepharose, and GST was used in control reactions. As shown in Fig. 6E, the in vitro translated LHCP coeluted with Cr-GST-cpSRP43 in all samples and was not present in the GST controls. Contrary to LHCP, neither Cr-cpSRP54-His nor stromal Cr-cpSRP54 was detected in the eluate fractions. In summary, these data show that Cr-cpSRP43 alone binds Cr-LHCP to form the transit complex in Chlamydomonas and indicate that this holds true for all organisms without a heterodimeric cpSRP.
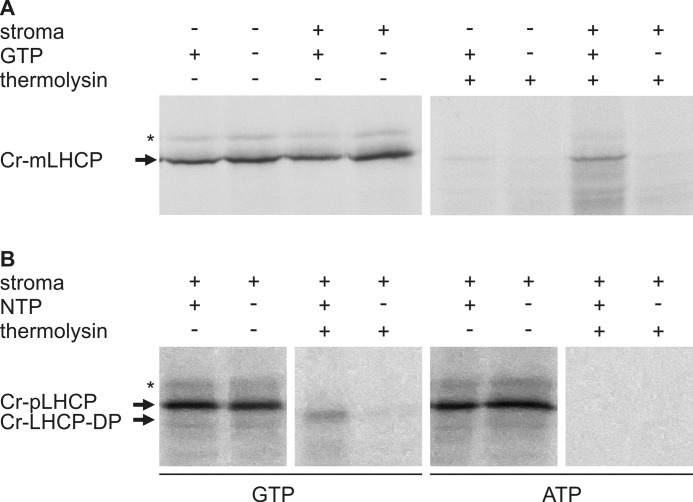
Integration of Cr-LHCP into the Thylakoid Membrane of Chlamydomonas Is GTP-dependent
In higher plants, the in vitro integration of LHCP into the thylakoid membrane requires the Alb3 insertase, the heterodimeric cpSRP43·cpSRP54 complex, the SRP receptor cpFtsY, and GTP, which is essential to trigger the GTPase cycle of the SRP GTPases. As our data demonstrated that in Chlamydomonas Cr-cpSRP54 has no role in transit complex formation, we aimed to investigate whether integration of Cr-LHCP into the thylakoid membrane requires the cpSRP GTPases. Experiments to reconstitute Cr-LHCP integration into isolated thylakoids with purified recombinant proteins were not successful. Therefore, integration assays using radiolabeled in vitro translated mature Cr-LHCP and thylakoids in the presence and absence of a stromal extract and GTP were performed. As shown in Fig. 7A, protease-resistant Cr-LHCP was only detected in assays containing both the stromal extract and GTP. To verify that the Cr-LHCP was integrated into the thylakoid membrane and to confirm the GTP dependence of the reaction, integration assays were conducted using radiolabeled in vitro translated precursor Cr-LHCP in the presence of stroma and GTP or ATP. Only in the presence of GTP was a protease-protected degradation fragment of the precursor Cr-LHCP detected (Fig. 7B). As the precursor sequence is predicted to comprise 15 residues (ChloroP prediction server), the size shift of about 2 kDa between the precursor Cr-LHCP and the degradation product suggests that the protease treatment led to efficient removal of the precursor sequence, whereas the other parts of the Cr-LHCP are largely protected from digestion by the thylakoid membrane. These data show clearly that Cr-LHCP integration in Chlamydomonas is GTP-dependent.
FIGURE 7.
GTP dependence of LHCP insertion into thylakoid membranes of Chlamydomonas. Thylakoids isolated from Chlamydomonas were incubated with in vitro translated, radiolabeled mature Cr-LHCP (Cr-mLHCP) (A) or precursor Cr-LHCP (Cr-pLHCP) (B) in the presence or absence of a stromal extract from Chlamydomonas and GTP or ATP as indicated. After incubation, thylakoids were protease-treated and washed with NaOH to remove non-integrated Cr-LHCP. Cr-LHCP-DP indicates a degradation product of precursor Cr-LHCP (Cr-pLHCP) generated by thermolysin treatment of thylakoids after the insertion reaction. The asterisk (*) indicates an unspecific background translation product.
