Abstract
A small pool of NK1.1+ CD8+ T cells is harbored among the “conventional” CD8+ T cell compartment. Conclusions driven from analysis of immune responses mediated by cytotoxic CD8+ T cells are often made on the total population, which includes these “contaminating” NK1.1+ CD8+ T cells. An as yet unresolved question is whether NK1.1+ CD8+ cells are conventional T cells that acquire NK1.1 expression upon activation or delineation into memory phenotype, or whether they are a distinct cell population that induces immune responses in a different manner than conventional T cells. To address this question, we used the Listeria monocytogenes model of infection and followed CD8+ NK1.1+ T cells alongside NK1.1− CD8+ T cells in each phase of the immune response: innate, effector and memory. Our central finding is that CD8+ NK1.1+ cells and conventional NK1.1− CD8+ T cells both contribute to the adaptive immune response to Listeria, but only CD8+ NK1.1+ cells were equipped with the ability to provide a rapid innate immune response, as demonstrated by early and antigen-independent IFNγ production, granzyme B expression, and degranulation. More importantly, purified conventional CD8+ T cells alone in the absence of any “contaminating” CD8+ NK1.1+ cells were not sufficient to provide early protection to lethally infected mice. These results highlight the role of CD8+ NK1.1+ T cells in mounting early innate responses important for host defense and support the therapeutic potential of this subset to improve the effectiveness of protective immunity.
Keywords: innate immunity, adaptive immunity, NKT cells, CD8+ T cells, Listeria monocytogenes, IFNγ, cytotoxicity, degranulation
INTRODUCTION
The immune system is comprised of a rapid, antigen-independent innate arm, and an antigen-specific delayed adaptive arm. CD8+ T cells are an integral part of classical antigen-dependent immunity against a variety of viral and bacterial pathogens. Development of cytotoxic T lymphocyte (CTL) activity is a process that takes days (1, 2). To acquire full functional competence, CD8+ T cells must first be activated by the innate immune system, enter a phase of replication, and then differentiate into effector cytotoxic T cells that provide long-lasting protection. However, while the adaptive CD8+ T cell response is developing, innate immune cells, especially NK cells, play a critical role in eradicating infected cells, often utilizing similar strategies as CD8+ T cells, namely the production of IFNγ, granzymes and perforins (3–5).
Recent reports have suggested innate capabilities within the CD8+ T cell population (6, 7). Specifically, memory CD8+ T cells were shown to secrete IFNγ in response to IL12/IL18 stimulation in an antigen-independent manner (8). Memory CD8+ T cells were also shown to contribute to innate immune responses and early protection from pathogen re-encounter (9–11). In these studies Listeria-primed memory CD8+ T cells were able to secrete IFNγ and granzyme B upon reinfection with Listeria in an antigen-independent manner (9, 10, 12–14). Ruiz et al identified this population of memory CD8+ T cells as NK1.1+ CD8+ T cells (13). Similarly lymphocytic choriomeningitis virus (LCMV)-primed memory CD8+ T cells were able to rapidly produce innate IFNγ upon infection with murine cytomegalovirus (MCMV) infection in an antigen-independent manner (15). However, these innate capabilities were identified in the context of prior pathogen exposure.
Although the existence of CD8+ NK1.1+ cells (most often termed CD8+ NKT cells) has been reported for more than two decades, their legitimacy as a distinct T cell subset is still under debate (16–23). To compare and contrast its function with that of the conventional CD8+ T cell (NK1.1−) compartment, we used Listeria monocytogenes (LM) infection model and examined the kinetics of responses by both populations during infection. This model of infection has a well-established pattern of antigen-specific CD8+ T cell adaptive immune responses in mice required for bacterial clearance, but also allows the study of innate immune responses to control bacterial burden during the early phase of infection (24–27). In this study, we show that CD8+ NKT and conventional NK1.1− CD8+ T cells both contribute to the adaptive response to Listeria infection; however, only CD8+ NKT cells and not NK1.1− CD8+ T cells had the ability to produce rapid innate immune responses, as demonstrated by early and antigen-independent proliferation, IFNγ production, granzyme B expression, and degranulation. Importantly, when conventional CD8+ NK1.1− T cells were adoptively transferred into immunodeficient mice, these cells were inferior to NKT cells in protecting mice against early infection. Thus, we propose that in naïve mice, a subset of CD8+ T cells that express NK1.1 have innate capabilities critically important for early host defense against initial infection. Accordingly, we propose that the pattern of NK1.1 expression in CD8+ T cells is similar to the pattern of CD25 expression in CD4+ T cells (28) with both constitutive and acquired expression yielding two different subsets of CD8+ T cells that have distinct functions during the course of an immune response.
MATERIAL AND METHODS
Animal procedures
Adult C57BL/6 WT, Rag2−/−, Rag2−/−γc−/−, CD1d−/− mice were purchased from Taconic. All mice were housed in a specific pathogen free room; all Listeria-infected mice were housed in specific ABSL-2 facility. For i.v. infections, mice were anesthetized with Ketamine 80 mg/kg and Xylazine 10 mg/kg (i.p., in 200 μl PBS). Intravenous infections were performed retro-orbitally. Blood and tissue samples were collected and processed at the indicated time points in accordance with University of Michigan Animal Care and Use Committee and approval to use mice was granted by the University of Michigan in accordance with the US National Institute of Health requirements for the care and use of animals. Care for mice was provided in accordance with PHS and AAALAC standards.
Listeria monocytogenes infection
Listeria monocytogenes expressing Ovalbumin (LM-Ova) strain 10403s (29) was a kind gift from Mary O’Riordan (University of Michigan). LM-Ova was grown in BHI or LB media with 5 μg/ml Erythromycin (30). Dose and route of LM-Ova infection for priming and prime/boost regimen have been previously established (29, 31, 32). We collected bacteria in a mid-log phase and injected intravenously 103, 104, 105 or 2x105 CFU/mouse. The infection dose was determined based on the following formula: OD600 of 1 = 1.2x109 bacteria/ml; the dose was validated retrospectively on BHI or LB agar plates + 5 μg/ml Erythromycin (Erm). LM-Ova burden was determined using colony forming unit determination as previously detailed by culturing serially diluted homogenized spleen and liver on BHI/Erm or LB/Erm agar plates (27, 33).
In vivo treatment
Where indicated, mice were treated with 2 mg/mouse of BrdU (Sigma) for 3 days (once a day) or with 4 mg/kg poly I:C (GE Healthcare) once (intraperitoneally, in 200 μl PBS).
Lymphocyte isolation
Single cell suspensions of spleen, liver and PBLs were prepared in RPMI supplemented with 5% FCS. Cells were passed through a nylon mesh (70 μm), red blood cells were lysed and cells were counted and stained. Liver lymphocytes were prepared by perfusion and then crushed through a nylon mesh. Liver cells were then passed through a 40%/70% percoll gradient and centrifuged at 2000 rpm for 20 min at room temperature. Cells were harvested from the interface and then counted and stained.
