Abstract
Biosynthesis of the phytohormone ethylene is under tight regulation to satisfy the need for appropriate levels of ethylene in plants in response to exogenous and endogenous stimuli. The enzyme 1-aminocyclopropane-1-carboxylic acid synthase (ACS), which catalyzes the rate-limiting step of ethylene biosynthesis, plays a central role to regulate ethylene production through changes in ACS gene expression levels and the activity of the enzyme. Together with molecular genetic studies suggesting the roles of post-translational modification of the ACS, newly emerging evidence strongly suggests that the regulation of ACS protein stability is an alternative mechanism that controls ethylene production, in addition to the transcriptional regulation of ACS genes. In this review, recent new insight into the regulation of ACS protein turnover is highlighted, with a special focus on the roles of phosphorylation, ubiquitination, and novel components that regulate the turnover of ACS proteins. The prospect of cross-talk between ethylene biosynthesis and other signaling pathways to control turnover of the ACS protein is also considered.
Keywords: 14-3-3, ACS, ethylene, phosphorylation, protein turnover
INTRODUCTION
The simple gas ethylene has been recognized as a plant growth regulator for a century (Crocker and Knight, 1908; Knight et al., 1910; Neljubov, 1901). Ethylene influences many aspects of plant growth and developmental processes, including germination, fruit ripening, flower senescence, leaf abscission, nodulation, lateral root initiation, and the response to a variety of abiotic and biotic stresses (Abeles et al., 1992; Mattoo and Suttle, 1991). The Russian scientist Neljubov first demonstrated that ethylene is the responsible component caused the early defoliation of the tree nearby a leaking illuminating gas main in a small German town in the late1800’s (Neljubov, 1901). Neljubov used pea seedlings to determine that ethylene is the active component of the illuminating gas by exposing the filtered illuminating gas to pea seedlings. The pea seedlings exhibited distinctive morphology changes which include shortening of hypocotyl and root, swelling and thickening of hypocotyl, and formation of exaggerated hook. This seedling phenotype was later defined as the triple response, which is a hallmark of the ethylene response of dark-grown seedlings (Knight et al., 1910). 30 years later, Gane showed that ethylene is naturally produced by plants (Gane, 1934).
Through the efforts of Yang and co-workers, the biosynthesis of ethylene was fully elucidated in late 1980’s (Fig. 1) (Kende, 1993; Yang and Hoffman, 1984; Zarembinski and Theologis, 1994). The pathway of ethylene biosynthesis is simple and straightforward. Ethylene is synthesized from the amino acid methionine via two intermediates, S-adenosyl methionine (SAM) and 1-aminocyclopropane-1-carboxylic acid (ACC) (Adams and Yang, 1977; Lieberman and Mapson, 1964). In the first step, methionine is converted to SAM by SAM synthases. Conversion of SAM to ACC, which is catalyzed by a family of ACC synthase (ACS) enzymes, is the first committed and rate-limiting step in ethylene biosynthesis (Boller et al., 1979; Yang and Hoffman, 1984). During this conversion, 5-methylthioadenosine (MTA) is generated as a by-product which is readily recycled back to the Yang cycle to conserve the methlythio group of MTA into the methionine (Murr and Yang, 1975). This salvage step enables plants to maintain a constant level of cellular methionine, which can be utilized during rapid ethylene production (Sauter et al., 2013). ACC is then converted to ethylene by ACC oxidase (ACO), a member of the oxygenase/oxidase superfamily of enzymes (Dong et al., 1992). The levels of free ACC are regulated by formation of ACC derivatives, malonyl-ACC, γ-glutamyl-ACC, and jasmonyl-ACC, which likely affect the pools of free ACC for ethylene biosynthesis (Van de Poel and Van Der Straeten, 2014).
Fig. 1.
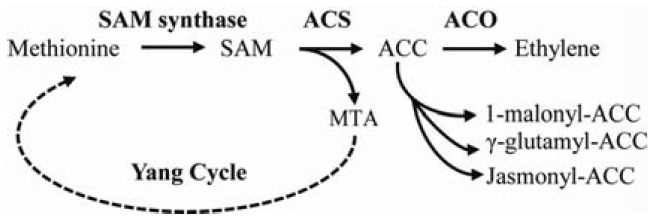
The ethylene biosynthetic pathway and its intermediates. S-adenosyl methionine (SAM) synthase uses methionine as a precursor for SAM. SAM is subsequently converted to 1-aminocyclopropane-1-carboxylic acid (ACC) in the first committed step in the ethylene biosynthesis by a family of ACC synthase (ACS) proteins, resulting in the release of methylthioadenosine (MTA). ACC is then finally converted to ethylene by ACC oxidase (ACO). MTA is recycled back to Yang cycles. Dashed line indicates additional enzymatic steps in the Yang cycle.
As gaseous ethylene is diffusible and not degraded in plant cells, strict regulation of ethylene biosynthesis is necessary to its diverse function in plant growth and development. Due to its role in the rate-limiting step of the ethylene biosynthesis pathway, ACS has been considered as a major point of the regulation in the pathway. Regulation of the transcript levels of ACS genes appears to be a key mechanism to control changes in ethylene production in plants (Argueso et al., 2007; Harpaz-Saad et al., 2012). However, recent studies suggest that post-translational modifications, such as phosphorylation and ubiquitination, serve as an important mechanism to regulate the stability of the ACS proteins, thus controlling the levels of ethylene in plants (Argueso et al., 2007; Chae and Kieber, 2005; Chae et al., 2003).
The family of ACS proteins can be grouped into 3 types, type-1, type-2, and type-3, based on the presence of distinct consensus sequences including phosphorylation target sites near the non-catalytic C-termini (Fig. 2) (Chae and Kieber, 2005; McClellan and Chang, 2008; Yoshida et al., 2005). Type-1 ACS proteins contain a relatively long C-terminal domain that shares highly conserved sequences and the target sites for a mitogen-activated protein kinase (MAPK) and calcium-dependent protein kinase (CDPK) (Hernández Sebastià et al., 2004; Kim et al., 2003; Liu and Zhang, 2004). Type-2 ACS proteins have only the predicted CDPK phosphorylation site. However, type-2 ACS proteins contain a unique regulatory motif called a target of ethylene overproducer 1 (ETO1) (TOE) which overlaps with the CDPK target site. TOE motif mediates interaction with ETO1 E3 ligase and its two paralogs, ETO1-Like (EOL1 and EOL2) and is required for degradation of type-2 ACS (Chae et al., 2003; Christians et al., 2009; Wang et al., 2004; Yoshida et al., 2005; 2006); type-3 ACS contains no target sites for a CDPK and MAPK, with only a short stretch of amino acids in the C-terminal extension (Chae and Kieber, 2005).