In Arabidopsis, the Complex Formation between cpSRP43 and cpSRP54 Is Required for the Formation of Low Molecular Weight Transit Complexes
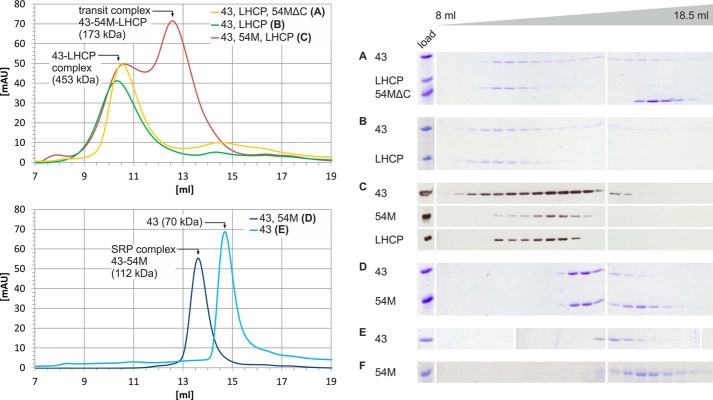
The finding that cpSRP54 has no role in transit complex formation in chlorophytes but was recruited as a high affinity binding partner of cpSRP43 during the evolution of the green lineage prompted the question of a physiological driving force for this adaptation. Studies analyzing the cpSRP system of higher plants initially reported that both cpSRP subunits are required to keep LHCP in a soluble insertion-competent form (8, 14, 15, 30). However, more recent results showed that At-cpSRP43 alone acts as a chaperone for LHCPs and keeps them soluble, challenging the role of At-cpSRP54 in transit complex formation (23, 24). Here we analyzed the formation of soluble LHCP-containing complexes as described above in the presence of At-cpSRP43 alone and in the presence of additional At-cpSRP54M or At-cpSRP54MΔ530–564, which lacks the cpSRP43-binding motif at its extreme C terminus (Fig. 8). In the presence of equimolar amounts of At-cpSRP43 and At-cpSRP54MΔ530–564 or At-cpSRP43 alone, LHCP formed a soluble complex with At-cpSRP43 with an apparent molecular mass of ∼453 ± 36 kDa, whereas At-cpSRP54MΔ530–564 eluted separately (Fig. 8, A and B). The addition of At-cpSRP43 in a 5-fold molar excess resulted in the formation of an At-cpSRP43·LHCP complex with the same apparent molecular weight, and excess non-complexed At-cpSRP43 was separated.3 Notably, in the presence of At-cpSRP43 and At-cpSRP54M, the dominant soluble complex containing LHCP and both cpSRP subunits eluted at a significantly lower apparent molecular mass of 174 ± 9 kDa (Fig. 8C). As controls, the elution profiles of the At-cpSRP43·cpSRP54M complex and the single proteins At-cpSRP43 and At-cpSRP54M were analyzed. The cpSRP complex and the single proteins eluted at 112 (Fig. 8D), 70 (Fig. 8E), and 40 kDa (Fig. 8F) and were therefore significantly smaller than the At-cpSRP43·cpSRP54M·LHCP complex. These results confirm the previous finding that At-cpSRP43 alone can form a soluble complex with LHCP (23, 24). Interestingly, however, our data show that At-cpSRP54M is additionally required for the formation of a transit complex with a drastically reduced molecular weight (>2-fold) compared with the At-cpSRP43·LHCP complex. Furthermore, our data show that At-cpSRP54M can fulfil this function only when it is able to form a complex with At-cpSRP43.
FIGURE 8.
