Significance
The EPIGEN Brazil Project is the largest Latin-American initiative to study the genomic diversity of admixed populations and its effect on phenotypes. We studied 6,487 Brazilians from three population-based cohorts with different geographic and demographic backgrounds. We identified ancestry components of these populations at a previously unmatched geographic resolution. We broadened our understanding of the African diaspora, the principal destination of which was Brazil, by revealing an African ancestry component that likely derives from the slave trade from Bantu/eastern African populations. In the context of the current debate about how the pattern of deleterious mutations varies between Africans and Europeans, we use whole-genome data to show that continental admixture is the main and complex determinant of the amount of deleterious genotypes in admixed individuals.
Keywords: Latin America, population genetics, Salvador SCAALA, Bambuí Cohort Study of Ageing, Pelotas Birth Cohort Study
Abstract
While South Americans are underrepresented in human genomic diversity studies, Brazil has been a classical model for population genetics studies on admixture. We present the results of the EPIGEN Brazil Initiative, the most comprehensive up-to-date genomic analysis of any Latin-American population. A population-based genome-wide analysis of 6,487 individuals was performed in the context of worldwide genomic diversity to elucidate how ancestry, kinship, and inbreeding interact in three populations with different histories from the Northeast (African ancestry: 50%), Southeast, and South (both with European ancestry >70%) of Brazil. We showed that ancestry-positive assortative mating permeated Brazilian history. We traced European ancestry in the Southeast/South to a wider European/Middle Eastern region with respect to the Northeast, where ancestry seems restricted to Iberia. By developing an approximate Bayesian computation framework, we infer more recent European immigration to the Southeast/South than to the Northeast. Also, the observed low Native-American ancestry (6–8%) was mostly introduced in different regions of Brazil soon after the European Conquest. We broadened our understanding of the African diaspora, the major destination of which was Brazil, by revealing that Brazilians display two within-Africa ancestry components: one associated with non-Bantu/western Africans (more evident in the Northeast and African Americans) and one associated with Bantu/eastern Africans (more present in the Southeast/South). Furthermore, the whole-genome analysis of 30 individuals (42-fold deep coverage) shows that continental admixture rather than local post-Columbian history is the main and complex determinant of the individual amount of deleterious genotypes.
Latin Americans, who are classical models of the effects of admixture in human populations (1, 2), remain underrepresented in studies of human genomic diversity, notwithstanding recent studies (3, 4). Indeed, no large genome-wide study on admixed South Americans has been conducted so far. Brazil is the largest and most populous Latin-American country. Its over 200 million inhabitants are the product of post-Columbian admixture between Amerindians, Europeans colonizers or immigrants, and African slaves (1). Interestingly, Brazil was the destiny of nearly 40% of the African diaspora, receiving seven times more slaves than the United States (nearly 4 million vs. 600,000).
Here, we present results of the EPIGEN Brazil Initiative (https://epigen.grude.ufmg.br), the most comprehensive up-to-date genomic analysis of a Latin-American population. We genotyped nearly 2.2 million SNPs in 6,487 admixed individuals from three population-based cohorts from different regions with distinct demographic and socioeconomic backgrounds and sequenced the whole genome of 30 individuals from these populations at an average deep coverage of 42× (Fig. 1B and SI Appendix, sections 1, 2, and 8). By leveraging on a population-based approach, we (i) identified and quantified ancestry components of three representative Brazilian populations at a previously unmatched geographic resolution; (ii) developed an approximate Bayesian computation (ABC) approach and inferred aspects of the admixture dynamics in Northeastern, Southeastern, and Southern Brazil; (iii) elucidated how aspects of the ancestry-related social history of Brazilians influenced their genetic structure; and (iv) studied how admixture, kinship, and inbreeding interact and shape the pattern of putative deleterious mutations in an admixed population.
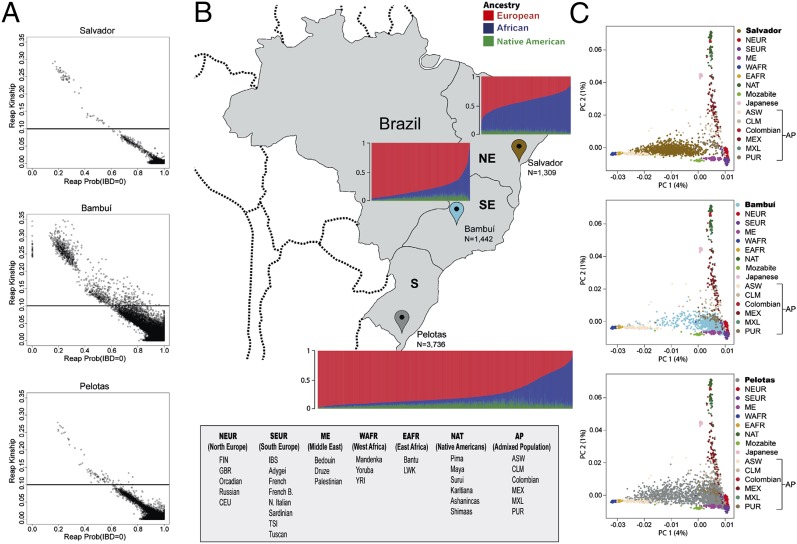
Fig. 1.
