Abstract
Background & Aims
Over-expression of FoxM1 correlates with poor prognosis in hepatocellular carcinoma (HCC). Moreover, the Ras-signaling pathway is found to be ubiquitously activated in HCC through epigenetic silencing of the Ras-regulators. We investigated the roles of FoxM1 in Ras-driven HCC, and on HCC cells with stem-like features.
Methods
We employed a transgenic mouse model that expresses the oncogenic Ras in the liver. That strain was crossed with a strain that harbor floxed alleles of FoxM1 and the MxCre gene that allows conditional deletion of FoxM1. FoxM1 alleles were deleted after development of HCC, and the effects on the tumors were analyzed. Also, FoxM1-siRNA was used in human HCC cell lines to determine its role in the survival of the HCC cells with stem cell features.
Results
Ras-driven tumors over-express FoxM1. Deletion of FoxM1 inhibits HCC progression. There was increased accumulation of reactive oxygen species (ROS) in the FoxM1-deleted HCC cells. Moreover, FoxM1-deletion caused a disproportionate loss of the CD44+ and EpCAM+ HCC cells in the tumors. We show that FoxM1 directly activates expression of CD44 in human HCC cells. Moreover, the human HCC cells with stem cell features are addicted to FoxM1 for ROS-regulation and survival.
Conclusion
Our results provide genetic evidence for an essential role of FoxM1 in the progression of Ras-driven HCC. In addition, FoxM1 is required for the expression of CD44 in HCC cells. Moreover, FoxM1 plays a critical role in the survival of the HCC cells with stem cell features by regulating ROS.
Keywords: FoxM1, Ras-driven liver cancer, liver cancer cells with stem cell features
Introduction
Hepatocellular carcinoma (HCC), the most common form of liver cancer, is the third most leading cause for cancer related deaths (1). Etiologically, HCC is linked to mainly viral hepatitis and alcohol abuse. However, in the developed countries, nonalcoholic fatty liver disease or nonalcoholic steatohepatitis (NASH) also has become a significant contributing factor towards the development of HCC (1). Recent studies identified ubiquitous activation of the Ras signaling pathway in HCC. For example, the Ras effector proteins RASSF1A, NORE1A/B are epigenetically silenced in HCC (2). One study reported epigenetic silencing of RASSF1A and NORE1B in 97% of HCC (3). More recent studies characterized epigenetic silencing of the Ras GTPase-activating proteins (GAPs) in HCC. For example, over 70% of HCC patients exhibit silencing of RASAL1 and DAB2IP (4). Also, NF1, another Ras GTPase activating protein, is silenced in HCC. PITX1, a transcription factor that activates expression of RASAL1, is also suppressed by promoter methylation in about 42% of HCC, and in greater than 80% of HCC with poor prognosis. Reduced expression or silencing of the GAPs causes increase in the levels of the GTP-bound active form of the Ras protein, leading to activated Ras-signaling in HCC. Interestingly, another study with 69 Chinese patients demonstrated mutations in H-Ras, codons 40 and 61, in about 71% of the patients (5).
Mounting evidence also indicates the presence of cells with stem cell properties in HCC (6). These cells were characterized based on their ability to generate tumors in immune-deficient mice. For example, CD90+, CD44+, CD133+, CD13+ and EpCAM+ cells in HCC are considered to possess features of stem cells (6, 7). A recent study, using a chemical carcinogenesis protocol that gives rise to HCC in mice, identified and characterized HCC precursor cells (HcPCs) in mouse liver (8). The HcPCs appear several months before appearance of HCC. These cells, which are not fully transformed HCC cells, give rise to tumor only when suitable microenvironment is provided, such as CCl4-damaged liver (8). Interestingly, these precursor cells include EpCAM+ and CD44+ cells, suggesting the possibility that at least some of the liver cancer cells with stem-like properties are derived from the HcPCs.
The fork-head box family transcription factor FoxM1 is important in liver cancer. It is over-expressed in HCC and its over-expression coincides with poor prognosis for liver cancer patients (9). FoxM1 stimulates expression of genes required for G1 to S progression and G2 to M progression, and thus, supports cell proliferation (10, 11). Interestingly, FoxM1 also stimulates expression of the anti-oxidant genes, such as MnSOD, Catalase and PRDX3 (12). That function of FoxM1 plays important role in attenuating oxidative stress in tumor cells especially the ones that generate a lot of reactive oxygen species (ROS). Several studies also indicated a role of FoxM1 in the activation of the “stemness” genes expression, including Oct4, Nanog, c-Myc, Sox2 and Bmi1 (13-15). Interestingly, in glioma, FoxM1 was shown to collaborate with the Wnt-signaling pathway to increase the levels of b-catenin in the nucleus (16, 17).
In mouse model, FoxM1 is essential for the development of liver cancer. For example, in a chemical carcinogenesis protocol, diethylnitrosamine (DEN) induces liver cancer in the male mice with a penetrance of nearly 100%. Male mice lacking FoxM1 expression in the adult liver, when exposed to DEN, did not develop liver cancer (18). Deregulated FoxM1, on the other hand, drives aggressive liver cancer progression (19). The transcriptional activity of FoxM1 is inhibited by the tumor suppressor ARF (18). Mice over-expressing FoxM1 in the absence of ARF (Arf-null background) when subjected to the DEN liver carcinogenesis protocol develop highly aggressive metastatic liver cancer (19). In the absence of ARF, over-expression of FoxM1 activates genes that are involved in all steps of metastasis, including those involved in the colonization of a distal organ. In that regard, it is noteworthy that FoxM1 has been implicated in the metastasis of other tumors (20-22).
In this study, we provide genetic evidence that targeting FoxM1 inhibits progression of liver cancer induced by activated Ras. Moreover, targeting FoxM1 preferentially eliminates cells with stem cell features in human HCC cell lines as well as CD44+ and EpCAM+ HCC cells in Ras-induced mouse liver tumors.
