Highlight
Two additional members of the Arabidopsis AINTEGUMENTA-LIKE (AIL) transcription factor gene family, AIL5 and AIL7, are shown to have overlapping functions with ANT in floral organ development.
Key words: AINTEGUMENTA-LIKE, Arabidopsis thaliana, carpel patterning, flower development, organ growth, organ initiation, petal, sepal fusion, unequal genetic redundancy.
Abstract
AINTEGUMENTA (ANT) is an important regulator of Arabidopsis flower development that has overlapping functions with the related AINTEGUMENTA-LIKE6 (AIL6) gene in floral organ initiation, identity specification, growth, and patterning. Two other AINTEGUMENTA-LIKE (AIL) genes, AIL5 and AIL7, are expressed in developing flowers in spatial domains that partly overlap with those of ANT. Here, it is shown that AIL5 and AIL7 also act in a partially redundant manner with ANT. The results demonstrate that AIL genes exhibit unequal genetic redundancy with roles for AIL5, AIL6, and AIL7 only revealed in the absence of ANT function. ant ail5 and ant ail7 double mutant flowers show alterations in floral organ positioning and growth, sepal fusion, and reductions in petal number. In ant ail5, petals are often replaced by filaments or dramatically reduced in size. ant ail7 double mutants produce increased numbers of carpels, which have defects in valve fusion and a loss of apical tissues. The distinct phenotypes of ant ail5, ant ail7 and the previously characterized ant ail6 indicate that AIL5, AIL6, and AIL7 make unique contributions to flower development. These distinct roles are also supported by genetic analyses of ant ail triple mutants. While ant ail5 ail6 triple mutants closely resemble ant ail6 double mutants, ant ail5 ail7 triple mutants exhibit more severe deviations from the wild type than either ant ail5 or ant ail7 double mutants. Furthermore, it is shown that AIL5, AIL6, and AIL7 act in a dose dependent manners in ant and other mutant backgrounds.
Introduction
Flowers arise on the flanks of the inflorescence meristem at the sites of auxin maxima (Reinhardt et al., 2003). Within these flower primordia, floral organ primordia are initiated at characteristic positions within concentric rings called whorls. The sites of floral organ initiation within a flower appear to correspond to auxin maxima, although it is not clear if auxin specifies the founder cell population or accumulates after these cells are specified and promotes organ outgrowth (Chandler et al., 2011). After initiation, these primordia adopt a sepal, petal, stamen or carpel fate as a consequence of the activities of distinct combinations of four classes of floral organ identity genes (reviewed in Krizek and Fletcher, 2005). In whorl one, class A and E genes specify sepal identity. In whorl two, class A, B, and E genes specify petal identity. In whorl three, class B, C, and E genes specify stamen identity, while in whorl four, class C and E genes specify carpel identity. Most of the class A, B, C, and E genes encode MADS domain transcription factors that form tetrameric complexes regulating distinct target genes in each whorl of the flower (Smaczniak et al., 2012). Furthermore, these MADS domain protein complexes appear to act throughout floral organ development activating both early and later targets during floral organogenesis (Ito et al., 2004, 2007; Kaufmann et al., 2010; Wuest et al., 2012; O’Maoleidigh et al., 2013).
Members of the AINTEGUMENTA-LIKE/PLETHORA (AIL/PLT) family of transcription factors play important roles in many developmental processes in plants including flower development (reviewed in Horstman et al., 2014). AILs are a small subgroup of eight proteins within the large AP2/ERF transcription factor family (see Supplementary Fig. S1 at JXB online). The founding member of the family, AINTEGUMENTA (ANT), is a key regulator of floral organ growth. Mutations in AINTEGUMENTA (ANT) result in flowers with smaller floral organs while ectopic expression of ANT results in floral organs that reach a larger final size (Elliott et al., 1996; Klucher et al., 1996; Krizek, 1999; Mizukami and Fischer, 2000). In addition, ant mutants have ovule defects and are female sterile (Elliott et al., 1996; Klucher et al., 1996). Three other AIL genes: AIL5, AIL6, and AIL7 are expressed in developing flowers at lower levels than ANT but in spatial domains that partially overlap that of ANT (Nole-Wilson et al., 2005) (see Supplementary Fig. S1 at JXB online). Loss of AIL5, AIL6 or AIL7 function by itself has no phenotypic consequences on flower development (Nole-Wilson et al., 2005; Krizek, 2009; Prasad et al., 2011). In the case of AIL6, this is due to genetic redundancy with ANT (Krizek, 2009). ant ail6 flowers exhibit severe defects in floral organ positioning, identity specification, growth, and patterning. These flowers consist primarily of small sepals, filaments, and unfused carpel valves; they lack petals and stamens and a normal gynoecium (Fig. 1F). The floral organ identity defects in ant ail6 flowers are likely to be a consequence of reduced expression of class B and C genes during the early stages of flower development.
Fig. 1.
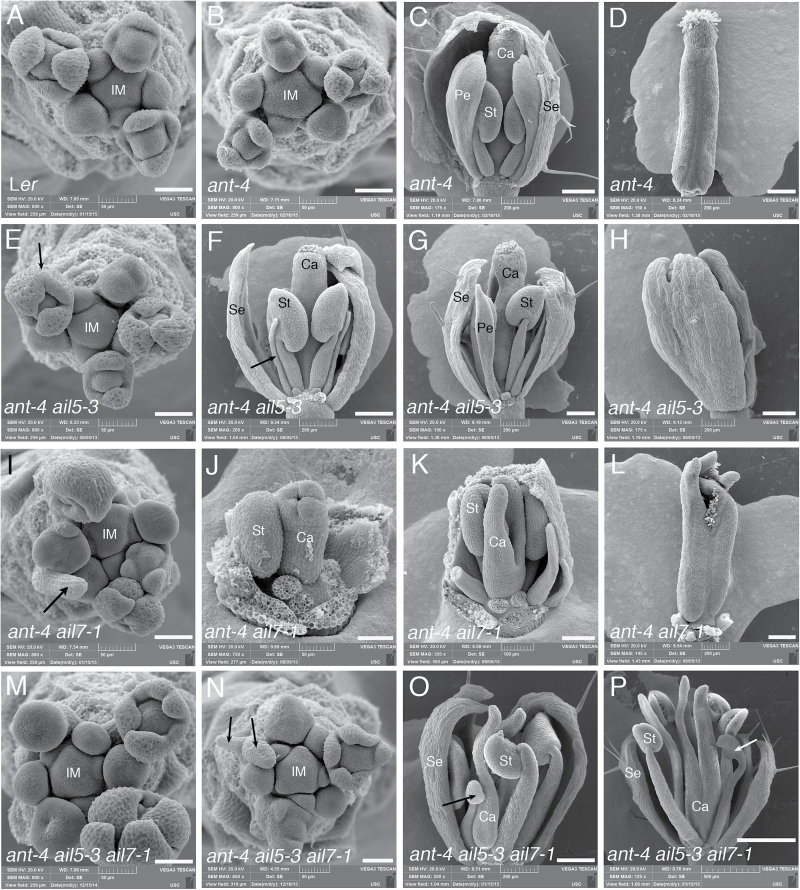
AIL5 and AIL7 have partially overlapping functions with ANT in flower development. (A) Ler flower. (B) ant-4 flower. (C) ant-4 ail5-2 flower. (D) ant-4 ail5-3 flower. (E) ant-4 ail7-1 flower. (F) ant-4 ail6-2 flower. (G) ant-4 ail5-3 ail6-2 flower. (H) Early-arising (left) and later-arising (right) ant-4 ail5-3 ail7-1 flowers. (I) ant-4 ail6-2 ail7-1/+ flowers. (J) ant-4 ail6-2 ail7-1/+ inflorescence. (K) Early-arising (left) and later-arising (right) ant-4 ail6-2/+ ail7-1 flowers. (L) ant-4 ail6-2/+ ail7-1 inflorescence.