Continental admixture and kinship analysis of the EPIGEN Brazil populations. (A) Kinship coefficient for each pair of individuals and the probability that they share zero identity by descent (IBD) alleles (IBD = 0). Horizontal lines represent a kinship coefficient threshold used to consider individuals as relatives. (B) Brazilian regions, the studied populations, and their continental individual ancestry bar plots. N represents the numbers of EPIGEN individuals in the Original Dataset (including relatives; detailed in SI Appendix, section 6). (C) PCA representation, including worldwide populations and the EPIGEN populations, using only unrelated individuals (Dataset U; explained in SI Appendix, section 6). The three graphics derive from the same analysis and are different only for the plotting of the EPIGEN individuals. AP, admixed population; ASW, Americans of African ancestry in USA; CEU, Utah residents with Northern and Western European ancestry; CLM, Colombians from Medellin, Colombia; EAFR, east Africa; FIN, Finnish in Finland; French B, Basque; GBR, British in England and Scotland; IBS, Iberian population in Spain; LWK, Luhya in Webuye, Kenya; ME, Middle East; MXL/MEX, Mexican ancestry from Los Angeles; N., (North) Italian; NAT, Native American; NE, northeast; NEUR, north Europe; PC, principal component; PUR, Puerto Ricans from Puerto Rico; S, south; SE, southeast; SEUR, south Europe; TSI, Toscani in Italia; YRI, Yoruba in Ibadan, Nigeira; WAFR, west Africa.
Results and Discussion
Populations, Continental Ancestry, and Population Structure.
We studied the following three population-based cohorts (Fig. 1B). (i) SCAALA (Social Changes, Asthma and Allergy in Latin America Program) (5) (1,309 individuals) from Salvador, a coastal city with 2.7 million inhabitants in Northeastern Brazil that harbors the most conspicuous demographic and cultural African contribution (6). We inferred (7) that this population has the largest African ancestry (50.8%; SE = 0.35) among the EPIGEN populations, with 42.9% (SE = 0.35) and 6.4% (SE = 0.09) of European and Amerindian ancestries, respectively. Notably, this African ancestry is lower than that usually observed in African Americans (8, 9). (ii) The Bambuí Aging Cohort Study (10), ongoing in the homonymous city of ∼15,000 inhabitants, in the inland of Southeastern Brazil (1,442 individuals who were 82% of the residents older than 60 y old at the baseline year). We estimated that Bambuí has 78.5% (SE = 0.4) of European, 14.7% (SE = 0.4) of African, and 6.7% (SE = 0.1) of Amerindian ancestries. (iii) The 1982 Pelotas Birth Cohort Study (11) (3,736 individuals; 99% of all births in the city at the baseline year). Pelotas is a city in Southern Brazil with 214,000 inhabitants. Ancestry in Pelotas is 76.1% (SE = 0.33) European, 15.9% (SE = 0.3) African, and 8% (SE = 0.08) Amerindian.
By comparing autosomal mtDNA and X-chromosome diversity, we found across the three populations the signature of a historical pattern of sex-biased preferential mating between males with predominant European ancestry and women with predominant African or Amerindian ancestry (12) (SI Appendix, sections 6.6 and 6.9, Fig. S12, and Table S18). We determined (13) that individuals from Salvador and Pelotas were, with few exceptions, unrelated and have low consanguinity (Fig. 1A and SI Appendix, Figs. S1 and S2). Conversely, the Bambuí cohort has the highest family structure and inbreeding [Fig. 1A and SI Appendix, section 4.1 (discussion about the age structure of this cohort) and Figs. S1 and S2]. Bambuí includes several families with more than five related individuals showing at least one second-degree (or closer) relative. Bambuí mean inbreeding coefficient (0.010; SE = 0.0008) (SI Appendix, Fig. S2) is comparable with estimates observed in populations with 15–25% of consanguineous marriages from India (14). Interestingly, inbreeding in Bambuí was correlated with European ancestry (ρSpearman = 0.20; P < 10−15). These higher inbreeding and kinship structures are consistent with Bambuí being the smallest and the most isolated of the EPIGEN populations.
Continental genomic ancestry in Latin America (and specifically, in Brazil) is correlated with a set of phenotypes, such as skin color and self-reported ethnicity, and social and cultural features, such as socioeconomic status (15–17). We observed a positive correlation across the three EPIGEN populations between SNP-specific Africans/Europeans FST (a measurement of informativeness of ancestry) and SNP-specific FIT (a measurement of departure from Hardy–Weinberg equilibrium) (SI Appendix, Fig. S3). This finding indicates that, after five centuries of admixture, Brazilians still preferentially mate with individuals with similar ancestry (and its correlated morphological phenotypes and socioeconomic characteristics), a trend also observed in Mexicans and Puerto Ricans (18). Interestingly, the highest correlations were found in Pelotas and Bambuí, consistent with their higher proportion of individuals with a clearly predominant ancestry (European or African) compared with Salvador (Fig. 1 B and C). Conversely, in Salvador, despite its highest mean African ancestry, individuals are more admixed (Fig. 1 B and C), probably because of a combination of a longer history of admixture (see below) and the lower and more homogeneous socioeconomic status of this cohort (5).