Materials and methods
Animal Studies
All animal experiments were preapproved by the UIC institutional animal care and use committee. Previously described H-ras12V mice (23) were crossed with FoxM1 fl/fl MxCre C57/BL6 mice (24) to obtain FoxM1 fl/fl MxCre H-ras12V and FoxM1 +/+ MxCre H-ras12V mice. For deletion studies, 8 months old male mice (FoxM1 +/+ MxCre H-ras12V and FoxM1 fl/fl MxCre H-ras12V) were subjected to five or ten ip injections (every other day) with 250 μg of synthetic synthetic polyinosinic polyinosinic-polycytidylic acid (polyIpolyC) (Sigma-Aldrich, St. Louis, MO) to induce expression of the Mx-Cre transgene. For xenograft tumor, male Nu/Nu strain mice were purchased from Charles River Laboratories (USA). Huh7 cells were transfected with control or FoxM1 siRNA. Twenty-four hours post transfection, cells (total of 1×106) were subcutaneously injected.
Immunohistochemistry
Immunohistochemical stainings were performed following standard procedure. Antigen retrieval was done using sodium citrate buffer and sections were then treated with antibodies overnight. Additional blocking step was performed using Avidin/biotin Vectastain kit following manufacturer's protocol. Antibody information can be found in Table S1. Visualization was done using DAB and counterstained using Hematoxylin (Polyscientific, Bay Shore, NY). For antibodies of mouse origin, mouse on mouse (MOM) kit was used. All used reagents are from Vector Labs (Burlingame, CA) unless otherwise indicated.
RT-PCR, Western Blot, and Chromatin Immunoprecipitation
RNA was Trizol extracted (Invitrogen, Carlsbad, CA) and cDNA was synthesized using Bio-Rad reverse transcriptase (Bio-Rad, Hercules, CA). cDNA was amplified using SYBR Green (Bio-Rad, Hercules, CA) and analyzed via iCycler software. All primer sequences are shown in Table S2. Western blots and chromatin-IPs were performed following previously described procedures (25). Antibodies are listed in Table S1.
FACS analysis and cell sorting
Antibodies used for FLOW analysis are listed in Table S1. Cells (re-suspended in PBS with 2%FBS and 2mM EDTA) were incubated with PE- and/or FITC-conjugated antibodies for 20 min on ice. More detailed description of FACS analysis and cell sorting can be found in Supplemental Material.
Statistical analysis
Statistical significance was calculated by the Student's t test (2 tailed). Statistically significant changes were indicated with asterisks (*, p < 0.05; **, p < 0.01, ***, p<0.001).
Results
Targeting FoxM1 inhibits Ras-induced HCC progression
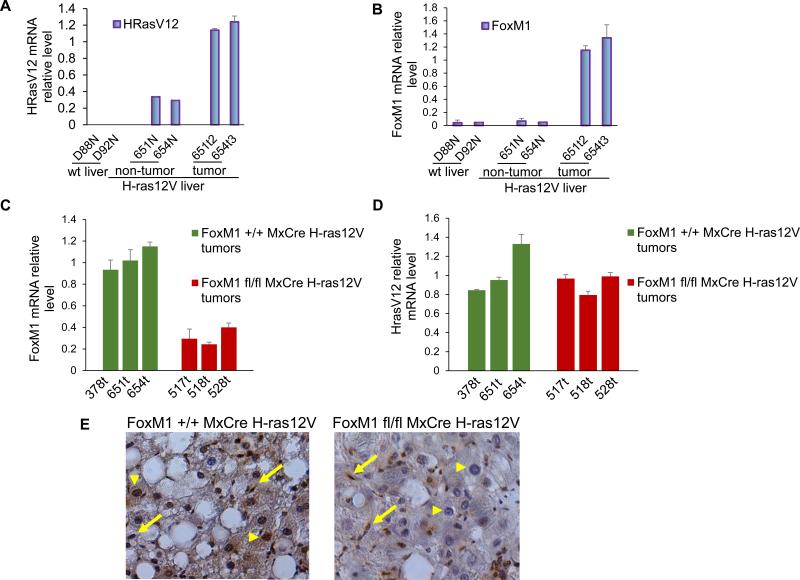
The Ras-signaling pathway is frequently activated in HCC (4). A transgenic mouse strain expressing H-ras12V oncogene specifically in the liver, in which expression is driven by the albumin promoter, has been described (23). The male mice of this transgenic strain develop hepatocellular adenomas by 6 to 8 months followed by HCC at 9 months of age with high penetrance (>88%). Also, there is evidence for micro- and macrovesicular steatosis in both female and male mice (26). To investigate the roles of FoxM1 in HCC progression in that model, we generated a tri-transgenic strain that in addition to H-ras12V contained floxed alleles of FoxM1 and the MxCre gene. MxCre allows conditional deletion of the FoxM1 alleles. Consistent with the previous findings (27), we observed higher expression of the H-ras12V mRNA in the tumor tissue of the transgenic mice in comparison to the non-tumor tissue sections (Fig. 1A). The H-ras12V induced HCC exhibited much higher expression of FoxM1 mRNA (Fig. 1B). To delete FoxM1 in the tumor we injected the male mice at 8 months of age with polyIpolyC (five injections every other day) that activates expression of Cre recombinase from the Mx promoter. Total RNAs from the tumor tissues of FoxM1 deleted (FoxM1 fl/fl) and undeleted (FoxM1+/+) were compared. We detected a significant reduction of the FoxM1-mRNA in the tumors from the FoxM1-deleted samples. However, the reduction of the FoxM1 mRNA was not very quantitative (Fig. 1C). The reason for a partial loss of FoxM1- mRNA became clear from immunohistochemical staining for FoxM1. In the undeleted samples (FoxM1+/+), clear FoxM1 expression in the nucleus was detected in the HCC cells as well as in the fibroblast-like cells in the tumor nodules (See arrow in Fig. 1E). In the FoxM1 deleted samples (FoxM1 fl/fl), we observed clear loss of FoxM1 signals from the HCC cells (arrow head), but not from the other cells. Therefore, it appears that there is some specificity in the expression of Cre in this system that allows specific deletion of FoxM1 in the HCC cells. Interestingly, loss of FoxM1 expression had negligible effect on the expression of H-ras12V mRNA in the tumors (Fig. 1D).