While roles for AIL5 and AIL7 in floral organ development have not been described, these two genes have been shown to regulate shoot phyllotaxy and lateral root initiation in combination with AIL6. In plt3 plt5 plt7 (i.e. ail5 ail6 ail7) triple mutants, flower initiation deviates from spiral phyllotaxy with two adjacent flowers often separated by 90° or 180° rather than 137.5° (Prasad et al., 2011). The positioning of lateral roots is also disrupted in this triple mutant with clusters of lateral roots arising in the triple mutant rather than being distributed along the root longitudinal axis (Hofhuis et al., 2013). In addition to these roles in primordium positioning, AIL5 also regulates germination and seedling growth (Yamagishi et al., 2009; Yano et al., 2009). Furthermore, ectopic expression of AIL5 results in the formation of embryo-like structures on seedlings (Tsuwamoto et al., 2010). This phenotype is similar to that resulting from over-expression of BABY BOOM (BBM), another member of the AIL family (Boutilier et al., 2002). AIL5 misexpression also results in the production of larger flowers, similar to the phenotype of plants over-expressing ANT (Nole-Wilson et al., 2005).
To investigate the possible roles of AIL5 and AIL7 in flower development further, double and triple mutants were made. ant ail5 and ant ail7 double mutants show more severe phenotypes than ant single mutants indicating that AIL5 and AIL7 have partially redundant functions with ANT during flower development. While ant ail5, ant ail6 and ant ail7 double mutants share some phenotypic similarities, each double mutant has a unique appearance. This suggests that AIL5, AIL6, and AIL7 make distinct contributions to flower development in the absence of ANT. This conclusion is further supported by the unique phenotypes of ant ail triple mutants. Furthermore, it was found that AIL5, AIL6, and AIL7 act in a dose-dependent manner in the ant background. Dose dependent behaviour is also observed for AIL6 and AIL7 in the ant ail7 and ant ail6 backgrounds, respectively. Floral organ development is, for the most part, normal in ail5 ail6 ail7 plants, although the petals are slightly smaller in size. These results indicate that these three genes only contribute to flower development in the absence of ANT. These results demonstrate unequal genetic redundancy among members of the AIL gene family.
Materials and methods
Plant materials and growth conditions
Mutants used in the study were ant-4 (Baker et al., 1997; Nole-Wilson and Krizek, 2006), ail5-2 (Prasad et al., 2011), ail5-3 (identified in this study), ail6-2 (Krizek, 2009), and ail7-1 (Krizek, 2009). Plants were grown on a soil mixture of Metro-Mix 360:perlite:vermiculite (5:1:1 by vol.) in 16h days (100–150 μmol m–2 s–1) at a temperature of 20–22 °C.
Genetics and PCR genotyping
ant-4 is in the Landsberg erecta (er) background while the ail5-2, ail5-3, ail6-2, and ail7-1 alleles were originally in the Columbia background. Double and triple mutants were identified in the F2 or later generations by PCR genotyping. All double and triple mutants characterized were those carrying the er allele. ant-4 was PCR genotyped as described previously (Krizek, 2009). Genotyping of ail5-2, ail5-3, ail6-2, and ail7-1 was performed using the primers listed in Supplementary Table S1 at JXB online. The ail5-2 ail6-2 ail7-1 and ail5-3 ail6-2 ail7-1 triple mutants phenotypically characterized were those containing er from crosses of double mutants carrying er.
SEM
Tissue for SEM was fixed, dried, dissected, and coated as described previously (Krizek, 1999). SEM analyses were performed on a Tescan Vega 3 SBU variable pressure SEM.
Petal measurements
Petal measurements were performed on at least 12 petals from flower at positions 1–10 on an inflorescence from at least four different plants. Petal measurements were performed essentially as described previously (Trost et al., 2014). Petals from approximately stage 13 flowers (at the time when the long stamens were at the same height as the carpel) were removed with forceps and placed on Sellotape. Once all petals were collected, the tape was stuck to a piece of black plexiglass and scanned at a resolution of 3600 dpi in 8-bit greyscale. Petal area, length, and width were determined using Image J software.
Gynoecium clearing
Gynoecium clearing was performed as described previously (Wynn et al., 2014). Briefly, the tissue was fixed in ethanol:acetic acid (9:1 v/v) for 2h at room temperature and rinsed twice in 90% ethanol. The tissue was transferred to Hoyer’s solution (70% chloral hydrate, 5% gum arabic, 4% glycerol) for several hours, dissected, and mounted.
RNA extraction, RT-PCR and real time PCR
RNA was extracted from inflorescences using Trizol following the manufacturer’s instructions with cleanup on an RNeasy column (Qiagen). The RNA was DNased while on the column. First strand cDNA synthesis was performed using Quanta qScript cDNA SuperMix (Quanta BioSciences) following the manufacturer’s instructions. RT-PCR and real-time PCR was performed using the primers listed in Supplementary Table S1 at JXB online. The RT-PCR experiment usd the following PCR conditions: 40 cycles of 92 °C for 30 s, 55 °C for 30 s, and 72 °C for 2min followed by 1 cycle of 72 °C for 5min. Real-time PCR was performed on a BioRad iCycler as described previously (Krizek and Eaddy, 2012).
Results
Mutations in AIL5 have no effect on flower development but enhance ant single mutants
Several ail5 alleles have been described previously. ail5-1 corresponds to a transgenic inverted repeat AIL5 knockdown line (Nole-Wilson et al., 2005). ail5-2 (i.e. plt5-2) contains a T-DNA insertion within the eighth exon of the gene (Prasad et al., 2011) (see Supplementary Fig. S2A at JXB online). In addition, three ail5 alleles (cho1-1, cho1-2, and cho1-3) have been described (Nambara et al., 2002; Yamagishi et al., 2009). Another allele (ail5-3) that contains a T-DNA insertion within the first intron of the gene was identified (see Supplementary Fig. S2A at JXB online) (Alonso et al., 2003). Like the previously identified ail5-1 and ail5-2 alleles, ail5-3 produces normal flowers (see Supplementary Fig. S2B–D at JXB online). No full-length transcript was produced in ail5-3, although partial transcripts corresponding to sequences downstream of the T-DNA insertion site were detected (see Supplementary Fig. S2E, F at JXB online). Transcripts corresponding to sequences upstream of ail5-2 were also detected, indicating that neither ail5-2 nor ail5-3 are RNA null alleles (see Supplementary Fig. S2F at JXB online).