Three outcomes illustrate how population subdivision and inbreeding (both partly ancestry-dependent) interact to shape population structure in admixed populations with different sizes (SI Appendix, Figs. S1 and S3). First, Bambuí (the smallest city) has the strongest departure from Hardy–Weinberg equilibrium (FIT =0.016; SE = 0.00003) because of both inbreeding (FIS = 0.010; SE = 0.0008) and ancestry-based population subdivision (ρFIT-FST = 0.18; P < 10−16). Second, Pelotas (a medium-sized city; FIT = 0.012; SE = 0.00002) has negligible inbreeding (FIS = −0.001; SE = 0.0002) but the strongest ancestry-based population subdivision (ρFIT-FST = 0.38; P < 10−16). Third, the large city of Salvador shows the lowest inbreeding and ancestry-based population subdivision (FIT = −0.003; SE = 0.00002; FIS = −0.001; SE = 0.0003; ρFIT-FST = 0.08; P < 10−16).
Overall, the EPIGEN populations studied by a population-based approach exemplify how ancestry, kinship, and inbreeding may be differently structured in small (Bambuí), medium (Pelotas), and large (Salvador) admixed Latin-American populations. These populations fairly represent the three most populated Brazilian regions (Northeast, Southeast, and South) with their geographic distribution and continental ancestry (Fig. 1) and are good examples of the Latin-American genetic diversity with their ethnic diversity.
Differences in Admixture Dynamics.
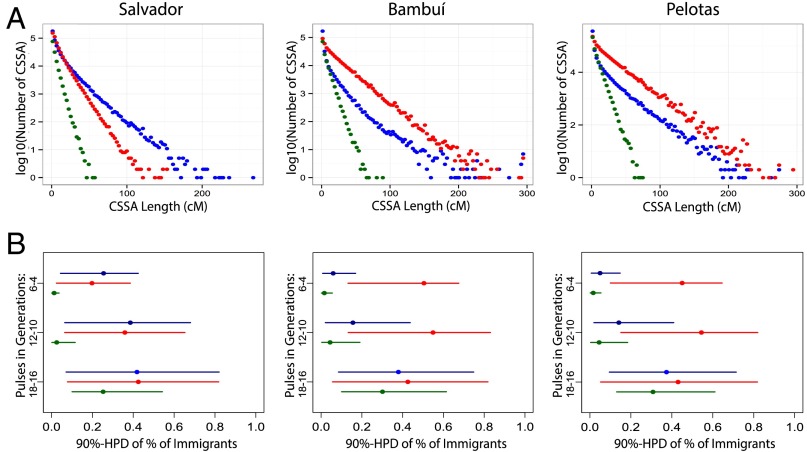
We estimated the continental origin of each allele for each SNP along each chromosome of the EPIGEN individuals (19) (SI Appendix, section 6.7) and calculated the lengths of chromosome segments of continuous specific ancestry (CSSA) (Fig. 2A), with distribution that informs how admixture occurred over time. By leveraging on the model by Liang and Nielsen (20) of CSSA, we developed an ABC framework to infer admixture dynamics (SI Appendix, section 6.8). We simulated CSSA distributions generated by a demographic history of three pulses of trihybrid admixture that occurred 18–16, 12–10, and 6–4 generations ago, conditioning on the observed current admixture proportions of each of the EPIGEN populations. This demographic model conciliates statistical complexity and the real history of admixture. We inferred the posterior distributions of nine parameters mn,P, where m is the proportion of immigrant individuals entering in the admixed population from the n ancestral population (African, European, or Native-American ancestry) in the P admixture pulse.
Fig. 2.
Distributions of lengths of chromosomal segments of (A) CSSA and (B) admixture dynamics inferences estimated for three EPIGEN Brazilian populations. (A) CSSA lengths were distributed in 50 equally spaced bins per population. Red, blue, and green dots represent a European, an African, and a Native-American CSSA, respectively. (B) We inferred the posterior densities of the proportions of immigrants (with respect to the admixed population) from each origin, and we show their 90% highest posterior density (HPD) intervals. Inferences are based on a model of three pulses of admixture (vertical axis) simulated based on the model of CSSAs evolution by Liang and Nielsen (20). Inferences are based on approximate Bayesian computation. Ancestry color codes are red for European, blue for African, and green for Native American.
Interestingly, ABC results (Fig. 2B) show that the observed low Native-American ancestry was mostly introduced in different regions of Brazil soon after the European Conquest of the Americas, which is consistent with the posterior depletion of the Native-American population in Brazil. Also, we inferred a predominantly earlier European colonization in the Northeast (Salvador) vs. a more recent immigration in Southeastern and Southern Brazil (Bambuí and Pelotas), consistent with historical records (brasil500anos.ibge.gov.br/). Conversely, African admixture showed a decreasing temporal trend shared by the three EPIGEN populations (21). Complementary explanations are continuous local immigration into the admixed populations from communities with high African ancestry already settled in Brazil [for example, quilombos (i.e., Afro-Brazilian slave-derived communities in Brazil) (22)].
Dissecting European Ancestry.