Fig. 1. Loss of FoxM1 expression in the HCC cells following MxCre induced deletion in transgenic mice expressing H-ras12V.
Total RNA from (FoxM1+/+) normal liver, transgenic non-tumor and tumor tissues were assayed for expression of H-ras12V (A) and FoxM1 (B). FoxM1-mRNA (C) and H-ras12V-mRNA (D) levels without (FoxM1+/+) and with deletion of FoxM1 (FoxM1fl/fl) were compared. The RNAs were assayed by quantitative RT-PCR. (E) Tumor sections from FoxM1 undeleted (FoxM1+/+) and FoxM1 deleted (FoxM1fl/fl) samples were compared by immunohistochemcal staining for FoxM1. Arrows indicate fibroblast-like cells in tumor sections and arrowheads indicate the HCC cells.
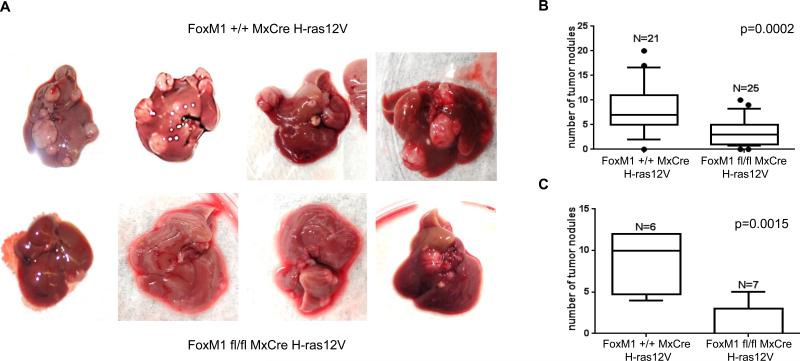
In the H-ras12V model, the number of HCC nodules and their sizes varied significantly. Therefore, to measure an effect of FoxM1 deletion we analyzed the tumor nodules from large cohorts of mice following 5 ip injections of polyIpolyC. We counted the total number of the HCC nodules irrespective of size. Deletion of FoxM1 caused a significant decrease in the number of tumor nodules (Fig. 2A, B). We also compared mice following 10 ip injections of polyIpolyC. The decrease in the number of nodules was significantly greater in that latter set of FoxM1 fl/fl animals. Some of the mice (2 out of 7) had no tumor nodule to be detected (Fig. 2C, and Fig. 1S).
Fig. 2. Decrease in the number of tumor nodules following deletion of FoxM1.
Transgenic male mice (Alb-H-ras12V FoxM1+/+ MxCre and Alb-H-ras12V FoxM1fl/fl MxCre) at 8 months of age were injected with 5 dosages of polyIpolyC (panels A and B) or 10 dosages (panel C) of polyIpolyC. The numbers of the tumor nodules were compared 4 weeks after the last injection. Box plots for the numbers are shown along with p values.
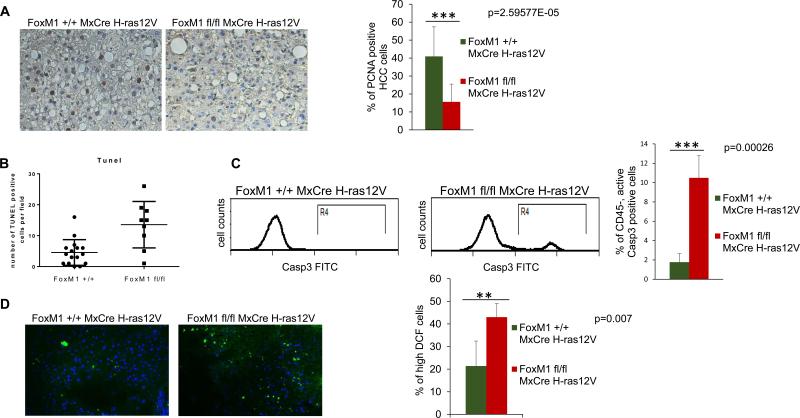
To further analyze the consequence of FoxM1 loss, sections from approximately equal size surviving tumor nodules were analyzed for PCNA expression by immunohistochemical staining. We quantified only the HCC cells for PCNA expression. There was a significant decrease in the PCNA positive HCC cells in the tumor sections from the FoxM1-deleted samples (Fig. 3A), indicating a decrease in proliferation. To analyze the mechanism of reduced proliferation in FoxM1-deleted tumors, we performed immunohistochemistry staining for Cyclin E and Plk1 (Supplementary Fig. 2), pro-proliferation factors that are known transcriptional targets of FoxM1. As expected, we observed decrease in the percentage of Cyclin E and Plk1 positive cells in the tumors following FoxM1 deletion. Moreover, we analyzed RNA isolated from the FoxM1 +/+ and FoxM1 fl/fl tumors, and observed decrease in the expression of Cdc25B, Aurora B and Plk1 in the FoxM1-deleted tumors (Supplementary Fig. 3). These results would explain the reduced proliferation observed in the FoxM1-deleted tumors.
Fig. 3. Decreased proliferation, increased apoptosis and increased accumulation of ROS in tumors following deletion of FoxM1.
Tumor sections from mice (Alb-H-ras12V FoxM1+/+ MxCre and Alb-H-ras12V FoxM1fl/fl MxCre) following 5 injections of polyIpolyC were compared for PCNA expression by immunohistochemical staining (A). The tumor sections were also subjected to TUNEL assays (B); Single cell suspensions of the tumor tissues were compared also for active caspase3+ cells by flow-cytometry (C). Frozen sections of the tumor tissues were compared for accumulation of ROS following treatments with DCF-DA (D). The sections were treated with 10 μM 5-(6)-chloromethyl-2-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) (Invitrogen, Carlsbad, CA) for 30 min at 37 °C and counterstained with DAPI. Images were taken using a fluorescent microscope at ×20 magnification.