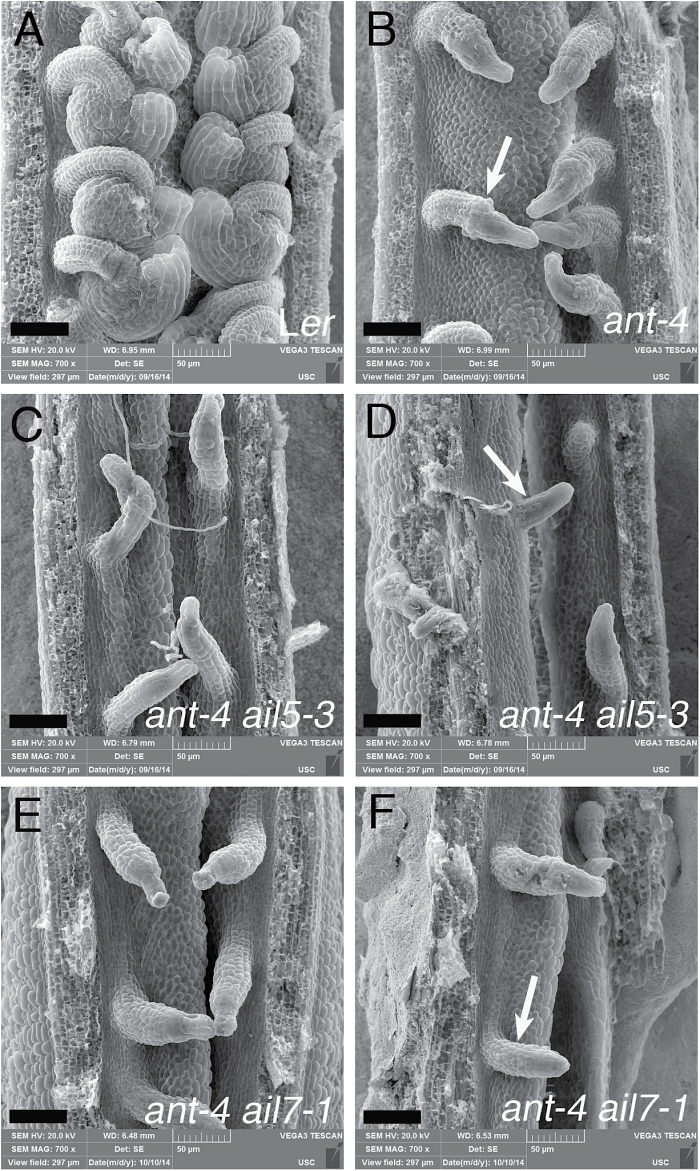
To investigate whether AIL5 acts redundantly with ANT, ant-4 ail5-2 and ant-4 ail5-3 double mutants were constructed. Both double mutants show a more severe second whorl phenotype compared with ant-4. ant-4 ail5-2 flowers produce similar numbers of petals as ant-4 but these petals are smaller in size (Fig. 1A–C; Tables 1, 2). ant-4 ail5-3 flowers produce many fewer petals than ant-4; most second whorl organs are missing or replaced with filaments (Figs 1D, 2C, F; Table 1). The petals that are present are very thin (Fig. 2G; Table 2). The ant-4 ail5-3 double mutant was characterized in more detail.
Table 1.
Floral organ counts in Ler, ant, ant ail5, ant ail7, and ant ail5 ail7 flowers at positions 1–30 on the inflorescence
| Ler 1–30 | ant-4 1–30 | ant-4 ail5-2 1–30 | ant-4 ail5-3 1–30 | ant-4 ail7-1 1–30 | ant-4 ail5-3 ail7-1 1–30 | |
|---|---|---|---|---|---|---|
| Whorl 1 | ||||||
| Se | 4.0 | 3.99 | 4.02 | 3.78 | 4.01 | 4.41 |
| Filament | 0.01 | |||||
| Curled white | 0.03 | 0.01 | ||||
| Total | 4.0 | 3.99 | 4.02 | 3.82 | 4.01 | 4.41 |
| Whorl 2 | ||||||
| Pe | 4.0 | 3.74 | 3.91 | 0.56 | 1.72 | 0.93 |
| Filament | 0.02 | 0.91 | 0.05 | 0.49 | ||
| St/Pe | 0.01 | 0.01 | 0.05 | |||
| Total | 4.0 | 3.77 | 3.92 | 1.47 | 1.77 | 1.47 |
| Whorl 3 | ||||||
| St | 5.77 | 4.38 | 4.87 | 4.24 | 4.82 | 4.56 |
| St-like | 0.24 | |||||
| Filament | 0.01 | 0.02 | 0.28 | |||
| Pe/St | 0.01 | 0.06 | 0.18 | |||
| Total | 5.77 | 4.38 | 4.89 | 4.26 | 4.88 | 5.26 |
| Whorl 4 | ||||||
| Ca | 2.0 | 2.0 | 2.0 | 2.0 | 2.35 | 2.32 |
| Total | 2.0 | 2.0 | 2.0 | 2.0 | 2.35 | 2.32 |
| Total of all whorls | 15.77 | 14.14 | 14.83 | 11.55 | 13.01 | 13.46 |
Fig. 2.
Scanning electron micrographs of Ler, ant-4 and ant-4 ail mutant combinations. (A) Ler inflorescence meristem. (B) ant-4 inflorescence meristem. (C) ant-4 flower. (D) ant-4 carpels. (E) ant-4 ail5-3 inflorescence meristem. Arrow points to the reduced boundary between two adjacent sepal primordia. (F) ant-4 ail5-3 flower with filaments in the second whorl in place of petals. Arrow points to one filament. (G) ant-4 ail5-3 flower with a thin petal in the second whorl. (H) ant-4 ail5-3 flower with fused sepals. (I) ant-4 ail7-1 inflorescence meristem. Arrow points to filament-like structure that arises in place of a flower. (J) Young ant-4 ail7-1 flower showing abnormal development of the fourth whorl carpels. (K) ant-4 ail7-1 flower with two unfused carpels in the fourth whorl. (L) ant-4 ail7-1 carpels in which the valves are unfused at their apex and some stigmatic tissue arises at their edges. (M, N) ant-4 ail5-3 ail7-1 inflorescence meristems. Arrows in (N) point to filament-like structures that arise in place of flowers. (O, P) ant-4 ail5-3 ail7-1 flowers with stamenoid organs (arrows). IM, inflorescence meristem; Se, sepal; Pe, petal; St, stamen; Ca, carpel. Scale bars, 50 μm (A, B, E, I, J, M, N), 100 μm (K) 200 μm (C, D, F–H, L, O), 500 μm (P).
Table 2.
Petal area, width, and length in various genotypes
Genotypes grouped together were grown and measured at the same time.
| Petal area (mm2) | Petal length (mm) | Petal width (mm) | |
|---|---|---|---|
| Ler | 1.82±0.18 | 2.95±0.19 | 0.97±0.09 |
| ant-4 | 0.94±0.11 | 2.47±0.18 | 0.59±0.07 |
| ant-4 ail5-2 | 0.48±0.07 | 2.14±0.13 | 0.28±0.04 |
| ant-4 ail5-3 | 0.40±0.10 | 2.24±0.22 | 0.20±0.05 |
| ant-4 ail7-1 | 1.02±0.21 | 2.40±0.21 | 0.66±0.09 |
| ant-4 ail5-3 ail7-1 | 0.59±0.09 | 2.33±0.18 | 0.32±0.05 |
| ant-4 | 0.83±0.09 | 2.38±0.14 | 0.58±0.05 |
| ant-4 ail5-3/+ | 0.68±0.09 | 2.28±0.18 | 0.49±0.06 |
| Ler | 1.77±0.27 | 2.85±0.23 | 1.03±0.09 |
| ail5-2 ail6-2 ail7-1 | 1.38±0.19 | 2.31±0.11 | 0.94±0.09 |
| ail5-3 ail6-2 ail7-1 | 1.31±0.18 | 2.27±0.17 | 0.93±0.06 |
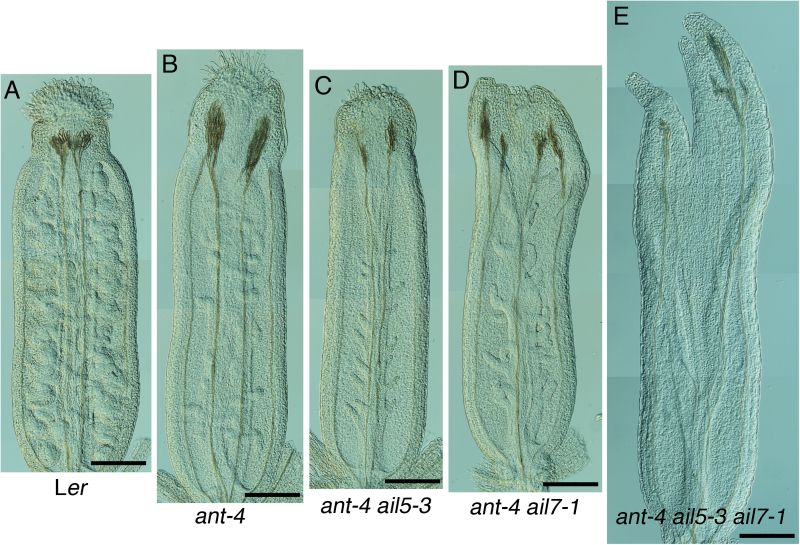
In addition to petal defects, ant-4 ail5-3 flowers exhibit more severe defects in whorls one and four compared with ant-4. ant-4 ail5-3 flowers show an increase frequency of sepal fusion (Table 3) and scanning electron microscopy of ant-4 ail5-3 flowers reveals altered patterns of sepal initiation and growth (Fig. 2A, B, E). Sepal primordia do not always arise in the characteristic adaxial and abaxial positions separated by 180 °C (Fig. 2E). Boundaries between adjacent sepals are not established in some flowers leading to the growth of two adjacent sepals as a fused structure (Fig. 2E, H). Examination of a cleared ant-4 ail5-3 gynoecium suggested that the double mutant might exhibit a more severe defect in ovule development than ant-4 (Fig. 3A–C). While ant-4 ovules do not initiate integuments (Baker et al., 1997), they often show a slight swelling in the chalazal region (Fig. 4A, B); this swelling was not observed in ant-4 ail5-3 ovules (Fig. 4C, D). In addition, some ant-4 ail5-3 ovule primordia were very short with no cellular distinction of regional identities (nucellus, chalaza, funiculus) along the apical-basal axis of the ovule (Fig. 4D).