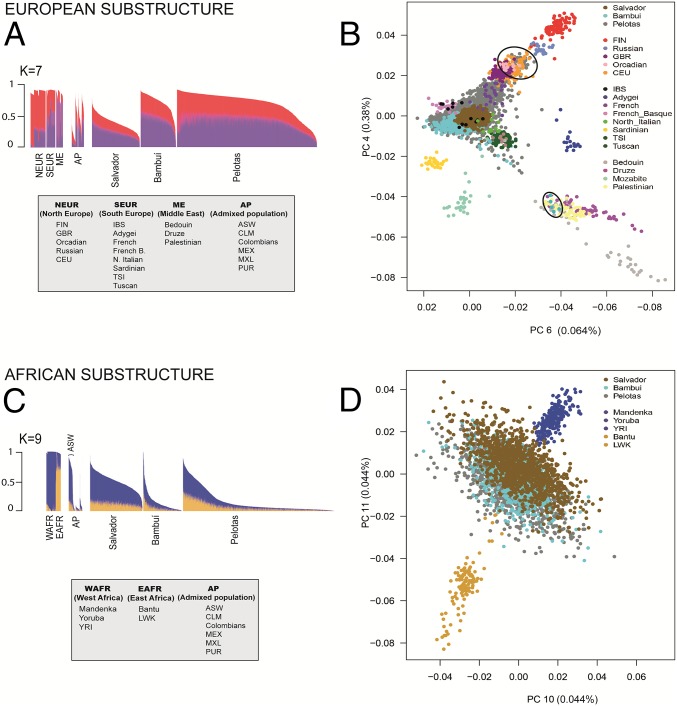
To dissect the ancestry of Brazilians at a subcontinental level, we applied (i) the ADMIXTURE method (7) by increasing the number of ancestral clusters (K) that explains the observed genetic structure (SI Appendix, Figs. S4 and S5) and (ii) the Principal Component Analysis (PCA) (23) (Figs. 1C and 3 B and D and SI Appendix, Fig. S6). To study biogeographic ancestry, we excluded sets of relatives that could affect our inferences at the within-continent level (24). We developed a method based on complex networks to reduce the relatedness of the analyzed individuals by minimizing the number of excluded individuals (SI Appendix, section 6.1). Using this method, we created the Dataset Unrelated (Dataset U), including 5,825 Brazilians, 1,780 worldwide individuals, and no pair of individuals closer than second-degree relatives. Hereafter, PCA and ADMIXTURE results are relative to Dataset U.
Fig. 3.
European and African ancestry clusters in the Brazilian populations. We show (A and C) relevant ADMIXTURE individual ancestry bar plots and (B and D) plots of principal components (PCs) that dissect ancestry within (A and B) Europe and (C and D) Africa. We performed the analyses using Dataset U (unrelated Brazilians and worldwide individuals). We only plot individuals from relevant ancestral populations. Complete ADMIXTURE and PCA results are represented in SI Appendix, section 6 and Figs. S4–S6. Black ellipses in B show some individuals from Pelotas (Southern Brazil) clustering with northern European individuals toward the top and individuals from Bambuí (Southeastern Brazil) clustering with Middle Eastern individuals toward the bottom. AP, admixed population; ASW, Americans of African ancestry in USA; CEU, Utah residents with Northern and Western European ancestry; CLM, Colombians from Medellin, Colombia; EAFR, east Africa; FIN, Finnish in Finland; French B, Basque; GBR, British in England and Scotland; IBS, Iberian population in Spain; LWK, Luhya in Webuye, Kenya; ME, Middle East; MXL/MEX, Mexican ancestry from Los Angeles; N., (North) Italian; NAT, Native American; NE, northeast; NEUR, north Europe; PUR, Puerto Ricans from Puerto Rico; S, south; SE, southeast; SEUR, south Europe; TSI, Toscani in Italia; YRI, Yoruba in Ibadan, Nigeira; WAFR, west Africa.
Brazil received several immigration waves from diverse European origins during the last five centuries (brasil500anos.ibge.gov.br/): Portuguese (the first colonizers), who also arrived in large numbers during the last 150 y; Italians (mostly to the South and Southeast); and Germans (mostly to the South). In our PCA representation (Fig. 3B), the European component of the genomes of most Brazilians is similar to individuals from the Iberian Peninsula and neighboring regions. The resemblance in within-European ancestry of individuals from Pelotas (South) and Bambuí (Southeast) to central North Europeans and Middle Easters, respectively (Fig. 3B), reflects a geographically wider European ancestry of these two populations with respect to Salvador. Considering the total European ancestry estimated by ADMIXTURE, we inferred a higher proportion of North European-associated ancestry in Pelotas (40.2%) than in Bambuí (35.8%) and Salvador (36.7%; P < 10−15; Wilcoxon tests) (Fig. 3A, red cluster in K = 7). We confirmed these results by analyzing a reduced number of SNPs with a larger set of European individuals and populations (25, 26) (SI Appendix, section 6.2).
Brazil, the Main Destination of the African Diaspora.
African slaves arrived to Brazil during four centuries, whereas most arrivals to the United States occurred along two centuries, and the geographic and ethnic origin of Brazilian slaves differ from Caribbeans and African Americans (27). In fact, the Portuguese Crown imported slaves to Brazil from western and central west Africa (the two are the major sources of the slave trade to all of the Americas) as well as Mozambique. We detected two within-Africa ancestry clusters in the current Brazilian population (Fig. 3C, K = 9 and SI Appendix, section 6.3): one associated with the Yoruba/Mandenka non-Bantu western populations (Fig. 3C, blue) and one associated with the Luhya/HGDP (Human Genome Diversity Project) Bantu populations from eastern Africa (Fig. 3C, mustard). Interestingly, the proportions of these ancestry clusters, which are present across all of the analyzed African and Latin-American populations, differ across them. The blue cluster in Fig. 3C predominates in African Americans and in Salvador, accounting for 83% and 75% of the total African ancestry, respectively (against 17% and 25%, respectively, of the mustard cluster in Fig. 3C) (SI Appendix, Table S17). Comparatively, the mustard cluster in Fig. 3C is more evident in Southeastern and Southern Brazil (36% and 44% of African ancestry in Bambuí and Pelotas, respectively). These results are consistent with the fact that a large proportion of Yoruba slaves arrived in Salvador, whereas the Mozambican Bantu slaves disembarked primarily in Rio de Janeiro in Southeastern Brazil (21). These results show for the first time, to our knowledge, that the genetic structure of Latin Americans reflects a more diversified origin of the African diaspora into the continent. Interestingly, the two within-African ancestry clusters in the Brazilian populations (showing an average FST of 0.02) are characterized by 3,318 SNPs, with the 10% top FST values higher than 0.06, and include 38 SNPs that are hits of genome-wide association studies (SI Appendix, section 7 and Table S25).
