ABSTRACT
Thermococcus kodakarensis grows optimally at 85°C and possesses two chaperonins, cold-inducible CpkA and heat-inducible CpkB, which are involved in adaptation to low and high temperatures, respectively. The two chaperonins share a high sequence identity (77%), except in their C-terminal regions. CpkA, which contains tandem repeats of a GGM motif, shows its highest ATPase activity at 60°C to 70°C, whereas CpkB shows its highest activity at temperatures higher than 90°C. To clarify the effects of changes in ATPase activity on chaperonin function at lower temperatures, various CpkA variants were constructed by introducing single point mutations into the C-terminal region. A CpkA variant in which Glu530 was replaced with Gly (CpkA-E530G) showed increased ATPase activity, with its highest activity at 50°C. The efficacy of the CpkA variants against denatured indole-3-glycerol-phosphate synthase of T. kodakarensis (TrpCTk), which is a CpkA target, was then examined in vitro. CpkA-E530G was more effective than wild-type CpkA at facilitating the refolding of chemically unfolded TrpCTk at 50°C. The effect of cpkA-E530G on cell growth was then examined by introducing cpkA-E530G into the genome of T. kodakarensis KU216 (pyrF). The mutant strain, DA4 (pyrF cpkA-E530G), grew as well as the parental KU216 strain at 60°C. In contrast, DA4 grew more vigorously than KU216 at 50°C. These results suggested that the CpkA-E530G mutation prevented cold denaturation of proteins under cold-stress conditions, thereby enabling cells to grow in cooler environments. Thus, a single base pair substitution in a chaperonin gene allows cells to grow vigorously in a new environment.
IMPORTANCE Thermococcus kodakarensis possesses two group II chaperonins, cold-inducible CpkA and heat-inducible CpkB, which are involved in adaptation to low and high temperatures, respectively. CpkA might act as an “adaptive allele” to adapt to cooler environments. In this study, we compared the last 20 amino acids within the C termini of the chaperonins and found a clear correlation between the CpkA-type chaperonin gene copy number and growth temperature. Furthermore, we introduced single mutations into the CpkA C-terminal region to clarify its role in cold adaptation, and we showed that a single base substitution allowed the organism to adapt to a lower temperature. The present data suggest that hyperthermophiles have evolved by obtaining mutations in chaperonins that allow them to adapt to a colder environment.
INTRODUCTION
Chaperonin, also known as heat shock protein 60 (HSP60), belongs to an evolutionarily conserved protein family that enables host cells to survive under stressful conditions, including restricted temperature, high/low salinity, and high/low hyperosmotic pressure. Chaperonins are classified into two subfamilies, group I and group II, on the basis of sequence homology and structural differences (1). The thermosome is a group II chaperonin found in hyperthermophilic archaea, in which it plays a key role in thermal adaptation. Unlike bacterial chaperonins, which function as homotetradecamers, archaeal chaperonins form a heterohexadecamer comprising two back-to-back rings; each of these rings comprises eight heterogeneous chaperonin subunits. Like bacterial chaperonins, archaeal chaperonin monomers comprise three domains: the apical domain (which binds unfolded proteins), the intermediate domain (which acts as a hinge, thereby allowing the movement of the apical domain as well as the transition between trans and cis conformations needed for chaperonin function), and the equatorial domain (which is responsible for the ATPase and refolding activities in the central cavity of the ringed complex). However, thermosomes do not have cochaperonins, such as GroES. Rather, there is a conserved protrusion from the apical domain, which may play a role in guiding unfolded proteins. Phylogenetic analyses revealed that bacterial chaperonin genes have undergone many rounds of duplication to generate these diverse structures (2). The hyperthermophilic archaeon Thermococcus kodakarensis possesses two gene copies of a chaperonin: cpkA and cpkB (3, 4). CpkB is a classic thermosome, which is an essential heat-inducible factor that enables the cells to survive at high temperatures (>85°C). A relative of T. kodakarensis, Pyrococcus sp., has only one chaperonin, which is an orthologue of CpkB; therefore, this organism has a narrow but high growth temperature range (80 to 100°C). However, CpkA is an atypical archaeal group II chaperonin, which is cold inducible at the stationary phase of cell growth and is not found in Pyrococcus sp. A cpkA disruptant strain showed poor cell growth at 60°C but no significant growth defect at 85°C and 93°C (5). T. kodakarensis indole-3-glycerol-phosphate synthase (TrpCTk or InGPS) was identified as one of the specific target proteins for CpkA in T. kodakarensis. The refolding of partially cold-denatured TrpCTk was accelerated by the addition of CpkA but not by adding CpkB (6).
CpkA and CpkB are highly homologous (77%), except for their C-terminal regions. The C-terminal region of CpkA harbors tandem repeats of a GGM motif. Such coding tandem repeats are usually involved in protein-protein interactions (7). For example, in Escherichia coli, the C-terminal region of GroEL (which is also rich in GGM motifs) is involved in ATP hydrolysis and protein binding (8–12). Langer et al. found that removing the 50 C-terminal amino acid residues of GroEL resulted in a loss of ATPase activity (9). McLennan et al. found that although the 16 C-terminal residues of GroEL were dispensable for E. coli cell growth, the truncated protein could not complement the groEL(Ts) mutation in E. coli (13). Machida et al. reported that the 23 C-terminal residues of GroEL (which are hydrophilic) were not required for chaperonin function (10). However, when the last 6 residues (526-KNDAAD-531) were replaced by a neutral (neither hydrophobic nor hydrophilic) sequence (526-GGGAAG-531) or a hydrophobic sequence (526-IGIAAI-531), chaperonin function was defective both in vitro and in vivo. Suzuki et al. used a special microscopic method, called zero-mode waveguides, to show that the integrity of the C terminus of the chaperonin region facilitates the transition from the first to the second rate-limiting step (14).
Studies of the C-terminal regions of group II chaperonins have just begun. Luo and Robb mutated the 15 to 25 C-terminal residues of the Pyrococcus furiosus chaperonin, which is highly homologous to CpkB, in an attempt to modify its thermostability and activity (15). An EK-rich motif (528-EKEKEKEGEK-537) was the key domain responsible for increased stability at around 100°C. Zhang et al. found that flexible interwoven termini determined the thermal stability of thermosomes, especially in the β subunit, which has more charged amino acids than the α subunit (16). However, the function of the CpkA C-terminal region remains unknown. In the present study, we found a clear correlation between the CpkA-type chaperonin gene copy number and growth temperature. We introduced single mutations into the CpkA C-terminal region to increase the number of GGM motifs to clarify its role during the evolution of T. kodakarensis.
MATERIALS AND METHODS
Microorganisms, media, and growth conditions.
The microorganisms (strains) used in this study are listed in Table 1. T. kodakarensis strains KU216 (ΔpyrF) and DA4 (ΔpyrF; carries a cpkA variant which encodes Gly in place of Glu530 [cpkA-E530G]) were precultivated anaerobically overnight (18 h) at 85°C in 100 ml of MA-YT medium containing the following (per liter): 30.4 g of Marine Art SF reagent, 5 g of yeast extract (Nacalai Tesque, Kyoto, Japan), 5 g of tryptone (Nacalai Tesque), and 5 g of pyruvate (Wako Pure Chemical Industries, Ltd., Osaka, Japan). The culture was added to 20 ml of MA-2YTP medium (30.4 g/liter Marine Art SF reagent, 10 g/liter yeast extract, 10 g/liter tryptone, and 10 g/liter pyruvate) and incubated at a given temperature. All growth studies were carried out three times. For solid medium, 1% Gelrite (Wako, Osaka, Japan) was mixed with 2 ml/liter polysulfide solution (10 g of Na2S · 9H2O and 3 g of sulfur flowers in 15 ml of H2O). E. coli DH5α strains were routinely cultivated at 37°C in lysogeny broth (LB) medium. Ampicillin (50 μg/ml) was added to the medium to select transformants.
TABLE 1.
