Abstract
Immunoassays are widely utilized due to their ability to quantify a vast assortment of biomolecules relevant to biological research and clinical diagnostics. Recently, immunoassay capabilities have been improved by the development of multiplex assays that simultaneously measure multiple analytes in a single sample. However, these assays are hindered by high costs of reagents and relatively large sample requirements. For example, in vitro screening systems currently dedicate individual wells to each time point of interest and this limitation is amplified in screening studies when the investigation of many experimental conditions is necessary; resulting in large volumes for analysis, a correspondingly high cost and a limited temporal experimental design. Microfluidics based immunoassays have been developed in order to overcome these drawbacks. Together, previous studies have demonstrated on-chip assays with either a large dynamic range, high performance sensitivity, and/or the ability to process samples in parallel on a single chip. In this report, we develop a multiplex immunoassay possessing all of these parallel characteristics using commercially available reagents, which allows the analytes of interest to be easily changed. The device presented can measure 6 proteins in 32 samples simultaneously using only 4.2 µL of sample volume. High quality standard curves are generated for all 6 analytes included in the analysis, and spiked samples are quantified throughout the working range of the assay. In addition, we demonstrate a strong correlation (R2=0.8999) between in vitro supernatant measurements using our device and those obtained from a bench-top multiplex immunoassay. Finally, we describe cytokine secretion in an in vitro inflammatory hippocampus culture system, establishing proof-of-concept of the ability to use this platform as an in vitro screening tool. The low-volume, multiplexing abilities of the microdevice described in this report could be broadly applied to numerous situations where sample volumes and costs are limiting.
Introduction
The immunoassay is one of the most versatile and widely used assays. Highly selective antibody-antigen interactions allow the measurement of any analyte for which specific antibodies are available. The flexibility of this technique permits the analysis of a variety of biomolecules, including cytokines, viruses, antibodies, drugs, hormones, and bacteria1. This has led to its utilization in a variety of applications both in the clinic and in basic research2. In recent years, the development of methods for multiplex analysis, i.e. the measurement of a panel of analytes within a single sample, has further improved the capabilities of the assay. Luminex has developed one of the most popular multiplexing platforms using optically encoded, antibody-conjugated microbeads. This technology allows for the simultaneous quantification of up to 100 proteins in a single sample3, and microbeads pre-conjugated with antibodies specific for many different molecules are commercially available from a variety of manufacturers. Due to its broad applicability and flexibility, thousands of studies have been published utilizing this technology4. However, these assays can cost several thousand dollars per kit due to the high cost of monoclonal antibodies and assay reagents and typically require at least 50µL of volume per sample.
In typical in vitro studies, one well is dedicated to each time point of interest due to the relatively high sample consumption of conventional immunoassays. Therefore, the number of time points under investigation is often limited due to the amount of cells, culture reagents, and supplies required as the number of experimental conditions increases. This limitation is exacerbated in high-throughput screening studies investigating the effect of drug candidates5 or soluble factors6 on cell secretion. A large reduction in immunoassay sample requirements could facilitate repeated sampling from individual wells. This would result in the ability to perform studies with far fewer cells, greatly increased temporal resolution, and decreased experimental costs.
Several microfluidic based multiplex immunoassays have been developed to address the drawbacks of conventional bench-top methods. These devices have utilized a variety of different approaches in order to facilitate multiplexing. These include, but are not limited to, DNA encoded antibody libraries7–9, the aforementioned Luminex microbeads10, 11, performing parallel single protein immunoassays in a CD format12, patterning antibodies at known positions within microchannels13–16, and quantum dot barcodes17. Taken together, previous approaches have demonstrated the ability to perform multiplex immunoassays in microfluidic devices with low sample volumes, high performance sensitivity, large dynamic ranges, commercial reagent compatibility, and a high sample throughput. However, some reports only demonstrate the ability to generate standard curves without assessing quantification accuracy across the working range of the assay or comparing measurements to those obtained from conventional immunoassays.
In this report, we build on previous publications to demonstrate all of aforementioned advantages in a single device. We present a microfluidic multiplex immunoassay device capable of analyzing 32 samples simultaneously in a small sample volume (<5 µL). The device utilizes commercially available Luminex reagents, which allows this device to be used to multiplex virtually any panel of analytes for which minimally cross-reactive specific antibodies can be generated. In addition, we test and demonstrate the accuracy of the device over a large dynamic range with sensitivity comparable to the standard bench-top assay. Finally, we measure supernatants generated with our established co-culture model of lipopolysaccharide (LPS) treated rat hippocampal slices18. LPS treatment of hippocampal slices induces a cytokine response, while co-culture with alginate encapsulated human mesenchymal stem cells (eMSC) provides immunomodulation. Using our device, we quantify this inflammatory response using small sample volumes, demonstrating the ability of the device to be used as an analysis tool for in vitro screening studies.
Materials and Methods
Immunoassay Reagents, Spiked Sample Preparation, and Conventional Immunoassay
Bio-Plex Pro immunoassay reagents including cytokine standards, Luminex microbeads conjugated to antibodies specific to rat IL-6, TNF-α, IL-13, IL-1β, IL-10, and MCP-1, the respective biotinylated detection antibodies, streptavidin-phycoerytrin, assay buffer, and wash buffer were used as received in both on-chip and benchtop immunoassays (Bio-Rad Laboratories, Hercules, CA). Bovine serum albumin solution (BSA) was prepared at 0.05% w/v in phosphate buffered saline (PBS) (Life Technologies, Carlsbad, CA). The lyophilized standards containing a cocktail of 24 inflammatory markers were reconstituted and diluted in BSA and media in device validation and supernatant analysis studies, respectively. For the validation study, known samples were prepared by spiking cytokine standards into BSA solutions. The antibody conjugated beads and streptavidin-phycoerythrin were prepared at 34X and 100X in assay buffer, respectively. Detection antibodies were diluted 20X in detection antibody diluent. The benchtop multiplex assay was performed as per manufacturer recommendations.
Organotypic Hippocampal Slice Culture
All animal procedures were approved by the Rutgers University Institutional Animal Care and Use Committee (Piscataway, NJ). Organotypic hippocampal slice cultures (OHSC) were prepared according to established methods19. Briefly, Sprague-Dawley rat pups (Taconic Biosciences Inc., Rensselaer, NY) at postnatal day 8–10 were decapitated, the hippocampus rapidly dissected, sliced into 400µm sections with a McIllwain tissue chopper (Vibratome, St. Louis, MO), and immersed in ice-cold Gey's balanced salt solution (Sigma-Aldrich, St. Louis, MO) supplemented with 4.5mg/ml glucose (Sigma-Aldrich, St. Louis, MO). Slices were separated and plated onto Millicell CM culture inserts (EMD Millipore, Billerica, MA) and maintained at 37°C in 5% CO2 for 14 days. Maintenance medium consisted of 25% heat-inactivated horse serum (Life Technologies, Carlsbad, CA), 25% Hank's balanced salt solution (HBSS) (Sigma-Aldrich, St. Louis, MO) and 50% minimum essential medium (MEM) with added Earle's salts (Sigma-Aldrich, St. Louis, MO), supplemented with 1mM glutamine (Sigma-Aldrich, St. Louis, MO) and 4.5mg/ml glucose (Sigma-Aldrich, St. Louis, MO). Medium was changed every 3 to 4 days.