Pattern of Deleterious Variants: Effect of Continental Admixture, Kinship, and Inbreeding.
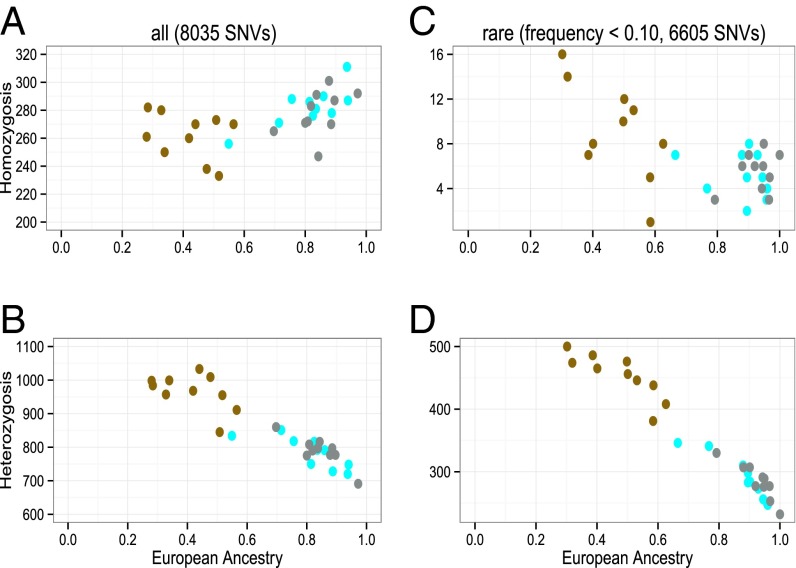
Based on whole-genome data from 30 individuals (10 from each of three EPIGEN populations), we identified putative deleterious nonsynonymous variants (28) (SI Appendix, section 8). There are recent interest in and apparently conflicting results on whether Europeans have proportionally more deleterious variants in homozygosis than Africans (29–32). Lohmueller et al. (29) explained these differences as an effect of the Out of Africa bottleneck on current non-African populations. Out of Africa would have enhanced the effect of genetic drift and attenuated the effect of purifying natural selection, preventing, in many instances, the extinction of (mostly weakly) deleterious variants in non-Africans.
We investigated how European ancestry shapes the amount of deleterious variants in homozygosis (a more likely genotype for common/weakly deleterious variants) and heterozygosis in admixed Latin-American individuals. We observed three patterns (Fig. 4). (i) Considering all (i.e., weakly and highly) deleterious variants, for a class of individuals with high European ancestry (>65%; from Bambuí and Pelotas), the individual number of deleterious variants in homozygosis is correlated with European ancestry, but importantly, this correlation is not observed among individuals with intermediate European ancestry (from Salvador) (Fig. 4A). (ii) The individual number of deleterious variants (both all and rare classes) in heterozygosis (Fig. 4 B and D) decreases linearly with European ancestry, regardless the cohort of origin. This result is also observed for rare deleterious variants in homozygosis, although the pattern is not very clear in this case (Fig. 4C). (iii) There are no differences in the amount of deleterious variants between individuals from Bambuí and Pelotas. These populations have similar continental admixture proportions and dynamics, but different post-Columbian population sizes and histories of isolation, assortative mating, kinship structure, and inbreeding. Taken together, our results are consistent with the results and evolutionary scenario proposed by Lohmueller et al. (29) and Lohmueller (31), and suggest that, in Latin-American populations, the main determinant of the amount of deleterious variants is the history of continental admixture, although in a more complex fashion than previously thought (pattern i). Comparatively, the role of local demographic history seems less relevant.
Fig. 4.
Individual numbers of genotypes with nonsynonymous deleterious variants in homozygosis and heterozygosis vs. European ancestry based on the whole-genome sequence (42×) of 30 individuals (10 from each population): Salvador (Northeast; brown), Bambuí (Southeast; cyan), and Pelotas (South; gray). Deleterious variants were identified using CONDEL (28) and corrected for the bias reported by Simons et al. (30). Spearman correlation between European ancestry and the number of all deleterious variants in homozygosis for Bambuí and Pelotas individuals was 0.57 (P = 0.009). The numbers of genotypes considering all deleterious variants in homozygosis or heterozygosis are in A and B, respectively, and considering only rare deleterious variants are in C (in homozygosis) and D (in heterozygosis). SNVs, single nucleotide variants.