At-cpSRP54M is required for the formation of low molecular weight transit complexes in Arabidopsis. To analyze the ability of cpSRP components to form soluble complexes with LHCP, the following combinations of recombinant proteins were used, and protein mixtures were separated by size exclusion chromatography: At-His-cpSRP43 (43), LHCP, and At-His-cpSRP54MΔ530–564 (54MΔC) (A); At-His-cpSRP43 and LHCP (B); and At-His-cpSRP43, At-His-cpSRP54M (54M), and LHCP (C). As controls, the combination of At-His-cpSRP43 and At-His-cpSRP54M (D) and the single proteins At-His-cpSRP43 (E) and At-His-cpSRP54M (F) were analyzed. The single proteins were treated in the same way as in assays analyzing complex formation. As At-cpSRP54M has a very low absorbance at 280 nm, the chromatogram is not shown. Elution fractions ranging from 8 to 18.5 ml were analyzed by SDS-PAGE and Coomassie staining (A, B, D, E, and F) or Western blotting (C) to distinguish between LHCP and At-His-cpSRP54M as both proteins have the same running behavior in SDS-PAGE. The complex formation assays shown in A, B, and D contained equimolar amounts of the indicated recombinant proteins, whereas in the assay shown in C At-His-cpSRP54M, At-His-cpSRP43, and LHCP were incubated at a molar ratio of 1:2:2. mAU, milliabsorbance units.
Discussion
In eukaryotic organisms of the green lineage, the antenna systems of photosystems I and II are composed of LHC proteins, which consist of three transmembrane-spanning regions. As LHC proteins are encoded in the nucleus, the chloroplast needs to provide an efficient system for the posttranslational passage of these hydrophobic proteins through the aqueous stromal environment to the thylakoid membrane. During evolution, chloroplasts retained the evolutionarily conserved 54-kDa GTPase cpSRP54 from the ancient SRP-dependent protein transport machinery that mediates the cotranslational transport of membrane proteins in prokaryotes. As the importance of cpSRP54 for LHCP transport has been demonstrated in higher plants, it was reasonable to initially assume that the posttranslational cpSRP pathway evolved using this component as a key player and cpSRP43 as an additional component that replaced the ancient SRP RNA. However, later research highlighted the crucial role of cpSRP43 in this transport system. First, At-cpSRP43 appeared to be sufficient for mediating LHCP transport in Arabidopsis plants lacking both At-cpSRP54 and At-cpFtsY (31). Second, At-cpSRP43 alone has been demonstrated to act in vitro as a chaperone for LHCP and is able to keep LHCP in a soluble form (23, 24). Third, apart from acting as a chaperone, At-cpSRP43 can function as a targeting factor because it is able to interact directly with the translocase Alb3 within the thylakoid membrane (19–22). In higher plants, the complete pool of stromal cpSRP43 is associated with cpSRP54, forming a stable cpSRP complex (7, 8) the subunits of which interact with high affinity as a Kd value of 2.5 nm was described for the interaction between At-cpSRP43 and At-cpSRP54M (32). Likewise, the ability of the cpSRP proteins from the moss Physcomitrella to form stable complexes at least in vitro has been recently described (25). These findings have led to the view that the posttranslational cpSRP43·cpSRP54 complex-dependent transport system coevolved with the LHC proteins, and it has been speculated that further evolution might lead to a novel targeting system that is dependent on cpSRP43 alone (33) as cpSRP43 seems to have all requisite functions for LHCP delivery as described above. Surprisingly, however, we now show that the cpSRP complex did not coevolve with the appearance of LHC proteins in green algae but occurred later during the transition from aquatic to terrestrial life. Instead, non-complexed cpSRP43 coevolved with the LHC proteins to keep them in a soluble state as we demonstrated for the Chlamydomonas proteins. The importance of Cr-cpSRP43 for Cr-LHCP transport in Chlamydomonas in vivo has been reported recently as a cpSRP43-null mutant exhibited a drastic reduction of chlorophyll content and light-harvesting complexes (34). It can be speculated that the function of cpSRP43 and cpSRP54/cpFtsY is restricted to posttranslational LHCP delivery and cotranslational transport of chloroplast-encoded membrane proteins, respectively, in green algae. However, this notion is probably not correct because (a) we demonstrated a clear GTP dependence of Cr-LHCP integration, which points strongly to an involvement of the cpSRP GTPases Cr-cpSRP54 and Cr-cpFtsY in the integration reaction, and (b) Cr-cpSRP43 appears to be insufficient for Cr-LHCP delivery in Chlamydomonas as the loss of the cpftsy gene also significantly reduced the levels of LHC proteins (35). Therefore, it is highly likely that Cr-cpSRP54 functions cooperatively with Cr-cpFtsY in posttranslational LHCP transport in Chlamydomonas, but in contrast to the posttranslational cpSRP system of higher plants, Cr-cpSRP54 acts downstream of Cr-cpSRP43.