The HCC cells in the tumor sections were analyzed also for TUNEL. In addition, we looked at the active caspase3 positive cells. Clearly, deletion of FoxM1 caused a significant increase in the TUNEL and active caspase3 positive cells, indicating an increased apoptosis (Fig. 3B and C). To investigate the cause of increased apoptosis in the FoxM1-deleted tumors, we stained tumors for the anti-apoptotic protein Survivin whose expression is regulated by FoxM1. The results clearly show decrease in the percentage of Survivin expressing cells in FoxM1-deleted tumor samples (Supplemental Figure 2). Previous cell culture studies indicated that the oncogenic Ras expressing cells depends upon FoxM1 to regulate the levels of the reactive oxygen species (ROS) (12). Consistent with that, we observed increased ROS in the tumor sections from the FoxM1-deleted samples compared to the undeleted samples (Fig. 3D). Together, these observations on decrease in the number of tumor nodules and increased apoptosis along with loss of proliferation in the remaining tumors upon deletion of FoxM1 provide genetic evidence that targeting FoxM1 inhibits H-ras12V-induced HCC progression. It is possible that MxCre might have deleted FoxM1 in other cells in the tumor nodules, such as the macrophages, that contributed to the apoptosis of HCC cells. But, that is expected also from a therapeutic agent that would target FoxM1 in liver cancer. In that regard, the inhibition of HCC progression is significant because there was no noticeable side effects of the MxCre mediated deletion of FoxM1 in the adult mice.
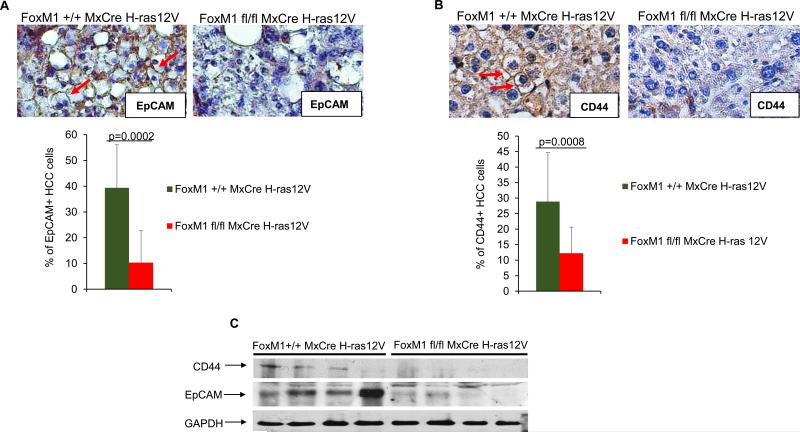
Deletion of FoxM1 causes a disproportionate loss of the EpCAM+ and the CD44+ HCC cells in the H-ras12V model
A recent study characterized precursors of HCC cells (8). These cells are not fully transformed and give rise to HCC only in specific context, and they contain cells that are positive for EpCAM as well as cells positive for CD44. Interestingly, these markers are found also in the liver cancer cells with stem cell features (6). We analyzed for the presence of those cells in the H-ras12V-induced HCC with and without deletion of FoxM1. Immunohistochemical staining of tumor section, derived from comparable size nodules, with EpCAM antibody indicated that a significant population of the HCC cells, about 40%, were positive for EpCAM. Interestingly, deletion of FoxM1 caused a severe reduction in the EpCAM expressing HCC cells (Fig. 4A). Next we examined tumors for the CD44+ cells and observed significant loss of these cells in FoxM1-deleted tumors compared to the control tumors (Fig. 4B). We compared 6 tumors of each genotype by immunohistochemical staining, and counted CD44 expressing HCC cells. While percentage of CD44+ cells in control tumor sections averaged to 28.9%, the percentage of those cells in the FoxM1-deleted tumor sections was significantly lower, with an average of 12.2%. The reduction in EpCAM and CD44 was evident also in a western blot assay (Fig. 4C). Together, these observations on decrease in the percentages of the EpCAM+ and CD44+ HCC cells over the total number of the HCC cells in the FoxM1-deleted tumors suggest that those cells are more dependent upon FoxM1 in comparison to the other HCC cells. We speculate that these cells originated from the liver cancer progenitor cells known to be CD44 and EpCAM positive (8), or these cells are bona fide liver cancer cells with stem cell features. Interestingly, the FoxM1-deleted tumor specimens also exhibited decrease in expression of “stemness” genes Bmi1, Nanog and c-Myc (Supplemental Fig. 3).
Fig. 4. Loss of the EpCAM+ and CD44+ HCC cells in the tumors following deletion of FoxM1.
Tumor sections from mice (Alb-H-ras12V FoxM1+/+ MxCre and Alb-H-ras12V FoxM1fl/fl MxCre) following 5 injections of polyIpolyC were compared for HCC cells that are positive for EpCAM (A) and CD44 (B). Quantification of the percentages of the EpCAM+ and CD44+ HCC cell over the total number of HCC cells are shown. For EpCAM+ cells, we quantified HCC cells from 9 different fields. For CD44+ cells, we quantified HCC cells from 16 different fields and 6 different tumors per genotype. (C) Protein extracts (150 ug) from tumor fragments were assayed by western blot.