Conclusion
A thread of historical facts has modeled the genetic structure of Brazilians. Our population-based and fine-scale analyses revealed novel aspects of the genetic structure of Brazilians. In 1870, blacks were the major ethnic group in Brazil (21), but this scenario changed after the arrival of nearly 4 million Europeans during the second one-half of the 19th century and the first one-half of the 20th century. This immigration wave was encouraged by Brazilian officials as a way of “whiting” the population (33), and it transformed Brazil into a predominantly white country, particularly in the Southeast and South. Consistently, (i) we observed that larger chromosomal segments of continuous European ancestry in the southeast/south are the signature of this recent European immigration, and (ii) we traced the European ancestry in the Southeast/South of Brazil to a wider geographical region (including central northern Europe and the Middle East) than in Salvador (more restricted to the Iberian Peninsula). However, neither this massive immigration nor the internal migration of black Brazilians have concealed two components of their African ancestry from the genetic structure of Brazilians: one associated with the Yoruba/Mandenka non-Bantu populations, which is more evident in the Northeast (Salvador), and one associated with central east African/Bantu populations, which is more present in the Southeast/South. This result broadens our understanding of the genetic structure of the African diaspora. Furthermore, we showed that positive assortative mating by ancestry is a social factor that permeates the demographic history of Brazilians and also, shapes their genetic structure, with implications for the design of genetic association studies in admixed populations. For instance, because mating by ancestry produces Hardy–Weinberg disequilibrium, filtering SNPs for genome-wide association studies based on the Hardy–Weinberg equilibrium conceals real aspects of the genetic structure of these populations. Finally, in Latin-American populations, the history of continental admixture rather than local demographic history is the main determinant of the burden of deleterious variants, although in a more complex fashion than previously thought. We speculate that future studies on populations from Northern Brazil (including large cities, such as Manaus, next to the Amazon forest) or the Central-West may reveal larger and different dynamics of Amerindian ancestry. Also, fine-scale studies on large urban centers from the Southeast and South of Brazil, such as Rio de Janeiro or Sao Paulo, that have been the destination of migrants from all over the country during the last decades, may show an even more diversified origin of Brazilians, including Japanese ancestry components, for instance, that we did not identify in our study. The EPIGEN Brazil initiative is currently conducting studies to clarify how the genetic variation and admixture interact with environmental and social factors to shape the susceptibility to complex phenotypes and diseases in the Brazilian populations.
Methods
Genotyping and Data Curation.
Genotyping was performed by the Illumina facility using the HumanOmni2.5–8v1 array for 6,504 individuals and the HumanOmni5-4v1 array for 270 individuals (90 randomly selected from each cohort). After that, we performed quality control analysis of the data using Genome Studio (Illumina), PLINK (34), GLU (code.google.com/p/glu-genetics/), Eigenstrat (35), and in-house scripts. This study was approved by the Brazilian National Research Ethics Committee (CONEP, resolution 15895).
Whole-Genome Sequencing and Functional Annotation.
We randomly selected 10 individuals from each of the three EPIGEN populations. The Illumina facility performed whole-genome sequencing of these individuals from paired-end libraries using the Hiseq 2000 Illumina platform. CASAVA v.1.9 modules were used to align reads and call SNPs and small INDELs (insertion or deletion of bases). Each genome was sequenced, on average, 42 times, with the following quality control parameters: 128 Gb (Gigabase) of passing filter aligned to the reference genome (HumanNCBI37_UCSC), 82% of bases with data quality (QScore) ≥30, 96% of non-N reference bases with a coverage ≥10×, a HumanOmni5 array agreement of 99.53%, and a HumanOmni2.5 array agreement of 99.27%. Functional annotation was performed with ANNOVAR (August 2013 release) with the refGene v.hg19_20131113 reference database in April of 2014. The nonsynonymous variants were predicted to be deleterious using CONDEL v2.0 (cutoff = 0.522) (28), which calculates a consensus score based on MutationAssessor (36) and FatHMM (37). These results were corrected for the bias reported in the work by Simons et al. (30), which evidenced that, when the human reference allele is the derived one, methods that infer deleterious variants tend to underestimate its deleterious effect (SI Appendix, section 8).
Relatedness and Inbreeding Analysis.
We estimated the kinship coefficients for each possible pair of individuals from each of the EPIGEN populations using the method implemented in the Relatedness Estimation in Admixed Populations (REAP) software (13). It estimates kinship coefficients solely based on genetic data, taking into account the individual ancestry proportion from K parental populations and the K parental populations allele frequencies per each SNP. For these analyses, we calculated individual ancestry proportion and K parental populations allele frequencies per each SNP using the ADMIXTURE software (7) in unsupervised mode assuming three parental populations (K = 3). Inbreeding coefficients were also estimated for each individual using REAP. We represented families by networks, which were defined as groups of individuals (vertices) linked by kinship coefficient higher than 0.1 (edges).
F Statistics.
The FIS statistic for each population is estimated as the average of the REAP inbreeding coefficients across individuals. For each SNP i and each population, we estimated the departure from Hardy–Weinberg equilibrium as FIT(i) = (Hei − Hoi)/Hei, where Hoi and Hei are the observed and the expected heterozygosities under Hardy–Weinberg equilibrium for the SNP i, respectively. We estimated the population FIT by averaging FIT(i) across SNPs. We estimated the FST for each SNP between the YRI and CEU populations using the R package hierfstat (38). The correlation between YRI vs. CEU FST and FIT values for each SNP was calculated by the Spearman's rank correlation-ρ using the R cor.test function.
Population Structure Analyses.