Human Mesenchymal Stem Cell Culture
Human mesenchymal stem cells (MSCs) were purchased from Texas A&M at passage one and cultured as previously described20. Briefly, cells were cultured in MEM-α medium (Life Technologies, Carlsbad, CA), supplemented with 10% FBS (Atlanta Biologicals, Lawrenceville, GA), 1ng/ml basic fibroblast growth factor (bFGF) (Peprotech, Rocky Hill, NJ), 100 units/ml penicillin and 100µg/ml streptomycin (Life Technologies, Carlsbad, CA). Cells were plated at 5000 cells per cm2 and allowed to proliferate to 70% confluence before passaging, and were only used at passages 2 through 5. All cultures were incubated at 37°C in 5% CO2.
Alginate Microencapsulation
Alginate Poly-L-Lysine microencapsulation of MSCs was performed as previously described21, using a 2.2% alginate (Sigma-Aldrich, St. Louis, MO) and cell solution of 4 million cells/mL. Alginate beads were generated using an electrostatic bead generator (Nisco, Zurich, Switzerland), resuspended in MEM-α (Life Technologies, Carlsbad, CA), and transferred to 25 cm2 tissue culture flasks, and used for experiments one day post-encapsulation.
LPS Injury & Co-culture
Organotypic slices cultured on transwell membrane inserts were added to 24-well plates and either cultured alone or co-cultured with eMSC at 1×105cells/well. Maintenance medium was exchanged for serum-free medium (75% MEM with added Earle's salts, 25% HBSS, 1mM glutamine, and 4.5mg/mL glucose). Cultures were stimulated with 1µg/ml LPS (Escherichia coli 055:B5, Sigma-Aldrich, St. Louis, MO)22, 23 and media supernatants were collected at 6, 12, 24, or 48 hours and immediately frozen at −20°C. All collected supernatants were then thawed simultaneously and diluted at 1:10 in media. Diluted samples were aliquotted for on-chip and benchtop immunoassays and then frozen at −80°C until they were thawed on ice for analysis.
Device Fabrication and Operation
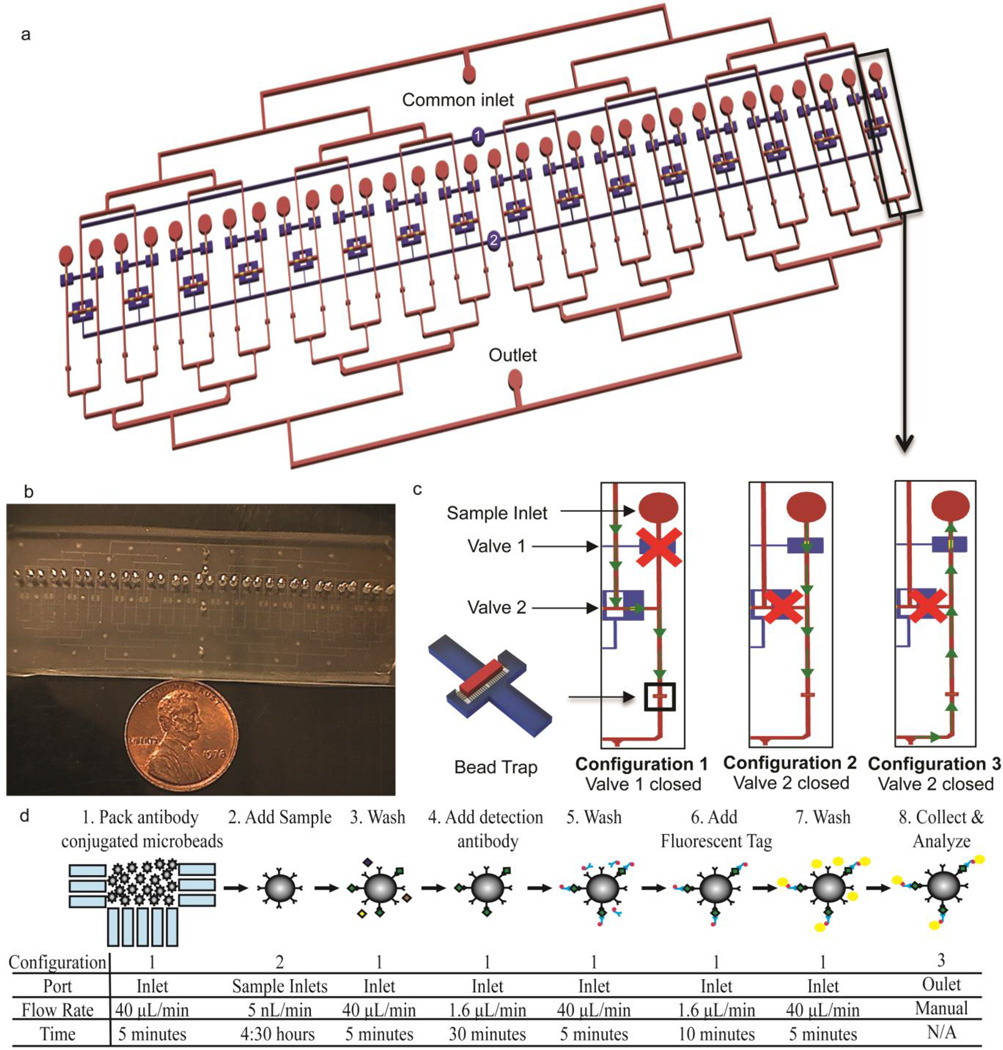
The device is an expanded version of our previous design24, which allows for 32 samples to be assayed simultaneously. A schematic and picture of the device are shown in Fig. 1a and Fig. 1b, respectively. The assay operates using a packed bed of capture antibody conjugated microbeads, allowing sequential reagent introduction over a compact space resulting in a minimal device footprint. While steric issues are a possibility in packed bed formats, eliminating this concern requires a much larger reaction area. For example, an immunoassay device has been developed with free flowing beads in solution rather than in a packed bed format25. With this approach, very long reaction channels are necessary, prohibiting large numbers of samples to be processed simultaneously and consuming a large amount of sample. In addition, operation is more complex as the beads need to be transferred between different reagent solutions and precise flow balancing is required. Therefore, we chose the packed bed format to reduce sample volume, simplify device operation, and process samples in parallel.
Fig. 1.
Device layout and assay principle a) Overall schematic of the device. The blue channels represent the pneumatic control layer and the red channels and gold valve seats denote the fluidic network b) Photograph of the device. The entire device measures 3 inches wide by 1 inch long, designed to fit on a standard glass slide c) Schematic of the valve states and fluidic paths through individual channels during different assay steps. The valves are actuated by pressurizing the inlets of the respective pneumatic channels, labeled 1 and 2 in Fig. 1a d) Overview of the assay workflow. The valve configuration and fluidic path (shown in panel c), port used for the introduction of the solution, flow rate, and duration is specified for each assay step
All assay reagents are introduced through a single common inlet. Individual sample inlets are positioned upstream of bead traps. The bead traps consist of an array of small features with a width of 3µm spaced 7µm apart, which blocks the passage of 6.5µm beads while allowing the passage of fluid. The pneumatic valves divert fluid flow to the proper regions of the device throughout the assay26. Devices were fabricated as described in our previous report24, except that the valve seats were fabricated at a height of 30µm and photomasks for the expanded design were used. Briefly, the bead traps were first patterned at a height of 7µm with SU-8 2007 (Microchem Corp., Newton, MA) using a reduced exposure dose relative to the manufacturer’s recommendations to improve resolution27. The wafer was flood exposed, hard baked, and coated with two 15µm layers of AZ Electronic Material AZ9260 photoresist (Capitol Scientific Inc., Austin, TX). The valve seats were patterned, and then reflowed to round the channel cross-section to allow for complete valve closure. The remaining fluidic network was then fabricated at a height of 41µm using SU-8 2025 (Microchem Corp., Newton, MA). The pneumatic channels were patterned on a separate layer, also at a height of 41µm. The fluidic wafer was reproduced in polyurethane, which was used in subsequent soft lithography steps to extend the life of the mold28. Poly (dimethylsiloxane) was prepared at a 10:1 pre-polymer to curing agent ratio, poured thick (3–4mm) on the fluidic mold, and spin coated onto the pneumatic wafer. The devices were then assembled using multilayer soft lithography techniques29. Prior to use, devices were primed overnight submerged in deionized water under vacuum in a desiccator.