Table 3.
Floral organ fusion in Ler, ant, ant ail5, ant ail7, and ant ail5 ail7 flowers at positions 1–30 on the inflorescence
| Ler 1–30 | ant-4 1–30 | ant-4 ail5-3 1–30 | ant-4 ail7-1 1–30 | ant-4 ail5-3 ail7-1 1–30 | |
|---|---|---|---|---|---|
| % of flowers with Se fusion | 0 | 2.0 | 46.3 | 43.6 | 39.4 |
| % of flowers with St fusion | 0 | 10.1 | 8.05 | 11.4 | 7.34 |
| % of flowers with unfused Ca | 0 | 3.0 | 2.0 | 96.7 | 95.5 |
| % of flowers with Ca more than halfway unfused | 0 | 0 | 0 | 37.3 | 66.6 |
| % of flowers with Ca lacking stigmatic tissue | 0 | 2.2 | 0 | 69.3 | 74.8 |
Fig. 3.
AIL5 and AIL7 contribute to gynoecium development. (A) Ler gynoecium. (B) ant-4 gynoecium. (C) ant-4 ail5-3 gynoecium. (D) ant-4 ail7-1 gynoecium. (E) ant-4 ail5-3 ail7-1 gynoecium. Scale bars, 200 μm.
Fig. 4.
AIL5 and AIL7 contribute to ovule development. (A) Ler ovules. (B) ant-4 ovules. Arrow points to swelling in chalazal region. (C, D) ant-4 ail5-3 ovules. Arrow in (D) points to small ovule primordium that lacks regional distinctions. (E, F) ant-4 ail7-1 ovules. Arrow in (F) points to small ovule primordium that lacks regional distinctions. Scale bars, 50 μm.
Mutations in AIL7 enhance ant single mutants
Previously, mutations in AIL7 were shown to have no phenotypic consequence on flower development (Krizek, 2009). In addition, it was shown that AIL7 did not act redundantly with its closest homologue AIL6. To investigate possible genetic redundancy between ANT and AIL7, the ant-4 ail7-1 double mutant was made. ant-4 ail7-1 flowers show an enhanced phenotype in whorls one, two, and four compared with ant-4 (Fig. 1B, E). Like ant-4 ail5-3, the ant-4 ail7-1 double mutant displays an increased frequency of sepal fusion in the first whorl (Table 3) and altered initiation and growth of sepal primordia (Fig. 2I). In the second whorl, ant-4 ail7-1 double mutants produce fewer petals than ant-4 (Table 1). These petals are similar in area to ant-4 petals (Table 2). In the fourth whorl, ant-4 ail7-1 carpels are unfused to varying extents (Table 3; Fig. 2D, K, L). In approximately one-third of flowers, the fusion defects extend more than halfway down the length of the carpel values (Table 3; Fig. 2K). Stigmatic tissue is often reduced or absent in ant-4 ail7-1 carpels (Figs 2L, 3D; Table 3). Defects in carpel growth are evident early in development of the fourth whorl (Fig. 2J). ant-4 ail7-1 mutants also appear to have slightly more severe ovule defects than ant-4. ant-4 ail7-1 ovule primordia exhibit less pronounced swelling of the chalazal region and are sometimes quite short and lack regional distinctions, similar to ant-4 ail5-3 ovules (Fig. 4E, F). In addition to the floral organ defects mentioned above, flower initiation from the inflorescence meristem was occasionally disrupted with some flowers being replaced by filament-like structures in which floral organs were not initiated (Fig. 2I).
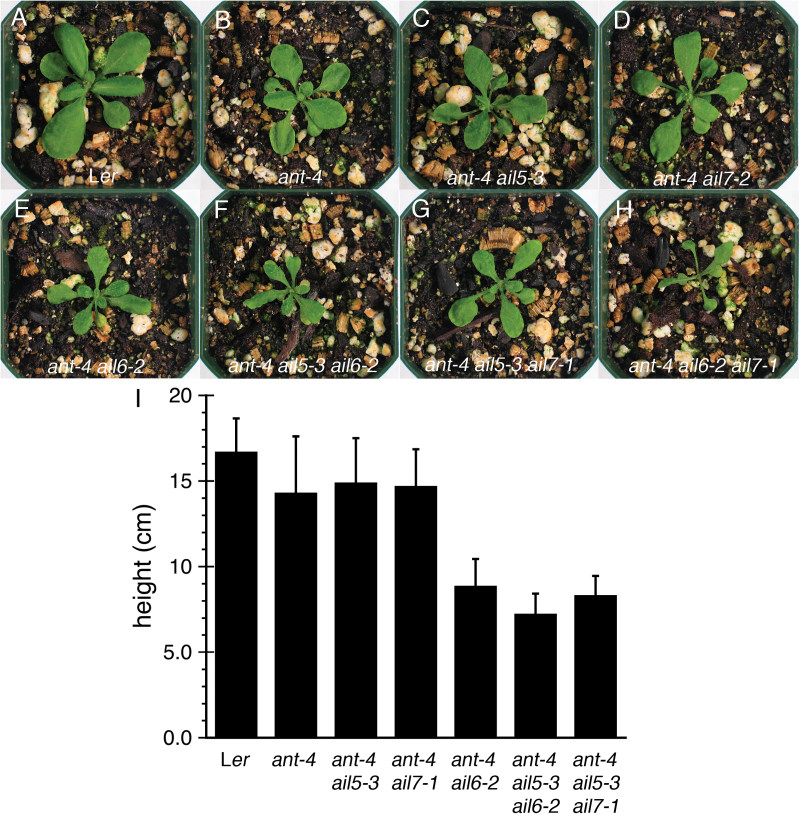
The vegetative phenotypes of ant-4 ail5-3 and ant-4 ail7-1 double mutants were also investigated. No dramatic differences in rosette leaf size or plant height were observed in either of these double mutants compared with ant-4 (Fig. 5A–D, I). This is in contrast to ant-4 ail6-2 double mutants that produce smaller rosette leaves and shorter plants compared with ant-4 (Fig. 5E, I).