To study population structure, we applied (i) the ADMIXTURE method (7), increasing the number of ancestral clusters (K) that explains the observed genetic structure from K = 3, and (ii) PCA (35) (Figs. 1C and 3 and SI Appendix, section 6 and Figs. S4–S6). To study biogeographic ancestry, we have to exclude sets of relatives that could affect our inferences at within-continental level (24). We conceived and applied a method based on complex networks to reduce the relatedness of the analyzed individuals by minimizing the number of excluded individuals (SI Appendix, section 6.1). Applying this method, we created Dataset U, with 5,825 Brazilians, 1,780 worldwide individuals, and no pairs of individuals closer than second-degree relatives (REAP kinship coefficient >0.10) (SI Appendix, Table S13). We performed ADMIXTURE analyses with both the Original Dataset and Dataset U (SI Appendix, section 6 and Figs. S4 and S5).
PCA and ADMIXTURE analyses were performed with integrated datasets comprising the three cohort-specific EPIGEN working datasets and the public datasets populations described in SI Appendix, section 5. For the PCA and ADMIXTURE analyses, we used the SNPs shared by all of these populations, comprising a total of 8,267 samples and 331,790 autosomal SNPs (called the Original Dataset).
Analyses with X-chromosome data used only female samples from the Original Dataset. To perform such analyses, we integrated genotype data of shared SNPs from the X chromosome of EPIGEN female samples (from all three cohorts) and the X chromosome of female samples from the public datasets populations described in SI Appendix, section 5. This data integration yielded genotyping data with 5,792 SNPs for 4,192 females.
Local Ancestry Analyses.
We inferred chromosome local ancestry using the PCAdmix software (19) and ∼2 million SNPs shared by EPIGEN (Original Dataset) and the 1000 Genomes Project (SI Appendix, section 5.2). Considering our SNPs density, we defined a window length of 100 SNPs following the work by Moreno-Estrada et al. (27). PCAdmix infers the ancestry of each window. Local ancestry inferences were performed after linked markers (r2 > 0.99) were pruned to avoid ancestry misestimating caused by overfitting (4). We considered only the windows in which ancestry was inferred by the forward–backward algorithm with a posterior probability >0.90.
After local ancestry inferences, we calculated the lengths of the chromosomal segments of CSSA for each haplotype from each chromosome from each individual. The distribution of CSSA length was organized in 50 equally spaced bins defined in centimorgans and plotted for each population (Fig. 2A).
For the local ancestry analyses, we used phased data from the 1000 Genomes Project populations YRI and LWK (Africans) as well as CEU, FIN, GBR, TSI, and IBS (Europeans), Native-American populations Ashaninka and Shimaa [from the Tarazona–Santos group LDGH (Laboratory of Human Genetic Diversity) dataset], and the three EPIGEN populations (Original Dataset). The SHAPEIT software (39) was used to generate phased datasets.
We estimated admixture dynamics parameters using ABC. We used the model by Liang and Nielsen (20) to simulate CSSA distributions generated by a demographic history of three pulses of trihybrid admixture occurring 18–16, 12–10, and 6–4 recent generations ago conditioned on the observed admixture proportions of the EPIGEN populations. We inferred the posterior distributions of nine parameters mn,P (SI Appendix, section 6.8).
Lineage Markers Haplogroups Inferences.
We performed mtDNA haplogroup assignments using HaploGrep (40), a web tool based on Phylotree (build 16) for mtDNAhaplogroup assignment. For Y-chromosome data, we inferred haplogroups using an automated approach called AMY tree (41). For Y-chromosome haplogroups, we considered the Karafet tree (42) and more recent studies to describe additional subhaplogroups. By these means, an updated tree was considered based on the information given by The International Society of Genetic Genealogy (ISOGG version 9.43; www.isogg.org).
Supplementary Material
Acknowledgments
The authors thank David Alexander and Fernando Levi Soares for technical help and discussion and Rasmus Nielsen and Mason Liang for sharing their software for continuous specific ancestry simulations and feedback on its use. Centro Nacional de Processamento de Alto Desempenho em MG/Financiadora de Estudos e Projetos–Ministério da Ciência, Tecnologia e Inovação, Centro Nacional de Super Computação, and Programa de Desenvolvimento Tecnológico em Insumos para Saúde-Bioinformatics Platform at Fundação Oswaldo Cruz-Minas Gerais provided computational support. The EPIGEN Brazil Initiative is funded by the Brazilian Ministry of Health (Department of Science and Technology from the Secretaria de Ciência, Tecnologia e Insumos Estratégicos) through Financiadora de Estudos e Projetos. The EPIGEN Brazil investigators received funding from the Brazilian Ministry of Education (CAPES Agency), Brazilian National Research Council (CNPq), Pró-Reitoria de Pesquisa from the Universidade Federal de Minas Gerais, and the Minas Gerais State Agency for Support of Research (FAPEMIG).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (PRJEB9080 (ERP010139) Genomic Epidemiology of Complex Diseases in Population-Based Brazilian Cohorts), accession no. EGAS00001001245, under EPIGEN Committee Controlled Access mode.
4A complete list of the Brazilian EPIGEN Project Consortium can be found in SI Appendix.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1504447112/-/DCSupplemental.