Cell staining and Flow Cytometry
All cell suspensions were treated with 2.4G2 and then surface stained with the following fluorochrome-conjugated antibodies: CD3 (145-2C11 or 500A2), CD8 (53-6.7), CD4 (RM4-5), NK1.1 (PK136), CD49b (DX5), CD127 (A7R34), CD132 (4G3), CD19 (1d3), CD244 (m2B4), CD27 (LG.7F9), CD44 (IM7), CD62L (MEL-14), CD94 (18d3), MHC class II (M5/114.15.2), Ly49A (YE1/48.10.6 or A1), Ly49A/D (12A8), Ly49C/I (5E6), Ly49D (4E5), Ly49G (AT-8), Ly49H (3D10), Ly49I (YLI-90), NKG2A/C/E (20d5), NKG2D (CX5), NKp46 (29A1.4), CD69 (H1.2F3), and CD107a (1D4B). For intracellular cytokine staining, cells were first incubated for 4 hours at 37°C in the presence of protein transporter inhibitor Golgi stop (BD Bioscience). Subsequently, cells were surface stained, then treated with Cytofix/Cytoperm buffer (BD Biosciences), followed by incubation with fluorochrome-conjugated antibodies against IFNγ (XMG1.2) and Granzyme B (GB11). BrdU staining was performed using BD BRDU staining kit (BD Biosciences) according to manufacturer’s instructions. All antibodies were purchased from BD Biosciences, eBiosciences, or Biolegend. APC conjugated CD1d-PBS57-tetramer and PE-conjugated H2M3-fMIGWII (LemA)-tetramer were obtained from NIH Tetramer Core Facility. PE-conjugated H2Kb-SIINFEKL (OVA)-tetramer was from MBL International. Cells were acquired on FACSCanto or AriaIII flow cytometers (BD Bioscience) using FACSDiVa software, and data were analyzed using FlowJo software (Tree Star). All FACS analyses were performed after excluding contamination of doublets. Briefly, samples were first gated by comparing FSC-W versus FSC-H; to exclude doublets, events with a high FSC-W profile were gated out of the total sample population. The remaining population was then examined based on its SSC-W/SSC-H profile, and any SSC-Whigh events were similarly excluded. When indicated, cells were sorted on a FACS AriaIII at the flow cytometry core facility at the University of Michigan.
IFNγ secretion assay
Total splenocytes from uninfected or LM-Ova infected mice were isolated at the indicated time points, and stimulated ex-vivo in the presence of Golgi stop with a combination of recombinant mouse IL-12 (5 ng/mL; Peprotech) and IL-18 (25 ng/mL; R&D systems) for 4 hours, or for 24 hours in the presence of Ova (4 μg/ml) or LemA (4 μg/ml) peptides. Following incubation cells were surface stained, fixed and permeabilized, and stained for intracellular IFNγ.
Adoptive transfer experiments
Sorted populations of conventional CD8+ T cells (CD3+NK1.1−CD1d-tetramer− CD8+), NKT cells (CD3+NK1.1+CD1d-tetramer−), or CD8+ NKT cells CD8+CD3+NK1.1+CD1d-tetramer− or CD8−CD3+NK1.1+CD1d-tetramer−) were intravenously injected into Rag2−/−/γc−/− recipient mice (100,000 cells per mouse). Three days post-transfer, Rag2−/−/γc−/− recipient mice were infected with 100,000 CFU of LM- and monitored for survival for 10 consecutive days.
Statistical analysis
Statistically significant differences were determined using Two Way ANOVA with a Bonferroni post-hoc test (time x cell population) or by One Way ANOVA with a Student-Newman-Keuls post-hoc test (p value < 0.05). Survival curve comparison was performed by Mantel-Cox log-rank test. Graphs in this paper are presented as Mean of the average ± s.e.m. GraphPad Prism6 software was utilized for statistical analysis.
RESULTS
Conventional CD8+ NK1.1− T cells and CD8+ NKT cells both contribute to the adaptive response to Listeria infection
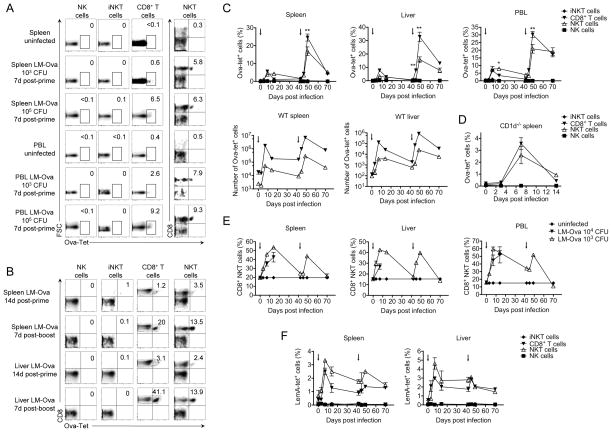
CD8+NK1.1+ cells comprise a subset of cells from the NKT compartment that are CD1d-tet− and therefore distinct from iNKT cells (Supplemental Figures 1–2). To determine whether this cell population (termed CD8+ NKT cells) has functions distinct from that of conventional CD8+NK1.1− T cells (termed conventional CD8+ T cells), we studied both cell populations (CD8+ NK1.1+ and CD8+ NK1.1−) during the host response against Listeria monocytogenes (LM) infection, which requires both innate and adaptive immunity for effective bacterial clearance. To examine both effector and memory T cell responses, we used a typical prime/boost infection regimen using OVA-expressing LM (LM-Ova) (29, 31, 32) with the first dose administered at day 0 (103–104 CFU) followed by a second high dose (2x105 CFU) administered at day 42 (Supplemental Figure 3). Kinetic studies of conventional CD8+NK1.1− T cell numbers in the spleen, liver, and blood from LM-Ova infected mice showed peak antigen-specific responses consistent with the development of effector (day 7 post-prime) and memory (day 7 post-boost) antigen specific CD8+ T cell responses as indicated by H2Kb-SIINFEKL tetramer (Ova-tetramer) positivity (Figure 1A–E). Likewise, we found that CD8+NK1.1+ T cells were capable of mounting OVA-specific T cell responses with similar kinetics as their counterparts from the conventional CD8+ T cell compartment, with ~6% of conventional CD8+ T cells and ~8% of CD8+ NKT cells positive for Ova-tetramer staining on day 7 (peak of the effector response) (Figure 1A–E). These results were observed in the spleen, liver, and blood of infected wild type mice (Figure 1C, E), as well as in CD1d−/− mice lacking iNKT cells (Figure 1D). Of interest, we found that a high dose of infection (i.e., 105 CFU) similarly primed the two CD8+ T cell compartments, while a low dose of infection (i.e 103 CFU) efficiently primed CD8+ NKT cells but not conventional CD8+ T cells, suggesting an increased sensitivity to priming in CD8+ NKT cells (Figure 1A, E). However, after re-challenge, the pool size of Ova-tetramer+ CD8+ T cells increased similarly in both T cell populations indicating similar outcomes of memory response (Figure 1C, E). To further confirm these observations, we used another tetramer, LemA (Listeria specific N-formylated peptide LemA; f-MIGWII), known to detect Listeria-specific T cell responses (34) (Figure 1F). Although this tetramer was not ideal to study Listeria-specific T cell memory response, results at the effector phase confirmed the efficient priming of both CD8+ T cell subsets with a slightly higher frequency of LemA-specific T cells among CD8+NKT cells compared to conventional CD8+ T cells (Figure 1F). Of note, the absolute numbers of tetramer-positive (e.g. OVA or LemA) cells were higher for conventional CD8+ T cells with a similar profile for both tetramers (Figure 1C, F and Supplementary Figure 4), likely reflecting the much larger pool size of conventional CD8+ T cell compartment as compared to that of the NKT cell compartment [~50 times, (Supplemental Figure 1)].