As described above, cpSRP54 of higher plants forms a stable heterodimer with cpSRP43, and we show in this study that the At-cpSRP43·cpSRP54M heterodimer is required for the formation of a transit complex with a significantly smaller molecular mass (∼170 kDa) than that observed for the soluble At-cpSRP43·LHCP complex (∼450 kDa). It should be noted that the molecular weight of the At-cpSRP43·LHCP complex was independent of various experimental conditions such as salt type, salt concentration (e.g. up to 1 m NaCl), and different detergents in various concentrations or molar ratios of the recombinant proteins.3 Furthermore, the molecular mass of 170 kDa of the small transit complex is very close to the 120 kDa of the LHCP soluble intermediate that is formed in the stroma (36). Previously, both cpSRP subunits At-cpSRP43 and At-cpSRP54 were reported to be required to facilitate LHCP insertion into thylakoid membranes in vitro, and mutations in At-cpSRP54 that prevent cpSRP complex formation drastically reduced LHCP insertion in vitro (8, 12, 14, 15, 30). Therefore, it is highly likely that the low molecular weight transit complex enables efficient LHCP insertion in higher plants. The precise molecular constraints that triggered the recruitment of cpSRP54 for posttranslational LHCP transport in complex with cpSRP43 during evolution remain to be clarified but might be attributed to structural and functional changes of the light-harvesting antennae during the transition to life on land (37, 38).
Acknowledgments
We thank the Kazusa DNA Research Institute (Japan) for providing the cDNA clone AV640228 and H. Paulsen (University of Mainz, Germany) for the kind gift of pea LHCP inclusion bodies, the expression vector pDSII, and the LHCP antibody.
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grants SFB 642 and SCHU 1163/5-1 and by the Ruhr University Research School funded by Germany's Excellence Initiative (DFG Grant GSC 98/1).
B. Dünschede and D. Schünemann, unpublished observation.
- SRP
- signal recognition particle
- LHCP
- light-harvesting chlorophyll-a/b-binding protein
- cp
- chloroplast
- CD
- chromodomain
- −LTH
- medium lacking leucine, tryptophan, and histidine
- At
- A. thaliana
- Cr
- C. reinhardtii
- Rubisco
- ribulose-bisphosphate carboxylase/oxygenase
- Pp
- P. patens
- LHC
- light-harvesting complex.