A Rheodyne MXP7970-000 switching valve (Idex Health & Science LLC, Oak Harbor, WA) was connected to the common inlet to allow for switching of reagent solutions without introducing bubbles into the device. Assay buffer was flowed at 40µL/min for 10 minutes to block non-specific binding to the channel walls with all valves open. Pneumatic valve 1 was then closed causing fluid to flow from the common inlet through the bead traps. A mixed solution of the 6 antibody conjugated microbeads was introduced at 40µL/min for 5 minutes in order to pack the bead beds (Fig. 1c, Configuration 1). Wash buffer was then flowed at 40µL/min for 5 minutes to further pack the bead beds and ensure all beads were in the traps. Pneumatic valve 2 was then closed and pipette tips containing each standard or sample were inserted into the sample ports. With this valve closed, fluid is prohibited from mixing between adjacent channels must flow from the sample inlet over the packed bed of beads located directly downstream (Fig. 1c, Configuration 2). A syringe pump was connected to the outlet and set to withdraw at 500nL/min, corresponding to a flow rate of 15.6nL/min/channel. Sample incubation was carried out for 4.5 hours, which was determined in our previous study to provide an assay sensitivity of 10pg/mL (358fM for IL624. Under these conditions, 4.2µL of sample was consumed per channel.
At the conclusion of the sample incubation, pneumatic valve 2 was opened, pneumatic valve 1 closed, and the beads were washed at 40µL/min for 5 minutes (Configuration 1, Fig. 1c). Secondary antibodies and streptavidin-phycoerythrin were flowed at 1.6µL/min for 30 and 10 minutes, respectively, each followed by a wash step performed for 5 minutes at 40µL/min. The flow rate and duration of each reagent incubation step was selected in order to obtain the dynamic range presented in this work. At the completion of the assay, new pipette tips were inserted into the sample ports, pneumatic valve 2 was closed, and wash buffer was flowed into the common outlet (Fig. 1c, Configuration 3). The beads were collected from the sample inlet ports and transferred to a vacuum filter 96-well plate included with the immunoassay kit, the wash buffer was removed, and the beads were resuspended in assay buffer. The plate was then transferred to a Bio-Plex 200 equipped with Bio-Plex Manager 5.0 (Bio-Rad, Hercules CA). The assay workflow is summarized in Fig. 1d, and a picture of the experimental system is shown in Fig. 2.
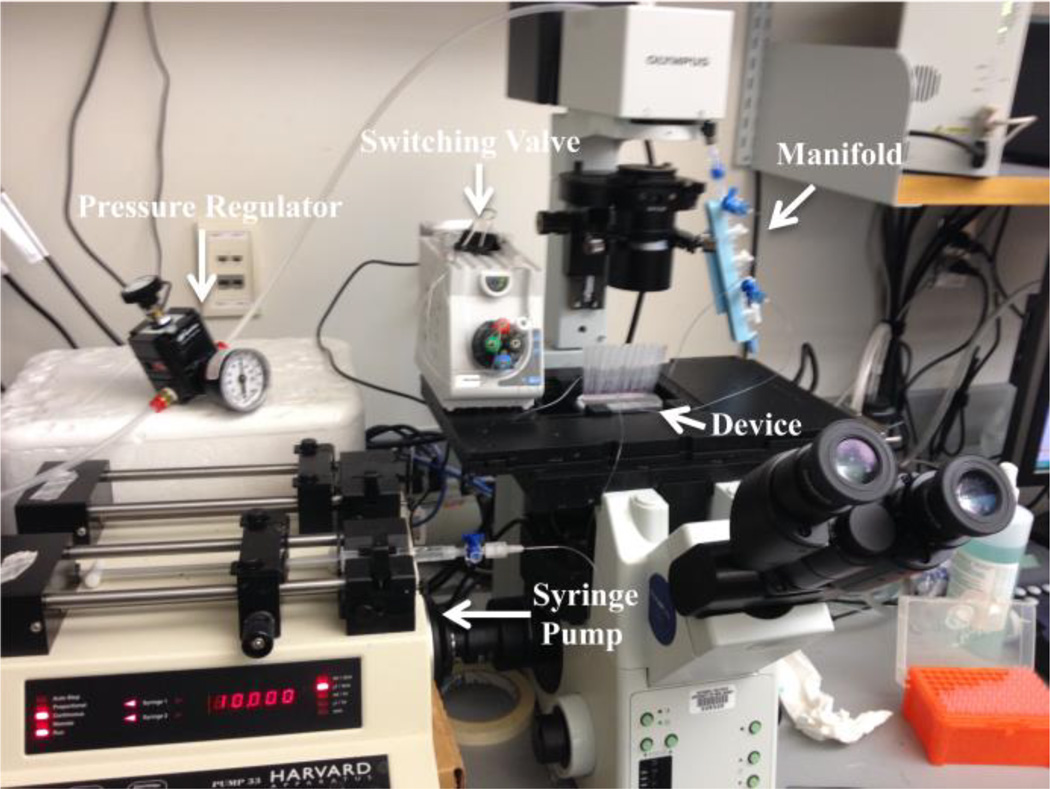
Fig 2.
Experimental system. The device is mounted on a microscope stage. The syringe pump is connected to a switching valve which is also connected to the device inlet for all assay steps in Configuration 1 (see Fig. 1c), facilitating the switching of solutions introduced into the device without removing tubing. In Configuration 2, the syringe pump is connected to the outlet of the device and set to withdraw fluid, pulling samples from the pipette tips inserted in the sample ports over the bead beds. In Configuration 3, the syringe is removed from the syringe pump and solution is manually infused through the outlet to collect the beads in pipette tips inserted into the sample ports. A pressure regulator provides compressed air to the manifold connected to each individual pneumatic channel allowing for each pneumatic valve to be individually opened and closed.
Data Analysis
Bio-Plex Manager 5.0 software was used to obtain the median fluorescence intensities of the beads and calculate the sample concentrations. Normalized standard curves were generated by dividing each fluorescence reading by the intensity of the highest standard for that analyte and fitting a 5-parameter logistic regression30 using MasterPlex ReaderFit software (Hitachi Solutions, San Bruno, CA). For the validation studies, the measured concentration of the spiked samples were compared with their expected concentrations calculated from the dilutions used to prepare the samples. For the in vitro supernatant studies, the measurements taken using the microfluidic immunoassay were compared to the concentration obtained using the conventional benchtop multiplex immunoassay. A linear regression and 95% confidence interval of the fit was constructed for the individual measurement of each sample in Matlab (Mathworks, Natick, MA) (n=10 for each of the 7 known samples in the validation studies, n=6 for the 7 samples in the in vitro supernatant studies). The confidence intervals were calculated for the linear fit itself, not to be confused with wider confidence limits for linear regressions that are to be used for the prediction of new observations31. For the in vitro supernatant analysis, measurements that fell below the limit of detection (LOD) of the microfluidic immunoassay were omitted from the analysis.