Strains and plasmids used for this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) phoA supE44 λ− thi-1 gyrA96 relA1 | |
| BL21(DE3)pLysE | F− ompT hsdSB(rB− mB−) gal dcm (DE3)pLysE Camr | |
| T. kodakarensis | ||
| KU216 | pyrF | 24 |
| DA4 | pyrF cpkA-E530G | This study |
| Plasmids | ||
| pUD2-cpkAfr | Plasmid used to replace the cpkA gene in T. kodakarensis | 5 |
| pUD2-cpkA-E530G | pUD2-cpkAfr derivative; cpkA-E530G | This study |
| pCPAE | Expression vector for cpkA in E. coli | 20 |
| pCPAE-E530G | pCPAE derivative; carries cpkA-E530G mutation; expression vector for E. coli | This study |
| pCPAE-E530M | pCPAE derivative; carries cpkA-E530M mutation; expression vector for E. coli | This study |
| pCPAE-Q533G | pCPAE derivative; carries cpkA-Q533G mutation; expression vector for E. coli | This study |
| pCPAE-Q533M | pCPAE derivative; carries cpkA-Q533M mutation; expression vector for E. coli | This study |
| pCPAE-P538G | pCPAE derivative; carries cpkA-P538G mutation; expression vector for E. coli | This study |
| pCPAE-P538M | pCPAE derivative; carries cpkA-P538M mutation; expression vector for E. coli | This study |
| pCPAE-D545G | pCPAE derivative; carries cpkA-D545G mutation; expression vector for E. coli | This study |
| pCPAE-D545M | pCPAE derivative; carries cpkA-D545M mutation; expression vector for E. coli | This study |
Alignment of protein sequences and modeling of protein structure.
All chaperonin (amino acid) sequences used for sequence alignment were identified and retrieved from the public National Center for Biotechnology Information (NCBI) database (17). The 20 C-terminal amino acids of each chaperonin were then aligned using ClustalW2, supported by the European Bioinformatics Institute.
A three-dimensional (3D) structural octamer of full-length CpkA was built in MODELLER 9.12 (18), using the chaperonin α subunit of Thermococcus strain KS-1 (PDB identifier [ID] 1Q2V) as the template. The protein sequence of CpkA was retrieved from the NCBI sequence database. The construction generated three models, and the one with the lowest discrete optimized protein energy score was selected.
DNA manipulation.
DNA was isolated and handled according to the standard techniques described by Sambrook and Russell (19). A small-scale preparation of plasmid DNA was prepared from E. coli cells by using a GenElute plasmid miniprep kit (Sigma-Aldrich, St. Louis, MO). DNA sequencing was performed using a BigDye Terminator cycle sequencing ready reaction kit, version 3.1, and a model 3130 capillary DNA sequencer (Applied Biosystems, CA). The plasmids and primers used are listed in Tables 1 and 2, respectively.
TABLE 2.
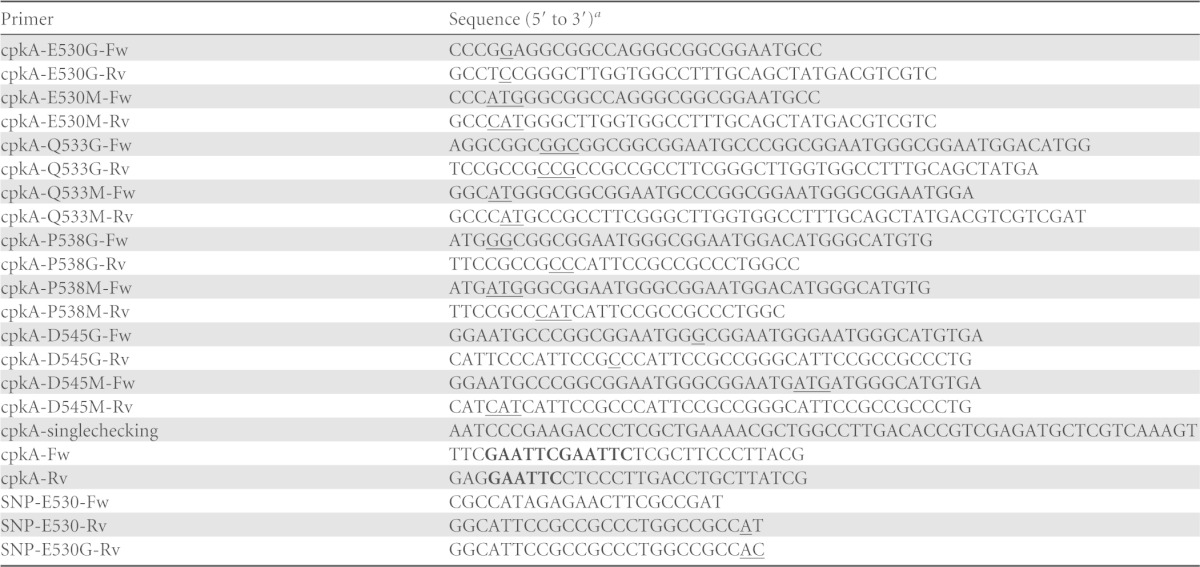
The mutated nucleotides within the cpkA gene are indicated by underlining. Introduced restriction sites are indicated in bold.
Construction of expression plasmids encoding cpkA variants.
pCPAE (20) was used as a template. Briefly, expression plasmids for the cpkA variants (encoding E530G, E530M, Q533G, Q533M, P538G, P538M, D545G, and D545M mutations) were generated by site-directed mutagenesis. To construct pCPAE-E530G, PCR was performed using primers cpkA-E530G-Fw and cpkA-E530G-Rv and the amplified DNA was ligated into pCPAE. E. coli DH5α cells were transformed with the ligated DNA, and plasmid DNA was extracted from positive transformants. Plasmid construction was confirmed by nucleotide sequencing with the primer cpkA-singlechecking (Table 2). Mutant plasmids pCPAE-E530M, -Q533G, -Q533M, -P538G, -P538M, -D545G, and -D545M were constructed using primers cpkA-E530M-Fw and cpkA-E530M-Rv, cpkA-Q533G-Fw and cpkA-Q533G-Rv, cpkA-Q533M-Fw and cpkA-Q533M-Rv, cpkA-P538G-Fw and cpkA-P538G-Rv, cpkA-P538M-Fw and cpkA-P538M-Rv, cpkA-D545G-Fw and cpkA-D545G-Rv, and cpkA-D545M-Fw and cpkA-D545M-Rv, respectively. The plasmids were then transformed into E. coli BL21(DE3) RIL cells.
Protein expression and purification.
Recombinant CpkA and its variants were expressed in E. coli BL21(DE3) RIL at 25°C as previously described (20). CpkA and its variants remained in the soluble fraction after heat treatment at 85°C for 30 min. Further purification was carried out by anion-exchange chromatography (MonoQ HR 5/5; GE Healthcare, Tokyo, Japan) followed by gel filtration on a Superdex 200 column (GE Healthcare). Recombinant TrpCTk (T. kodakarensis indole-3-glycerol-phosphate synthase; InGPS) was expressed and purified as previously described (21).
Thermostability of CpkA and its variants.
The secondary structures of chaperonins were measured using a far-UV circular dichroism (CD) spectroscopy method in wavelength scan mode (220 nm to 260 nm) at 20°C in a J-820 spectrometer (Jasco, Japan), as described previously (6). The thermostability of CpkA and its variants was measured by far-UV CD spectroscopy using a J-820 spectrometer (Jasco, Japan) set in temperature scan mode. Chaperonins were dissolved in 25 mM Tris-HCl (pH 7.8) buffer containing 300 mM KCl (final concentration, 50 μg ml−1), and the change in absorption at 222 nm was monitored from 30 to 100°C. The temperature scan rate was 1°C min−1.
Enzyme assay to examine ATPase activity.
ATPase activity was measured by monitoring the amount of phosphate released from ATP by chaperonins. The temperature dependency of the ATPase activity of CpkA and its variants was examined by incubating each chaperonin at 80 nM in HKM buffer (25 mM HEPES-NaOH, 100 mM KCl, 5 mM MgCl2, pH 7.5) at different temperatures (20, 40, 60, 80, and 93°C). The reaction mixtures were preheated to the test temperature for 5 min prior to the addition of 2 mM ATP to trigger the reaction. After 10 min, the reaction was terminated by cooling on ice, and the amount of released phosphate was measured using a Biomol Green kit (Enzo Life Science, NY) (22, 23). One unit of ATPase activity was defined as the amount of enzyme that liberated 1 nmol of inorganic phosphate from ATP in 1 min.