Figure 1. CD8+ NKT cells mount antigen-specific T cell response to Listeria infection with kinetics similar to that of conventional CD8+ T cells.
(A) Spleen and blood (PBL) of LM-Ova-infected mice (103 or 105 CFU) were analyzed on day 7 post-infection. Plots show the frequency of Ova-tetramer positive cells among gated NK, iNKT, and CD8+ T cells. For NKT cells, we show the distribution of Ova-tetramer versus CD8. (B–F) Mice were infected with 104 CFU of LM-Ova on day 0 and re-challenged with 2x105 CFU on day 42, as indicated by arrows (C, E-F). (B) Plots show the distribution of CD8 versus Ova-tetramer staining among gated NK, iNKT, T, and NKT cell subsets. (C–D) Graphs show kinetics of frequency (C–D) and numbers (C) of Ova-tetramer positive cells among NK, iNKT, and CD8+ T cells from infected wild type (C) and CD1d−/− (D) mice. (E) Graphs show the frequency of CD8+ NKT cells in spleen, liver, and blood from mice infected with 104 or 105 CFU LM-Ova. (F) Graphs show kinetics of frequency of LemA-tetramer positive cells among NK, iNKT, and CD8+ T cells in spleen and liver. Data are representative of two independent experiments, n=4, mean ± s.e.m (D). Data are representative from one of three independent experiments, n = 3 for each (for days 0, 7, 49), data are representative from one of two independent experiments n = 3 for each (for days 3, 14, 42, 45, 70), mean ± s.e.m (C, E–F). *p < 0.05, **p < 0.01 between NKT and conventional CD8+ T cells.
Likewise, the pattern of memory T cell responses (35, 36) as indicated by the central (CD62L+CD127+) and effector (CD62L−CD127+) memory phenotypes were not dramatically different in conventional CD8+ NK1.1− T cells and CD8+ NKT cells (Figure 1B, 2A–B) with some minor, but statistically significant differences in the conversion of CD8+ NKT cells to central memory phenotype at day 7 post infection. CD8+ NKT cells, however, were of similar phenotype as conventional CD8+ T cells at later time points (Figure 2A–B). Consistently, we found that IFNγ release in response to ex-vivo stimulation with Ova-peptide was equally robust in both CD8+ T cells CD8+ NKT cell compartments during the effector and memory phases (Figure 2C). Likewise, equivalent levels of granzyme B expression were observed in both CD8+ T cells subsets at both phases of the adaptive response mounted against Listeria (Figure 2D). Collectively, our data suggest that conventional CD8+ NK1.1− T cells and CD8+ NKT cells have similar adaptive responses to Listeria infection.
Figure 2. Similar patterns of CD8+ T cell memory generation in response to Listeria infection from both NKT and conventional T cell compartments.
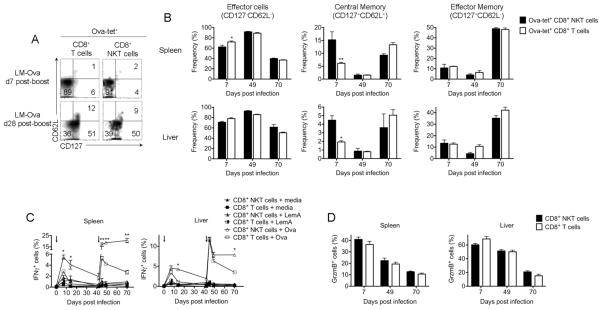
Mice were infected with 104 CFU of LM-Ova on day 0 and re-challenged with 2x105 CFU on day 42, as indicated by arrows in (C). (A) Plots show the distribution of CD62L versus CD127 among gated Ova-tetramer+ CD8+ NKT or CD8+ T cells 7 and 28 days post-boost. (B) Graphs summarize the frequency of effector, central memory, and effector memory cells among Ova-tetramer+ CD8+ NKT or CD8+ T cells on day 7 post-prime and days 7 and 28 post-boost. (C) At the indicated timepoints, cells from spleen and liver were re-stimulated ex-vivo with Ova or LemA peptides. Graph show the kinetics of the frequency of IFNγ-expressing cells among CD8+ NKT versus CD8+ T cells. (D) At the indicated timepoints, granzyme B expression was measured in CD8+ NKT or CD8+ conventional T cells. Data are representative from one of two independent experiments, n = 3 for each, mean ± s.e.m (A–D). *p < 0.05, **p < 0.01 between NKT and conventional CD8+ T cells.