Fig. 5.
AIL5 and AIL7 contribute to leaf development and shoot growth. (A). Ler plant. (B) ant-4 plant. (C) ant-4 ail5-3 plant. (D) ant-4 ail7-1 plant. (E) ant-4 ail6-2 plant. (F) ant-4 ail5-3 ail6-2 plant. (G) ant-4 ail5-3 ail7-1 plant. (H) ant-4 ail6-2 ail7-1 plant. All plants in (A–H) were 20-d-old. (I) Graph of plant heights. All plants were 42-d-old.
ant ail5 ail6 triple mutant flowers resemble ant ail6 double mutant flowers
To investigate the functions of AIL5 and AIL7 further, ant-4 ail5-3 ail6-2 and ant-4 ail5-3 ail7-1 triple mutants were made. ant-4 ail6-2 ail7-1 triple mutants were shown previously not to make flowers due to the termination of the shoot apical meristem during vegetative development (Fig. 5H) (Mudunkothge and Krizek, 2012). ant-4 ail5-3 ail6-2 triple mutant flowers resemble ant-4 ail6-2 flowers with regard to floral organ types with no petals or true stamens present in either genotype (Fig. 1F, G; Table 4). Instead both triple mutants produce flowers that primarily consist of sepals, stamenoid organs, and unfused carpel valves (Table 4). ant-4 ail5-3 ail6-2 triple mutants do show enhanced leaf and shoot phenotypes compared with ant-4 ail6-2. The triple mutant produces smaller leaves and the plants are shorter in height (Fig. 5E, F, I). This indicates that while AIL5 does not make additional contributions to flower development in the absence of ANT and AIL6, it does contribute to leaf and shoot growth in the absence of these genes.
Table 4.
Floral organ counts in ant-4 ail6-2 and ant-4 ail5-3 ail6-2 flowers at positions 1–20 on the inflorescence
| ant-4 ail6-2 1–20 | ant-4 ail5-3 ail6-2 1–20 | |
|---|---|---|
| Se | 4.24 | 3.92 |
| Flat white | 0.02 | |
| Filament | 0.78 | 1.56 |
| Swollen filament | 0.60 | 0.37 |
| Flat green | 0.02 | 0.06 |
| Stamenoid organs | 1.30 | 2.15 |
| St/Va | 0.35 | 0.17 |
| Ca Valve like (unfused) | 2.04 | 2.56 |
| Total | 9.35 | 10.79 |
ant ail5 ail7 triple mutants show more severe vegetative and reproductive phenotypes than either double mutant
Overall, ant-4 ail5-3 ail7-1 triple mutant flowers show more severe deviations from the wild type than either ant-4 ail5-3 or ant-4 ail7-1 double mutants (Fig. 1D, E, H). The triple mutant flower phenotype is complex with aspects that are novel, additive, epistatic or synergistic compared with either double mutant. ant-4 ail5-3 ail7-1 flowers contain more sepals than either double mutant (Table 1) but with similar amounts of sepal fusion as the two double mutants (Table 3). Similar to both ant-4 ail5-3 and ant-4 ail7-1 double mutants, the pattern and growth of sepal primordia is also altered in ant-4 ail5-3 ail7-1 triple mutants (Fig. 2M, N). In the second whorl of the triple mutant, the numbers of petals and filaments are intermediate between the two double mutants (Table 1). Petal size in ant-4 ail5-3 ail7-1 is also intermediate between ant-4 ail5-3 and ant-4 ail7-1 (Table 2). In the third whorl, there is more variation in organ type than observed in either double mutant with some filaments, stamen-like and petaloid stamens present in addition to normal stamens (Table 1; Fig. 2O, P). With regard to carpel number in the fourth whorl, the triple mutant resembles ant-4 ail7-1 (Table 1); however, there is a more dramatic loss of carpel valve fusion in the triple mutant (Table 3). Approximately two-thirds of ant-4 ail5-3 ail7-1 flowers contain carpels that are unfused for more than half of their length; this is almost twice the number observed in ant-4 ail7-1. These gynoecia only rarely produce ovule primordia (Fig. 3E).
In addition, the overall morphology of the floral organs and organization of the triple mutant flowers degrade with developmental time such that later-arising flowers appear increasingly disorganized (Fig. 1H). Stamens in later-arising flowers often do not make pollen, exhibit altered morphology (Fig. 2O, P), and are reduced in size. As described for ant-4 ail7-1 inflorescences, ant-4 ail5-3 ail7-1 inflorescences also show the replacement of some flowers with thick filamentous structures (Fig. 2N). ant-4 ail5-3 ail7-1 plants possess narrower leaves and are reduced in height compared with ant-4 ail5-3 and ant-4 ail7-1 double mutants (Fig. 5C, D, G, I).
Dose-dependent behaviour of AIL6 and AIL7 in the ant ail7 and ant ail6 backgrounds
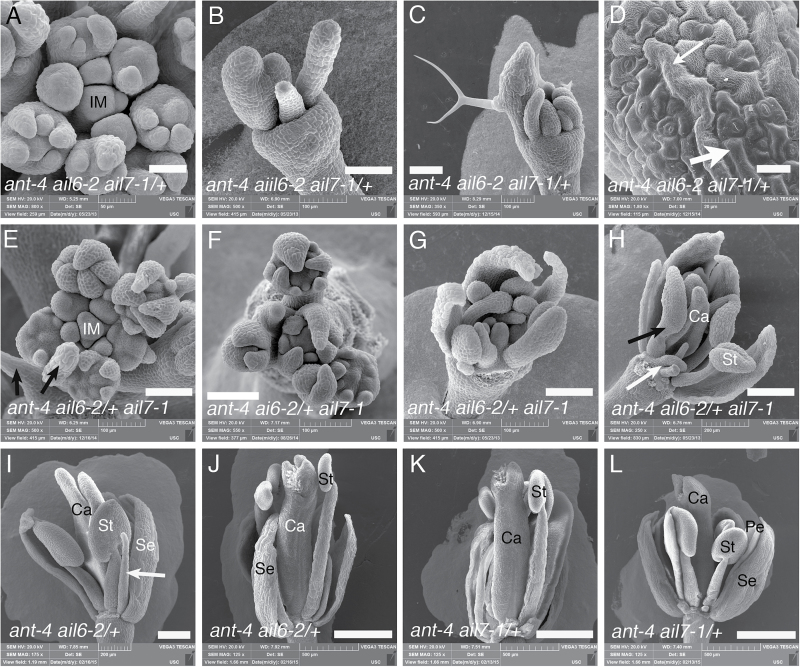
Because ant-4 ail6-2 ail7-1 plants do not possess shoot apical meristems that persist long enough to produce flowers, the effect of the combined loss of ANT, AIL6, and AIL7 on flower development cannot be examined. It was decided to investigate the consequences of the loss of a single copy of AIL6 or AIL7 in a background compromised for ANT and either AIL7 or AIL6 activity, respectively. ant-4 ail6-2 ail7-1/+ plants produce flowers that are more severe than ant-4 ail6-2 with fewer and smaller organs (Figs 1I, 6 A–C). ant-4 ail6-2 ail7-1/+ flowers produced 5.5±1.5 organs per flower while ant-4 ail6-2 flowers produced 9.5±1.9 organs. Most ant-4 ail6-2 ail7-1/+ floral organs are green and filamentous with overall morphologies that lack a resemblance to a normal floral organ (Figs 1I, 6B, C). Some of the organs arising in the outer region of these flowers contain giant cells characteristic of sepals as well as leaf-like epidermal cells (Fig. 6D). ant-4 ail6-2 ail7-1/+ inflorescences terminate with the production of filaments in place of flowers (Fig. 1J).