Fig. 2.
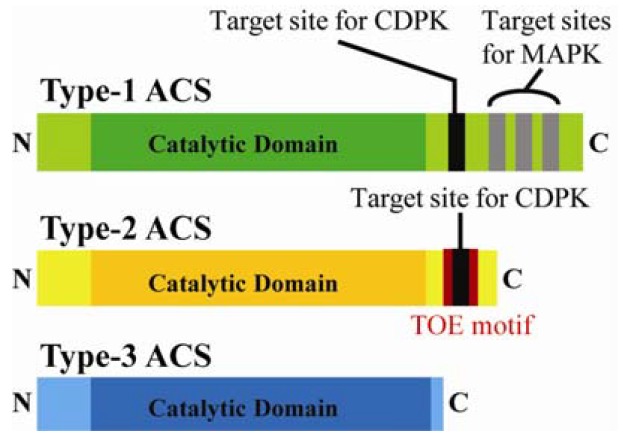
Structures of the different subgroups of ACC synthases and their regulatory motifs. Each type of ACS protein consists of a short N-terminus, followed by a conserved catalytic domain and a C-terminal domain with regulatory motifs, including phosphorylation sites. The target serine residue for calcium-dependent protein kinase (CDPK) is shown as a black bar and the target serine residues for mitogen-activated protein kinase (MAPK) are shown as grey bars. Target of ETO1 (TOE) motif in type-2 ACS is highlighted in red box. Type-3 ACS does not contain any regulatory motifs. The cartoon does not accurately represent the scale between the domains of ACS.
Here, I focus on recent advances and current knowledge on the protein turnover regulation of ACS in ethylene biosynthesis. New insights into the role of phosphorylation, ubiquitination and recently identified novel components governing the stability of ACS proteins are discussed. Finally, I conclude with the prospects regarding the cross-talk between ethylene and other cellular biosynthetic/signaling pathways. Transcriptional regulation in ethylene biosynthesis has been well documented, therefore not discussed in this review.
MOLECULAR GENETIC EVIDENCE FOR POST-TRANSLATIONAL MODIFICATION OF ACC SYNTHASES
Treatment of Arabidopsis etiolated seedlings with ethylene results in the triple response (Guzman and Ecker, 1990). The triple response has been extensively used to identify Arabidopsis mutants with defects in ethylene perception and signaling, as well as mutants affecting ethylene biosynthesis. The ethylene biosynthesis mutants can be further categorized into two groups: (1) cytokinin-insensitive (cin) mutants which are impaired in response to cytokinin, resulting in reduced levels of ethylene production in response to cytokinin; (2) ethylene-overproducer (eto) mutants which show a constitutive ethylene response due to increased ethylene biosynthesis (Chae et al., 2003; Vogel et al., 1998). The recessive cin5 mutant was identified as the first example of the cin mutants, and further characterization of the mutant revealed that the corresponding cin5 mutation is a loss of function allele of the ACS5 gene. The cin5 mutant is severely insensitive to exogenous cytokinin, and as a result, it fails to display the triple response. However, it shows normal triple response to ethylene, suggesting ACS5 is the primary target of cytokinin-mediated ethylene induction in etiolated seedlings (Vogel et al., 1998). Other phytohormones, such as auxin, brassinosteroids and ABA are also known hormonal triggers that increase ethylene production (Arteca and Arteca, 2008; Woeste et al., 1999a; Yi et al., 1999; Zhang et al., 2009). Auxin promotes ethylene production mainly through the increase of mRNA levels of specific ACS genes in various plant species. In Arabidopsis, the most of ACS genes are transcriptionally induced in response to auxin, and auxin treatment also alters the spatial expression pattern of the ACS genes (Tsuchisaka and Theologis, 2004). ABA has been shown to regulate ethylene production in apples, tomato and various plant tissues (Lara and Vendrell, 2000; Tari and Nagy, 1996; Zhang et al., 2009). In tomato, ethylene levels increase remarkably after ABA treatment and this coincides with the increase in the expression of LeACS2, whereas application of NDGA, an inhibitor of a key enzyme in ABA biosynthesis, suppresses the expression of LeACS2 (Zhang et al., 2009). Brassinosteroid is another phytohormone that enhances ethylene production by increasing the transcript abundance of ACS genes, but brassinosteroid in part promotes ethylene production by stabilizing ACS protein (Hansen et al., 2009; Yi et al., 1999). Cytokinin, however, stimulates ethylene production by acting on the stability of ACS proteins, thereby increasing the ethylene production in plants (Chae et al., 2003; Hansen et al., 2009; Vogel et al., 1998).
Analysis of eto mutants has provided further evidence that the stability of ACS proteins is regulated. Three eto mutants have been identified via genetic screens based on the constitutive triple response phenotype due to ethylene overproduction: eto1, eto2, and eto3 (Chae et al., 2003). Etiolated eto mutants exhibit the constitutive triple response and this phenotypes is rescued by treatment of mutant seedlings with ethylene biosynthesis inhibitor aminoethoxyvinylglycine (AVG). This suggests that the eto mutants are affected in ethylene biosynthesis. The dominant eto2 and eto3 mutations alter the C-terminal domain of ACS5 and ACS9, both type-2 ACS proteins, as the result of a single base insertion and a missense mutation, respectively. The eto2 and eto3 mutants significantly produce more ethylene than wild-type seedlings, but this increase in ethylene production is not correlated to the ACS5 or ACS9 gene expression, thus suggesting that the mutants control the ACS function at the post-translational level similar to the action of cytokinin. These results reveal that the C-terminal domain of both ACS proteins is a target for post-translational modification for degradation (Chae and Kieber, 2005).