FoxM1 activates expression of CD44 in HCC cells
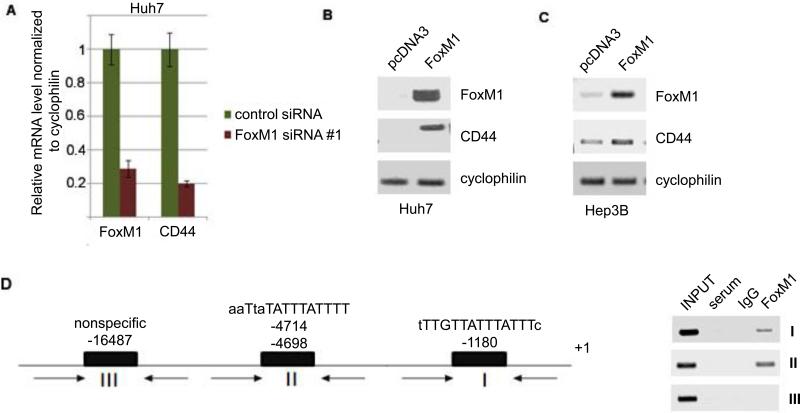
CD44 was shown to be important for the liver cancer cells with stem cell features (7, 28). Inhibition of CD44 leads to apoptosis of those cells in HCC (7). We observed that both mouse and human CD44 promoters contain FoxM1-binding sites. Therefore, we considered the possibility that CD44 might be a downstream transcriptional target of FoxM1. To test that, we analyzed human HCC cell lines Huh7 and Hep3B. The Ras-signaling pathway is active in these cell lines, as inhibition of that pathway reduces viability of the cells (29). We employed siRNA to deplete FoxM1. Depletion of FoxM1 caused a significant loss in the levels of CD44 mRNA (Fig. 5A). In a reciprocal experiment, we transfected a plasmid expressing FoxM1 into both Huh7 and Hep3B cell lines. Expression of FoxM1 caused an increase in the expression of CD44, indicating that FoxM1 regulates the mRNA levels of CD44 (Fig. 5B and C). Also, we performed chromatin immunoprecipitation (ChIP) in Huh7 cells to see an interaction of FoxM1 with the putative binding sites in the CD44 promoter (Fig. 5D). The results show that FoxM1 could associate with the CD44 promoter in the HCC cells and stimulate its expression. Surprisingly, in breast cancer cell line MDA-MB-231, we did not see a requirement for FoxM1 in the expression of CD44 (not shown). It is likely that FoxM1 stimulates CD44 expression in a cell type specific fashion.
Fig. 5. FoxM1 binds to the promoter of CD44 and activates its expression.
(A) Human HCC cells line Huh7 was transfected with control or FoxM1-siRNA #1. Seventy-two hours after transfection, total RNAs were assayed for FoxM1 and CD44 by quantitative RT-PCR. (B, C) Huh7 cells and Hep3B cells were transfected with control plasmid (pcDNA3) or plasmid expressing FoxM1. Forty-eight hours after transfection, total RNAs were compared for FoxM1 and CD44 mRNAs by semi-quantitative PCR assays. Cyclophilin-mRNA was assayed as loading control. (D) Upper panel, Putative FoxM1-binding sites in the human CD44 promoter are shown. The schematic also includes the location of the PCR primers (arrows) used in the chromatin-IP assays. Chromatin-IP assays indicating FoxM1 binding to the two proximal sites are shown (D).
Inhibition of FoxM1 preferentially eliminates the cancer cells with stem cell features
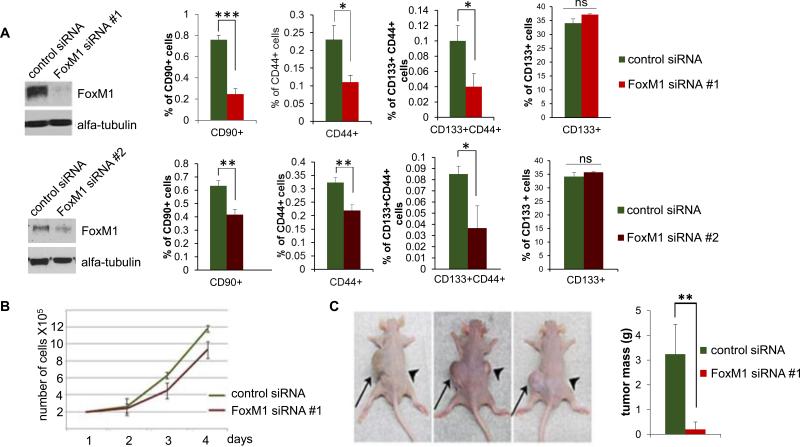
FoxM1 has been shown to stimulate expression of the “stemness” genes, including Sox2, Oct4, Nanog, c-Myc, Bmi1 and others (13-15). We investigated whether expression of those genes in HCC cells involve FoxM1. Using siRNA-mediated knockdown of FoxM1, as well as by over-expression of FoxM1, we observed evidence that expression of the “stemness” genes in HCC involves FoxM1 (Fig. 4S). Interestingly, several studies characterized cell surface markers for cells with stem cell properties in HCC (30). To investigate the effects of FoxM1-depletion on those cells, Huh7 cells were transfected with 2 independent siRNA against FoxM1 (FoxM1 siRNA #1 or FoxM1 siRNA #2) or control siRNA (Fig. 6A). The depletion of FoxM1 had only a marginal effect on cell growth (Fig. 6B). The transfected cells were then analyzed for the CD90+, CD133+ and CD44+ cells by flow cytometry. We observed significant decreases in the CD90+, CD44+ and CD133+CD44+ cells, while FoxM1 deletion did not have a statistically significant effect on the total CD133+ cell population. Consistent with the loss of the stem-like cancer cells, the FoxM1-depleted cells failed to generate tumor when injected subcutaneously in athymic nude mice (Fig. 6C). Lack of tumorigenicity was observed also in soft agar colony formation assay using other HCC lines (Fig. 5S). Together these results provide evidence that FoxM1 is critical for the survival of the cells with stem cell features in HCC. It is noteworthy that EpCAM was suggested as another marker for human liver cancer cells with stem cell properties. However, the majority of the Huh7 cells are EpCAM+ and depletion of FoxM1 had very little effect on the population of the EpCAM+ cells. It remains possible that they underwent differentiation because the FoxM1-depleted Huh7 cells failed to generate xenograft tumors.