Fig. 6.
AIL6 and AIL7 act in a dose-dependent manner as shown by scanning electron micrographs. (A) ant-4 ail6-2 ail7-1/+ inflorescence meristem. (B, C) ant-4 ail6-2 ail7-1/+ flowers. (D) Surface of a flat organ arising in the periphery of an ant-4 ail6-2 ail7-1/+ flower. Both giant cells characteristic of sepals (thin white arrow) and leaf-like cells (thick white arrow) are present. (E) ant-4 ail6-2/+ ail7-1 inflorescence meristem. Arrows point to filaments that arise in place of flowers. (F) Inflorescence apex of ant-4 ail6-2/+ ail7-1 plant. No inflorescence meristem is visible. (G) ant-4 ail6-2/+ ail7-1 flower with many organ primordia that arise in altered positions and lack normal morphologies. (H) ant-4 ail6-2/+ ail7-1 flower with stamenoid organ (black arrow). Short filaments are present in this flower (white arrow). (I) ant-4 ail6-2/+ flower with unfused carpel valves and a second whorl filament (arrow). (J) ant-4 ail6-2/+ flower with carpels that are unfused at their apex. (K, L) ant-4 ail7-1/+ flowers showing loss of carpel fusion at the apex of the gynoecium. IM, inflorescence meristem; Se, sepal; Pe, petal; St, stamen; Ca, carpel. Scale bars, 20 μm (D), 50 μm (A), 100 μm (B, C, E–G), 200 μm (H, I), 500 μm in (J–L).
The phenotypic consequences of the loss of a single copy of AIL6 in the ant-4 ail7-1 background were investigated next. ant-4 ail6-2/+ ail7-1 plants produce flowers that have more severe defects than ant-4 ail7-1 (Fig. 1E, K). ant-4 ail6-2/+ ail7-1 flowers produce more sepals but fewer petals, stamens, and carpels, with some of these second, third, and fourth whorl organs replaced by filaments (Table 5). Floral organ initiation patterns and the growth of floral organ primordia are more severely disrupted than in ant-4 ail7-1 flowers (Fig. 6E–G). Like ant-4 ail5-3 ail7-1, the ant-4 ail6-2/+ ail7-1 floral phenotype becomes more severe with developmental age. Later-arising ant-4 ail6-2/+ ail7-1 flowers exhibit some loss of normal floral organ morphologies (Figs 1K, 6H). ant-4 ail6-2/+ ail7-1 plants also show several inflorescence meristem defects. Filaments are sometimes produced in place of flowers (Fig. 6E). In addition, the inflorescence meristem often terminates with the production of flowers (Fig. 6F). In other cases, the inflorescence meristems get wider, splitting in half with both halves of the meristem continuing to initiate new flowers (Fig. 1L).
Table 5.
Floral organ counts in ant-4 ail7-1 and ant-4 ail6-2/+ ail7-1 flowers at positions 1–20 on the inflorescence
| ant-4 ail7-1 1–20 | ant-4 ail6-2/+ ail7-1 1–20 | |
|---|---|---|
| Whorl 1 | ||
| Se and Se like | 3.97 | 5.36 |
| Whorl 2 | ||
| Pe and Pe-like | 2.00 | 0.51 |
| Filament | 0.04 | 0.62 |
| St/Pe | 0.04 | 0.01 |
| Total | 2.08 | 1.14 |
| Whorl 3 | ||
| St and St-like | 5.04 | 4.35 |
| Filament | 0.02 | 0.94 |
| Pe/St | 0.05 | 0.01 |
| Total | 5.11 | 5.30 |
| Whorl 4 | ||
| Ca and Va like | 2.60 | 1.10 |
| Filament | 0.31 | |
| St/Va | 0.12 | |
| Total | 2.60 | 1.53 |
| Total of all whorls | 13.76 | 13.33 |
Dose-dependent behaviour of AIL5, AIL6, and AIL7 in the ant background
Any possible dosage effects of AIL5 were also investigated by examining flower development in ant-4 ail5-3/+ flowers. ant-4 ail5-3/+ flowers show a reduction in petal numbers with the replacement of a few petals with filaments but these defects are less severe than ant-4 ail5-3 double mutants (Tables 1, 6). The decrease in petal number gets more severe in later-arising flowers (Table 6). In addition, petals are slightly smaller than ant-4 but larger than in ant-4 ail5-3 (Table 2) indicating that the AIL5 dose does matter in a background lacking ANT activity.
Table 6.
Floral organ counts in ant and ant ail5/+ flowers at positions 1–30 on the inflorescence
| ant-4 1–10 | ant-4 11–20 | ant-4 21–30 | ant-4 ail5-3/+ 1–10 | ant-4 ail5-3/+ 11–20 | ant-4 ail5-3/+ 21–30 | |
|---|---|---|---|---|---|---|
| Whorl 1 | ||||||
| Se | 4.00 | 3.98 | 3.98 | 4.05 | 4.00 | 3.95 |
| Filament | 0.03 | |||||
| Total | 4.00 | 3.98 | 3.98 | 4.08 | 4.00 | 3.95 |
| Whorl 2 | ||||||
| Pe | 4.00 | 3.73 | 3.50 | 3.38 | 3.13 | 2.58 |
| Filament | 0.05 | 0.05 | 0.28 | 0.28 | 0.45 | |
| Total | 4.00 | 3.78 | 3.55 | 3.66 | 3.41 | 3.03 |
| Whorl 3 | ||||||
| St | 4.23 | 4.25 | 4.30 | 4.38 | 4.58 | 4.38 |
| Total | 4.23 | 4.25 | 4.30 | 4.38 | 4.58 | 4.38 |
| Whorl 4 | ||||||
| Ca | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 |
| Total | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 | 2.00 |
| Total of all whorls | 14.23 | 14.01 | 13.83 | 14.12 | 13.99 | 13.36 |
It was also found that AIL6 and AIL7 exhibit dose-dependent behaviour in the ant mutant background. For both AIL6 and AIL7, the respective ant ail/+ combination shows a phenotype intermediate between that of ant and the corresponding ant ail double mutant. ant-4 ail6-2/+ flowers produce fewer petals and filaments in place of some petals (Fig. 6I, J). In addition, these flowers show more severe defects in carpel fusion compared with ant-4 (Fig. 6I, J). Carpel fusion defects are also more common in ant-4 ail7-1/+ flowers compared with ant-4. The lack of carpel fusion in ant-4 ail7-1/+ flowers is usually restricted to the apex of the carpels (Fig. 6K, L)
Floral organ development is largely normal in ail5 ail6 ail7 triple mutants
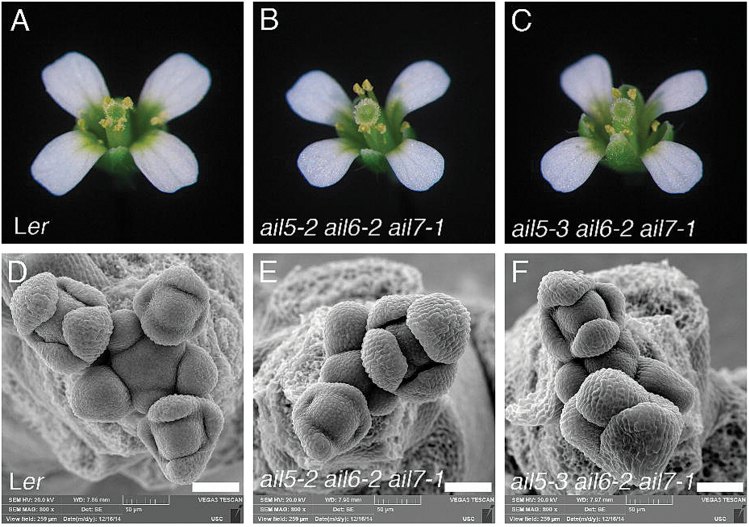
ail5-2 ail6-1 ail7-1 (i.e. plt3-1 plt5-2 plt7-1) triple mutants have previously been reported to exhibit defects in phyllotaxy, with two successive flowers often arising with divergence angles of 90 °C or 180 °C rather than 137.5 °C (Prasad et al., 2011). However, floral organ development has not been described in this triple mutant. Flower development was examined in the ail5-2 ail6-2 ail7-1 and ail5-3 ail6-2 ail7-1 triple mutants in the er background. These triple mutants did not exhibit dramatic differences in floral organ development compared with wild-type flowers (Fig. 7A–F), although the petals were slightly smaller in the triple mutant compared with Ler (Table 2). As described previously for ail5-2 ail6-1 ail7-1 ER, ail5-2 ail6-2 ail7-1 er and ail5-3 ail6-2 ail7-1 er triple mutants display alterations in the positioning of flower initiation within the inflorescence meristem (Prasad et al., 2011). The inflorescence meristems are surrounded by fewer flower primordia, which initiate at angles nearing 180 °C (Fig. 7D–F).