Results and Discussion
Device Validation
The ability of the device to accurately quantify multiple proteins in a single sample was first evaluated. In previous reports of microfluidic immunoassays, various degrees of assay testing have been performed ranging from simply generating standard curves11, 25, 32–39, comparing signals measured on- and off-chip7, 9, 10, 12, 14, or a direct comparison with the analogous bench-top assay15. In our studies, experiments were designed according to guidelines provided by the pharmaceutical industry for validating immunoassays for use in biomarker discovery studies40, 41. The guidelines require the use of at least 6 non-zero standards and 3 freshly spiked samples of known concentrations to construct standard curves and assess assay accuracy, respectively. Therefore, seven samples spiked with cytokines, eight standards at the manufacturer’s recommended concentrations, and a blank sample were prepared and processed in the device. The relatively large sample capacity allowed all 16 samples/standards to be assayed simultaneously in duplicate on a single chip.
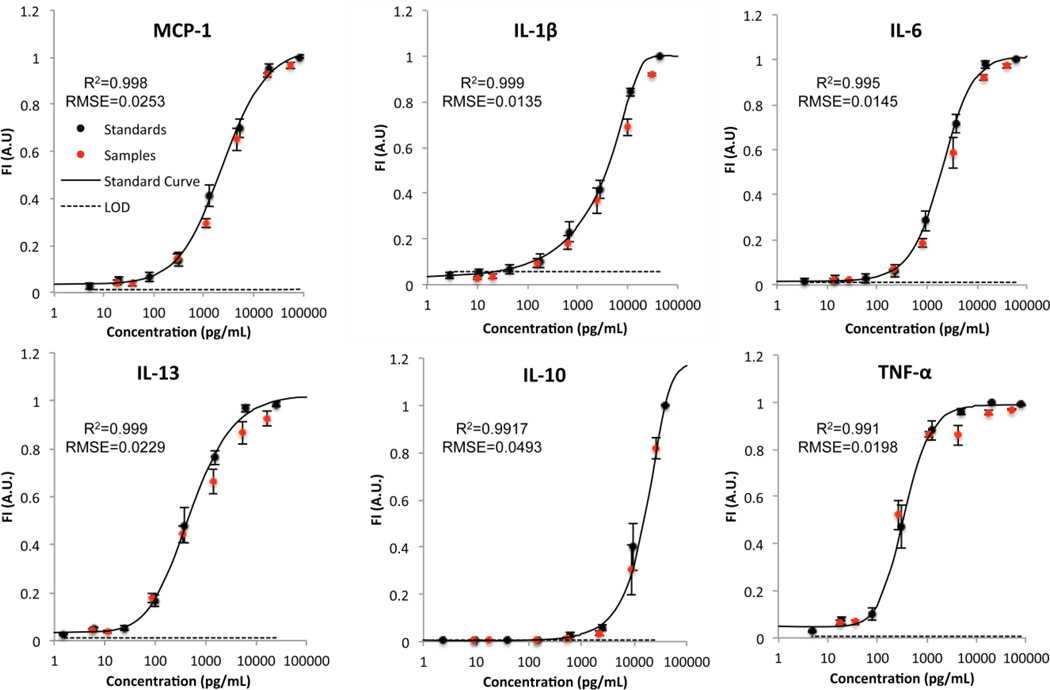
The standard curves generated from this study closely resemble the sigmoidal shape obtained when performing the benchtop assay, shown in Fig. 3. High quality standard curves for all 6 analytes were obtained, corroborated by a very high coefficient of determination and low root mean squared error (R2>0.99 and RMSE<0.05 for all analytes). The noise floor, calculated by the mean plus 3 standard deviations of the fluorescence of the blank sample, is also shown on the curves. The intersection of the standard curve and the noise floor was considered the LOD of each specific analyte. For analytes where the lower asymptote of the standard curve was higher than the noise floor (MCP-1, IL-6, TNF-α), the LOD was determined to be the concentration of the most dilute standard. Therefore, the LOD is influenced by the amount of non-specific binding and the sensitivity of the optical system. In a multiplex assay, non-specific binding can result from cross-reactivity between the different analytes included in the assay, antibodies of different species, and/or components of the sample matrix42. To achieve a desired sensitivity, the reaction must be allowed to continue until sufficient molecules have bound so that the signal can be differentiated from non-specific binding alone (i.e. above the noise floor).
Fig. 3.
Standard curves, sample measurements, and noise floor. Fluorescence intensities of the standards (black circles) and spiked samples of known concentrations (red circles) are shown overlaid on the standard curve generated using a 5-parameter logistic curve fit (black line). The limit of detection (LOD) is represented by the dotted black line. The error bars represent ± S.E.M., n=10 for each data point. RMSE=Root Mean Squared Error
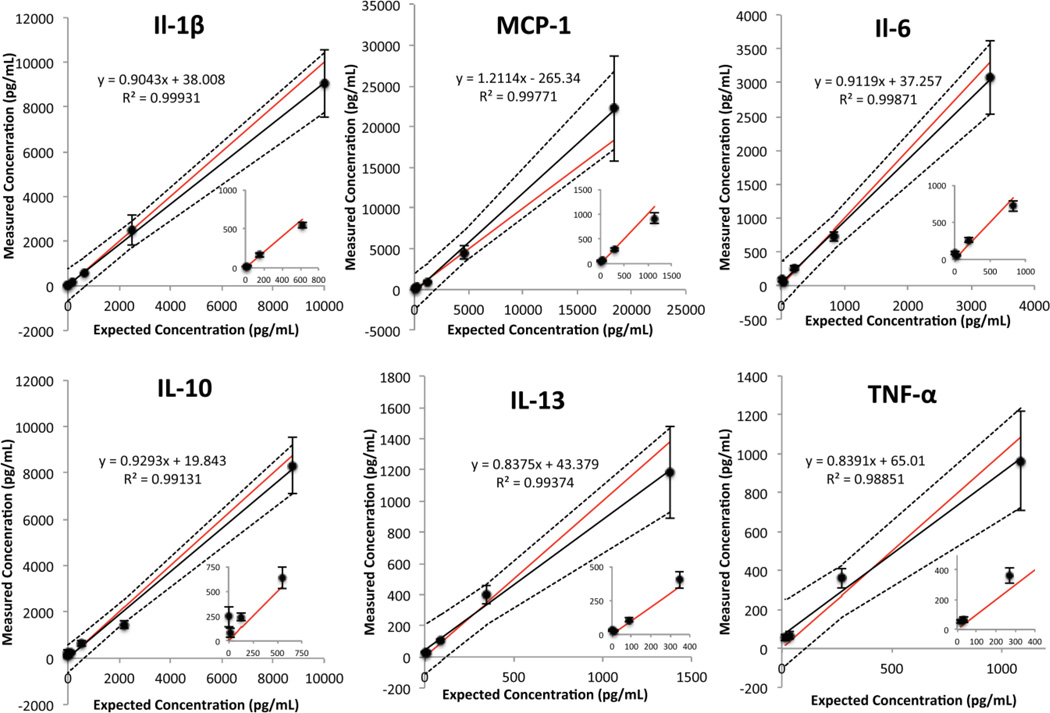
The fluorescence intensity of the spiked samples is plotted with respect to their expected concentrations and overlaid on the standard curves. Visual inspection reveals that the fluorescence measurements for the samples fall very close to their expected position on the standard curve. The ability of the device to quantify the concentration of the spiked samples was then evaluated. Fig. 4 shows the comparison of the expected and measured concentrations of the samples. The 95% confidence band of the linear regression straddles the perfect agreement between the expected and measured concentrations for all 6 analytes. In addition, the coefficient of determination of the linear regression for all six analytes was greater than 0.98. From this experiment, we defined the working range of the assay from the calculated LOD to the highest concentration shown for each analyte in Fig. 4. The working range of the assay is summarized in Table 1.