Examination of chaperonin activity.
Purified TrpCTk was chemically unfolded and used as a substrate for chaperonins in a refolding assay as previously described (6). TrpCTk (16 μM) was denatured on ice for 3 days in 50 mM Tris-HCl buffer (pH 8.0) containing 7 M urea. Next, 2 μl of unfolded TrpCTk (8 μM) was added to 198 μl of HKM buffer (100-fold dilution) containing 2 mM ATP in the presence or absence of wild-type or variant CpkA. The solutions were incubated for 30 min at 50 or 60°C, and the refolding rates were determined by monitoring the amount of InGPS activity at 50 or 60°C as previously described (6). One unit of InGPS activity was defined as the amount of activity that resulted in the formation of 1 μmol of InGPS in 1 min.
Plasmids used to construct mutant strain DA4.
The gene disruption strategy has been described previously (24). Briefly, plasmid pUD2-cpkA-E530G was first constructed by amplifying PCR fragments from the pUD2-cpkAfr plasmid by using primers cpkA-E530G-Fw and cpkA-E530G-Rv. The ligated PCR products were then used to transform E. coli DH5α cells. Plasmid DNA was extracted from transformants, and the nucleotide sequence of the cpkA region was confirmed by sequencing using the primer cpkA-singlechecking (Table 2). The resulting plasmid was named pUD2-cpkA-E530G and used to construct mutant strain DA4.
Transformation of T. kodakarensis.
To construct mutant strain DA4 (pyrF cpkA-E530G), KU216 was used as the parental strain, and plasmid pUD2-cpkA-E530G was introduced into this strain. Transformation was performed as described in our previous work (6). Genotypes of positive candidates were confirmed by sequencing. The correctly constructed mutant was named strain DA4.
Growth profiles of DA4 and KU216.
The growth of strains DA4 and KU216 was monitored by measuring the optical density at 600 nm (OD660) in an automatic Bioshaker (BR-43FH; Taitec, Japan). Cells were first precultured overnight (18 h) at 85°C in MA-YT medium followed by culture in MA-2YTP medium, with a 1% inoculation ratio. The cells were then cultured at the indicated test temperature (50, 60, or 85°C) for several days.
Competitive cell growth assay.
KU216 and DA4 were grown in MA-YT medium in separate cultures overnight (18 h) at 85°C. The two strains were then inoculated into fresh MA-2YTP medium (20 ml) at KU216/DA4 ratios of 1:1, 10:1, 102:1, and 104:1. The mixtures were then cultured at 50°C for 2 days. Each mixed culture was then inoculated into another 20 ml of fresh MA-2YTP medium and incubated at 50°C for another 2 days. Finally, the cells were spread on the MA-2YTP solid medium and incubated at 85°C overnight. Next, 106 colonies were picked at each KU216/DA4 ratio, and the genotypes of the cells were checked. The genotype of cells harboring wild-type cpkA was confirmed by PCR using primers SNP-E530-Fw and SNP-E530-Rv, whereas the genotype of cells harboring cpkA-E530G was confirmed using primers SNP-E530-Fw and SNP-E530G-Rv. To increase the specificity of the PCR, an extra mismatched nucleotide was introduced into the 3′ ends of primers SNP-E530-Rv and SNP-E530G-Rv (Table 2) according to previously described principles (25); this was done to avoid amplification of mismatches caused by the tandem repeats within the C-terminal region of the cpkA gene. The PCR conditions were as follows: initial denaturation for 2 min at 94°C followed by 20 cycles of denaturation at 94°C (15 s), annealing at 62°C (30 s), and extension at 68°C (1 min). GoTaq Green master mix (Promega, WI) was applied to perform the PCR described above. Genomic DNAs were extracted from 10 colonies randomly selected at each KU216/DA4 ratio to test the validity of the PCR described above. Briefly, the cpkA gene region was enriched in the first PCR amplification by using primers cpkA-Fw and cpkA-Rv. The genotypes were then confirmed by sequence analysis with the primer cpkA-singlechecking (Table 2).
RESULTS
Sequence alignment and temperature-dependent distribution of chaperonins.
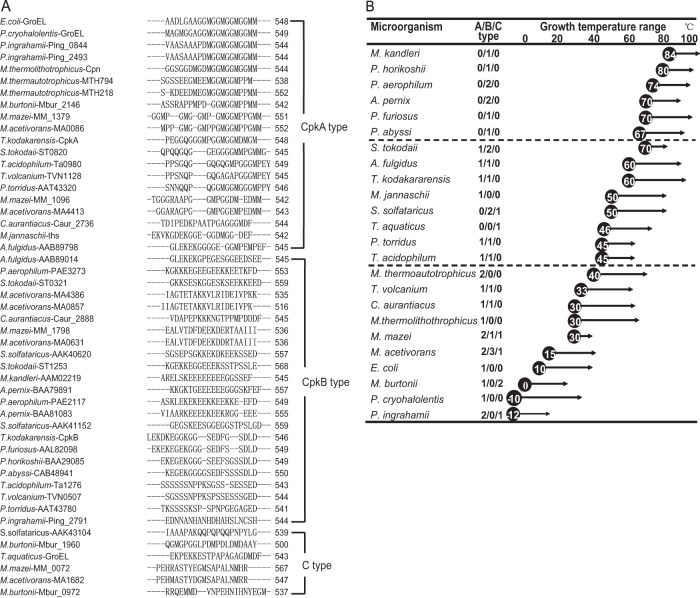
Although CpkA and CpkB share high overall sequence identity (77%), they harbor unique differences within the C-terminal region. To examine the role of the C-terminal region, we compared the last 20 amino acids within the C termini of chaperonins expressed by microorganisms at different growth temperatures (Fig. 1). The aligned sequences are shown in Fig. 1A. The C-terminal regions of chaperonins fell into three general clusters: CpkA type, CpkB type, and C type. The CpkA-type chaperonin had a mildly hydrophobic and flexible C terminus comprised primarily of GGM motifs, whereas the CpkB-type chaperonin had a more hydrophilic C-terminal region comprising glutamic acid (Glu [D]), serine (Ser [S]), lysine (Lys [K]), and aspartic acid (Asp [E]). More interestingly, there were some “promiscuous” chaperonins that fell between these two clusters (C type) (Fig. 1A). C-type chaperonins contained more glycine (Gly [G]), alanine (Ala [A]), and methionine (Met [M]) residues but also retained a considerable number of charged or polar amino acids. In other words, two major types of C-terminal region were identified: one showing increased hydrophobicity and flexibility (CpkA type) and the other showing an increased electrical charge (CpkB type). Some hyperthermophiles, such as Pyrococcus furiosus and Aeropyrum pernix, harbor only CpkB-type chaperonins. CpkB-type chaperonins seem to be indispensable for the preferred high growth temperature of these organisms. The copy number of the CpkA-type chaperonin tended to increase as the optimal cell growth temperature decreased. This tendency was not unique to archaea (Fig. 1B). The extremely psychrophilic bacterium Psychromonas ingrahamii, which shows maximal growth at 15°C and minimal growth at 0°C or lower, harbored two CpkA-type chaperonins and one C-type chaperonin, whereas Psychrobacter cryohalolentis, which grows optimally at 22°C, harbored only a CpkA-type chaperonin. Therefore, the appearance of CpkA-type chaperonins appears to be related to adaptation to low temperatures. Microorganisms harboring the CpkA-type chaperonin might adapt to lower growth temperatures more readily than those in which it is absent.
FIG 1.