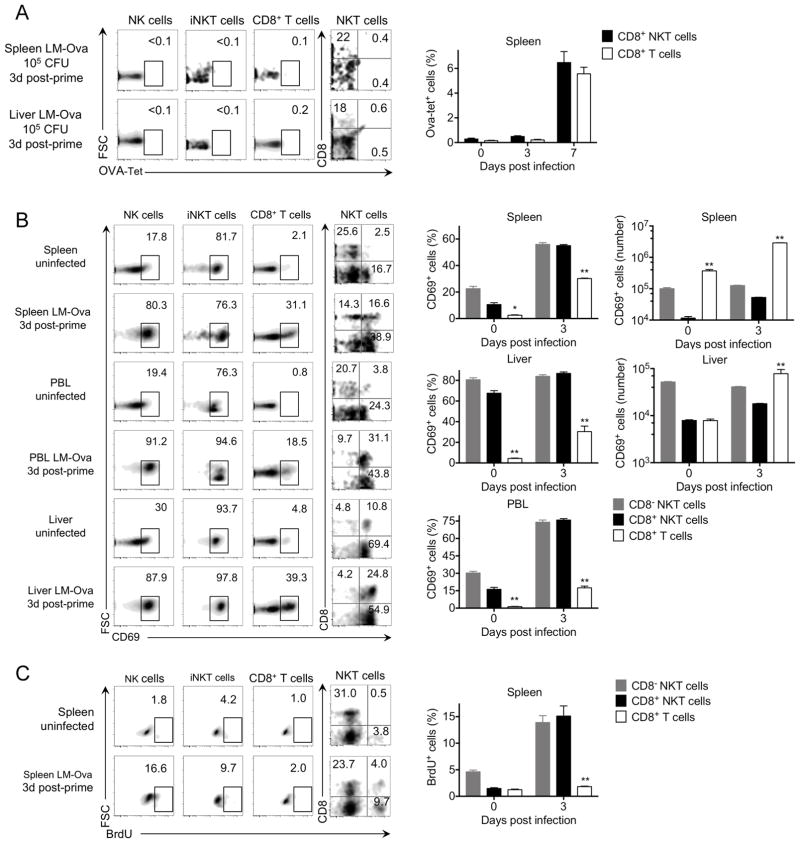
CD8+ NKT cells can provide innate responses against Listeria infection
The kinetics of Ova-tetramer staining is consistent with the absence of antigen-specific OVA-tetramer+ T cells early during LM-Ova infection of naïve mice in either conventional CD8+ T cells or CD8+ NKT cell populations (Figure 1, 3A). Accordingly, we chose day 3 post-infection to examine early responses to LM-Ova infection in both CD8+ T cell subsets (CD8+ NK1.1+ and CD8+ NK1.1−). This included analysis of early T cell activation (CD69), T cell expansion (BrdU), innate IFNγ production, cytotoxicity (granzyme B), and degranulation (CD107). Analysis of the spleen, liver, and blood showed expression of CD69 on all lymphocyte subsets on day 3 indicating early activation (Figure 3B). However, despite early activation of CD8+ T cells from both compartments, only CD8+ NKT cells exhibited robust expansion (BrdU+) at day 3 post-infection in the absence of previous Listeria exposure (Figure 3C), which correlated with more robust CD69 expression in CD8+ NKT cells (Figure 3B). Since the pool size of conventional CD8+ T cell compartment is ~50 times larger then the NKT cell compartment (Supplemental Figure 1), However, the absolute numbers of CD69+ conventional T cells on days 0–3 post infection were slightly higher than that of CD8+ NKT cells likely due to the increased pool size of the conventional CD8+ T cell compartment (~50 times, Supplemental Figure 1).
Figure 3. Activation and expansion of CD8+ NKT versus conventional CD8+ T cells in the early response to Listeria infection.
C57BL/6 mice were infected with 105 CFU LM-Ova and analyzed 3 days post-infection in indicated organs. Plots show the frequency of Ova-tetramer+ (A), CD69+ (B), and BrdU+ (C) cells among gated NK, iNKT, and CD8+ T cells. For NKT cells, we show the distribution of CD8 versus Ova-tetramer, CD69, and BrdU. Graphs summarize the absolute number and/or frequency of Ova-tetramer+ (A), CD69+ (B), and BrdU+ (C) cells among CD8− NKT cells, CD8+ NKT cells, and conventional CD8+ T cells. Data are representative from one of two independent experiments, n = 3 for each (A–B), from one of two independent experiments, n = 4 for each (C), mean ± s.e.m. *p < 0.05, **p < 0.01 between CD8+ NKT and conventional CD8+ T cells.
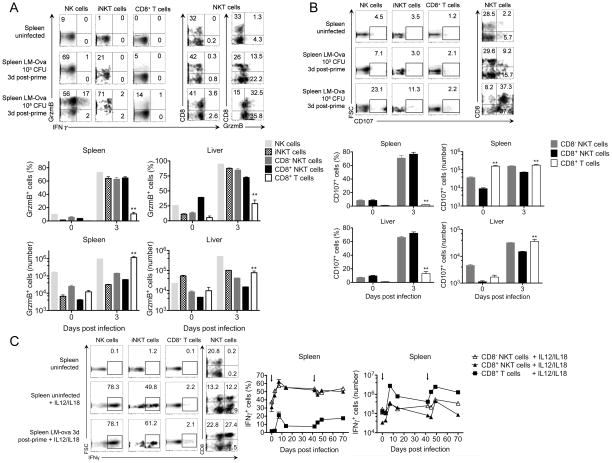
Next, we compared CD8+ NKT cells and conventional NK1.1− CD8+ T cells for their ability to exhibit rapid cytotoxicity against Listeria infection (Figure 4). Expression of granzyme B (Figure 4A) and the degranulation marker CD107 (Figure 4B) was examined at day 3 post-infection. Results revealed significantly greater expression of granzyme B in CD8+ NKT cells as compared to conventional CD8+ NK1.1− T cells, suggesting the ability of CD8+ NK1.1− T cells to rapidly acquire cytotoxic activity early during Listeria infection (Figure 4A). These results were further confirmed by efficient degranulation of CD8+ NKT cells as indicated by the significantly increased frequency of CD107+ cells on day 3 post-infection compared to conventional CD8+ NK1.1− T cells (Figure 4B), which typically require more time to acquire full functional competence (i.e., 6–7 days (1, 2)). Notably, the early potent cytotoxicity among CD8+ NKT cells is antigen-independent as indicated by the lack of OVA-tetramer staining 0–3 days post LM-Ova infection. Again, based on the conventional T cell compartment being up to 50 times larger than the NKT cell compartment (Supplemental Figure 1), the absolute number of CD107+ or GrzmB+ conventional CD8+ T cells on days 0–3 post infection was larger as compared to CD8+ NKT cells.
Figure 4. CD8+ NKT cells mount a robust innate immune response to Listeria infection.
C57BL/6 mice were infected with 103 or 105 CFU LM-Ova and analyzed 3 days post-infection in indicated organs. (A) Plots show the distribution of Granzyme B versus IFNγ expression among gated NK, iNKT, and CD8+ T cells. In NKT cells, we show the distribution of CD8 versus IFNγ and granzyme B expression. Graphs summarize the absolute number and frequency of granzyme B+ cell subsets. (B) Plots show the frequency of CD107 among gated NK, iNKT, and CD8+ T cells. In NKT cells, we show the distribution of CD8 versus CD107. Graphs summarize the absolute number and frequency of CD107 in CD8− NKT, CD8+ NKT, and CD8+ T cells. (C) Spleens from uninfected or day 3-infected mice were stimulated ex-vivo with IL-12 plus IL-18 for 4 hours. Plots show the frequency of IFNγ expression among gated NK, iNKT, and CD8+ T cells. In NKT cells, we show the distribution of CD8 versus IFNγ. Graphs show the kinetics of IFNγ expression among CD8− NKT, CD8+ NKT, and CD8+ T cells at frequency and absolute number levels. Data are representative from one of two independent experiments, n = 3 for each (A–C), mean ± s.e.m. *p < 0.05, **p < 0.01 between CD8+ NKT and conventional CD8+ T cells. For the kinetics of IFNγ expression, p < 0.01 for all time points tested between CD8+ NKT and conventional CD8+ T cells.