Fig. 6. Depletion of FoxM1 in Huh7 cells leads to specific loss of the cancer cells with stem cell features.
Huh7 cells were transfected with control siRNA, FoxM1-siRNA #1 and FoxM1 siRNA #2. (A) Western blots show the extent of FoxM1 depletion. The siRNA-transfected cells were treated with PE-tagged CD90-ab, FITC-tagged CD44-ab and PE-tagged CD133-ab and analyzed using a cell sorter. (B) Cell-growth following FoxM1-depletion was assayed by direct cell counting at the indicated times. (C) Twenty-four hours after control or FoxM1-siRNA transfection, 106 cells were subcutaneously injected into nude mice. The control-siRNA transfected cells were injected on the left side and the FoxM1-siRNA #1 transfected cells on the right side of 5 different mice. A picture of 3 mice after 6 weeks is shown. Quantification of the tumor mass from all five mice is plotted. Statistically significant changes were indicated with asterisks (*, p < 0.05; **, p < 0.01, ***, p<0.001.)
FoxM1 supports survival of the cells with stem-like features by regulating the levels of ROS
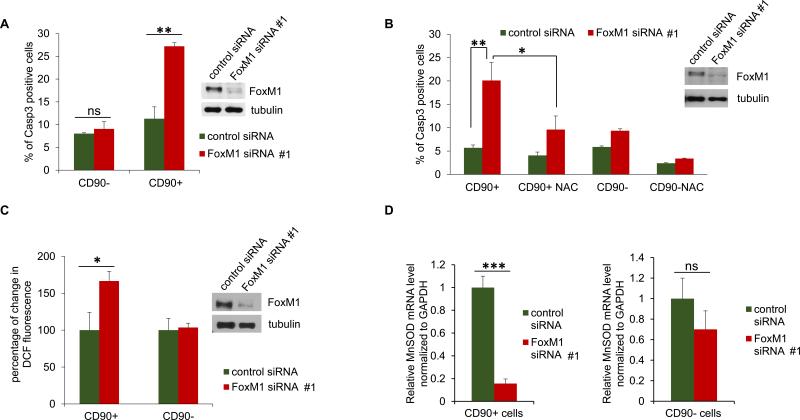
We investigated apoptosis of the CD90+ and the CD90- population in Huh7 cells. Forty-eight hours after transfection of siRNA, the cells were incubated with PE-tagged CD90-ab as well as a FITC-tagged active caspase3 detection reagent. The cells were separated using a cell sorter, gating for the CD90+/active caspase3+ and CD90-/active caspase3+ cells. Depletion of FoxM1 caused a significant increase in the CD90+/active caspase3+ cells (Fig. 7A). The CD90- population did not exhibit any significant increase in caspase3+ cells, indicating that the CD90+ cells undergo preferential apoptosis following depletion of FoxM1. Interestingly, the increase in the active caspase3+ CD90+ population was stunted in experiment where cells were incubated with the ROS scavenger N-acetyl cysteine (NAC) for 24h before analysis (Fig. 7B), suggesting that ROS accumulation plays a role in the apoptosis of the CD90+ cells. We measured the levels of ROS following depletion of FoxM1. The cells were incubated with PE-tagged CD90-ab as well as the ROS detection reagents DCF-DA, and the cells were then analyzed by flow cytometry. We observed increases in ROS mainly in the CD90+ population (Fig. 7C). Moreover, when the CD90+ and CD90- sorted populations were analyzed for MnSOD-mRNA, we observed a stronger dependence of the CD90+ cells on FoxM1 for MnSOD expression in comparison to that in the CD90- population (Fig. 7D). These observations indicate an important role of the FoxM1 induced expression of the antioxidant genes in the survival of the cells with stem cell features in HCC.
Fig. 7. Depletion of FoxM1 causes ROS-dependent apoptosis of CD90+ HCC cells.
Huh7 cells were transfected with control or FoxM1-siRNA #1 (all panels). Forty-eight hours after transfection, the cells were incubated with PE-tagged CD90-ab and FITC-tagged active caspase 3 detection reagent followed by separation of the CD90+ and active caspase 3 + cells using a cell sorter. Active Caspase 3+ CD90 + and the active caspase 3+ CD90- cells are plotted (A). (B) Twenty-four hours after siRNA transfection, the cells were treated with N-acetyl cysteine (NAC) for 24h followed by incubation with PE-tagged CD90-ab and FITC-tagged active caspase 3-detection reagent. The CD90+ and active caspase 3 + cells were quantified using a cell sorter. (C) Forty-eight hours after transfection, the cells were incubated with PE-tagged CD90-ab and DCF-DA followed by separation of the CD90+ and CD90- cells using a cell sorter. (D) Forty-eight hours after transfection, the cells were incubated with PE-tagged CD90-ab followed by separation of the CD90+ and the CD90- cells using a cell sorter. Total RNAs isolated from the fractionated cells were analyzed for MnSOD mRNA expression by quantitative RT-PCR. Statistically significant changes were indicated with asterisks (*, p < 0.05; **, p < 0.01, ***, p<0.001.)
Discussion
Work presented here is significant in several ways. First, we provide genetic evidence that FoxM1 is essential for H-ras12V-driven liver cancer (HCC) progression. Targeting FoxM1 leads to a disproportionate loss of the CD44+ and EpCAM+ HCC cells, in vivo. Moreover, FoxM1 plays important roles in the maintenance of the cancer cells with stem-like features, at least partly, through its ability to regulate the levels of ROS.