Three types of chaperonin and their distribution in prokaryotes. (A) Alignment of the C-terminal regions (last 20 amino acids) of chaperonins from Pyrococcus, Thermococcus, Methanococcus, Sulfolobus, Thermus, and E. coli. Chaperonins whose C-terminal regions do not belong to the CpkA or CpkB type are labeled “C type.” (B) Distribution of chaperonins according to growth temperature. Prokaryotes examined included Aeropyrum pernix (37), Archaeoglobus fulgidus (38), Chloroflexus aurantiacus (39), Escherichia coli (40), Methanosarcina acetivorans (41), Methanococcoides burtonii (38), Methanocaldococcus jannaschii (38), Methanopyrus kandleri (38), Methanococcus mazei (42), Methanothermobacter thermautotrophicus (43), Methanothermobacter thermolithotrophicus (44), Picrophilus torridus (45), Psychromonas ingrahamii (46), Pyrobaculum aerophilum (38), Pyrococcus abyssi (38), Psychrobacter cryohalolentis (47), Pyrococcus furiosus (48), Pyrococcus horikoshii (38), Sulfolobus solfataricus (38), Sulfolobus tokodaii (49), Thermus aquaticus (50), Thermococcus kodakarensis (51), Thermoplasma acidophilum (38), and Thermoplasma volcanium (38). The numbers in the middle column represent the numbers of CpkA-type, CpkB-type, and C-type genes. The circled numbers indicate the lowest growth temperatures, and the arrows indicate the growth temperature ranges.
Variant construction and protein stability.
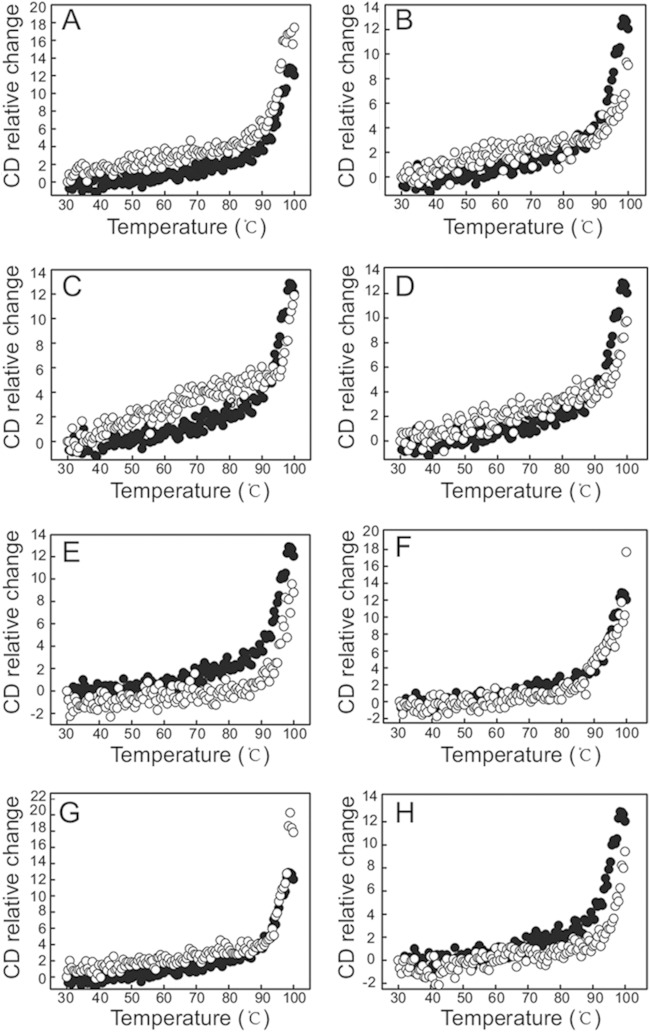
The C-terminal tail of GroEL is rich in GGM motifs; however, five of the residues in the tail region of CpkA are neither Gly nor Met (these residues are Pro529, Glu530, Gln533, Pro538, and Asp545). Therefore, to make this region resemble that of GroEL, we replaced Glu530, Gln533, Pro538, and Asp545 with Gly or Met. Pro529 was not replaced because mutation of this residue to Met or Gly would not result in successive GGM motifs. The variant proteins were then expressed in E. coli BL21(DE3) RIL and purified as described in Materials and Methods. The molecular masses of the chaperonins were around 60 kDa (see Fig. S1A in the supplemental material). CpkA and its variants showed almost identical CD profiles within the far-UV region (220 nm to 260 nm) at 20°C, suggesting that all possessed very similar secondary structures (see Fig. S1B). Next, we examined the thermostability of each purified protein. The thermal denaturation curves of the purified proteins were determined by monitoring the change in the CD value at 222 nm as the temperature increased from 20 to 100°C at a rate of 5°C min−1. All spectra were corrected to allow for the contribution of the buffer to the signal (Fig. 2A to H). All of the chaperonins showed a major unfolding event at around 90°C. The thermal unfolding plots for CpkA-E530M, -Q533M, -P538M, and -D545G were very similar to that for wild-type CpkA. CpkA-E530G and -Q533M showed slightly lower thermal stabilities, while CpkA-P538G and -D545M showed slightly higher stabilities than that of wild-type CpkA.
FIG 2.
Thermostability of CpkA and its variants. Each sample contained 50 μg ml−1 of chaperonin dissolved in 25 mM Tris-HCl (pH 7.8) containing 300 mM KCl. Sample denaturation was monitored by measuring the change in absorption at 222 nm at temperatures from 30°C to 100°C. Data for each sample were plotted after subtracting data obtained at the indicated temperatures. The denaturation of CpkA is indicated by black dots in each frame, whereas that of mutants is indicated by white dots. (A) CpkA-E530G. (B) CpkA-E530M. (C) CpkA-Q533G. (D) CpkA-Q533M. (E) CpkA-P538G. (F) CpkA-P538M. (G) CpkA-D545G. (H) CpkA-D545M.
Effects of mutations on ATPase activity.
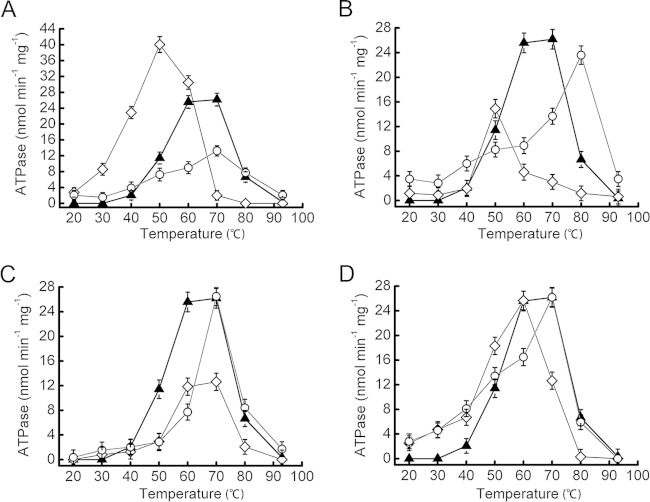
Next, we examined the temperature dependency of the ATPase activity of each variant. The results are shown in Fig. 3. CpkA showed the highest ATPase activity between 60 and 70°C. Figure 3A shows that CpkA-E530G showed optimal ATPase activity at 50°C; the value was almost 4-fold higher than that of CpkA at the same temperature. CpkA-E530M showed the most activity at 70°C, although the value was almost half that of wild-type CpkA. The ATPase activity of CpkA-E530M at 50°C was a little lower than that of wild-type CpkA. CpkA-Q533G showed the most activity at 50°C, although it was similar to that of wild-type CpkA. The optimum activity of CpkA-Q533M was displayed at 80°C and was 4-fold higher than that of wild-type CpkA at this temperature (Fig. 3B). CpkA-P538G showed a temperature-dependent profile similar to that of CpkA, although its ATPase activity was almost 2-fold lower. CpkA-P533M showed optimal activity at 70°C, with a value similar to that of wild-type CpkA (Fig. 3C). Finally, CpkA-D545G showed its highest activity at 60°C, with a value equal to that of wild-type CpkA. The optimal activity of CpkA-D545M was detected at 70°C, with a value equal to that of wild-type CpkA (Fig. 3D).
FIG 3.
ATPase activities of CpkA and its variants. The temperature-dependent ATPase activities of CpkA (triangles) and its variants (glycine substitutions are indicated by diamonds, and methionine substitutions are indicated by circles) are shown in panels A to D. (A) CpkA, CpkA-E530G, and CpkA-E530M. (B) CpkA, CpkA-Q533G, and CpkA-Q533M. (C) CpkA, CpkA-P538G, and CpkA-P538M. (D) CpkA, CpkA-D545G, and CpkA-D545M.