Finally, we assessed the contribution of conventional CD8+ T cells versus CD8+ NKT cells in the early production of IFNγ (Figure 4A, C). In this case, cells from untreated or infected (day 3 post-infection) mice were examined for intracellular expression of IFNγ in response to the cytokine stimulus IL-12/IL-18. We found that CD8+ NKT cells were as potent as iNKT and NK cells in producing IFNγ in response to IL-12/IL-18 alone (Figure 4C). In sharp contrast, conventional CD8+ T cells exhibited poor IFNγ production in response to IL-12/IL-18 stimulation (Figure 4C). Similarly, CD8+ NKT cells, but not CD8+ NK1.1− T cells, harvested from 3 day-infected mice and re-stimulated ex-vivo with IL-12/IL-18, produced significant levels of IFNγ early during Listeria infection (Figure 4C). After the early innate phase, the expression of IFNγ in both CD8+ NKT and conventional CD8+ T cell populations peaks at day 7 post-Listeria priming or boost. Importantly, the absolute number of IFNγ-producing conventional CD8+ T cells was still significant, likely due to the much larger CD8+ T cell compartment size compared to the NKT cell compartment (Supplemental Figure 1), and confirms previous results that demonstrate the responsiveness of conventional CD8+ T cells to IL12/IL18 stimuli (6). Based on the ability of CD8+ NKT cells to produce IFNγ and granzyme B in an antigen-independent manner as well as degranulate at a very early stage in LM-Ova infection, our data strongly suggest that CD8+ NK1.1+ cells can provide innate functions during Listeria infection.
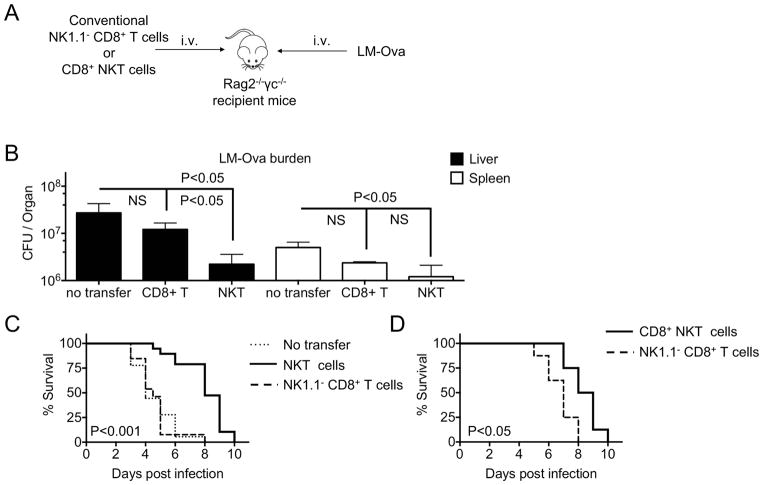
Conventional NK1.1− CD8+ T cells alone are not sufficient to provide early protection to lethally infected Rag−/−γc−/− host mice
Based on the above results, we questioned whether conventional NK1.1− CD8+ T cells alone (i.e., without contaminating CD8+ NKT cells) are capable of providing protection against Listeria infection. To this aim, we transferred similar numbers (105 cells) of sorted conventional CD8+ NK1.1− T cells (Supplemental Figure 1E) or CD1d-tetramer− NKT cells (Supplemental Figure 1C) into Rag2−/−γc−/− recipient mice, which lack functional T, B and NK cells, and assessed their effects on mice survival for 10 days post-infection (Figure 5A). Because the early response to Listeria infection (as measured by T cell activation and proliferation, IFNγ production, cytotoxicity and degranulation) was not significantly different between CD8+ NKT and CD8− NKT cell subsets (Figures 3–4), we first chose to transfer total NKT cells to overcome the limiting numbers of CD8+ NKT cells despite pooling of mice donors. LM-infected Rag−/−γc−/− mice adoptively transferred with NKT cells showed a 12-fold reduction of bacterial burden in the livers compared to infected mice that received no cells. However, infected Rag−/−γc−/− mice adoptively transferred with conventional CD8+ NK1.1− T cells showed no reduction in bacterial titers (Figure 5B). Similarly, we observed a 4-fold versus 2-fold reduction of Listeria CFU titers in spleens of recipient mice transferred with NKT cells versus conventional CD8+ T cells, respectively (Figure 5B). Importantly, the reduction in bacterial burden in mice adoptively transferred with NKT cells was associated with prolonged survival compared to infected Rag−/−γc−/− mice, which received conventional CD8+ T cells alone (Figure 5C). In fact, analysis of Rag−/−γc−/− mice adoptively transferred with conventional CD8+ NK1.1− T cells showed no difference in survival compared to recipient mice that received no cells after challenge with a lethal dose of LM-Ova (Figure 5C). To directly confirm that the CD8+ sub-population of NKT cells can provide early innate immunity that can protect from early lethality from Listeria infection, we adoptively transferred either CD8+ T or CD8+ NKT cells into Rag−/−γc−/− mice (Figure 5D). Consistent with Figure 5C, the adoptive transfer of CD8+ NKT cells delayed mortality from Listeria infection. While the role of conventional CD8+ T cells in providing long-lasting protection has been well documented, altogether our data suggest innate responses by CD8+ NKT cells may be critical in the early defense against LM infection.
Figure 5. Transfer of conventional CD8+ T cells without CD8+ NKT cells impairs survival of lethally Listeria-challenged Rag2−/−γc−/− recipient mice.
(A) Rag2−/−γc−/− mice were left untreated (no cells transferred) or injected with 100,000 of sorted NKT, CD8+ NKT, or conventional CD8+ T cells. At day 3 post-transfer, mice were challenged with lethal dose of 100,000 CFU/mouse of LM-Ova. (B) Graphs show Listeria burden (CFU/organ) in liver and spleen on day 3 post-infection. (C) Graph shows percentage of mice survival as indicated by Kaplan–Meyer survival curves. (B) Data are pooled from two independent experiments, n = 8 for no cells and NKT cells transfer, n = 4 for CD8+ T cells transfer, mean ± s.e.m. (C) Data are pooled from at least two independent experiments, n = 18 for no cells transfer, n = 19 for NKT cells transfer, n = 13 for CD8+ T cells transfer. (D) Graph shows percentage of mice survival as indicated by Kaplan–Meyer survival curves, n = 8 per group.
DISCUSSION
With the recent development of both the NK and NKT cell fields, it has become increasingly recognized that T cells expressing the NK1.1 marker form a distinct T cell compartment comprising several subsets as diverse as the heterogeneity of the conventional T cell compartment. In this study, we have distinguished the conventional T cell compartment lacking NK1.1 from the NKT cell compartment that express NK1.1. Although the size pool of the conventional T cell compartment is ~50 times larger than the NKT cell compartment, both cell compartments harbor CD4+ and CD8+ T cell subsets. A phenotypic comparison of surface marker expression between the conventional CD8+ T cell compartment and the CD8+ NKT cell compartment showed distinct profiles that may reflect differential roles in immunity (Supplementary Figure 2). In this study, we showed that this minor population of CD8+ NKT cells (that is easily overlooked) can rapidly expand during the innate phase of the immune response against Listeria infection, produce copious amounts of IFNγ and granzyme B, and contributes to improved early survival. Using LM-Ova tetramer, we confirmed that the early response mediated by CD8+ NKT cells is antigen-independent. Interestingly, similar to the conventional CD8+ T cells, this cell population was able to mount adaptive immunity late during infection and generate memory response after re-exposure to Listeria.