THE ROLE OF UBIQUITINATION IN ETHYLENE BIOSYNTHESIS
Characterization of the eto1 revealed that ubiquitination via the 26S proteasome pathway is involved in regulating ethylene biosynthesis by modulating the protein stability of type-2 ACS proteins. Recessive eto1 produces a nearly 10-fold excess of ethylene compared to wild-type etiolated Arabidopsis seedlings and exhibits the constitutive triple response (Woeste et al., 1999b). Epistasis analysis demonstrates that ACS5 acts downstream of ETO1; the eto1cin5 double mutant produces significantly reduced amount of ethylene compared to eto1 itself (Chae et al., 2003), indicating ETO1 plays a role as a negative regulator by acting through ACS5 in ethylene biosynthesis. (Wang et al., 2004). ETO1 encodes a novel plant-specific protein that contains a Broad-complex, Tramtrack, Bric-á-brac (BTB) and Tetratrico-peptide repeat (TPR) with a coiled-coil domain (Wang et al., 2004). Proteins containing BTB motifs have been shown to participate in substrate recognition via their protein-protein interaction motifs and bridge substrates to Cullin 3 (CUL3)-based ubiquitin ligase complexes for degradation (Albagli et al., 1995; Pintard et al., 2004). The TPR motifs are involved in diverse protein-protein interactions, and also serve as a scaffold for the assembly of high-order protein complexes (Blatch and Lassle, 1999). ETO1 interacts with ACS5 and CUL3, and in the case of ACS5, this interaction is dependent on the C-terminus of ACS5, as ETO1 fails to interact with ACS5eto (Pintard et al., 2004; Wang et al., 2004). Consequently, these studies suggest that ETO1 serves as a substrate-specific adaptor to bridge ACS5 to the CUL3 to regulate ACS5 protein degradation. Analysis of ETO1 function in plants shows that ETO1 suppresses cytokinin-induced ACS protein stabilization via the C-terminus of ACS5. Overexpression of ETO1 inhibits cytokinin-induced ethylene biosynthesis, and overexpression of ACS5 results in a partial constitutive triple response which is suppressed by co-expression of ETO1 (Wang et al., 2004). However, additional data indicate that cytokinin-mediated stabilization and ETO1-mediated destabilization are at least partially independent effects; exogenous cytokinin treatment still increases ACS5 protein stability in etiolated eto2 and eto3 seedlings, suggesting cytokinin partially acts via an alternative mechanism that is independent of ETO1 and ACS5 C-terminus (Hansen et al., 2009).
Other factors that modify E3 ligase function involved in ethylene biosynthesis are the Related to the Ubiquitin (RUB) and RUB1 Conjugating Enzyme (RCE1) (Bostick et al., 2004; Larsen and Cancel, 2004). Like ubiquitin, RUB functions through a covalent attachment to target proteins. In Arabidopsis, RUB attaches to the cullins, thereby promoting the activity of the SCF (for Skp, Cdc53p/Cul1, and F-box protein) ubiquitin ligase complex for polyubiquitination of target proteins. Interestingly, RNA interference lines of RUB exhibit the partial triple response in etiolated seedlings due to the increase in ethylene biosynthesis, implying that conjugation of RUB to CUL3 is required for the activation of ETO1 containing CUL3 E3 ligase complex (Bostick et al., 2004). Analysis of the rce1 mutant also demonstrated that the modification of RUB is required for regulating ethylene biosynthesis (Larsen and Cancel, 2004). RCE1 encodes a RUB conjugating enzyme, and has been shown to conjugate RUB to the SCF complex to modify the activity of the complex. The recessive rce1 mutant displays the constitutive triple response and overproduces ethylene. Interestingly, the basis of ethylene overproduction phenotype of rce1 is not due to enhanced ACS activity, but due to an increase of ACO activity. ACO is generally not a rate-limiting step in ethylene biosynthesis during vegetative Arabidopsis tissues; however, ethylene biosynthesis in R. palustris is limited by ACO activity during submergence. Elevated ethylene levels have been shown to increase ACO activity, which could explain the increased ACO activity in rce1 (Vriezen et al., 1999).
XBAT32, a RING domain-containing ankyrin repeat subfamily of E3 ligases, also plays a role in ethylene biosynthesis by controlling the turnover of ACS4 (type-2) and ACS7 (type-3) (Lyzenga et al., 2012). XBAT32 was previously identified as a positive regulator of lateral root development and the xbat32 mutant produces significantly less ethylene than wild type (Nodzon et al., 2004; Prasad et al., 2010). XBAT32 interacts with the ACS proteins and it catalyzes the attachment of ubiquitin to the ACS proteins (Prasad et al., 2010), suggesting XBAT32 negatively regulates the ethylene biosynthesis by regulating the stability of ACS proteins.
THE ROLES OF PHOSPHORYLATION IN ETHYLENE BIOSYNTHESIS
Phosphorylation is one of the most abundant post-translational modifications which affect many important aspects of protein function, including activity, stability, subcellular localization and protein-protein interaction (Holt et al., 2009). Mounting evidence suggests that ethylene biosynthesis is regulated by phosphorylation events which likely influence ACS protein turnover. Studies from the application of kinase and phosphatase inhibitors to tomato suspension cell cultures and tissues indicated that phosphorylation regulates the activity and/or turnover of ACS (Kamiyoshihara et al., 2010). A CDPK present in the extracts of wounded tomato fruits phosphorylates LeACS2 (Mayfield et al., 2007). The extract containing CDPK activity phosphorylates the C-terminal domain of LeACS2 in vitro, but the activity of the LeACS2 does not show a significant increase, suggesting phosphorylation regulates the turnover of ACS rather than affecting the activity. The C-terminus of LeACS2 contains a consensus phosphorylation target site for a CDPK, and this CDPK recognition site is present in a subset of ACS isoforms (Fig. 2) (Kamiyoshihara et al., 2010). Unlike the target sites of MAPK in the C-terminal domain of the type-1 ACS proteins, phosphorylation of the CDPK target site, which lies immediately upstream from the target site for MAPK has not been shown to be phosphorylated in vivo and in vitro.
Among three types of ACS, protein stability of the type-1 and type-2 ACS proteins has been shown to be directly regulated by phosphorylation. Genetic and biochemical studies of a MAPK pathway have revealed that pathogen-activated Arabidopsis MPK6 phosphorylates the type-1 ACS2 and ACS6, which leads to increased accumulation of these ACS proteins and, hence, increases ethylene production (Joo et al., 2008; Liu and Zhang, 2004). MPK6 phosphorylates 3 serine residues residing within the consensus MAPK target site in the C-terminus in vitro, suggesting MPK6-mediated phosphorylation of ACS2 and ACS6 prevents their degradation, resulting in an increase in ethylene biosynthesis in response to pathogen attack. Li et al. (2014) report a similar result that rice MPK3 and 6 are involved in ethylene production via the Salt-Intolerance 1 receptor-like kinase (SIT1), but ACS stability was not discussed in their work.