Nonalcoholic fatty liver disease (NAFLD), including nonalcoholic steatohepatitis (NASH) is a significant risk factor for liver cirrhosis, and becoming a major cause of hepatocellular carcinoma (1). Recent studies also indicated almost ubiquitous activation of the Ras-signaling pathway in hepatocellular carcinoma through silencing expression of the Ras regulatory GAP proteins (4). A transgenic mouse model that expresses H-ras12V in the liver was shown to develop steatosis, linking activated Ras-signaling to steatosis (26). Moreover, the male mice in that strain develop hepatocellular carcinoma. Interestingly, we observed that FoxM1 is expressed at a high level in those tumors (HCCs), and it is critical for the survival of the tumors. The genetically engineered mouse model for HCC used in this study is significant because of its relatedness to human HCC. Our observation that inhibition of FoxM1 inhibits HCC progression in that model further supports the notion that FoxM1 is a key molecular target for HCC. It is noteworthy that deletion of FoxM1 also inhibits initiation of lung cancer (31).
We employed the MxCre transgene and double stranded RNA to induce expression of Cre recombinase conditionally to delete FoxM1 after tumor development. Surprisingly, we observed loss of FoxM1 mainly in the HCC cells, but not in other fibroblast-like cells within the tumor nodules. Nevertheless, the reduction in FoxM1 expression caused a decrease in cell proliferation and an increase in apoptosis of the HCC cells, which coincided with a reduction in the number of tumor nodules. Some of the mice with longer induction of the Cre recombinase exhibited a total loss of the tumor nodules. It is noteworthy that we observed inhibition of HCC progression without any significant loss of expression of the H-ras12V driver oncogene. There was a significant increase in the levels of ROS in the HCC nodules following deletion of FoxM1. It is likely that the increased ROS might be an important factor in increasing apoptosis of the HCC cells that is likely related to the loss of the tumor nodules.
The disproportionate loss of the EpCAM+ and CD44+ HCC cells in H-ras12V induced HCC is interesting because it is likely related to the mechanism by which FoxM1 deletion inhibits HCC progression. These markers were shown to be present in the HCC precursor cells (8). Moreover, the liver cancer cells with stem cell features were shown to express these markers. Importantly, CD44 has been shown to protect cancer cells with stem cell properties from ROS-induced apoptosis by increasing the cellular levels of anti-oxidants (32). Also, it was shown that CD44 is important for survival of the liver cancer cells with stem cell features (7). We observed that both human and mouse CD44 promoters contain FoxM1 binding sites, and that FoxM1 interacts with those sites in the human gene promoter to activate expression of CD44. That could potentially explain why FoxM1 is essential for HCC development because the HCC precursor cells express CD44 (8). However, we did not observe dependence of CD90 expression on FoxM1 in the human liver cancer cell lines.
Our observations on the liver cancer cells with stem cell features provide further insights into the mechanism by which FoxM1 supports HCC progression. The CD90+, CD44+ and the CD133+/CD44+ cells in culture were induced to apoptosis by transient depletion of FoxM1, which had only marginal effect on the majority of the cells in culture. Moreover, FoxM1-depleted cells were not tumorigenic, and that would be consistent with a loss of the cancer stem-like cells. We observed increased ROS accumulation following depletion of FoxM1 in those cells. Moreover, inhibition of ROS decreased the apoptosis of the CD90+ cells. While some, but not all, of the CD90+ cells were also positive for CD44, we observed that expression of the anti-oxidant gene MnSOD in the CD90+ cells was dependent upon FoxM1. Loss of the FoxM1-activated anti-oxidant gene expression could explain the loss of the CD90+ cells. However, since FoxM1 deletion also inhibits the “stemness” genes in these HCC cells, it is possible that a significant population of those also undergo differentiation upon transient loss of FoxM1.
Our data suggest that MnSOD expression strongly depends upon FoxM1 in the CD90+ cells, but not in the CD90- cell population. MnSOD expression is activated by a variety of transcription factors, including p53 (33). It is likely that in the CD90- cells one or more of those transcription factor(s) participate in the expression of MnSOD. Together, these observations are highly significant with regards to development of new therapeutic strategies against HCC focusing on the FoxM1 pathway.
Supplementary Material
Acknowledgment
The work was supported by a grant (CA175380) from the National Cancer Institute to PR. PR is supported also by a Merit Review Grant (BX000131) from the Veteran Administration.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author's contributions: Dragana Kopanja: study concept and design, acquisition of data, analysis and interpretation of data, preparation of manuscript; Akshay Pandey: acquisition and analysis of data, Megan Kiefer, Zebin Wang, Neha Chandan, Janai R. Carr, Roberta Franks, Dae-Yeul Yu, Grace Guzman and Ajay Maker: acquisition of data; Pradip Raychaudhuri: study concept and design, interpretation of data, study supervision and manuscript preparation.
No conflict of interest with regards to this manuscript.
Financial support: NCI (no conflict of interest).