The question of whether these CD8+NK1.1+ T cells that have innate properties represent a distinct T cell lineage or arise from conventional CD8+ T cells that have acquired NK1.1 expression remained largely controversial (16–20, 22). On the one hand, results showing the up-regulation of NK1.1 on CD8+ T cells upon in vitro and in vivo stimulation suggest a conventional T cell origin (17–20, 22). On the other hand, the observation that CD8+ NK1.1+ T cells can develop in thymectomized mice supports rather a distinct lineage for these CD8+ NKT cells (16). Recent reports suggested an innate component within memory CD8+ T cells that express NK1.1; however, these cells were elicited only after transfer of memory Listeria-primed CD8+ T cells and are therefore functionally significant in the context of pathogen re-infection (10, 13). In contrast, in the present study, we examined CD8+NK1.1+ T cells from naïve specific pathogen-free mice, and demonstrated that these cells are endowed with both innate and adaptive responses after Listeria infection. While we cannot rule out the possibility that some CD8+NK1.1+ T cells in the current study arise from a pool of memory T cells, the distinct phenotypic profile and the specialized innate function of NK1.1+CD8+ T cells supports a different lineage that warrants further study.
The notion that T cells can perform as innate cells has been previously proposed and debated in a number of reports (6, 7, 14, 15, 37–39). While the role of NK cells producing IFNγ during the early phase of Listeria infection is indisputable, Anderson et al showed evidence of early IFNγ production in the absence of NK cells (37). Specifically, their results showed that infected γc−/− mice, which lack NK cells, but not Rag−/−γc−/− (which lack T, B, and NK cells), were able to mount early resistance to Listeria, indicating that T cells can functionally replace NK cells for early IFNγ production, which is necessary for activating the innate immune system upon Listeria infection (37). When dissecting the source of IFNγ during the early response to Listeria, Thale et al provided evidence that T cells can produce innate IFNγ (38). Of interest, only a small population (2–3%) of T cells was able to produce early IFNγ, and among those T cells, CD8+ T cells were described as the major source (38). D’Orazio et al confirmed that only few CD8+ T cells were capable of producing IFNγ in the early response to Listeria and reported that these cells expressed CD44hi (39). Similar observations were made by Ghanem et.al. in humans, where a small population of CD8+ T cells (< 3%) was able to secrete IFNγ in response to Listeria (40). Recently, Schenkel et al provided evidence that CD8+ T cells can trigger innate responses to increase immunity against unrelated pathogens (14). Moreover, Suarez-Ramirez et al showed that CD8+ T cells can acquire innate functions and produce IFNγ independently of TCR stimulation (15). In the same line, Berg et.al showed that memory CD8+ T cells are able to reduce Listeria burden in an antigen-independent and IFNγ-dependent manner (8). Naïve CD8+ T cells, however, had reduced (as compared to memory CD8+ T cells) expression of IL12Rβ2, IL18Rα and IL18Rβ subunits, which negatively affect their ability to secrete IFNγ and reduce LM-Ova burden at early (day 3) time points (8). In addition, memory CD8+ T cells but not naïve CD8+ T have been shown to rapidly co-localize with LM-Ova-infected cells during the early phase of re-infection (11). Here, we show evidence of innate responses against the first exposure to Listeria infection confined within the subset of CD8+ NKT cells.
Evidence of innate responses within CD8+ NKT cells are provided by the rapid expression of CD69, CD107, IFNγ, and Granzyme B which develop in an antigen-independent manner. In the absence of any NK1.1+ CD8+ T cells, conventional CD8+ NK1.1− T cells were inferior to NKT cells in providing protection against early lethality to Listeria infection. Whether these cells are capable of rapid co-localization with LM-Ova-infected cells, like the case of memory CD8+ T cells during re-infection, remains to be determined. Also, the exact mechanisms by which CD8+ NKT cells recognize Listeria-infected cells remain unclear. We show that CD8+ NKT cells express activating and inhibitory NK-cell receptors at high levels (e.g., NKG2D, NKG2A) in contrast to conventional CD8+ T cells, which have minimal to no expression of these receptors (Supplemental Figure 2). Thus, it is possible that CD8+ NKT cells use the NK-cell receptor machinery to recognize and kill Listeria-infected cells in an antigen-independent, nonspecific manner. Interestingly, a recent study reported that vectors expressing the NKG2D ligand RAE-1γ dramatically enhanced the effectiveness of CD8+ T-cell response suggesting a promising approach in the development of CD8+ T-cell–based vaccines (41). Accordingly, one possible candidate for the origin of innate CD8+ NKT cell function is the subset of MHC-unrestricted NKT cells (42, 43). These NKT cells have been recently described as sharing many characteristics of NK cells including the expression of the killer cell lectin-like receptors (Klr), the rapid production of IFNγ in response to cytokine stimuli (IL-12/IL-18), and the potent cytotoxic program (granzyme B) in response to innate signals (Poly:IC) (43). Notably, the identity of these T cells as MHC-unrestricted T cells makes them excellent candidates to serve as professional innate T cells while maintaining adaptive functions. Based on this, we speculate that NK1.1 in CD8+ T cells is the equivalent of CD25 in CD4+ T cells with NK1.1 constitutively expressed on professional innate CD8+ T cells and up-regulated on activated CD8+ T cells in the same manner as CD25 is constitutively expressed on regulatory CD4+ T cells and up-regulated on activated CD4+ T cells (28).
Of note, the role of NK1.1+ cells in the early phase of Listeria (and other bacterial) infection has been a subject of a large debate. Several studies that used the NK1.1-depleting antibody (PK136 mAbs) showed improved Listeria clearance (44–46). Further, Berg et. al showed that NK cells were not protective from Listeria infection, in contrast to memory CD8+ T cells (47). However, these studies did not differentiate between different populations of immune cells that express NK1.1 (CD8+ or CD8− NKT). Here, we have carefully investigated the role of CD8+ NKT cells and identified their protective roles at early stages of Listeria infection. It is possible that robust activation of IFNγ production by the more abundant NK cell population is detrimental to the host, while the specialized population of NKT cells can limit Listeria burden in a more controlled manner without triggering excessive activation of the immune system of the host in antigen-independent manner.
The notion that the heterogeneity of CD8+ T cell responses can provide both innate resistance and adaptive immunity will certainly impact our current understanding of the cellular requirements for generating systemic immunity against infection perhaps in the same manner that the heterogeneity of CD4+ T cells with both effector and suppressive functions has impacted our understanding of autoimmunity.