Until recently, the effect of phosphorylation on type-2 ACS protein stability was not clear; neither direct phosphorylation nor responsible kinase has been identified. However, a recent study demonstrated that a Casein Kinase isoform 1.8 (CK 1.8) phosphorylates the type-2 ACS5 protein, which in turn promotes the interaction between ETO1 and ACS5, resulting in the degradation of ACS5 protein (Tan and Xue, 2014). The ck1.8 mutant displays the constitutive triple response due to overproduction of ethylene similar to the eto mutant. Interestingly, the triple response of ck1.8 seedlings is only observed in the hypocotyl and hook, not in the roots, implying that CK1.8 affects specific aspects of ethylene-mediated seedling growth responses.
NEW PLAYERS AND THEIR ROLES IN ACS PROTEIN TURNOVER REGULATION IN ETHYLENE BIOSYNTHESIS
Several recent studies have identified novel regulatory factors which target multiple ACS isoforms belonging in different types of ACS (Fig. 3). This regulatory feature is distinct from that of previous known regulatory proteins with a type-specific targeting (e.g. ETO1/EOL E3 ligase for type-2 and MAPK3/6 for type-1 ACS). 14-3-3 proteins, novel regulator of ethylene biosynthesis, target all three types of ACS (Fig. 3) (Yoon and Kieber, 2013a). 14-3-3 proteins are a family of evolutionarily well-conserved dimer proteins that specifically interact with phosphoproteins and are involved in a diverse array of physiological processes (Darling et al., 2005; Dougherty and Morrison, 2004; Freeman and Morrison, 2011). Upon interaction with target proteins, 14-3-3 proteins change their localization, stability, and activity, resulting in changes in physiological responses (Freeman and Morrison, 2011). There are 13 functional 14-3-3 genes in Arabidopsis and their encoded proteins possess a highly conserved target binding domain, which can recognize a short stretch of peptide containing phosphoserine or phosphothreonin on target proteins (Aitken et al., 1992; De Boer et al., 2013; Denison et al., 2011). This could allow different 14-3-3 isoforms to function redundantly by recognizing similar sets of target proteins. However, increasing results suggest that a defined subset of 14-3-3 isoforms display specific functions, such as the regulation of stomatal opening, flowering time, and phytochrome signaling in Arabidopsis (Mayfield et al., 2007; Paul et al., 2008; Purwestri et al., 2009; Tseng et al., 2012). It is unclear whether this is a result of biochemical specificity or simply differences in expression patterns of 14-3-3 isoforms in plants.
Fig. 3.
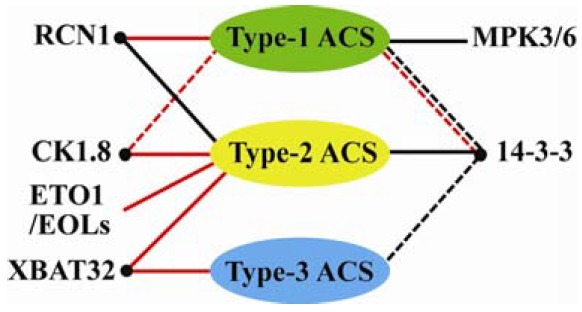
Regulatory elements determine the stability of ACS in Arabidopsis. Mitogen-activated protein kinase 3/6 (MPK3/6) and Ethylene Overproducer 1/ETO1-Like (ETO1/EOL) E3 ligases target a specific type of ACS, whereas 14-3-3, Roots curl in 1-N-naphthyl-phthalamic acid 1 (RCN1), XBAT32, and casein kinase 1.8 (CK1.8) could regulate the protein stability of more than one type of ACS. Positive or negative regulation of the different types of ACS protein stability is shown as black or red lines, respectively. Solid lines indicate interactions for which there is direct experimental evidence. Dashed lines indicate hypothetical interactions suggested by indirect evidence.
14-3-3 interacts with all three types of ACS proteins via a non-C-terminal domain of the proteins and there is no specificity in the interaction between 14-3-3 isoforms and ACS proteins in bimolecular fluorescence complementation assay (Yoon and Kieber, 2013a). 14-3-3 stabilizes ACS protein by direct interaction and by negatively regulating the stability of the E3 ligases, ETO1/EOLs, which specifically target the type-2 ACS proteins for degradation (Yoon and Kieber, 2013a). Studies from mammalian and yeast systems have suggested that the stability of F-box proteins which promote ubiquitination in the ubiquitinproteasome pathway, is regulated based on the availability of substrates through an autocatalytic process (Ho et al., 2008; Wee et al., 2005). It is possible that 14-3-3 proteins preferentially interact with type-2 ACS proteins, which in turn leads to the depletion of the ACS proteins for the ETO1/EOLs, thus regulating the turnover of both sets of proteins. Alternatively, the interaction with dimeric 14-3-3 proteins may cause the ETO1/EOLs to dimerize, thereby promoting self-ubiquitination and subsequent degradation. Finally, the interaction with 14-3-3 proteins could enhance the interaction with distinct E3 ligases, such as XBAT32, leading to the ubiquitination and subsequent degradation of the ETO1/EOLs. Intriguingly, in mammalian cells, 14-3-3σ interacts with and regulates the protein stability of a short-lived p53 tumor suppressor protein and its cognate E3 ligases COP1 and MDM2 (Su et al., 2011; Yang et al., 2007). 14-3-3σ stabilizes p53 by down-regulation of MDM2 and COP1 protein stability. This 14-3-3-mediated inverse stability regulation on p53 and MDM2 and COP shows a similar regulatory mechanism by which Arabidopsis 14-3-3 ω control the protein stability of ACS5 and ETO1/EOLs, suggesting that the function of a subset of 14-3-3 isoforms in protein stability regulation is evolutionarily conserved between mammalian and plants. Several findings imply that 14-3-3 also regulates ACS stability independently of ETO1/EOLs (Yoon and Kieber, 2013a; 2013b). First, 14-3-3 interacts with ACS5eto2, a type-2 ACS with a lack of TOE motif for ETO1/EOL interaction. Secondly, 14-3-3 directly interacts and stabilizes the sole type-3 ACS7 and type-1 ACS2, whose protein stability is not regulated by the ETO1/EOLs. Finally, 14-3-3 increases ACS stability in eto1;eol1;eol2 triple mutant. Thus, there is at least one other system acting to degrade type-2 ACS proteins in addition to the ETO1/EOLs, but in the manner dependent on 14-3-3 function. This is consistent with the observation that cytokinin and brassinosteroid increase type-2 ACS function partly through a TOE-independent mechanism (Hansen et al., 2009).