Reference
- 1.Page JM, Harrison SA. NASH and HCC. Clinics in liver disease. 2009;13:631–647. doi: 10.1016/j.cld.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Calvisi DF, Ladu S, Gorden A, Farina M, Conner EA, Lee JS, Factor VM, et al. Ubiquitous activation of Ras and Jak/Stat pathways in human HCC. Gastroenterology. 2006;130:1117–1128. doi: 10.1053/j.gastro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Macheiner D, Heller G, Kappel S, Bichler C, Stattner S, Ziegler B, Kandioler D, et al. NORE1B, a candidate tumor suppressor, is epigenetically silenced in human hepatocellular carcinoma. Journal of hepatology. 2006;45:81–89. doi: 10.1016/j.jhep.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. Journal of hepatology. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sui G, Ma X, Liu S, Niu H, Dong Q. Study of the correlation between H-ras mutation and primary hepatocellular carcinoma. Oncology letters. 2012;4:779–782. doi: 10.3892/ol.2012.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S, Chan KW, Guan XY. In search of liver cancer stem cells. Stem Cell Rev. 2008;4:179–192. doi: 10.1007/s12015-008-9035-z. [DOI] [PubMed] [Google Scholar]
- 7.Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 8.He G, Dhar D, Nakagawa H, Font-Burgada J, Ogata H, Jiang Y, Shalapour S, et al. Identification of liver cancer progenitors whose malignant progression depends on autocrine IL-6 signaling. Cell. 2013;155:384–396. doi: 10.1016/j.cell.2013.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun H, Teng M, Liu J, Jin D, Wu J, Yan D, Fan J, et al. FOXM1 expression predicts the prognosis in hepatocellular carcinoma patients after orthotopic liver transplantation combined with the Milan criteria. Cancer Lett. 2011;306:214–222. doi: 10.1016/j.canlet.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 12.Park HJ, Carr JR, Wang Z, Nogueira V, Hay N, Tyner AL, Lau LF, et al. FoxM1, a critical regulator of oxidative stress during oncogenesis. EMBO J. 2009;28:2908–2918. doi: 10.1038/emboj.2009.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gemenetzidis E, Elena-Costea D, Parkinson EK, Waseem A, Wan H, Teh MT. Induction of human epithelial stem/progenitor expansion by FOXM1. Cancer Res. 2010;70:9515–9526. doi: 10.1158/0008-5472.CAN-10-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Park HJ, Carr JR, Chen YJ, Zheng Y, Li J, Tyner AL, et al. FoxM1 in tumorigenicity of the neuroblastoma cells and renewal of the neural progenitors. Cancer Res. 2011;71:4292–4302. doi: 10.1158/0008-5472.CAN-10-4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Z, Tan G, Ding M, Dong D, Chen T, Meng X, Huang X, et al. Foxm1 transcription factor is required for maintenance of pluripotency of P19 embryonal carcinoma cells. Nucleic Acids Res. 2010;38:8027–8038. doi: 10.1093/nar/gkq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang N, Wei P, Gong A, Chiu WT, Lee HT, Colman H, Huang H, et al. FoxM1 promotes beta-catenin nuclear localization and controls Wnt target-gene expression and glioma tumorigenesis. Cancer cell. 2011;20:427–442. doi: 10.1016/j.ccr.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gong A, Huang S. FoxM1 and Wnt/beta-catenin signaling in glioma stem cells. Cancer research. 2012;72:5658–5662. doi: 10.1158/0008-5472.CAN-12-0953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, et al. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park HJ, Gusarova G, Wang Z, Carr JR, Li J, Kim KH, Qiu J, et al. Deregulation of FoxM1b leads to tumour metastasis. EMBO Mol Med. 2011;3:21–34. doi: 10.1002/emmm.201000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang C, Du J, Xie K. FOXM1 and its oncogenic signaling in pancreatic cancer pathogenesis. Biochimica et biophysica acta. 2014;1845:104–116. doi: 10.1016/j.bbcan.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue J, Lin X, Chiu WT, Chen YH, Yu G, Liu M, Feng XH, et al. Sustained activation of SMAD3/SMAD4 by FOXM1 promotes TGF-beta-dependent cancer metastasis. The Journal of clinical investigation. 2014;124:564–579. doi: 10.1172/JCI71104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balli D, Ustiyan V, Zhang Y, Wang IC, Masino AJ, Ren X, Whitsett JA, et al. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. The EMBO journal. 2013;32:231–244. doi: 10.1038/emboj.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang AG, Moon HB, Lee MR, Hwang CY, Kwon KS, Yu SL, Kim YS, et al. Gender-dependent hepatic alterations in H-ras12V transgenic mice. Journal of hepatology. 2005;43:836–844. doi: 10.1016/j.jhep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 24.Gusarova GA, Wang IC, Major ML, Kalinichenko VV, Ackerson T, Petrovic V, Costa RH. A cell-penetrating ARF peptide inhibitor of FoxM1 in mouse hepatocellular carcinoma treatment. J Clin Invest. 2007;117:99–111. doi: 10.1172/JCI27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy N, Stoyanova T, Dominguez-Brauer C, Park HJ, Bagchi S, Raychaudhuri P. DDB2, an essential mediator of premature senescence. Mol Cell Biol. 2010;30:2681–2692. doi: 10.1128/MCB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26.Wang AG, Moon HB, Chae JI, Kim JM, Kim YE, Yu DY, Lee DS. Steatosis induced by the accumulation of apolipoprotein A-I and elevated ROS levels in H-ras12V transgenic mice contributes to hepatic lesions. Biochemical and biophysical research communications. 2011;409:532–538. doi: 10.1016/j.bbrc.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 27.Wang AG, Moon HB, Lee MR, Hwang CY, Kwon KS, Yu SL, Kim YS, et al. Gender-dependent hepatic alterations in H-ras12V transgenic mice. J Hepatol. 2005;43:836–844. doi: 10.1016/j.jhep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 28.Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, et al. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694–707. doi: 10.1016/j.stem.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Charette N, De Saeger C, Lannoy V, Horsmans Y, Leclercq I, Starkel P. Salirasib inhibits the growth of hepatocarcinoma cell lines in vitro and tumor growth in vivo through ras and mTOR inhibition. Molecular cancer. 2010;9:256. doi: 10.1186/1476-4598-9-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu LL, Fu D, Ma Y, Shen XZ. The power and the promise of liver cancer stem cell markers. Stem cells and development. 2011;20:2023–2030. doi: 10.1089/scd.2011.0012. [DOI] [PubMed] [Google Scholar]
- 31.Wang IC, Ustiyan V, Zhang Y, Cai Y, Kalin TV, Kalinichenko VV. Foxm1 transcription factor is required for the initiation of lung tumorigenesis by oncogenic Kras. Oncogene. 2013 doi: 10.1038/onc.2013.475. [DOI] [PubMed] [Google Scholar]
- 32.Lin CH, Hung PH, Chen YJ. CD44 Is Associated with the Aggressive Phenotype of Nasopharyngeal Carcinoma through Redox Regulation. International journal of molecular sciences. 2013;14:13266–13281. doi: 10.3390/ijms140713266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dhar SK, Tangpong J, Chaiswing L, Oberley TD, St Clair DK. Manganese superoxide dismutase is a p53-regulated gene that switches cancers between early and advanced stages. Cancer research. 2011;71:6684–6695. doi: 10.1158/0008-5472.CAN-11-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.