Effects of mutations on chaperonin activity.
InGPS from T. kodakarensis (TrpCTk) is a target protein for CpkA (6). Because the refolding of unfolded TrpCTk requires the assistance of CpkA, we performed an in vitro refolding assay using chemically unfolded TrpCTk as the substrate. Purified TrpCTk (25.4 kDa) (see Fig. S1A in the supplemental material) was denatured in 7 M urea on ice and then refolded at either 50 or 60°C in the presence or absence of wild-type or mutant CpkA. The recovered InGPS activity is shown in Fig. 4. At 60°C, the InGPS activities recovered by CpkA and CpkA-E530G were almost the same (0.66 and 0.67 U/mg, respectively), but they were only 28% of the native activity (2.39 U/mg). The InGPS activity recovered by CpkA-D545G (0.63 U/mg) was a little lower than that recovered by CpkA and CpkA-E530G. The InGPS activity of refolded TrpCTk (assisted by these three chaperonins) was almost 7-fold higher than that of spontaneously refolded TrpCTk (0.1 U/mg). CpkA-E530M, -Q533G, -Q533M, -P538G, -P538M, and -D545M were also able to assist in the refolding of TrpCTk, but the recovered InGPS activities were lower than those obtained with CpkA, CpkA-E530G, and CpkA-D545G. There were significant differences between the chaperonin activities at 50°C. When TrpCTk was refolded in the absence of CpkA, the specific activity of the refolded TrpCTk was 0.06 U/mg (about 10% of the native activity [0.58 U/mg]). This increased to 0.25 U/mg (43% of native activity) in the presence of CpkA. Moreover, refolded TrpCTk showed 1.5-fold higher activity (0.38 U/mg) in the presence of CpkA-E530G than in the presence of CpkA. Refolded TrpCTk also showed a slightly higher activity (0.3 U/mg) in the presence of CpkA-E530M than in the presence of CpkA. In general, the chaperonin activities of the variants were consistent with their ATPase activities (Fig. 3); i.e., a higher ATPase activity equated to a higher chaperonin activity and vice versa. The InGPS activity of refolded TrpCTk was not significantly higher in the presence of CpkA-Q533G, -Q533M, -D545G, and -D545M than in the presence of wild-type CpkA. Moreover, CpkA-P538G and -P538M were less effective than CpkA in terms of refolding of TrpCTk; their ATPase activities were also much lower than that of CpkA.
FIG 4.
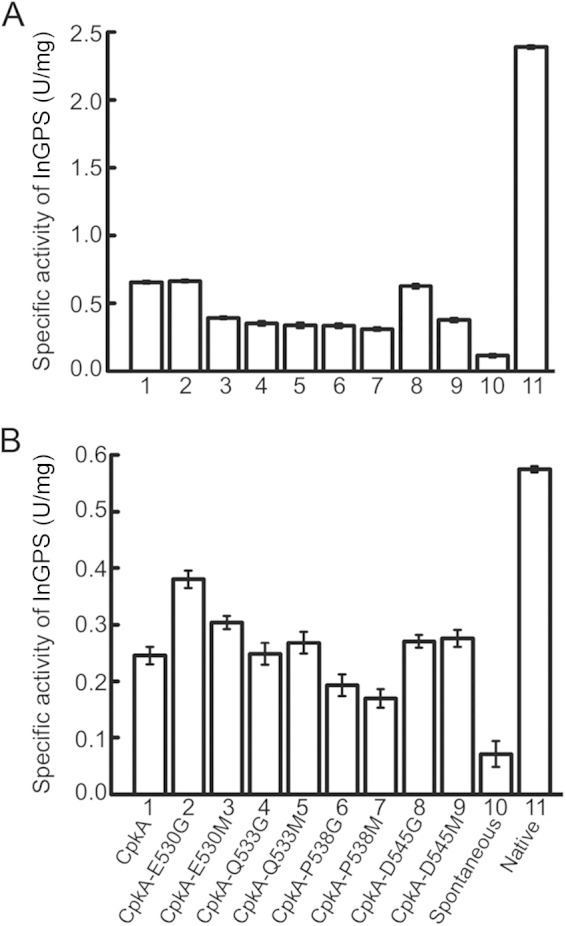
Effects of chaperonins on TrpCTk refolding. The InGPS activity of refolded TrpCTk was measured at 60°C (A) and 50°C (B) in the absence or presence of chaperonins. Columns 1 to 9 show the InGPS activities of refolded TrpCTk in the presence of CpkA, CpkA-E530G, CpkA-E530M, CpkA-Q533G, CpkA-Q533M, CpkA-P538G, CpkA-P538M, CpkA-D545G, and CpkA-D545M, respectively. Column 10 shows the InGPS activity of spontaneously refolded TrpCTk in the absence of chaperonin. Column 11 shows the activity of native TrpCTk. One unit of specific activity was defined as the amount of activity resulting in the formation of 1 μmol of InGPS in 1 min. All analyses were conducted in triplicate, and mean values are shown.
Growth profiles of strains KU216 and DA4 and growth competition assay results.
To observe the effect of the E530G mutation on T. kodakarensis in vivo, we constructed strain DA4 (pyrF cpkA-E530G) and compared its growth profile with that of the parental strain KU216. At 85°C, DA4 showed a little higher growth rate during log phase than that of KU216 (Fig. 5A). At 60°C, DA4 showed a slightly shortened lag period in the growth curve (Fig. 5B). However, at 50°C, the lag period of DA4 was much shorter than that of KU216. Strain DA4 started to propagate 30 h earlier and reached the stationary growth phase nearly 80 h earlier than strain KU216 (Fig. 5C). These results indicate that strain DA4 has a growth advantage in a cold-stressed environment.
FIG 5.
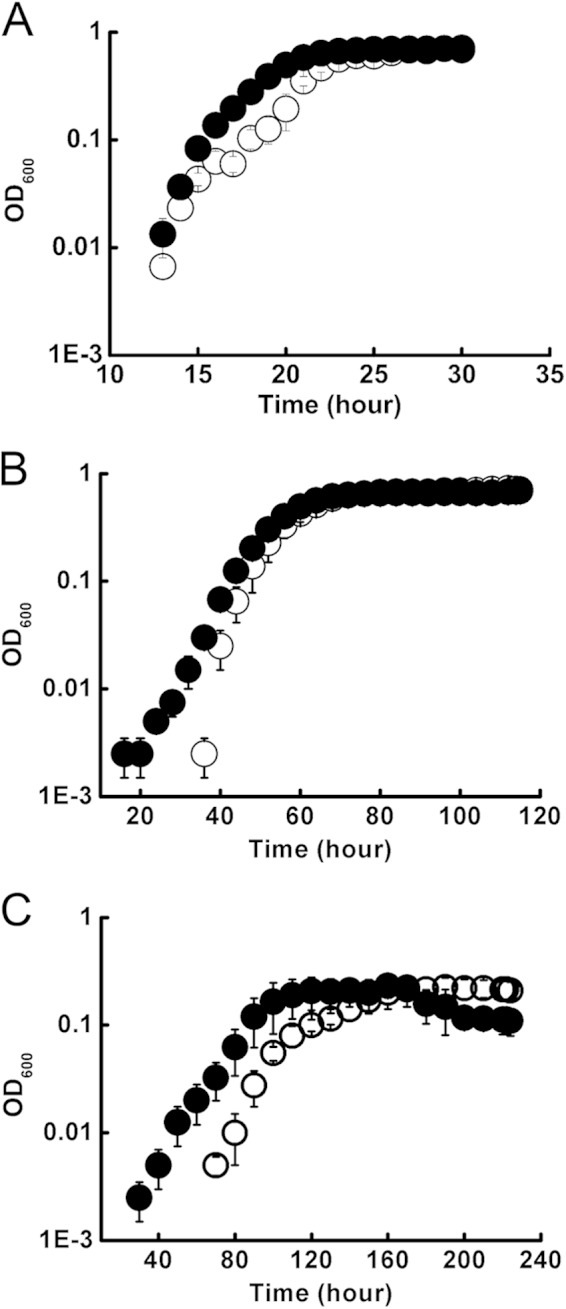
Growth profiles of KU216 and DA4. KU216 (○) and DA4 (●) were cultured in MA-2YTP medium at 85°C (A), 60°C (B), and 50°C (C), and their growth profiles were monitored by measuring the OD660.