Supplementary Material
Acknowledgments
We thank Dr. Mary O’Riordan for providing Listeria monocytogenes (10403s LM-ova). We thank the NIH tetramer core facility for providing CD1d and LemA tetramers.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interests
References
- 1.Zelinskyy G, Kraft AR, Schimmer S, Arndt T, Dittmer U. Kinetics of CD8+ effector T cell responses and induced CD4+ regulatory T cell responses during Friend retrovirus infection. Eur J Immunol. 2006;36:2658–2670. doi: 10.1002/eji.200636059. [DOI] [PubMed] [Google Scholar]
- 2.Wong P, Pamer EG. CD8 T cell responses to infectious pathogens. Annual review of immunology. 2003;21:29–70. doi: 10.1146/annurev.immunol.21.120601.141114. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Terme M, Ullrich E, Delahaye NF, Chaput N, Zitvogel L. Natural killer cell-directed therapies: moving from unexpected results to successful strategies. Nature immunology. 2008;9:486–494. doi: 10.1038/ni1580. [DOI] [PubMed] [Google Scholar]
- 5.Fehniger TA, Cai SF, Cao X, Bredemeyer AJ, Presti RM, French AR, Ley TJ. Acquisition of murine NK cell cytotoxicity requires the translation of a pre-existing pool of granzyme B and perforin mRNAs. Immunity. 2007;26:798–811. doi: 10.1016/j.immuni.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Berg RE, Cordes CJ, Forman J. Contribution of CD8+ T cells to innate immunity: IFN-gamma secretion induced by IL-12 and IL-18. Eur J Immunol. 2002;32:2807–2816. doi: 10.1002/1521-4141(2002010)32:10<2807::AID-IMMU2807>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Lertmemongkolchai G, Cai G, Hunter CA, Bancroft GJ. Bystander activation of CD8+ T cells contributes to the rapid production of IFN-gamma in response to bacterial pathogens. J Immunol. 2001;166:1097–1105. doi: 10.4049/jimmunol.166.2.1097. [DOI] [PubMed] [Google Scholar]
- 8.Berg RE, Crossley E, Murray S, Forman J. Memory CD8+ T cells provide innate immune protection against Listeria monocytogenes in the absence of cognate antigen. J Exp Med. 2003;198:1583–1593. doi: 10.1084/jem.20031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soudja SM, Ruiz AL, Marie JC, Lauvau G. Inflammatory monocytes activate memory CD8(+) T and innate NK lymphocytes independent of cognate antigen during microbial pathogen invasion. Immunity. 2012;37:549–562. doi: 10.1016/j.immuni.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soudja SM, Chandrabos C, Yakob E, Veenstra M, Palliser D, Lauvau G. Memory-T-cell-derived interferon-gamma instructs potent innate cell activation for protective immunity. Immunity. 2014;40:974–988. doi: 10.1016/j.immuni.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bajenoff M, Narni-Mancinelli E, Brau F, Lauvau G. Visualizing early splenic memory CD8+ T cells reactivation against intracellular bacteria in the mouse. PLoS One. 2010;5:e11524. doi: 10.1371/journal.pone.0011524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kupz A, Guarda G, Gebhardt T, Sander LE, Short KR, Diavatopoulos DA, Wijburg OL, Cao H, Waithman JC, Chen W, Fernandez-Ruiz D, Whitney PG, Heath WR, Curtiss R, 3rd, Tschopp J, Strugnell RA, Bedoui S. NLRC4 inflammasomes in dendritic cells regulate noncognate effector function by memory CD8(+) T cells. Nature immunology. 2012;13:162–169. doi: 10.1038/ni.2195. [DOI] [PubMed] [Google Scholar]
- 13.Ruiz AL, Soudja SM, Deceneux C, Lauvau G, Marie JC. NK1.1+ CD8+ T cells escape TGF-beta control and contribute to early microbial pathogen response. Nat Commun. 2014;5:5150. doi: 10.1038/ncomms6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 2014 doi: 10.1126/science.1254536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suarez-Ramirez JE, Tarrio ML, Kim K, Demers DA, Biron CA. CD8 T Cells in Innate Immune Responses: Using STAT4-Dependent but Antigen-Independent Pathways to Gamma Interferon during Viral Infection. MBio. 2014;5 doi: 10.1128/mBio.01978-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond K, Cain W, van Driel I, Godfrey D. Three day neonatal thymectomy selectively depletes NK1.1+ T cells. International immunology. 1998;10:1491–1499. doi: 10.1093/intimm/10.10.1491. [DOI] [PubMed] [Google Scholar]
- 17.Ikarashi Y, Maruoka H, Shinohara K, Sugimura T, Terada M, Wakasugi H. Mouse NK1.1+ cytotoxic T cells can be generated by IL-2 exposure from lymphocytes which express an intermediate level of T cell receptor. Immunol Lett. 1998;61:165–173. doi: 10.1016/s0165-2478(98)00014-5. [DOI] [PubMed] [Google Scholar]
- 18.Assarsson E, Kambayashi T, Sandberg JK, Hong S, Taniguchi M, Van Kaer L, Ljunggren HG, Chambers BJ. CD8+ T cells rapidly acquire NK1.1 and NK cell-associated molecules upon stimulation in vitro and in vivo. J Immunol. 2000;165:3673–3679. doi: 10.4049/jimmunol.165.7.3673. [DOI] [PubMed] [Google Scholar]
- 19.Kambayashi T, Assarsson E, Michaelsson J, Berglund P, Diehl AD, Chambers BJ, Ljunggren HG. Emergence of CD8+ T cells expressing NK cell receptors in influenza A virus-infected mice. J Immunol. 2000;165:4964–4969. doi: 10.4049/jimmunol.165.9.4964. [DOI] [PubMed] [Google Scholar]
- 20.Slifka MK, Pagarigan RR, Whitton JL. NK markers are expressed on a high percentage of virus-specific CD8+ and CD4+ T cells. J Immunol. 2000;164:2009–2015. doi: 10.4049/jimmunol.164.4.2009. [DOI] [PubMed] [Google Scholar]
- 21.Pellicci DG, Hammond KJ, Coquet J, Kyparissoudis K, Brooks AG, Kedzierska K, Keating R, Turner S, Berzins S, Smyth MJ, Godfrey DI. DX5/CD49b-positive T cells are not synonymous with CD1d-dependent NKT cells. J Immunol. 2005;175:4416–4425. doi: 10.4049/jimmunol.175.7.4416. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Wang H, Xia J, Liang T, Wang G, Li X, Yang YG. Activated CD8 T cells acquire NK1.1 expression and preferentially locate in the liver in mice after allogeneic hematopoietic cell transplantation. Immunol Lett. 2013;150:75–78. doi: 10.1016/j.imlet.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond KJ, Pelikan SB, Crowe NY, Randle-Barrett E, Nakayama T, Taniguchi M, Smyth MJ, van Driel IR, Scollay R, Baxter AG, Godfrey DI. NKT cells are phenotypically and functionally diverse. Eur J Immunol. 1999;29:3768–3781. doi: 10.1002/(SICI)1521-4141(199911)29:11<3768::AID-IMMU3768>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 24.Busch DH, Pamer EG. T lymphocyte dynamics during Listeria monocytogenes infection. Immunol Lett. 1999;65:93–98. doi: 10.1016/s0165-2478(98)00130-8. [DOI] [PubMed] [Google Scholar]