Fig. 7.
Loss of AIL5, AIL6, and AIL7 together has little effect on floral organ development. (A) Ler flower, (B) ail5-2 ail6-2 ail7-1 er flower. (C) ail5-3 ail6-2 ail7-1 er flower. (D) Scanning electron micrograph of Ler inflorescence. (E) Scanning electron micrograph of ail5-2 ail6-2 ail7-1 er inflorescence. (F) Scanning electron micrograph of ail5-3 ail6-2 ail7-1 er inflorescence. Scale bars, 50 μm.
Discussion
AIL5, AIL6 and AIL7 make distinct contributions to vegetative and reproductive development
Flowers produced by ant ail5, ant ail6, and ant ail7 double mutants share some phenotypic similarities. Flowers from all three double mutants show either a reduction in the number of petals (ant ail5, ant ail7) or a complete absence of petals (ant ail6). Defects in petal initiation are observed even with the loss of a single copy of AIL5 or AIL6 in the ant mutant background. Thus petal initiation seems to be particularly sensitive to AIL activity. In addition, all three ant ail double mutants show defects in sepal positioning within the periphery of the flower primordium (Fig. 2; Krizek, 2009). In the case of ant ail5-3 and ant-4 ail7-1, this often leads to the fusion of adjacent sepals. Despite these similarities, the ant ail5, ant ail6, and ant ail7 double mutants also exhibit distinct phenotypes, particularly in the inner whorls. Stamen initiation and identity specification as well as carpel positioning, growth, and patterning is only dramatically altered in ant ail6 (Krizek, 2009). Besides ant ail6 flowers, defects in carpel fusion also occur in most ant ail7 flowers, but only rarely in ant and ant ail5 flowers.
These results indicate that AIL5, AIL6, and AIL7 make distinct contributions to flower development. Each of these contributions appears to overlap completely with ANT function in floral organ development, as no defects are observed in ail5, ail6 or ail7 single mutants. In the absence of ANT, AIL5 contributes primarily to sepal positioning and growth and petal initiation and growth. By contrast, AIL6 contributes to primordium positioning and growth throughout the flower, petal and stamen identity specification, and carpel growth and patterning (Krizek, 2009). AIL7 primarily contributes to sepal positioning, petal initiation, and carpel growth. Thus, AIL6 makes more important contributions to floral organ development than AIL5 or AIL7. The distinct roles of AIL5, AIL6, and AIL7 in flower development are not unexpected given the distinct expression pattern of these genes within developing flowers (see Supplementary Fig. S1 at JXB online; Nole-Wilson et al., 2005). However, the particular phenotypes are not necessarily correlated with the spatial expression patterns of each gene. For example, in wild-type flowers, AIL7 mRNA is present only in the centre of the floral primordium starting at stage 2 of flower development yet ant ail7 double mutants show defects in the positioning of sepal primordia in the periphery of stage 3 flowers. This could indicate a non-cell autonomous function of AIL7. A recent report suggests that PLT proteins can move between cells in the root (Mähünen et al., 2014). Alternatively, the expression pattern of AIL7 could be altered in the ant mutant background.
Increasing loss of AIL function in flowers is generally but not always associated with increasingly more severe deviations from wild-type flower development. An exception is the ant ail5 ail6 triple mutant, which closely resembles the ant ail6 double mutant suggesting that the AIL5 function completely overlaps the combined activities of ANT and AIL6. However, rather than acting in a partially redundant fashion, there are some suggestions of complex genetic interactions among AIL genes in flowers. For example, certain aspects of the ant ail5 ail7 triple mutant phenotype, such as an increased numbers of sepals and stamens, are novel with respect to either double mutant. Other aspects such as petal number and petal size are intermediate in value between the two doubles, suggesting that mutations in AIL7 partially rescue the petal defects of ant ail5 double mutants. It is possible that AIL5 and AIL7 have somewhat opposing actions in petal growth and may regulate common target genes in opposite directions. Petal area and petal width may be slightly increased in ant ail7 double mutants compared with ant single mutants (Table 1). Complex and possible antagonistic behaviours for ANT/AIL6 and AIL7 within the inflorescence meristem have been noted previously (Mudunkothge and Krizek, 2012). Opposing activities of AIL6 and AIL7 in the flower might contribute to the slight rescue of petal defects in the ant ail5 ail7 triple mutant if AIL6 activity is increased upon the loss of AIL7.
AIL5, AIL6, and AIL7 all contribute to vegetative growth in the absence of ANT. Generally, increasing loss of AIL function results in smaller rosette leaves and shorter plants, although AIL5, AIL6, and AIL7 do not make equivalent contributions to vegetative growth either. Once again, AIL6 appears to play a more important role than AIL5 or AIL7, as ant ail6 double mutant plants are the shortest of the ant ail double mutants.
Unequal genetic redundancy within the AIL family
Genetic redundancy occurs as a consequence of gene duplication. The initial full genetic redundancy of duplicated genes often disappears as one gene acquires mutations resulting in nonfunctionalization, neofunctionalization or subfunctionalization. Partial genetic redundancy results when neither gene is sufficient by itself to provide the ancestral function. In the case of unequal genetic redundancy, loss of one of the two genes results in a phenotype, loss of the other gene has no effect, while loss of both genes results in a more severe phenotype. This is the case with the AIL gene family in flowers. Loss of the ANT function results in a mutant floral phenotype, no mutant phenotype results from the loss of AIL5, AIL6 or AIL7, and an enhanced phenotype is observed in ant ail5, ant ail6, and ant ail7 double mutants. Such unequal genetic redundancy implies that the trait controlled by the genes depends in a quantitative manner on the sum of the activities of the two genes (Briggs et al., 2006). Although dispensable on its own, the remaining activity provided by the duplicated gene makes a significant contribution to the overall activity of the gene pair as shown by the severity of the phenotype upon the loss of both genes. Unequal genetic redundancy has been reported a number of times in Arabidopsis (Briggs et al., 2006).
The unequal redundancy of AIL genes in flower development may result from different expression levels and/or expression patterns of the genes. ANT is expressed at much higher levels than AIL5, AIL6 or AIL7 and in a broader domain within developing flowers (Nole-Wilson et al., 2005). Alternatively, the unequal genetic redundancies could be a consequence of different protein activities. It will be interesting to investigate the molecular basis for the unequal redundancies of AIL genes in flowers.