References
- 1. Akopian D., Shen K., Zhang X., Shan S. O. (2013) Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 82, 693–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Grudnik P., Bange G., Sinning I. (2009) Protein targeting by the signal recognition particle. Biol. Chem. 390, 775–782 [DOI] [PubMed] [Google Scholar]
- 3. Franklin A. E., Hoffman N. E. (1993) Characterization of a chloroplast homologue of the 54-kDa subunit of the signal recognition particle. J. Biol. Chem. 268, 22175–22180 [PubMed] [Google Scholar]
- 4. Nilsson R., Brunner J., Hoffman N. E., van Wijk K. J. (1999) Interactions of ribosome nascent chain complexes of the chloroplast- encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18, 733–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nilsson R., van Wijk K. J. (2002) Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; 'cpSRP54 caught in the act'. FEBS Lett. 524, 127–133 [DOI] [PubMed] [Google Scholar]
- 6. Groves M. R., Mant A., Kuhn A., Koch J., Dübel S., Robinson C., Sinning I. (2001) Functional characterization of recombinant chloroplast signal recognition particle. J. Biol. Chem. 276, 27778–27786 [DOI] [PubMed] [Google Scholar]
- 7. Klimyuk V. I., Persello-Cartieaux F., Havaux M., Contard-David P., Schuenemann D., Meiherhoff K., Gouet P., Jones J. D., Hoffman N. E., Nussaume L. (1999) A chromodomain protein encoded by the Arabidopsis CAO gene is a plant- specific component of the chloroplast signal recognition particle pathway that is involved in LHCP targeting. Plant Cell 11, 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schuenemann D., Gupta S., Persello-Cartieaux F., Klimyuk V. I., Jones J. D., Nussaume L., Hoffman N. E. (1998) A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. U.S.A. 95, 10312–10316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Richter C. V., Bals T., Schünemann D. (2010) Component interactions, regulation and mechanisms of chloroplast signal recognition particle-dependent protein transport. Eur. J. Cell Biol. 89, 965–973 [DOI] [PubMed] [Google Scholar]
- 10. Ouyang M., Li X., Ma J., Chi W., Xiao J., Zou M., Chen F., Lu C., Zhang L. (2011) LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat. Commun. 2, 277. [DOI] [PubMed] [Google Scholar]
- 11. Kogata N., Nishio K., Hirohashi T., Kikuchi S., Nakai M. (1999) Involvement of a chloroplast homologue of the signal recognition particle receptor protein, FtsY, in protein targeting to thylakoids. FEBS Lett. 447, 329–333 [DOI] [PubMed] [Google Scholar]
- 12. Tu C. J., Schuenemann D., Hoffman N. E. (1999) Chloroplast FtsY, chloroplast signal recognition particle, and GTP are required to reconstitute the soluble phase of light-harvesting chlorophyll protein transport into thylakoid membranes. J. Biol. Chem. 274, 27219–27224 [DOI] [PubMed] [Google Scholar]
- 13. Moore M., Harrison M. S., Peterson E. C., Henry R. (2000) Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275, 1529–1532 [DOI] [PubMed] [Google Scholar]
- 14. Funke S., Knechten T., Ollesch J., Schünemann D. (2005) A unique sequence motif in the 54-kDa subunit of the chloroplast signal recognition particle mediates binding to the 43-kDa subunit. J. Biol. Chem. 280, 8912–8917 [DOI] [PubMed] [Google Scholar]
- 15. Goforth R. L., Peterson E. C., Yuan J., Moore M. J., Kight A. D., Lohse M. B., Sakon J., Henry R. L. (2004) Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J. Biol. Chem. 279, 43077–43084 [DOI] [PubMed] [Google Scholar]
- 16. Holdermann I., Meyer N. H., Round A., Wild K., Sattler M., Sinning I. (2012) Chromodomains read the arginine code of post-translational targeting. Nat. Struct. Mol. Biol. 19, 260–263 [DOI] [PubMed] [Google Scholar]
- 17. DeLille J., Peterson E. C., Johnson T., Moore M., Kight A., Henry R. (2000) A novel precursor recognition element facilitates posttranslational binding to the signal recognition particle in chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 97, 1926–1931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stengel K. F., Holdermann I., Cain P., Robinson C., Wild K., Sinning I. (2008) Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43. Science 321, 253–256 [DOI] [PubMed] [Google Scholar]
- 19. Bals T., Dünschede B., Funke S., Schünemann D. (2010) Interplay between the cpSRP pathway components, the substrate LHCP and the translocase Alb3: an in vivo and in vitro study. FEBS Lett. 584, 4138–4144 [DOI] [PubMed] [Google Scholar]