Contributor Information
Collaborators: Neuza Alcantara-Neves, Nathalia M. Araújo, Márcio L.B. Carvalho, Jackson Santos Conceição, Josélia O.A. Firmo, Denise P. Gigante, Lindolfo Meira, Thais Muniz-Queiroz, Guilherme C. Oliveira, Isabel O. Oliveira, Sérgio V. Peixoto, Fernando A. Proietti, Domingos C. Rodrigues, Meddly L. Santolalla, Agostino Strina, and Camila Zolini
References
- 1.Salzano FM, Freire-Maia N. Populações Brasileiras; Aspectos Demográficos, Genéticos e Antropológicos. Companhia Editora Nacional; São Paulo, Brazil: 1967. [Google Scholar]
- 2.Giolo SR, et al. Brazilian urban population genetic structure reveals a high degree of admixture. Eur J Hum Genet. 2012;20(1):111–116. doi: 10.1038/ejhg.2011.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno-Estrada A, et al. Human genetics. The genetics of Mexico recapitulates Native American substructure and affects biomedical traits. Science. 2014;344(6189):1280–1285. doi: 10.1126/science.1251688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eyheramendy S, Martinez FI, Manevy F, Vial C, Repetto GM. Genetic structure characterization of Chileans reflects historical immigration patterns. Nat Commun. 2015;6:6472. doi: 10.1038/ncomms7472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreto ML, et al. Risk factors and immunological pathways for asthma and other allergic diseases in children: Background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (Salvador-SCAALA study) BMC Pulm Med. 2006;6:15. doi: 10.1186/1471-2466-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacelar J. A Hierarquia sas Raças. Negros e Brancos em Salvador. Pallas Editora; Rio de Janeiro: 2001. [Google Scholar]
- 7.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryc K, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA. 2010;107(2):786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lima-Costa MF, Firmo JO, Uchoa E. Cohort profile: The Bambui (Brazil) Cohort Study of Ageing. Int J Epidemiol. 2011;40(4):862–867. doi: 10.1093/ije/dyq143. [DOI] [PubMed] [Google Scholar]
- 11.Victora CG, Barros FC. Cohort profile: The 1982 Pelotas (Brazil) birth cohort study. Int J Epidemiol. 2006;35(2):237–242. doi: 10.1093/ije/dyi290. [DOI] [PubMed] [Google Scholar]
- 12.Salzano FM, Bortolini MC. The Evolution and Genetics of Latin American Populations. Cambridge Univ Press; New York: 2002. [Google Scholar]
- 13.Thornton T, et al. Estimating kinship in admixed populations. Am J Hum Genet. 2012;91(1):122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bittles AH. Endogamy, consanguinity and community genetics. J Genet. 2002;81(3):91–98. doi: 10.1007/BF02715905. [DOI] [PubMed] [Google Scholar]
- 15.Telles EE. Race in Another América: The Significance of Skin Color in Brazil. Princeton Univ Press; Princeton: 2006. [Google Scholar]
- 16.Lima-Costa MF, et al. Epigen-Brazil group Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative) Sci Rep. 2015;5:9812. doi: 10.1038/srep09812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruiz-Linares A, et al. Admixture in Latin America: Geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 2014;10(9):e1004572. doi: 10.1371/journal.pgen.1004572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Risch N, et al. Ancestry-related assortative mating in Latino populations. Genome Biol. 2009;10(11):R132. doi: 10.1186/gb-2009-10-11-r132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brisbin A, et al. PCAdmix: Principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 2012;84(4):343–364. doi: 10.3378/027.084.0401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang M, Nielsen R. The lengths of admixture tracts. Genetics. 2014;197(3):953–967. doi: 10.1534/genetics.114.162362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klein HS. Homo brasilis Aspectos Genéticos, Lingüísticos, Históricos e Socioantropológicos da Formação do Povo Brasileiro. 2nd Ed. FUNPEC-RP; Ribeirão Preto, Brasil: 2002. pp. 93–112. [Google Scholar]
- 22.Scliar MO, Vaintraub MT, Vaintraub PM, Fonseca CG. Brief communication: Admixture analysis with forensic microsatellites in Minas Gerais, Brazil: The ongoing evolution of the capital and of an African-derived community. Am J Phys Anthropol. 2009;139(4):591–595. doi: 10.1002/ajpa.21046. [DOI] [PubMed] [Google Scholar]
- 23.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11(7):459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson MR, et al. The Population Reference Sample, POPRES: A resource for population, disease, and pharmacological genetics research. Am J Hum Genet. 2008;83(3):347–358. doi: 10.1016/j.ajhg.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Botigué LR, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci USA. 2013;110(29):11791–11796. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreno-Estrada A, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9(11):e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González-Pérez A, López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88(4):440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmueller KE, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451(7181):994–997. doi: 10.1038/nature06611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46(3):220–224. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lohmueller KE. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Do R, et al. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet. 2015;47(2):126–131. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pena SD, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS ONE. 2011;6(2):e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Reva B, Antipin Y, Sander C. Determinants of protein function revealed by combinatorial entropy optimization. Genome Biol. 2007;8(11):R232. doi: 10.1186/gb-2007-8-11-r232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shihab HA, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34(1):57–65. doi: 10.1002/humu.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goudet J. Hierfstat, a package for r to compute and test hierarchical F-statistics. Mol Ecol Notes. 2005;5(1):184–186. [Google Scholar]
- 39.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 40.Kloss-Brandstätter A, et al. HaploGrep: A fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32(1):25–32. doi: 10.1002/humu.21382. [DOI] [PubMed] [Google Scholar]
- 41.Van Geystelen A, Decorte R, Larmuseau MHD. AMY-tree: An algorithm to use whole genome SNP calling for Y chromosomal phylogenetic applications. BMC Genomics. 2013;14(14):101–112. doi: 10.1186/1471-2164-14-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karafet TM, et al. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18(5):830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.