Unequal genetic redundancy is not observed in four AIL genes that function in the root: PLETHORA1 (PLT1), PLT2, PLT3/AIL6, and BBM/PLT4. These four genes contribute to root growth and root stem cell maintenance in an overlapping and largely additive manner (Galinha et al., 2007). Single mutations in each of these genes has modest effects on root development while higher order mutants show increasingly severe defects with plt1 plt2/+ plt3 bbm plants sometimes lacking root and hypocotyl (Aida et al., 2004; Galinha et al., 2007). The two most highly expressed PLT genes, PLT1 and PLT2, do make more significant contributions to root development than PLT3/AIL6 and BBM/PLT4 (Galinha et al., 2007). These different contributions result primarily from differences in expression and, to some extent, distinct protein activities (Galinha et al., 2007).
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. AIL family tree and summary of gene expression data.
Supplementary Fig. S2. ail5 mutants have a wild-type appearance.
Supplementary Table S1. Primers used in this study
Acknowledgements
I thank Soumitra Ghoshroy and the Electron Microscopy Centre staff for advice on the use of the SEM, Robert Franks for advice on the gynoecium clearing procedure, the Salk Institute Genomic Analysis Laboratory for providing the sequence-indexed Arabidopsis T-DNA insertion mutants, and the ABRC for Arabidopsis seeds. This work was supported by National Science Foundation (NSF) grants IOS 0922367 and IOS 1354452.
Glossary
Abbreviations:
- ANT
AINTEGUMENTA
- AIL5
AINTEGUMENTA-LIKE5
- AIL6
AINTEGUMENTA-LIKE6
- AIL7
AINTEGUMENTA-LIKE7
- RT-PCR
reverse transcription PCR.
References
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh Y-S, Amasino R, Scheres B. 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119, 109–120. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Baker SC, Robinson-Beers K, Villanueva JM, Gaiser JC, Gasser CS. 1997. Interactions among genes regulating ovule development in Arabidopsis thaliana . Genetics 145, 1109–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, et al. 2002. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell 14, 1737–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. 2006. Unequal genetic redundancies in Arabidopsis: a neglected phenomenon? Trends in Plant Sciences 11, 1360–1385. [DOI] [PubMed] [Google Scholar]
- Chandler JW, Jacobs B, Cole M, Cornelli P, Werr W. 2011. DORNROSHCEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Molecular Biology 76, 171–185. [DOI] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQJ, Gerentes D, Perez P, Smyth DR. 1996. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell 8, 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449, 1053–1057. [DOI] [PubMed] [Google Scholar]
- Hofhuis H, Laskowski M, Du Y, Prasad K, Grigg S, Pinon V, Scheres B. 2013. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Current Biology 23, 956–962. [DOI] [PubMed] [Google Scholar]
- Horstman A, Willemsen V, Boutilier K, Heidstra R. 2014. AINTEGUMENTA-LIKE proteins: hubs in a plethora of networks. Trends in Plant Sciences 19, 146–157. [DOI] [PubMed] [Google Scholar]
- Ito T, Ng K-H, Lim T-S, Yu H, Meyerowitz EM. 2007. The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis . The Plant Cell 19, 3516–3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Wellmer F, Yu H, Das P, Ito N, Alves-Ferreira M, Riechmann JL, Meyerowitz EM. 2004. The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS . Nature 430, 356–360. [DOI] [PubMed] [Google Scholar]
- Kaufmann K, Wellmer F, Muino JM, et al. 2010. Orchestration of floral initiation by APETALA1. Science 328, 85–89. [DOI] [PubMed] [Google Scholar]
- Klucher KM, Chow H, Reiser L, Fischer RL. 1996. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2 . The Plant Cell 8, 137–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA. 1999. Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs. Developmental Genetics 25, 224–236. [DOI] [PubMed] [Google Scholar]
- Krizek BA. 2009. AINTEGUMENTA and AINTEGUMENTA-LIKE6 act redundantly to regulate Arabidopsis floral growth and patterning. Plant Physiology 150, 1916–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek BA, Eaddy M. 2012. AINTEGUMENTA-LIKE6 regulates cellular differentiation in flowers. Plant Molecular Biology 78, 199–209. [DOI] [PubMed] [Google Scholar]
- Krizek BA, Fletcher JC. 2005. Molecular mechanisms of flower development: an armchair guide. Nature Review Genetics 6, 688–698. [DOI] [PubMed] [Google Scholar]
- Mähünen AP, ten Tusscher K, Siligato R, Smetana O, Díaz-Triviño S, Salojarvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. 2014. PLETHORA gradient formation mechanism separates auxin responses. Nature 515, 125–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y, Fischer RL. 2000. Plant organ size control: AINTEGUMENTA regulates growth and cell numbers during organogenesis. Proceedings of the National Academy of Sciences, USA 97, 942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudunkothge JM, Krizek BA. 2012. Three Arabidopsis AIL/PLT genes act in combination to regulate shoot apical meristem function. The Plant Journal 71, 108–121. [DOI] [PubMed] [Google Scholar]
- Nambara E, Suzuki M, Abrams S, McCarty DR, Kamiya Y, McCourt P. 2002. A screen for genes that function in abscisic acid signaling in Arabidopsis thaliana . Genetics 161, 1247–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Krizek BA. 2006. AINTEGUMENTA contributes to organ polarity and regulates growth of lateral organs in combination with YABBY genes. Plant Physiology 141, 977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nole-Wilson S, Tranby T, Krizek BA. 2005. AINTEGUMENTA-like (AIL) genes are expressed in young tissues and may specify meristematic or division-competent states. Plant Molecular Biology 57, 613–628. [DOI] [PubMed] [Google Scholar]
- O’Maoleidigh DS, Wuest SE, Rae L, et al. 2013. Control of reproductive floral organ identity specification in Arabidopsis by the C function regulator AGAMOUS. The Plant Cell 25, 2482–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad K, Grigg SP, Barkoulas M, et al. 2011. Arabidopsis PLETHORA transcription factors control phyllotaxis. Current Biology 21, 1123–1128. [DOI] [PubMed] [Google Scholar]
- Reinhardt D, Eva-Rachele P, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C. 2003. Regulation of phyllotaxis by polar auxin transport. Nature 426, 255–260. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Muino JM, et al. 2012. Characterization of MADS-domain transcription factor complexes in Arabidopsis flower development. Proceedings of the National Academy of Sciences, USA 109, 1560–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trost G, Vi SL, Czesnick H, Lange P, Holton N, Giavalisco P, Zipfel C, Kappel C, Lenhard M. 2014. Arabidopsis poly(A) polymerase PAPS1 limits founder-cell recruitment to organ primordia and suppresses the salicylic acid-independent immune response downstream of EDS1/PAD4. The Plant Journal 77, 688–699. [DOI] [PubMed] [Google Scholar]
- Tsuwamoto R, Yokoi S, Takahata Y. 2010. Arabidopsis EMBRYOMAKER encoding an AP2 domain transcription factor plays a key role in developmental change from vegetative to embryonic phase. Plant Molecular Biology 73, 481–492. [DOI] [PubMed] [Google Scholar]
- Wuest SE, O’Maoleidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F. 2012. Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proceedings of the National Academy of Sciences, USA 109, 13452–13457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn AN, Seaman AA, Jones AL, Franks RG. 2014. Novel functional roles for PERIANTHIA and SEUSS during floral organ identity specification, floral meristem termination, and gynoecial development. Frontiers in Plant Science doi: 10.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K, Tatematsu K, Yano R, Preston J, Kitamura S, Takahashi H, McCourt P, Kamiya Y, Nambara E. 2009. CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant and Cell Physiology 50, 330–340. [DOI] [PubMed] [Google Scholar]
- Yano R, Kanno Y, Jikumaru Y, Nakabayashi K, Kamiya Y, Nambara E. 2009. CHOTTO1, a putative double APETALA2 repeat transcripton factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis . Plant Physiology 151, 641–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.