Fig 4.
Comparison of the expected and measured concentration of the spiked samples. A linear regression (black line), 95% confidence interval of the fit (dotted lines), and line representing a perfect agreement (red line) are shown. The error bars represent ±S.E.M., n=10 for each data point.
Table 1.
Working range for each analyte in the multiplex assay. The dynamic range was calculated by dividing the highest quantified sample by the limit of detection and taking a logarithm (base 10).
| Analyte | Limit of Detection (pg/mL) |
Limit of Detection (fM) |
Highest Quantified Sample (pg/mL) |
Dynamic Range (Orders of Magnitude) |
|---|---|---|---|---|
| MCP-1 | <5.0 | 387 | 18,406 | 3.56 |
| Il-1β | 21.8 | 1211 | 10,000 | 2.66 |
| IL-6 | <3.6 | 151 | 3,310 | 2.96 |
| IL-13 | 15.4 | 963 | 1,380 | 1.95 |
| IL-10 | 103.1 | 5726 | 8,766 | 1.93 |
| TNF-α | <4.8 | 92 | 1,090 | 2.36 |
In Vitro Supernatant Quantification
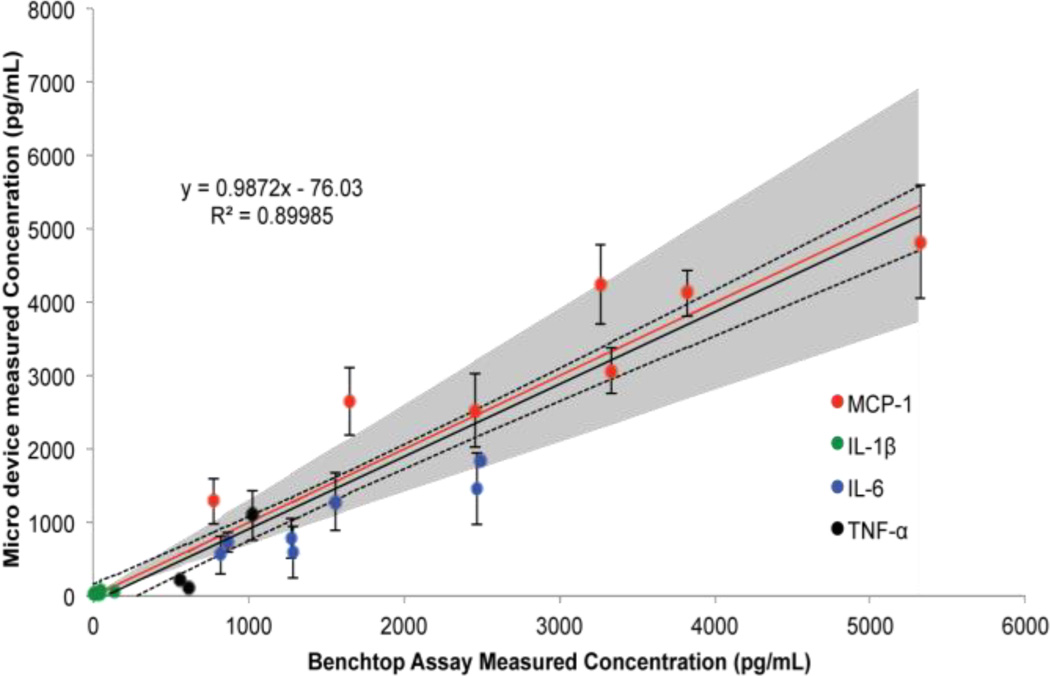
With the working range of the device established, the ability of the device to quantify in vitro samples was compared to a standard benchtop immunoassay performed as per the manufacturer’s protocol. Hippocampal slices extracted from neonatal rats were cultured on transwell inserts, treated with LPS, and cultured alone or co-cultured with eMSC. The supernatants were collected, aliquotted, and analyzed by the standard multiplex immunoassay and the microdevice in parallel. The same 6 analytes were analyzed as the validation studies; however, IL-13 was not measured in the samples. In addition, IL-10 was only measured in 3 samples in the conventional assay and was below the LOD for the microfluidic assay format. A comparison of quantification with the standard and microfluidic multiplex immunoassay is shown in Fig. 5, with the grey band denoting the 30% error allowed by the pharmaceutical industry for biomarker discovery immunoassays40. It is important to note that this acceptable error could affect the agreement of the data obtained from the two assay systems. Nonetheless, a good correlation exists between the bench-top and microfluidic immunoassay (R2=0.8999). However, IL-6 and TNF-α concentrations seem to be slightly under-predicted, while some MCP-1 measurements were over predicted, possibly biasing the linear fit. Moreover, the regression line possesses a slope very close to 1 and the 95% confidence interval of the linear fit encompasses the perfect agreement between the two assay formats. Overall, the microdevice provides comparable sample quantification of in vitro protein concentrations as a conventional assay using commercially available reagents.
Fig. 5.