To examine how dominant the growth of DA4 was at 50°C, we performed a cocultivation experiment. KU216 and DA4 were cultured separately to an OD660 of ∼0.6. The two cultures were then mixed at different KU216/DA4 ratios (1:1, 10:1, 102:1, and 104:1), and cell growth was monitored at 50°C. When growth entered log phase (after about 2 days), the cultures were inoculated into fresh medium and cultured for a further 2 days at 50°C. The cultures were then spread onto solid medium and incubated at 85°C overnight until colonies appeared. Finally, 106 colonies were picked and genotyped for each KU212/DA4 ratio. The final results are shown in Fig. 6. At a ratio of 1:1, DA4 was the dominant (∼87%) population. However, the percentage of DA4 gradually decreased as the DA4/KU216 ratio decreased (83.5% at a ratio of 10−1:1, 76.5% at 10−2:1, and 64.5% at 10−4:1, respectively). It is striking that DA4 was the dominant strain throughout, even when the starting population was 104-fold smaller than that of KU216.
FIG 6.
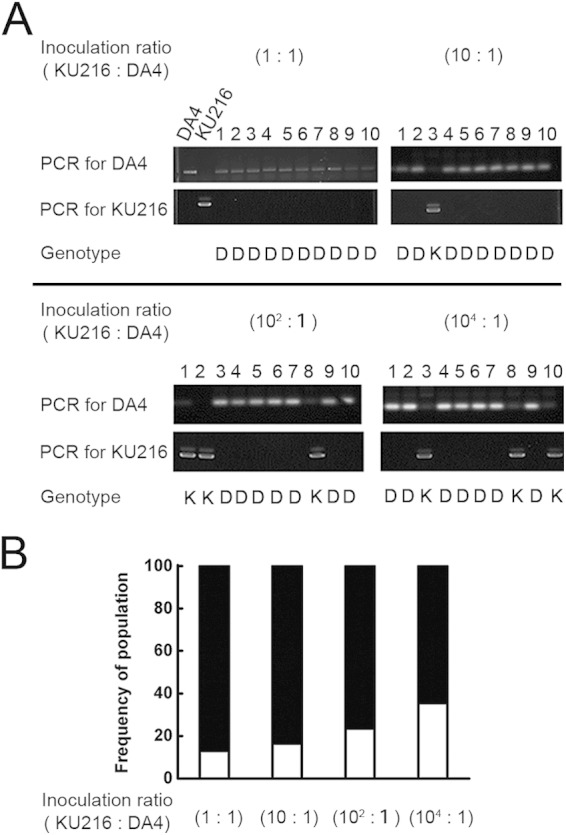
Dominant growth of DA4 at lower temperatures. KU216 and DA4 cells were precultured separately at 85°C. The two cultures were then mixed at different KU216/DA4 ratios (1:1, 10:1, 102:1, and 104:1) and cultured at 50°C for 2 days. The cultures were then added to fresh growth medium and cultured for another 2 days. The log-phase cultures were spread onto solid medium and cultured overnight at 85°C. Over 100 colonies for each KU216/DA4 ratio were randomly picked and genotyped by PCR, using primer pairs SNP-E530-Fw/SNP-E530-Rv and SNP-E530-Fw/SNP-E530G-Rv. (A) Genotypes of colonies confirmed by PCR and sequencing. The results for 10 representative colonies for each KU216/DA4 ratio are shown. The results were confirmed by sequencing (shown at the bottom of gels). K, KU216 genotype; D, DA4 genotype. (B) Proportions of DA4 and KU216 cells in cocultures at each KU216/DA4 ratio after culture at 50°C. DA4 and KU216 are indicated by black and white bars, respectively.
DISCUSSION
The duplication and divergence of α and β chaperonin subunit genes, such as cpkA and cpkB, in T. kodakarensis were thought to have occurred in a common ancestor; the α subunit was subsequently lost from the Pyrococcus lineage (26). However, the results of the present study suggest another possibility. The natural habitat of Thermococcus organisms is a solfatara, which fluctuates between intermediate (around the solfatara; ∼60°C) and high (at the center of the solfatara; ∼100°C) temperatures; therefore, these thermophiles have evolved an extra chaperonin that allows them to adapt to a colder environment. Living in an environment that moves between temperature extremes facilitates natural selection; therefore, such thermophiles can evolve quickly (27). Thus, the Thermococci adapted to lower temperatures by duplicating the chaperonin α subunit (CpkA). CpkA acts as an “adaptive allele” and enables T. kodakarensis to adapt to its changing habitat. In the absence of the cpkA gene, it is difficult to grow T. kodakarensis at 60°C (5).
Cold tolerance and cold adaptation are related but different behaviors. Cold tolerance usually occurs in response to a temporary drop in temperature, whereupon cold shock proteins (CSPs) enable their hosts to survive. Cold adaptation refers to a longer period of low temperature, triggering a response not only by CSPs but also by many nonstress proteins. However, these nonstress proteins are unstable and incorrectly folded under cold-stress conditions. Chaperonin plays an important role in protein folding and refolding; however, CpkB is not suited to this task under cold-stress conditions, because it has low ATPase activity at temperatures below 60°C (6). Thus, CpkA evolved as a unique cold-inducible chaperonin. CpkA and CpkB differ at the C terminus, which forms part of the equatorial domain. The C-terminal region of CpkA has characteristics similar to those of E. coli GroEL in terms of amino acid sequence. CpkA, a cold-inducible thermosome, was classified as a CpkA-type chaperonin belonging to group I chaperonins (which include GroEL of E. coli) (Fig. 1A). In contrast, MM_1798 encodes a mesophilic chaperonin that was classified as a CpkB-type chaperonin, along with thermosomes from Pyrococcus. Moreover, the growth temperature-dependent distribution of chaperonins (Fig. 1B) and the growth profile of the cpkA disruptant (T. kodakarensis strain DA1) suggest that CpkA-type chaperonins play a key role in cold adaptation. An extra C-type chaperonin in microorganisms might respond to other situations; an example is the TCP1 (CCT)-containing chaperonin in eukaryotic cells, which has up to eight different subunits that interact with different substrates (28). The wide variation in the C-terminal regions of chaperonins enables microorganisms to adapt to changing environments.