While 14-3-3 positively regulates ACS protein stability (Yoon and Kieber, 2013a; 2013b), studies from characterization of the RARE COLD INDUCIBLE 1A (RCI1A), which encodes a 14-3-3 φ isoform suggest that 14-3-3 φ negatively regulate the protein stability of ACS in response to cold stress (Catala et al., 2014). rci1a mutant displays increased levels of ACS6 protein in response to cold treatment, and this change is not due to the changes of the ACS6 mRNA levels. However, the direct effect of 14-3-3 on ACS protein stability was not demonstrated in the study. Catala et al. suggest that the discrepancy of two studies might be contributed to the functional specificities of 14-3-3 isoforms; Yoon and Kieber (2013) used 14-3-3 ω, while Catala et al. (2014) used 14-3-3 φ isoform (Bornke, 2005; Fu et al., 2000; Paul et al., 2012). A recent study also reported that single 14-3-3 isoform could have distinct functions depending on the binding sites on a given target protein (Ganguly et al., 2005).
Identification and characterization of the role of CK1.8 have also brought new insights into the post-translational regulation in ethylene biosynthesis (Tan and Xue, 2014). CK1.8 is a conserved serine/threonine protein kinase that plays role in various physiological processes, including blue light signaling, flowering, microtubule organization and brassinosteroid signaling in rice (Ben-Nissan et al., 2008; Dai and Xue, 2010; Liu et al., 2003; Tan et al., 2013). As briefly discussed in the previous section, the ck1.8 mutant overproduces ethylene resulting from accumulation of ACS proteins, suggesting CK1.8 is a negative regulator of ACS protein stability. CK1.8 phosphorylates ACS5 at threonine residue 463 (T463) which is located within the TOE motif, and phosphorylation on this site promotes the interaction with ETO1, indicating the phosphorylation of T463 on ACS5 plays a negative role in ACS5 protein stability. However, T463 is not highly conserved in type-2 ACS proteins. It can be found in only in a subset of Arabidopsis type-2 ACS (ACS5 and ACS9), type-1 (ACS6), and tomato type-1 (LeACS2), suggesting that CK1.8 could target different ACS types rather than committing to regulate a specific type of ACS (Fig. 3). The role of CK1.8 is somewhat in contrast to what has been observed from other studies indicating that phosphorylation promotes ACS protein stability.
The roots curl in 1-N-naphthylphthalamic acid 1 (RCN1) gene encodes one of three regulatory/scaffolding A subunits of Arabidopsis PP2A, and targets the type-1 protein ACS2 and ACS6 for regulating their stability (Skottke et al., 2011). Genetic studies revealed that the function of RCN1 requires ACS2 and ACS6 and the rcn1 mutant exhibits increased accumulation of the ACS6, suggesting phosphorylation promotes the protein stability of type-1 ACS. Strikingly, rcn1 shows different effects on type-2 ACS5 protein stability. In etiolated rcn1 seedlings, the accumulation of myc-tagged ACS5 is significantly reduced, whereas accumulation and turnover of the myc-ACS5eto2 is not affected, indicating that RCN1 plays a positive role in ACS5 stability through the TOE motif of ACS5. However, RCN1-directed dephosphorylation on ACS5 has not been evidenced, suggesting the possibility that RCN1 may dephosphorylate the ETO1 complex. Both RCN1 and CK1.8 regulate the stability of ACS5 through the TOE domain, but it is not clear whether the effects of RCN1 on ACS5 are dependent on CK1.8 or ETO1/ EOLs.
Due to the lack of regulatory motifs in the C-terminal domain, including phosphorylation sites, it was considered that ACS7, the sole type-3 ACS, may not be subjected to proteasome-mediated degradation pathway, and that it may be more stable than other ACS proteins (Chae and Kieber, 2005). However, Lyzenga et al., recently showed that the protein stability of ACS7 is also governed by the ubiquitin-mediated proteasomal degradation, and that degradation requires the Ring-type E3 ligase XBAT32 (Lyzenga et al., 2012). Interestingly, XBAT32 also confers protein instability to the type-2 ACS4, suggesting XBAT32-mediated degradation mechanism is not specific for the type-3 ACS. A cell-free degradation assay shows that changes in 4 lysine residues in the C-terminal domain of ACS4 results in accelerating degradation of ACS4 protein. This result is similar to the observation that K435R in the C-terminus of Flag-ACS7 promotes the turnover rate of the ACS7, suggesting the C-terminal lysine residues are not for ubiquitination, but for stabilization of ACS4 and ACS7. Shortly after, Xiong et al. raise interesting aspects of the protein stability regulation of ACS7. They showed that destabilization sequences of the ACS7 are located in the N-terminus of ACS7. The N-terminal 54 residues of the ACS7 confer significant instability to ACS71–54-GUS and first 14 amino acids are responsible for negative regulation of the ACS7 protein stability (Xiong et al., 2014). One possible explanation for this may be due to the nature of the C-terminal fusion of the ACS7-GUS used in the study. Traditionally, the N-terminal fusion of ACS has been routinely utilized for studying the turnover of ACS to avoid masking the C-terminal regulatory domain and this may be blamed for concealing the destabilization signals located at the N-termini and making only the C-terminal signals available to degradation machinery. It is interesting to further study the role of the N-terminal domain of other types of ACS whether they also contain putative degradation sequences in their N-termini.
IDENTIFICATION OF NOVEL REGULATORS IN ETHYLENE BIOSYNTHESIS: THE POINT OF CROSSTALK BETWEEN ETHYLENE AND OTHER BIOSYNTHETIC/SIGNALING PATHWAYS?