Comparison of quantification of in vitro supernatants using commercially available multiplex immunoassay reagents in the conventional and microfluidic assay formats. A linear regression (black line), 95% confidence interval of the fit (dotted lines), and line representing a perfect agreement (red line) are shown. The gray shaded region represents a ±30% error accepted by the pharmaceutical industry for biomarker discovery immunoassays. The error bars represent ±S.E.M., n=6 for each data point
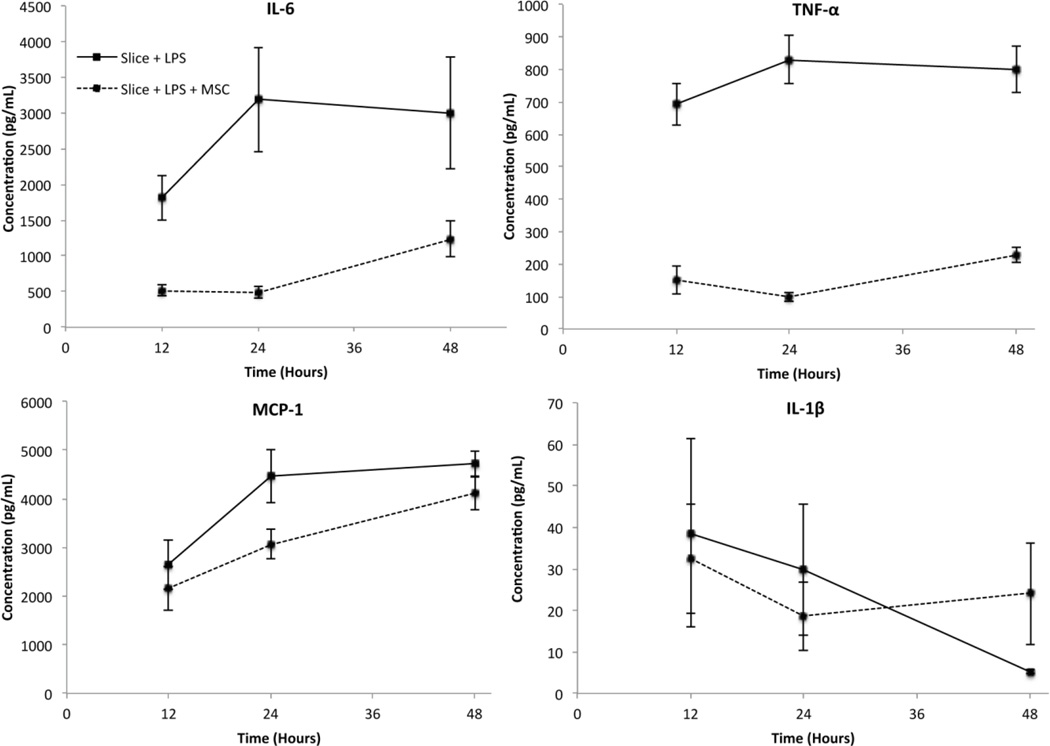
We then aimed to demonstrate proof-of-concept of this device as a low-volume in vitro screening tool. Using our group’s established co-culture model, we investigated the effect of eMSC on LPS activated hippocampal slices using our microfluidic device. The addition of LPS to the slices is known to elicit an inflammatory response resulting in cytokine secretion43, and co-culture with mesenchymal stem cells has been shown to modulate the immune response44. In addition, our previous studies have shown that alginate encapsulation allows the cells to remain viable for up to 60 days while permitting secreted proteins to diffuse through the capsule45. As indicated in Fig. 6, LPS induced increased secretion of TNF-α, IL-6 and MCP-1 from the slices during the 48 hour experimental time period, while IL-1β levels decreased. We also observed that the addition of eMSC modulated cytokine levels. TNF-α and IL-6 were markedly decreased at 12, 24 and 48 hours following LPS addition. The effect on MCP-1 secretion was less pronounced, with the largest decrease observed at 24 hours. The effect on IL-1β was much smaller and in fact, a slight increase was observed in the eMSC treated slices by 48 hours. Therefore, we were able to observe cytokine specific immunomodulatory effects of eMSC on activated hippocampus slices using our microdevice. More importantly, the temporal progression of cytokine secretion was quantified.
Fig. 6.
Effect of eMSC on rat hippocampal slices treated with LPS measured using the microdevice. All concentrations are shown at 10 fold dilutions. Error bars represent ± S.E.M., n=4 for TNF-a, Slice + LPS at 12 and 24 hours, n=6 for all other data points)
The decrease in TNF-α secretion observed from the hippocampal slice in the presence of MSC is consistent with our studies using conventional techniques18. While the biological effects of eMSC on hippocampal slices are discussed in detail in that report, the differences in the cytokines measured here demonstrates the ability of this device to be used as a screening tool in in vitro systems. Furthermore, we were able to perform this analysis using only 4.2µL of supernatant compared to the 50µL of sample volume required in the bench-top assay. With lowered sample consumption, one well can be used to perform a time course study by removing small volumes of supernatant at each time point of interest and allowing the culture to continue. Furthermore, the number of time points examined could be increased without requiring additional hippocampal slices, eMSC, and associated tissue culture reagents. These potential advantages demonstrate how reduced sample consumption provides additional cost and reagent savings beyond what is required to perform the assay.
In addition to lowering sample consumption, antibody conjugated microbeads consumption was reduced >13 fold (2.5µL to 0.184µL of each stock bead solution) and detection antibody requirement was reduced >16 fold (25µL to 1.5µL of each 1X detection antibody solution). Extrapolating the reduction of the limiting reagent (i.e. the antibody conjugated microbeads), we estimate that the assay kit used in this report, which contains sufficient reagents to analyze one 96 well plate and a 25% reagent excess, could potentially analyze 1560 samples using the on chip assay.
In addition to the significant reduction in required sample volume and accompanying cost savings provided by this device, its specifications make it well suited for a variety of applications. The device possesses sensitivity comparable to the benchtop assay (low pg/mL concentrations) with a dynamic range of ~2–3.5 orders of magnitude. This large working range allows for multiple analytes present at different concentrations to be measured simultaneously, as shown in our in vitro supernatant analysis. This mitigates the need to optimize dilutions prior to analysis in order to avoid saturation, which is necessary when using an assay with a smaller dynamic range.
Antibody conjugated microbeads have been used as an antibody immobilization surface in several microfluidic based immunoassays. This is due to the increase in surface area to volume ratio and decreased diffusion times compared to conventional immunoassays46. Many reports mechanically trap the beads, as was done in this study. Early reports employed mechanically trapping relatively large beads (45 um) and on-chip thermal lens microscopy33, 34. Another device recirculated fluid over a small number of beads immobilized behind a filter11 and performed detection using fluorescence microscopy. Beads have also been confined in a capillary tube, collected at the completion of the assay, and analyzed off-chip using flow cytometry10. In the absence of a permanent mechanical trap PDMS valves47 and magnetic trapping have also been utilized in conjunction with fluorescence imaging48. Lab-on-a-disc bead based approaches have also been applied, first developed for single protein assays using fluorescence detection49, and then expanded to multiplexing by performing assays for different analytes in parallel in conjunction with absorbance detection12. Moreover, some devices have performed the immunoassay with beads flowing in solution and utilized off-chip flow cytometry25, 50. While the overall assay principle in each of these technologies is similar, the major differences are marked by the methods used to immobilize the beads, introduce the samples and reagent solutions, and interrogate the beads at the completion of the assay.
The use of the Luminex multiplexing microbeads used in this study allows this device to measure many different analytes that are commercially available. These reagents are available for purchase pre-conjugated to capture antibodies specific for a wide variety of molecules in several species. Moreover, the concentrations of detection antibodies and streptavidin-PE used in this report are identical to those recommended by the manufacturer for the bench-top assay. Finally, this design allows for parallel processing of samples on a single chip. This device is an expanded version of our 8 channel device with the same basic layout24, suggesting scalability beyond the 32 sample capacity presented in this report. To the best of our knowledge, this is the first report simultaneously demonstrating low sample usage, high performance sensitivity, a large dynamic range, commercial reagent compatibility, quantification capabilities (confirmed with spiked samples and comparison to a conventional assay), and parallel sample processing. Furthermore, sacrificing assay sensitivity can significantly shorten the assay time. For example, we determined in a previous study that the sample incubation time could be reduced from 4.5 hours to less than one hour by increasing the limit of detection from 10pg/mL to 50pg/mL of IL6 (358fM to 1.79pM)24.
The combination of these characteristics allow for broad applications in both clinical and research settings. The immunoassay reagents used in this study can be used to measure samples in a variety of matrices, including media, cerebrospinal fluid (CSF), plasma, and serum. However, analysis of biological fluids would require sample preparation prior to analysis. Low protein samples such as CSF would require centrifugation to remove debris prior to introduction into the device, while whole blood would require treatment with an anticoagulant, separation, and dilution prior to analysis51.