In the present study, we constructed eight CpkA variants harboring single amino acid substitutions in the C-terminal region, leading to an increase in the number of GGM motifs. Purified CpkA-E530G showed increased ATPase and chaperonin activities at 50°C in vitro. Cold adaptation of T. kodakarensis is a multilocus-controlled phenomenon which is fine-tuned by chaperonins (5), transcription factors (29), the composition of the cell membrane (30), and DEAD-box RNA helicase (31). It is thought that beneficial mutations accumulate incrementally via each of these mechanisms and act synergistically to enable adaptation to cold environments. However, when we introduced cpkA-E530G into the genome of T. kodakarensis, we were surprised to see that a single base pair substitution enabled T. kodakarensis to adapt to a temperature of 50°C; indeed, it showed a shorter lag phase than that of its parental strain (KU216) (Fig. 5C). We believe that the increased ability to adapt to colder conditions correlates with the improved chaperonin activity of CpkA-E530G at 50°C. The C-terminal region of GroEL also plays a role in ATPase activity (9) and substrate binding (12). CpkA has a substrate preference similar to that of GroEL (6). Here we showed that the ATPase and chaperonin activities of CpkA were dependent on the C-terminal region (Fig. 3 and 4). However, because the high flexibility of the C-terminal region means that it cannot be observed within the crystal structure of the α subunit of the chaperonin from Thermococcus sp. strain KS-1 (PDB ID 1Q2V) (32), it is difficult to discuss the structure-function relationships of CpkA, which is an orthologue of the chaperonin in Thermococcus sp. KS-1 (99% identical). We did, however, construct a 3D structural model of a full-length CpkA octamer in MODELLER 9.12, using the 1Q2V structure as a template. The results showed that the C-terminal tail of CpkA is fully extended to form the floor of a cleft within the octamer structure (see Fig. S2A in the supplemental material) and that Glu530 faces a short loop between β1 and β2 which is adjacent to the catalytic residue Asp64 (see Fig. S2B). A cryo-electron microscopic study using chaperonins from Acidianus tengchongensis also revealed a flat-bottom shape of chaperonin (cpn-α complex) (16). According to the results presented herein, it is not arbitrary to say that the location of the amino acids within the C-terminal region is the primary factor affecting chaperonin function. Replacement of Glu530, which is ∼20 Å from the functional ATPase domain of CpkA (see Fig. S2B), by either Gly or Met plays an important role in increasing chaperonin activity at low temperatures. This distance was also an ideal mutation area in a directed evolution experiment (33). We also found that particular amino acids within the C-terminal region play another important role in cold adaptation. For example, Gly enhanced the ATPase activity at 50°C to a greater extent than that seen with Met. Pro, Ser, and Ala are the most common amino acids in tandem repeats, whereas Met, Ile, and Trp are largely absent (7). Replacing Gly with polar residues (E530G, Q538G, and D545G) increased the ATPase activity at 50°C, whereas replacing Met maintained the ATPase activity above 60°C. Functional movement of the catalytic core responsible for ATPase activity determines the catalytic rate and substrate binding of the chaperonin α subunit of Thermococcus KS-1 (32). The replacement of Glu530 with Gly in CpkA increased the ATPase activity and would remove any possible blemish between the intermediate domain and the equatorial domain, such that the functional movement would occur easily compared to that of the wild type (see Fig. S2B). Pro is usually thought of as a “turn maker” or “helix breaker,” and it dictates the local structure of a protein by providing extra rigidity (i.e., resulting in lower entropy). Replacing Pro538 with either Gly or Met (CpkA-P538G or CpkA-P538M) would alter the structure of the peptide backbone and push the C-terminal tail in the wrong direction, resulting in reduced ATPase activity. We also tried to replace both Glu530 and Asp545 with Gly (CpkA-E530G/D545G); however, this double mutant (CpkA-E530G/D545G) did not show any additional increase in ATPase activity at 50°C. An alternative evolutionary path would be to increase the copy number of CpkA-type chaperonin genes, thereby increasing chaperonin activity at lower temperatures; indeed, this is the evolutionary pathway taken by some mesophiles and psychrophiles (Fig. 1B).
The final cell yields of DA4 and KU216 were almost similar when the growth entered the stationary phase (Fig. 5). However, DA4 showed a shorter lag phase than that of KU216 at 50°C, suggesting that CpkA-E530G in DA4 could perform its function on target proteins from the very start of the growth cycle, whereas KU216 would take much longer to accumulate correctly folded proteins.
In summary, we identified two growth temperature-dependent trends (correlation between the CpkA-type chaperonin gene copy number and growth temperature) in group II chaperonins, both of which are driven by the initial duplication and subsequent mutation of a chaperonin gene. The tandem repeats within the C-terminal region of CpkA are a mutational hot spot (34). Here we showed that a single base pair substitution (E530G) in the C-terminal region of T. kodakarensis CpkA allowed the organism to adapt to a lower growth temperature (50°C). Because chaperonins regulate protein-protein interactions within cells, especially under stress conditions (35), changes in the activity of chaperonins will lead to changes in cell properties, such as cold adaptation. GroEL has a marked effect on the refolding of proteins called mutators, which are involved in DNA repair (36); thus, GroEL might indirectly increase the mutation rate of the host cell. Because GroEL and CpkA have similar substrate biases (6), we expect the role of CpkA in T. kodakarensis to resemble that of GroEL. Further in vivo studies should aim to derive a mesophilic lineage from hyperthermophiles to better understand cold-adaptive mechanisms in archaea.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported mainly by a grant from the Japan Society for the Promotion of Science (JSPS) (KAKENHI grant 26292045). Bioinformatic analysis was supported by a Grant for Individual Special Research, provided by Kwansei-Gakuin University.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00279-15.
REFERENCES
- 1.Frydman J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70:603–647. doi: 10.1146/annurev.biochem.70.1.603. [DOI] [PubMed] [Google Scholar]
- 2.Goyal K, Qamra R, Mande SC. 2006. Multiple gene duplication and rapid evolution in the groEL gene: functional implications. J Mol Evol 63:781–787. doi: 10.1007/s00239-006-0037-7. [DOI] [PubMed] [Google Scholar]
- 3.Fukui T, Atomi H, Kanai T, Matsumi R, Fujiwara S, Imanaka T. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res 15:352–363. doi: 10.1101/gr.3003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Izumi M, Fujiwara S, Takagi M, Fukui K, Imanaka T. 2001. Two kinds of archaeal chaperonin with different temperature dependency from a hyperthermophile. Biochem Biophys Res Commun 280:581–587. doi: 10.1006/bbrc.2000.4154. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara S, Aki R, Yoshida M, Higashibata H, Imanaka T, Fukuda W. 2008. Expression profiles and physiological roles of two types of molecular chaperonins from the hyperthermophilic archaeon Thermococcus kodakarensis. Appl Environ Microbiol 74:7306–7312. doi: 10.1128/AEM.01245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao L, Danno A, Fujii S, Fukuda W, Imanaka T, Fujiwara S. 2012. Indole-3-glycerol-phosphate synthase is recognized by a cold-inducible group II chaperonin in Thermococcus kodakarensis. Appl Environ Microbiol 78:3806–3815. doi: 10.1128/AEM.07996-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet 44:445–477. doi: 10.1146/annurev-genet-072610-155046. [DOI] [PubMed] [Google Scholar]
- 8.Farr GW, Fenton WA, Horwich AL. 2007. Perturbed ATPase activity and not “close confinement” of substrate in the cis cavity affects rates of folding by tail-multiplied GroEL. Proc Natl Acad Sci U S A 104:5342–5347. doi: 10.1073/pnas.0700820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langer T, Pfeifer G, Martin J, Baumeister W, Hartl FU. 1992. Chaperonin-mediated protein folding: GroES binds to one end of the GroEL cylinder, which accommodates the protein substrate within its central cavity. EMBO J 11:4757–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machida K, Kono-Okada A, Hongo K, Mizobata T, Kawata Y. 2008. Hydrophilic residues 526 KNDAAD 531 in the flexible C-terminal region of the chaperonin GroEL are critical for substrate protein folding within the central cavity. J Biol Chem 283:6886–6896. doi: 10.1074/jbc.M708002200. [DOI] [PubMed] [Google Scholar]