Several studies indicate that there are molecular components acing on the non-C-terminal domain of ACS proteins to regulate their stability. Cytokinin and brassinosteroid stabilize ACS5eto2 and ACS9eto3 and the effects of these two hormones on the protein stability are additive, suggesting cytokinin and brassinosteroid act through distinct TOE-independent mechanisms on these ACS proteins (Hansen et al., 2009). Genetic studies showed that cytokinin-mediated ACS stabilization requires a functional cytokinin signaling pathway (Hansen et al., 2009). Mutation in the signaling components, cytokinin receptors, AHPs, and type-A and type-B transcription factors, in the cytokinin signaling pathway produce reduced amounts of ethylene in response to exogenous cytokinin. The effect of brassinosteroid in ethylene biosynthesis is somewhat distinguished from the typical triple response that has been observed with cytokinin treatment; BR treatment results in a shortened and thickened hypocotyl formation; but it does not induce an exaggerated hook formation; and shortening of the root and hypocotyl is less severe than for cytokinin-treated seedlings. Interestingly, the ethylene-insensitive mutant ein2-5 still shows cytokinin and BR induced hypocotyl phenotypes, although the extent is not as severe as in wild type, indicating this process is independent of the ethylene signaling pathway. Although it has not been demonstrated whether gibberellin regulates the turnover of ACS proteins, studies from the characterization of a gai;eto2 (gibberellin insensitive;ethylene overproducing) double mutant showed that GA signaling is required for ACS stabilization via the TOE-independent manner, as the overproduction phenotype of eto2 is abolished in the gai;eto2 double mutant (De Grauwe et al., 2008a; 2008b). Furthermore, 14-3-3-mediated ETO1/EOL-independent stabilization of ACS proteins also indicates the existence of an alternative mechanism to stabilize ACS proteins (Yoon and Kieber, 2013a). It is not clear whether the ETO1/EOL-independent mechanism acts in the same pathway that is utilized by the other factors, but is it of great interest to further study to identify molecular elements involving in these regulatory pathways.
Together, these studies indicate that there are molecular components that act as the points of cross-talk between ethylene biosynthesis and other hormonal signaling pathways. Identification of these elements will bring new insights into understanding the mechanism by which protein turnover of ACS is regulated to coordinate and merge different hormonal inputs to regulate ethylene production which effects on many diverse ranges of plant growth and development.
Acknowledgments
The author thanks Alison Delong for critical reading of the manuscript and comments. This work was supported by a startup fund by Purdue University to GMY.
REFERENCES
- Abeles FB, Morgan PW, Saltveit MEJ. Ethylene in plant biology. San Diego, CA: Academic Press; 1992. [Google Scholar]
- Adams DO, Yang SF. Methionine metabolism in apple tissue – implication of S-adenosylmethionine as an intermediate in conversion of methionine to ethylene. Plant Physiol. 1977;60:892–896. doi: 10.1104/pp.60.6.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken A, Collinge DB, van Heusden BP, Isobe T, Roseboom PH, Rosenfeld G, Soll J. 14-3-3 proteins: a highly conserved, widespread family of eukaryotic proteins. Trends Biochem Sci. 1992;17:498–501. doi: 10.1016/0968-0004(92)90339-b. [DOI] [PubMed] [Google Scholar]
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: a new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Argueso CT, Hansen M, Kieber JJ. Regulation of ethylene biosynthesis. J. Plant Growth Regul. 2007;26:13. [Google Scholar]
- Arteca RN, Arteca JM. Effects of brassinosteroid, auxin, and cytokinin on ethylene production in Arabidopsis thaliana plants. J. Exp. Bot. 2008;59:3019–3026. doi: 10.1093/jxb/ern159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Nissan G, Cui W, Kim DJ, Yang Y, Yoo BC, Lee JY. Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiol. 2008;148:1897–1907. doi: 10.1104/pp.108.129346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lassle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Boller T, Herner RC, Kende H. Assay for and enzymatic formation of an ethylene precursor, 1-aminocyclopropane-1-carboxylic acid. Planta. 1979;145:293–303. doi: 10.1007/BF00454455. [DOI] [PubMed] [Google Scholar]
- Bornke F. The variable C-terminus of 14-3-3 proteins mediates isoform-specific interaction with sucrose-phosphate synthase in the yeast two-hybrid system. J. Plant Physiol. 2005;162:161–168. doi: 10.1016/j.jplph.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Bostick M, Lochhead SR, Honda A, Palmer S, Callis J. Related to ubiquitin 1 and 2 are redundant and essential and regulate vegetative growth, auxin signaling, and ethylene production in Arabidopsis. Plant Cell. 2004;16:2418–2432. doi: 10.1105/tpc.104.024943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R, Lopez-Cobollo R, Mar Castellano M, Angosto T, Alonso JM, Ecker JR, Salinas J. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell. 2014;26:3326–3342. doi: 10.1105/tpc.114.127605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Kieber JJ. Eto Brute? Role of ACS turnover in regulating ethylene biosynthesis. Trends Plant Sci. 2005;10:291–296. doi: 10.1016/j.tplants.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ. The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell. 2003;15:545–559. doi: 10.1105/tpc.006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hansen M, Binder BM, Kieber JJ, Vierstra RD. The BTB ubiquitin ligases ETO1, EOL1 and EOL2 act collectively to regulate ethylene biosynthesis in Arabidopsis by controlling type-2 ACC synthase levels. Plant J. 2009;57:332–345. doi: 10.1111/j.1365-313X.2008.03693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker W, Knight LI. Effect of illuminating gas and ethylene upon flowering carnation. Bot. Gaz. 1908;46:259–276. [Google Scholar]
- Dai C, Xue HW. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010;29:1916–1927. doi: 10.1038/emboj.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling DL, Yingling J, Wynshaw-Boris A. Role of 14-3-3 proteins in eukaryotic signaling and development. Curr. Top. Dev. Biol. 2005;68:281–315. doi: 10.1016/S0070-2153(05)68010-6. [DOI] [PubMed] [Google Scholar]