- 25.Pamer EG. Immune responses to Listeria monocytogenes. Nat Rev Immunol. 2004;4:812–823. doi: 10.1038/nri1461. [DOI] [PubMed] [Google Scholar]
- 26.Busch DH, I, Pilip M, Vijh S, Pamer EG. Coordinate regulation of complex T cell populations responding to bacterial infection. Immunity. 1998;8:353–362. doi: 10.1016/s1074-7613(00)80540-3. [DOI] [PubMed] [Google Scholar]
- 27.Tuma RA, Giannino R, Guirnalda P, Leiner I, Pamer EG. Rescue of CD8 T cell-mediated antimicrobial immunity with a nonspecific inflammatory stimulus. J Clin Invest. 2002;110:1493–1501. doi: 10.1172/JCI16356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Randolph DA, Fathman CG. Cd4+Cd25+ regulatory T cells and their therapeutic potential. Annu Rev Med. 2006;57:381–402. doi: 10.1146/annurev.med.57.121304.131337. [DOI] [PubMed] [Google Scholar]
- 29.Pope C, Kim SK, Marzo A, Masopust D, Williams K, Jiang J, Shen H, Lefrancois L. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- 30.Busch DH, Vijh S, Pamer EG. Animal model for infection with Listeria monocytogenes. Curr Protoc Immunol. 2001;Chapter 19(Unit 19):19. doi: 10.1002/0471142735.im1909s36. [DOI] [PubMed] [Google Scholar]
- 31.Kerksiek KM, Busch DH, Pamer EG. Variable immunodominance hierarchies for H2-M3-restricted N-formyl peptides following bacterial infection. J Immunol. 2001;166:1132–1140. doi: 10.4049/jimmunol.166.2.1132. [DOI] [PubMed] [Google Scholar]
- 32.Xu H, Chun T, Choi HJ, Wang B, Wang CR. Impaired response to Listeria in H2-M3-deficient mice reveals a nonredundant role of MHC class Ib-specific T cells in host defense. J Exp Med. 2006;203:449–459. doi: 10.1084/jem.20051866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung Y, Yamazaki T, Kim BS, Zhang Y, Reynolds JM, Martinez GJ, Chang SH, Lim H, Birkenbach M, Dong C. Epstein Barr virus-induced 3 (EBI3) together with IL-12 negatively regulates T helper 17-mediated immunity to Listeria monocytogenes infection. PLoS Pathog. 2013;9:e1003628. doi: 10.1371/journal.ppat.1003628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kerksiek KM, Ploss A, Leiner I, Busch DH, Pamer EG. H2-M3-restricted memory T cells: persistence and activation without expansion. J Immunol. 2003;170:1862–1869. doi: 10.4049/jimmunol.170.4.1862. [DOI] [PubMed] [Google Scholar]
- 35.Stemberger C, Huster KM, Koffler M, Anderl F, Schiemann M, Wagner H, Busch DH. A single naive CD8+ T cell precursor can develop into diverse effector and memory subsets. Immunity. 2007;27:985–997. doi: 10.1016/j.immuni.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 36.Huster KM, Stemberger C, Busch DH. Protective immunity towards intracellular pathogens. Curr Opin Immunol. 2006;18:458–464. doi: 10.1016/j.coi.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Andersson A, Dai WJ, Di Santo JP, Brombacher F. Early IFN-gamma production and innate immunity during Listeria monocytogenes infection in the absence of NK cells. J Immunol. 1998;161:5600–5606. [PubMed] [Google Scholar]
- 38.Thale C, Kiderlen AF. Sources of interferon-gamma (IFN-gamma) in early immune response to Listeria monocytogenes. Immunobiology. 2005;210:673–683. doi: 10.1016/j.imbio.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 39.D’Orazio SE, Troese MJ, Starnbach MN. Cytosolic localization of Listeria monocytogenes triggers an early IFN-gamma response by CD8+ T cells that correlates with innate resistance to infection. J Immunol. 2006;177:7146–7154. doi: 10.4049/jimmunol.177.10.7146. [DOI] [PubMed] [Google Scholar]
- 40.Bou Ghanem EN, D’Orazio SE. Human CD8+ T cells display a differential ability to undergo cytokine-driven bystander activation. Cell Immunol. 2011;272:79–86. doi: 10.1016/j.cellimm.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 41.Trsan T, Busche A, Abram M, Wensveen FM, Lemmermann NA, Arapovic M, Babic M, Tomic A, Golemac M, Brinkmann MM, Jager W, Oxenius A, Polic B, Krmpotic A, Messerle M, Jonjic S. Superior induction and maintenance of protective CD8 T cells in mice infected with mouse cytomegalovirus vector expressing RAE-1gamma. Proc Natl Acad Sci U S A. 2014;110:16550–16555. doi: 10.1073/pnas.1310215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda M, Shadeo A, MacFadyen AM, Takei F. CD1d-independent NKT cells in beta 2-microglobulin-deficient mice have hybrid phenotype and function of NK and T cells. J Immunol. 2004;172:6115–6122. doi: 10.4049/jimmunol.172.10.6115. [DOI] [PubMed] [Google Scholar]
- 43.Farr AR, Wu W, Choi B, Cavalcoli JD, Laouar Y. CD1d-unrestricted NKT cells are endowed with a hybrid function far superior than that of iNKT cells. Proc Natl Acad Sci U S A. 2014;111:12841–12846. doi: 10.1073/pnas.1323405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teixeira HC, Kaufmann SH. Role of NK1.1+ cells in experimental listeriosis. NK1+ cells are early IFN-gamma producers but impair resistance to Listeria monocytogenes infection. J Immunol. 1994;152:1873–1882. [PubMed] [Google Scholar]
- 45.Takada H, Matsuzaki G, Hiromatsu K, Nomoto K. Analysis of the role of natural killer cells in Listeria monocytogenes infection: relation between natural killer cells and T-cell receptor gamma delta T cells in the host defence mechanism at the early stage of infection. Immunology. 1994;82:106–112. [PMC free article] [PubMed] [Google Scholar]
- 46.Viegas N, Andzinski L, Wu CF, Komoll RM, Gekara N, Dittmar KE, Weiss S, Jablonska J. IFN-gamma production by CD27(+) NK cells exacerbates Listeria monocytogenes infection in mice by inhibiting granulocyte mobilization. Eur J Immunol. 2013;43:2626–2637. doi: 10.1002/eji.201242937. [DOI] [PubMed] [Google Scholar]
- 47.Berg RE, Crossley E, Murray S, Forman J. Relative contributions of NK and CD8 T cells to IFN-gamma mediated innate immune protection against Listeria monocytogenes. J Immunol. 2005;175:1751–1757. doi: 10.4049/jimmunol.175.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.