- 20. Dünschede B., Bals T., Funke S., Schünemann D. (2011) Interaction studies between the chloroplast signal recognition particle subunit cpSRP43 and the full-length translocase Alb3 reveal a membrane-embedded binding region in Alb3 protein. J. Biol. Chem. 286, 35187–35195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Falk S., Ravaud S., Koch J., Sinning I. (2010) The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J. Biol. Chem. 285, 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lewis N. E., Marty N. J., Kathir K. M., Rajalingam D., Kight A. D., Daily A., Kumar T. K., Henry R. L., Goforth R. L. (2010) A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J. Biol. Chem. 285, 34220–34230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Falk S., Sinning I. (2010) cpSRP43 is a novel chaperone specific for light-harvesting chlorophyll a,b-binding proteins. J. Biol. Chem. 285, 21655–21661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jaru-Ampornpan P., Shen K., Lam V. Q., Ali M., Doniach S., Jia T. Z., Shan S. O. (2010) ATP-independent reversal of a membrane protein aggregate by a chloroplast SRP subunit. Nat. Struct. Mol. Biol. 17, 696–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Träger C., Rosenblad M. A., Ziehe D., Garcia-Petit C., Schrader L., Kock K., Richter C. V., Klinkert B., Narberhaus F., Herrmann C., Hofmann E., Aronsson H., Schünemann D. (2012) Evolution from the prokaryotic to the higher plant chloroplast signal recognition particle: the signal recognition particle RNA is conserved in plastids of a wide range of photosynthetic organisms. Plant Cell 24, 4819–4836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Harris E. H. (1989) The Chlamydomonas Sourcebook: a Comprehensive Guide to Biology and Laboratory Use, p. 242, Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 27. Paulsen H., Finkenzeller B., Kühlein N. (1993) Pigments induce folding of light-harvesting chlorophyll a/b-binding protein. Eur. J. Biochem. 215, 809–816 [DOI] [PubMed] [Google Scholar]
- 28. Mason C. B., Bricker T. M., Moroney J. V. (2006) A rapid method for chloroplast isolation from the green alga Chlamydomonas reinhardtii. Nat. Protoc. 1, 2227–2230 [DOI] [PubMed] [Google Scholar]
- 29. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 30. Yuan J., Kight A., Goforth R. L., Moore M., Peterson E. C., Sakon J., Henry R. (2002) ATP stimulates signal recognition particle (SRP)/FtsY-supported protein integration in chloroplasts. J. Biol. Chem. 277, 32400–32404 [DOI] [PubMed] [Google Scholar]
- 31. Tzvetkova-Chevolleau T., Hutin C., Noël L. D., Goforth R., Carde J. P., Caffarri S., Sinning I., Groves M., Teulon J. M., Hoffman N. E., Henry R., Havaux M., Nussaume L. (2007) Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19, 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hermkes R., Funke S., Richter C., Kuhlmann J., Schünemann D. (2006) The alpha-helix of the second chromodomain of the 43 kDa subunit of the chloroplast signal recognition particle facilitates binding to the 54 kDa subunit. FEBS Lett. 580, 3107–3111 [DOI] [PubMed] [Google Scholar]
- 33. Nussaume L. (2008) Chloroplast SRP takes another road. Nat. Chem. Biol. 4, 529–531 [DOI] [PubMed] [Google Scholar]
- 34. Kirst H., Garcia-Cerdan J. G., Zurbriggen A., Ruehle T., Melis A. (2012) Truncated photosystem chlorophyll antenna size in the green microalga Chlamydomonas reinhardtii upon deletion of the TLA3-CpSRP43 gene. Plant Physiol. 160, 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kirst H., García-Cerdán J. G., Zurbriggen A., Melis A. (2012) Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene. Plant Physiol. 158, 930–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Payan L. A., Cline K. (1991) A stromal protein factor maintains the solubility and insertion competence of an imported thylakoid membrane protein. J. Cell Biol. 112, 603–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Alboresi A., Caffarri S., Nogue F., Bassi R., Morosinotto T. (2008) In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: identification of subunits which evolved upon land adaptation. PLoS One 3, e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Neilson J. A., Durnford D. G. (2010) Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth. Res. 106, 57–71 [DOI] [PubMed] [Google Scholar]