- De Boer AH, van Kleeff PJ, Gao J. Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma. 2013;250:425–440. doi: 10.1007/s00709-012-0437-z. [DOI] [PubMed] [Google Scholar]
- De Grauwe L, Chaerle L, Dugardeyn J, Decat J, Rieu I, Vriezen WH, Baghour M, Moritz T, Beemster GT, Phillips AL, et al. Reduced gibberellin response affects ethylene biosynthesis and responsiveness in the Arabidopsis gai eto2-1 double mutant. New Phytol. 2008a;177:128–141. doi: 10.1111/j.1469-8137.2007.02263.x. [DOI] [PubMed] [Google Scholar]
- De Grauwe L, Dugardeyn J, Van Der Straeten D. Novel mechanisms of ethylene-gibberellin crosstalk revealed by the gai eto2-1 double mutant. Plant Signal. Behav. 2008b;3:1113–1115. doi: 10.4161/psb.3.12.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison FC, Paul AL, Zupanska AK, Ferl RJ. 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 2011;22:720–727. doi: 10.1016/j.semcdb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Dong JG, Fernandez-Maculet JC, Yang SF. Purification and characterization of 1-aminocyclopropane-1-carboxylate oxidase from apple fruit. Proc. Natl. Acad. Sci. USA. 1992;89:9789–9793. doi: 10.1073/pnas.89.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J. Cell Sci. 2004;117:1875–1884. doi: 10.1242/jcs.01171. [DOI] [PubMed] [Google Scholar]
- Freeman AK, Morrison DK. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011;22:681–687. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 2000;40:617–647. doi: 10.1146/annurev.pharmtox.40.1.617. [DOI] [PubMed] [Google Scholar]
- Gane R. Production of ethylene by some ripening fruits. Nature. 1934;134:1008–1008. [Google Scholar]
- Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc. Natl. Acad. Sci. USA. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman P, Ecker JR. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell. 1990;2:513–523. doi: 10.1105/tpc.2.6.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Chae HS, Kieber JJ. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009;57:606–614. doi: 10.1111/j.1365-313X.2008.03711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S, Yoon GM, Matto AK, Kieber JJ. The formation of ACC and competition between polyamines and ethylene for SAM. Annu. Plant Rev. 2012;44:53–81. [Google Scholar]
- Hernández Sebastià C, Hardin SC, Clouse SD, Kieber JJ, Huber SC. Identification of a new motif for CDPK phosphorylation in vitro that suggests ACC synthase may be a CDPK substrate. Arch. Biochem. Biophys. 2004;428:81–91. doi: 10.1016/j.abb.2004.04.025. [DOI] [PubMed] [Google Scholar]
- Ho MS, Ou C, Chan YR, Chien CT, Pi H. The utility F-box for protein destruction. Cell. Mol. Life Sci. 2008;65:1977–2000. doi: 10.1007/s00018-008-7592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo S, Liu Y, Lueth A, Zhang S. MAPK phosphorylation-induced stabilization of ACS6 protein is mediated by the non-catalytic C-terminal domain, which also contains the cis-determinant for rapid degradation by the 26S proteasome pathway. Plant J. 2008;54:129–140. doi: 10.1111/j.1365-313X.2008.03404.x. [DOI] [PubMed] [Google Scholar]
- Kamiyoshihara Y, Iwata M, Fukaya T, Tatsuki M, Mori H. Turnover of LeACS2, a wound-inducible 1-aminocyclopropane-1-carboxylic acid synthase in tomato, is regulated by phosphorylation/dephosphorylation. Plant J. 2010;64:140–150. doi: 10.1111/j.1365-313X.2010.04316.x. [DOI] [PubMed] [Google Scholar]
- Kende H. Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1993;44:283–307. [Google Scholar]
- Kim CY, Liu Y, Thorne ET, Yang H, Fukushige H, Gassmann W, Hildebrand D, Sharp RE, Zhang S. Activation of a stress-responsive mitogen-activated protein kinase cascade induces the biosynthesis of ethylene in plants. Plant Cell. 2003;15:2707–2718. doi: 10.1105/tpc.011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight LI, Rose RC, Crocker W. Effects of various gases and vapors upon etiolated seedlings of the sweet pea. Science. 1910;31:635–636. [Google Scholar]
- Lara I, Vendrell M. Development of ethylene-synthesizing capacity in preclimacteric apples: interaction between abscisic acid and ethylene. J. Am. Soc. Hortic. Sci. 2000;125:505–512. [Google Scholar]
- Larsen PB, Cancel JD. A recessive mutation in the RUB1-conjugating enzyme, RCE1, reveals a requirement for RUB modification for control of ethylene biosynthesis and proper induction of basic chitinase and PDF1.2 in Arabidopsis. Plant J. 2004;38:626–638. doi: 10.1111/j.1365-313X.2004.02068.x. [DOI] [PubMed] [Google Scholar]
- Li CH, Wang G, Zhao JL, Zhang LQ, Ai LF, Han YF, Sun DY, Zhang SW, Sun Y. The Receptor-Like Kinase SIT1 Mediates Salt Sensitivity by Activating MAPK3/6 and regulating ethylene homeostasis in rice. Plant Cell. 2014;26:2538–2553. doi: 10.1105/tpc.114.125187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman M, Mapson LW. Genesis and biogenesis of ethylene. Nature. 1964;204:343–345. [Google Scholar]
- Liu Y, Zhang S. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell. 2004;16:3386–3399. doi: 10.1105/tpc.104.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW. Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 2003;36:189–202. doi: 10.1046/j.1365-313x.2003.01866.x. [DOI] [PubMed] [Google Scholar]
- Lyzenga WJ, Booth JK, Stone SL. The Arabidopsis RING-type E3 ligase XBAT32 mediates the proteasomal degradation of the ethylene biosynthetic enzyme, 1-aminocyclopropane-1-carboxylate synthase 7. Plant J. 2012;71:23–34. doi: 10.1111/j.1365-313X.2012.04965.x. [DOI] [PubMed] [Google Scholar]
- Mattoo AK, Suttle JC. The Plant Hormone Ethylene. Boca Raton: CRC Press; 1991. [Google Scholar]
- Mayfield JD, Folta KM, Paul AL, Ferl RJ. The 14-3-3 Proteins mu and upsilon influence transition to flowering and early phytochrome response. Plant Physiol. 2007;145:1692–1702. doi: 10.1104/pp.107.108654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan CA, Chang CL. The role of protein turnover in ethylene biosynthesis and response. Plant Sci. 2008;175:24–31. doi: 10.1016/j.plantsci.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr DP, Yang SF. Conversion of 5′-methylthioadenosine to methionine by apple tissue. Phytochemistry. 1975;14:1291–1292. [Google Scholar]
- Neljubov D. Uber die horizontale Nutation der Stengel von Pisum sativum und einiger Anderer. Pflanzen Beih. Bot. Zentralb. 1901;10:128–139. [Google Scholar]
- Nodzon LA, Xu WH, Wang Y, Pi LY, Chakrabarty PK, Song WY. The ubiquitin ligase XBAT32 regulates lateral root development in Arabidopsis. Plant J. 2004;40:996–1006. doi: 10.1111/j.1365-313X.2004.02266.x. [DOI] [PubMed] [Google Scholar]