With proper sample preparation, the device could also be applied to in vivo studies. For example, analysis of cerebrospinal fluid (CSF) in rat models of central nervous system diseases has been prohibited by the small amount of available volume. This limitation has forced developmental Alzheimer’s disease therapies to be studied in larger animals, resulting in increased drug compound, veterinary costs, and ethical concerns when compared to rodent models52. In addition, potential spinal cord injury biomarker candidates identified in a human clinical trial53 were unable to be further explored in a rat model with controlled injury. This resulted in samples needing to be pooled from multiple animals54 or the analysis of spinal tissue rather than CSF55, which can not be analyzed in human patients and is therefore not directly clinically translatable.
A scaled down assay would also provide advantages in a clinical setting. The decreased cost of the assay could drastically decrease the cost of diagnostic procedures. In addition, samples from pediatric and neonatal patients that do not yield sufficient sample volumes could be analyzed56. Finally, the high-throughput, low volume, and low cost multiplexing characteristics of this device are perfectly suited for biomarker discovery studies57. Also, like most microfluidic assays, automation of this device would be fairly straightforward58, potentially simplifying the complex procedure required to operate this device. This could eliminate the need for highly trained technicians to perform the assay in clinical labs, reducing assay variability and the laborious workflow associated with immunoassays.
Advancement of the proof-of-concept device into a practical product would require further development. First, systematically optimizing the flow rates, duration, and concentrations used in the reagent incubation steps could improve sensitivity and expand the dynamic range of the assay. In addition, a manifold would be needed to operate the device. Samples could be introduced to the sample ports at the beginning of the assay and the sequential introduction of reagents, valve operation, and sample propulsion could be driven with minimal pressure sources. In addition, the requirement to interrogate the beads off-chip represents a limitation of this technology. One option to eliminate this step could be to perform detection on-chip. Due to the packed bed, it would be difficult to individually analyze each bead microscopically. Alternatively, the beads could be released and transferred to an on-chip flow cytometer, but this would increase the device footprint and complicate the on-chip valving system. A simpler approach may be to include the necessary optics and fluid handling components in the manifold to automate the process of extracting the beads from the device and performing detection.
Conclusions
Herein we present a multiplex immunoassay device capable of performing 32 simultaneous multiplex immunoassays in only 4.2 µL of sample volume. This design allows for further scalability beyond a 32 sample capacity and allows for the analysis of virtually any analyte for which immunoassay antibodies are available. The device is shown to have high performance sensitivity with a dynamic range of ~2–3.5 orders of magnitude depending on the analyte. Furthermore, we demonstrate the ability to quantify samples across the entire working range of the assay and compare on-chip quantification with a standard benchtop multiplex immunoassay. To the best of our knowledge, no device demonstrated to date possesses the combination of the aforementioned characteristics. These capabilities allow for utilization in numerous situations where sample volumes and costs are limiting.
Acknowledgements
This work was partially funded by the National Institute of Health Grants P41EB002503 and UH2TR000503, the National Institute of Health Rutgers Biotechnology Training Program (T32GM008339), the National Science Foundation Integrated Science and Engineering of Stem Cells Program (DGE0801620), The New Jersey Commission on Brain Injury Research Grant CBIR12IRG019, and Corning Inc. The authors would like to thank Dr. Bhaskar Mitra and Henry Yu for support in developing the fabrication protocols, and Prof. Lawrence Williams for productive discussion in designing the experiments.
Notes and references
- 1.Henares TG, Mizutani F, Hisamoto H. Anal. Chim. Acta. 2008;611:17–30. doi: 10.1016/j.aca.2008.01.064. [DOI] [PubMed] [Google Scholar]
- 2.Wild D. The immunoassay handbook. San Diego: Elsevier Ltd.; 2005. [Google Scholar]
- 3.Dunbar SA. Clinica Chimica Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houser B. Archives of physiology and biochemistry. 2012;118:192–196. doi: 10.3109/13813455.2012.705301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fung E, Sugianto P, Hsu J, Damoiseaux R, Ganz T, Nemeth E. Molecular pharmacology. 2013;83:681–690. doi: 10.1124/mol.112.083428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barminko JA, Nativ NI, Schloss R, Yarmush ML. Biotechnology and Bioengineering. 2014 doi: 10.1002/bit.25282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang J, Ahmad H, Ma C, Shi Q, Vermesh O, Vermesh U, Heath J. Lab Chip. 2010;10:3157–3162. doi: 10.1039/c0lc00132e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin L, Vermesh O, Shi Q, Heath JR. Lab Chip. 2009;9:2016–2020. doi: 10.1039/b821247c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan R, Vermesh O, Srivastava A, Yen BKH, Qin L, Ahmad H, Kwong GA, Liu C-C, Gould J, Hood L, Heath JR. Nature Biotechnology. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Hartmann M, Wang Q, Poetz O, Schneiderhan-Marra N, Stoll D, Kazmaier C, Joos TO. PLoS ONE. 2010;5:e13125. doi: 10.1371/journal.pone.0013125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diercks A, Ozinsky A, Hansen C, Spotts J, Rodriguez D, Aderem A. Analytical Biochemistry. 2009;386:30–35. doi: 10.1016/j.ab.2008.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park J, Sunkara V, Kim T-H, Hwang H, Cho Y-K. Anal. Chem. 2012;84:2133–2140. doi: 10.1021/ac203163u. [DOI] [PubMed] [Google Scholar]
- 13.Bernard A, Michel B, Delamarche E. Anal. Chem. 2001;73:8–12. doi: 10.1021/ac0008845. [DOI] [PubMed] [Google Scholar]
- 14.Song Y, Zhang Y, Bernard PE, Reuben JM, Ueno NT, Arlinghaus RB, Zu Y, Qin L. Nature communications. 2012;3:1283. doi: 10.1038/ncomms2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Cordero JL, Nembrini C, Stano A, Hubbell JA, Maerkl SJ. Integrative Biology. 2013;5:650–658. doi: 10.1039/c3ib20263a. [DOI] [PubMed] [Google Scholar]