- 11.Tang YC, Chang HC, Chakraborty Hartl FU, Hayer-Hartl M. 2008. Essential role of the chaperonin folding compartment in vivo. EMBO J 27:1458–1468. doi: 10.1038/emboj.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. 2006. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell 125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 13.McLennan NF, McAteer S, Masters M. 1994. The tail of a chaperonin: the C-terminal region of Escherichia coli GroEL protein. Mol Microbiol 14:309–321. doi: 10.1111/j.1365-2958.1994.tb01292.x. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki M, Ueno T, Iizuka R, Miura T, Zako T, Akahori R, Miyake T, Shimamoto N, Aoki M, Tanii T, Ohdomari I, Funatsu T. 2008. Effect of the C-terminal truncation on the functional cycle of chaperonin GroEL: implication that the C-terminal region facilitates the transition from the folding-arrested to the folding-competent state. J Biol Chem 283:23931–23939. doi: 10.1074/jbc.M804090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo H, Robb FT. 2011. A modulator domain controlling thermal stability in the group II chaperonins of Archaea. Arch Biochem Biophys 512:111–118. doi: 10.1016/j.abb.2011.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Zhang K, Wang L, Liu Y, Chan KY, Pang X, Schulten K, Dong Z, Sun F. 2013. Flexible interwoven termini determine the thermal stability of thermosomes. Protein Cell 4:432–444. doi: 10.1007/s13238-013-3026-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acland A, Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bryant SH, Canese K, Church DM, Clark K, DiCuccio M, Dondoshansky I, Federhen S, Feolo M, Geer LY, Gorelenkov V, Hoeppner M, Johnson M, Kelly C, Khotomlianski V, Kimchi A, Kimelman M, Kitts P, Krasnov S, Kuznetsov A, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Karsch-Mizrachi I, Murphy T, Ostell J, O'Sullivan C, Panchenko A, Phan L, Pruitt DP, Rubinstein W, Sayers EW, Schneider V, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Siyan K, Slotta D, Soboleva A, Starchenko G, Tatusova TA, Trawick B, Vakatov D, Wang Y, Ward M, Wilbur W, Yaschenko E, Zbicz K. 2014. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5:Unit 5.6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 20.Izumi M, Fujiwara S, Takagi M, Kanaya S, Imanaka T. 1999. Isolation and characterization of a second subunit of molecular chaperonin from Pyrococcus kodakaraensis KOD1: analysis of an ATPase-deficient mutant enzyme. Appl Environ Microbiol 65:1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang X, Ezaki S, Fujiwara S, Takagi M, Atomi H, Imanaka T. 1999. The tryptophan biosynthesis gene cluster trpCDEGFBA from Pyrococcus kodakaraensis KOD1 is regulated at the transcriptional level and expressed as a single mRNA. Mol Gen Genet 262:815–821. doi: 10.1007/s004380051145. [DOI] [PubMed] [Google Scholar]
- 22.Harder KW, Owen P, Wong LK, Aebersold R, Clark-Lewis I, Jirik FR. 1994. Characterization and kinetic analysis of the intracellular domain of human protein tyrosine phosphatase beta (HPTP beta) using synthetic phosphopeptides. Biochem J 298:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin B, Pallen CJ, Wang JH, Graves DJ. 1985. Use of fluorinated tyrosine phosphates to probe the substrate specificity of the low molecular weight phosphatase activity of calcineurin. J Biol Chem 260:14932–14937. [PubMed] [Google Scholar]
- 24.Sato T, Fukui T, Atomi H, Imanaka T. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl Environ Microbiol 71:3889–3899. doi: 10.1128/AEM.71.7.3889-3899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu J, Huang S, Sun M, Liu S, Liu Y, Wang W, Zhang X, Wang H, Hua W. 2012. An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 8:34–42. doi: 10.1186/1746-4811-8-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archibald JM, Blouin C, Doolittle WF. 2001. Gene duplication and the evolution of group II chaperonins: implications for structure and function. J Struct Biol 135:157–169. doi: 10.1006/jsbi.2001.4353. [DOI] [PubMed] [Google Scholar]
- 27.Diaz Arenas C, Cooper TF. 2013. Mechanisms and selection of evolvability: experimental evidence. FEMS Microbiol Rev 37:572–582. doi: 10.1111/1574-6976.12008. [DOI] [PubMed] [Google Scholar]
- 28.Bigotti MG, Clarke AR. 2008. Chaperonins: the hunt for the group II mechanism. Arch Biochem Biophys 474:331–339. doi: 10.1016/j.abb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Hidese R, Nishikawa R, Gao L, Katano M, Imai T, Kato S, Kanai T, Atomi H, Imanaka T, Fujiwara S. 2014. Different roles of two transcription factor B proteins in the hyperthermophilic archaeon Thermococcus kodakarensis. Extremophiles 18:573–588. doi: 10.1007/s00792-014-0638-9. [DOI] [PubMed] [Google Scholar]
- 30.Matsuno Y, Sugai A, Higashibata H, Fukuda W, Ueda K, Uda I, Sato I, Itoh T, Imanaka T, Fujiwara S. 2009. Effect of growth temperature and growth phase on the lipid composition of the archaeal membrane from Thermococcus kodakaraensis. Biosci Biotechnol Biochem 73:104–108. doi: 10.1271/bbb.80520. [DOI] [PubMed] [Google Scholar]
- 31.Nagaoka E, Hidese R, Imanaka T, Fujiwara S. 2013. Importance and determinants of induction of cold-induced DEAD RNA helicase in the hyperthermophilic archaeon Thermococcus kodakarensis. J Bacteriol 195:3442–3450. doi: 10.1128/JB.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shomura Y, Yoshida T, Iizuka R, Maruyama T, Yohda M, Miki K. 2004. Crystal structures of the group II chaperonin from Thermococcus strain KS-1: steric hindrance by the substituted amino acid, and inter-subunit rearrangement between two crystal forms. J Mol Biol 335:1265–1278. doi: 10.1016/j.jmb.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 33.Arnold FH. 1998. When blind is better: protein design by evolution. Nat Biotechnol 16:617–618. doi: 10.1038/nbt0798-617. [DOI] [PubMed] [Google Scholar]
- 34.Fonville NC, Ward RM, Mittelman D. 2011. Stress-induced modulators of repeat instability and genome evolution. J Mol Microbiol Biotechnol 21:36–44. doi: 10.1159/000332748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palotai R, Szalay MS, Csermely P. 2008. Chaperones as integrators of cellular networks: changes of cellular integrity in stress and diseases. IUBMB Life 60:10–18. doi: 10.1002/iub.8. [DOI] [PubMed] [Google Scholar]
- 36.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. 2005. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell 122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 37.Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, Hoaki T, Maruyama T. 1996. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int J Syst Bacteriol 46:1070–1077. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 38.Garrity GM. 2001. Bergey's manual of systematic bacteriology, 2nd ed, vol 1 Springer-Verlag, New York, NY. [Google Scholar]
- 39.Pierson BK, Castenholz RW. 1974. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus aurantiacus, gen. and sp. nov. Arch Microbiol 100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 40.Jones PG, VanBogelen RA, Neidhardt FC. 1987. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol 169:2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sowers KR, Baron SF, Ferry JG. 1984. Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl Environ Microbiol 47:971–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mah RA. 1980. Isolation and characterization of Methanococcus mazei. Curr Microbiol 3:321–326. doi: 10.1007/BF02601895. [DOI] [Google Scholar]
- 43.Zeikus JG, Wolfe RS. 1972. Methanobacterium thermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J Bacteriol 109:707–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huber H, Thomm M, König H, Thies G, Stetter KO. 1982. Methanococcus thermolithotrophicus, a novel thermophilic lithotrophic methanogen. Arch Microbiol 132:47–50. doi: 10.1007/BF00690816. [DOI] [Google Scholar]
- 45.Schleper C, Puehler G, Holz I, Gambacorta A, Janekovic D, Santarius U, Klenk HP, Zillig W. 1995. Picrophilus gen. nov., fam. nov.: a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around pH 0. J Bacteriol 177:7050–7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Auman AJ, Breezee JL, Gosink JJ, Kampfer P, Staley JT. 2006. Psychromonas ingrahamii sp. nov., a novel gas vacuolate, psychrophilic bacterium isolated from Arctic polar sea ice. Int J Syst Evol Microbiol 56:1001–1007. doi: 10.1099/ijs.0.64068-0. [DOI] [PubMed] [Google Scholar]
- 47.Bakermans C, Ayala-del-Rio HL, Ponder MA, Vishnivetskaya T, Gilichinsky D, Thomashow MF, Tiedje JM. 2006. Psychrobacter cryohalolentis sp. nov. and Psychrobacter arcticus sp. nov., isolated from Siberian permafrost. Int J Syst Evol Microbiol 56:1285–1291. doi: 10.1099/ijs.0.64043-0. [DOI] [PubMed] [Google Scholar]
- 48.Fiala G, Stetter KO. 1986. Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch Microbiol 145:56–61. doi: 10.1007/BF00413027. [DOI] [Google Scholar]
- 49.Suzuki T, Iwasaki T, Uzawa T, Hara K, Nemoto N, Kon T, Ueki T, Yamagishi A, Oshima T. 2002. Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6:39–44. [DOI] [PubMed] [Google Scholar]
- 50.Brock TD, Freeze H. 1969. Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J Bacteriol 98:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Atomi H, Fukui T, Kanai T, Morikawa M, Imanaka T. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263–267. doi: 10.1155/2004/204953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.