- Paul AL, Folta KM, Ferl RJ. 14-3-3 proteins, red light and photoperiodic flowering: a point of connection? Plant Signal. Behav. 2008;3:511–515. doi: 10.4161/psb.3.8.5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AL, Denison FC, Schultz ER, Zupanska AK, Ferl RJ. 14-3-3 phosphoprotein interaction networks - does isoform diversity present functional interaction specification? Front. Plant Sci. 2012;3:190. doi: 10.3389/fpls.2012.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad ME, Schofield A, Lyzenga W, Liu H, Stone SL. Arabidopsis RING E3 ligase XBAT32 regulates lateral root production through its role in ethylene biosynthesis. Plant Physiol. 2010;153:1587–1596. doi: 10.1104/pp.110.156976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purwestri YA, Ogaki Y, Tamaki S, Tsuji H, Shimamoto K. The 14-3-3 protein GF14c acts as a negative regulator of flowering in rice by interacting with the florigen Hd3a. Plant Cell Physiol. 2009;50:429–438. doi: 10.1093/pcp/pcp012. [DOI] [PubMed] [Google Scholar]
- Sauter M, Moffatt B, Saechao MC, Hell R, Wirtz M. Methionine salvage and Sadenosylmethionine: essential links between sulfur, ethylene and polyamine biosynthesis. Biochem. J. 2013;451:145–154. doi: 10.1042/BJ20121744. [DOI] [PubMed] [Google Scholar]
- Skottke KR, Yoon GM, Kieber JJ, DeLong A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011;7:e1001370. doi: 10.1371/journal.pgen.1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su CH, Zhao R, Zhang F, Qu C, Chen B, Feng YH, Phan L, Chen J, Wang H, Wang H, et al. 14-3-3sigma exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res. 2011;71:884–894. doi: 10.1158/0008-5472.CAN-10-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan ST, Xue HW. Casein kinase 1 regulates ethylene synthesis by phosphorylating and promoting the turnover of ACS5. Cell Rep. 2014;9:1692–1702. doi: 10.1016/j.celrep.2014.10.047. [DOI] [PubMed] [Google Scholar]
- Tan ST, Dai C, Liu HT, Xue HW. Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant Cell. 2013;25:2618–2632. doi: 10.1105/tpc.113.114322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tari I, Nagy M. Abscisic acid and ethrel abolish the inhibition of adventitious root formation of pacrobutrazol-treated bean primary leaf cuttings. Biol. Plant. 1996;38:369–375. [Google Scholar]
- Tseng TS, Whippo C, Hangarter RP, Briggs WR. The role of a 14-3-3 protein in stomatal opening mediated by PHOT2 in Arabidopsis. Plant Cell. 2012;24:1114–1126. doi: 10.1105/tpc.111.092130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchisaka A, Theologis A. Unique and overlapping expression patterns among the Arabidopsis 1-amino-cyclopropane-1-carboxylate synthase gene family members. Plant Physiol. 2004;136:2982–3000. doi: 10.1104/pp.104.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Poel B, Van Der Straeten D. 1-aminocyclopropane-1-carboxylic acid (ACC) in plants: more than just the precursor of ethylene! Front. Plant Sci. 2014;5:640. doi: 10.3389/fpls.2014.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Woeste KE, Theologis A, Kieber JJ. Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA. 1998;95:4766–4771. doi: 10.1073/pnas.95.8.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vriezen WH, Hulzink R, Mariani C, Voesenek LA. 1-aminocyclopropane-1-carboxylate oxidase activity limits ethylene biosynthesis in Rumex palustris during submergence. Plant Physiol. 1999;121:189–196. doi: 10.1104/pp.121.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KL, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature. 2004;428:945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- Wee S, Geyer RK, Toda T, Wolf DA. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- Woeste KE, Vogel JP, Kieber JJ. Factors regulating ethylene biosynthesis in etiolated Arabidopsis thaliana seedlings. Physiol. Plant. 1999a;105:478–484. [Google Scholar]
- Woeste KE, Ye C, Kieber JJ. Two Arabidopsis mutants that overproduce ethylene are affected in the posttranscriptional regulation of 1-aminocyclopropane-1-carboxylic acid synthase. Plant Physiol. 1999b;119:521–530. doi: 10.1104/pp.119.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Xiao D, Xu X, Guo Z, Wang NN. The non-catalytic N-terminal domain of ACS7 is involved in the post-translational regulation of this gene in Arabidopsis. J. Exp. Bot. 2014;65:4397–4408. doi: 10.1093/jxb/eru211. [DOI] [PubMed] [Google Scholar]
- Yang SF, Hoffman NE. Ethylene biosynthesis and its regulation in higher plants. Ann. Rev. Plant Physiol. 1984;34:34. [Google Scholar]
- Yang HY, Wen YY, Lin YI, Pham L, Su CH, Yang H, Chen J, Lee MH. Roles for negative cell regulator 14-3-3 sigma in control of MDM2 activities. Oncogene. 2007;26:7355–7362. doi: 10.1038/sj.onc.1210540. [DOI] [PubMed] [Google Scholar]
- Yi HC, Joo S, Nam KH, Lee JS, Kang BG, Kim WT. Auxin and brassinosteroid differentially regulate the expression of three members of the 1-aminocyclopropane-1-carboxylate synthase gene family in mung bean (Vigna radiata L.) Plant Mol. Biol. 1999;41:443–454. doi: 10.1023/a:1006372612574. [DOI] [PubMed] [Google Scholar]
- Yoon GM, Kieber JJ. 14-3-3 regulates 1-aminocyclopropane-1-carboxylate synthase protein turnover in Arabidopsis. Plant Cell. 2013a;25:1016–1028. doi: 10.1105/tpc.113.110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon GM, Kieber JJ. ACC synthase and its cognate E3 ligase are inversely regulated by light. Plant Signal. Behav. 2013b;8:e26478. doi: 10.4161/psb.26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagata M, Saito K, Wang KL, Ecker JR. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005;5:14. doi: 10.1186/1471-2229-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Wang KL, Chang CM, Mori K, Uchida E, Ecker JR. The ACC synthase TOE sequence is required for interaction with ETO1 family proteins and destabilization of target proteins. C. 2006;62:427–437. doi: 10.1007/s11103-006-9029-7. [DOI] [PubMed] [Google Scholar]
- Zarembinski TI, Theologis A. Ethylene biosynthesis and action: a case of conservation. The. 1994;26:1579–1597. doi: 10.1007/BF00016491. [DOI] [PubMed] [Google Scholar]
- Zhang M, Yuan B, Leng P. The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit. J. Exp. Bot. 2009;60:1579–1588. doi: 10.1093/jxb/erp026. [DOI] [PMC free article] [PubMed] [Google Scholar]