- 16.Stern E, Vacic A, Rajan NK, Criscione JM, Park J, Ilic BR, Mooney DJ, Reed MA, Fahmy TM. Nature nanotechnology. 2009;5:138–142. doi: 10.1038/nnano.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klostranec JM, Xiang Q, Farcas GA, Lee JA, Rhee A, Lafferty EI, Perrault SD, Kain KC, Chan WC. Nano letters. 2007;7:2812–2818. doi: 10.1021/nl071415m. [DOI] [PubMed] [Google Scholar]
- 18.Stucky EC, Schloss RS, Yarmush ML, Shreiber DI. Cytotherapy. 2014 doi: 10.1016/j.jcyt.2015.05.002. (In Press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoppini L, Buchs P-A, Muller D. Journal of neuroscience methods. 1991;37:173–182. doi: 10.1016/0165-0270(91)90128-m. [DOI] [PubMed] [Google Scholar]
- 20.Parekkadan B, Van Poll D, Suganuma K, Carter EA, Berthiaume F, Tilles AW, Yarmush ML. PloS one. 2007;2:e941. doi: 10.1371/journal.pone.0000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maguire T, Novik E, Schloss R, Yarmush M. Biotechnology and bioengineering. 2006;93:581–591. doi: 10.1002/bit.20748. [DOI] [PubMed] [Google Scholar]
- 22.Lieberman AP, Pitha PM, Shin HS, Shin ML. Proceedings of the National Academy of Sciences. 1989;86:6348–6352. doi: 10.1073/pnas.86.16.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung IY, Benveniste EN. The Journal of Immunology. 1990;144:2999–3007. [PubMed] [Google Scholar]
- 24.Ghodbane M, Kulesa A, Yu H, Maguire T, Schloss R, Ramachandran R, Zahn J, Yarmush M. Microfluid. Nanofluid. 2015;18:199–214. doi: 10.1007/s10404-014-1416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sasso LA, Johnston IH, Zheng M, Gupte RK, Ündar A, Zahn JD. Microfluid. Nanofluid. 2012;13:603–612. doi: 10.1007/s10404-012-0980-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Unger MA, Chou H-P, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113–116. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 27.Chan-Park MB, Zhang J, Yan Y, Yue C. Sensors and Actuators B: Chemical. 2004;101:175–182. [Google Scholar]
- 28.Desai SP, Freeman DM, Voldman J. Lab Chip. 2009;9:1631–1637. doi: 10.1039/b822081f. [DOI] [PubMed] [Google Scholar]
- 29.Fordyce PM, Diaz-Botia C, DeRisi JL, Gomez-Sjoberg R. Lab Chip. 2012;12:4287–4295. doi: 10.1039/c2lc40414a. [DOI] [PubMed] [Google Scholar]
- 30.Gottschalk PG, Dunn JR. Analytical biochemistry. 2005;343:54–65. doi: 10.1016/j.ab.2005.04.035. [DOI] [PubMed] [Google Scholar]
- 31.Montgomery DC, Runger GC. 4 edn. John Wiley & Sons; 2007. [Google Scholar]
- 32.White RJ, Kallewaard HM, Hsieh W, Patterson AS, Kasehagen JB, Cash KJ, Uzawa T, Soh HT, Plaxco KW. Anal. Chem. 2012;84:1098–1103. doi: 10.1021/ac202757c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato K, Yamanaka M, Takahashi H, Tokeshi M, Kimura H, Kitamori T. Electrophoresis. 2002;23:734–739. doi: 10.1002/1522-2683(200203)23:5<734::AID-ELPS734>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 34.Sato K, Tokeshi M, Odake T, Kimura H, Ooi T, Nakao M, Kitamori T. Anal. Chem. 2000;72:1144–1147. doi: 10.1021/ac991151r. [DOI] [PubMed] [Google Scholar]
- 35.Ko Y-J, Maeng J-H, Ahn Y, Hwang S-Y, Cho NG, Lee S-H. Sensors and Actuators B: Chemical. 2008;132:327–333. [Google Scholar]
- 36.Lee BS, Lee J-N, Park J-M, Lee J-G, Kim S, Cho Y-K, Ko C. Lab Chip. 2009;9:1548–1555. doi: 10.1039/b820321k. [DOI] [PubMed] [Google Scholar]
- 37.Gervais L, Delamarche E. Lab Chip. 2009;9:3330–3337. doi: 10.1039/b906523g. [DOI] [PubMed] [Google Scholar]
- 38.Yoo SK, Kim YM, Yoon SY, Kwon HS, Lee JH, Yang S. Artificial organs. 2011;35:E136–E144. doi: 10.1111/j.1525-1594.2011.01240.x. [DOI] [PubMed] [Google Scholar]
- 39.Kartalov EP, Zhong JF, Scherer A, Quake SR, Taylor CR, Anderson WF. BioTechniques. 2006;40:85. doi: 10.2144/000112071. [DOI] [PubMed] [Google Scholar]
- 40.Valentin M-A, Ma S, Zhao A, Legay F, Avrameas A. Journal of pharmaceutical and biomedical analysis. 2011;55:869–877. doi: 10.1016/j.jpba.2011.03.033. [DOI] [PubMed] [Google Scholar]
- 41.Lee JW, Hall M. Journal of Chromatography B. 2009;877:1259–1271. doi: 10.1016/j.jchromb.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 42.Elshal MF, McCoy JP. Methods. 2006;38:317–323. doi: 10.1016/j.ymeth.2005.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huuskonen J, Suuronen T, Miettinen R, Van Groen T, Salminen A. J Neuroinflammation. 2005;2:25. doi: 10.1186/1742-2094-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foraker JE, Oh JY, Ylostalo JH, Lee RH, Watanabe J, Prockop DJ. Journal of Neurochemistry. 2011;119:1052–1063. doi: 10.1111/j.1471-4159.2011.07511.x. [DOI] [PubMed] [Google Scholar]
- 45.Barminko J, Kim JH, Otsuka S, Gray A, Schloss R, Grumet M, Yarmush ML. Biotechnology and Bioengineering. 2011 doi: 10.1002/bit.23233. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Derveaux S, Stubbe BG, Braeckmans K, Roelant C, Sato K, Demeester J, Smedt SC. Anal. Bioanal. Chem. 2008;391:2453–2467. doi: 10.1007/s00216-008-2062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoo SK, Kim YM, Yoon SY, Kwon H-S, Lee JH, Yang S. Artificial Organs. 2011 doi: 10.1111/j.1525-1594.2011.01240.x. no-no. [DOI] [PubMed] [Google Scholar]
- 48.Lacharme F, Vandevyver C, Gijs M. Microfluid. Nanofluid. 2009;7:479–487. [Google Scholar]
- 49.He H, Yuan Y, Wang W, Chiou N-R, Epstein AJ, Lee LJ. Biomicrofluidics. 2009;3:022401. doi: 10.1063/1.3116665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasso LA, Undar A, Zahn JD. Microfluid. Nanofluid. 2010;9:253–265. doi: 10.1007/s10404-009-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang H, Weng X, Li D. Microfluid. Nanofluid. 2011;10:941–964. [Google Scholar]
- 52.Shapiro JS, Stiteler M, Wu G, Price EA, Simon AJ, Sankaranarayanan S. Journal of Neuroscience Methods. 2011;205:36–44. doi: 10.1016/j.jneumeth.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 53.Kwon BK, Casha S, Hurlbert RJ, Yong VW. Clin. Chem. Lab. Med. 2011;49:425–433. doi: 10.1515/CCLM.2011.068. [DOI] [PubMed] [Google Scholar]
- 54.Lubieniecka JM, Streijger F, Lee JHT, Stoynov N, Liu J, Mottus R, Pfeifer T, Kwon BK, Coorssen JR, Foster LJ, Grigliatti TA, Tetzlaff W. PLoS ONE. 2011;6:e19247. doi: 10.1371/journal.pone.0019247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stammers AT, Liu J, Kwon BK. J Neurosci Res. 2012;90:782–790. doi: 10.1002/jnr.22820. [DOI] [PubMed] [Google Scholar]
- 56.Phillips TM. Electrophoresis. 2004;25:1652–1659. doi: 10.1002/elps.200305873. [DOI] [PubMed] [Google Scholar]
- 57.Shoemaker LD, Achrol AS, Sethu P, Steinberg GK, Chang SD. Neurosurgery. 2012;70:518. doi: 10.1227/NEU.0b013e3182333a26. [DOI] [PubMed] [Google Scholar]
- 58.Bange A, Halsall H, Heineman W. Biosensors and Bioelectronics. 2005;20:2488–2503. doi: 10.1016/j.bios.2004.10.016. [DOI] [PubMed] [Google Scholar]